Introduction
Gastric cancer is the fourth most common cancer and
second leading cause of cancer-related deaths worldwide (1,2).
Although surgery is the most effective treatment modality for
patients with resectable tumors, due to the low rates of early
detection and diagnosis, many patients with gastric cancer in China
are diagnosed at an advanced and unresectable clinical stage. For
advanced gastric cancer and for patients with recurrent cancer who
are not suitable candidates for reoperation, chemotherapy remains
the preferred treatment method (3).
However, at present, intrinsic or acquired multidrug resistance
(MDR) represents a major obstacle for successful cancer
chemotherapy (4). The mechanism
underlying MDR is complex due to interactions between various
factors, including drug efflux, the alteration of drug targets, DNA
damage repair, cell proliferation and apoptosis, and cell cycle
changes (5–7).
Noncoding RNAs (ncRNAs) have drawn much attention in
the field of cancer research. Of particular interest are microRNAs
(miRNA/miRs), which are small ncRNAs that are able to
post-transcriptionally modulate gene expression, with the primary
functions of controlling physiological and pathological processes,
including cancer onset, growth, and progression (8). To date, >200 miRNAs have been found
to be associated with gastric cancer development, progression, and
therapeutic response, and numerous studies have indicated that
miRNAs may play an important role in the MDR of various types of
cancer, including gastric cancer (9–11). It
has previously been reported that miR-30a is significantly
downregulated in gastric cancer cells (12,13).
However, there is a lack of research on whether miR-30a regulation
can affect the chemosensitivity of resistant gastric cancer cells,
and the underlying mechanisms regarding the potential effects of
miR-30a on drug resistance and cell autophagy require further
investigation. In the present study, we assessed the potential
effect of miR-30a on the MDR of the SGC7901 gastric cancer cell
line. Additionally, we aimed to provide insight into the molecular
mechanism underlying the effect of miR-30a on MDR, with focus on
chemotherapy-induced autophagy.
Materials and methods
Cell culture
The human gastric cancer cell line SGC-7901 was
purchased from the Cell Bank of the Chinese Academy of Medical
Sciences (Shanghai, China) and maintained in RPMI-1640 medium
containing 10% FBS (both from Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) and 0.1% penicillin-streptomycin (Beijing
Solarbio Science & Technology Co., Ltd., Beijing, China) at
37°C in a humid atmosphere (5% CO2, 95% air).
Cisplatin (CDDP) was purchased from Qilu
Pharmace-utical Co., Ltd. (Jinan, China) and dissolved with normal
saline before use. A CDDP-resistant variant SGC-7901/CDDP cell line
was established by gradually increasing the drug concentration
(0.1, 0.2, 0.3, 0.4, 0.5, and 0.6 µg/ml CDDP each for two weeks).
The SGC-7901/CDDP phenotype was then maintained by supplementing
the cell medium with 0.6 µg/ml cisplatin, which was stopped 2 weeks
prior to experiments.
Cell transfection
miR-30a mimics, and scramble miRNA negative control
(NC miRNA) were synthesized by Shanghai GenePharma Co., Ltd.
(Shanghai, China). SGC-7901/CDDP cells were transfected with 100 nM
miR-30a mimics for miR-30a overexpression. Transfection with NC
miRNA was performed in parallel as a control. Transfection was
performed using Lipofectamine 2000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) according to the
manufacturer's protocol (13). The
transfection efficiency was measured via the reverse transcription
(RT)-PCR analysis of miR-30a expression described below.
Reverse
transcription-semi-quantitative polymerase chain reaction
(RT-sqPCR) analysis of miR-30a and MDR1 mRNA expression
A TRIzol kit was used to extract total RNA according
to the manufacturer's instructions (Invitrogen; Thermo Fisher
Scientific, Inc.), and the RNA samples were quantified by UV
spectrophotometry.
miRNA-specific cDNA was obtained by RT using
stem-loop primers and a TaqMan MicroRNA Reverse Transcription kit
(Applied Biosystems; Thermo Fisher Scientific, Inc.), the RT primer
used for miR-30a was 5′-CGTCGCTACATCCAGTGTAGCATATGCGACGCTTCCAGT-3′.
Subsequently, the level of mature miR-30a was detected using a
TaqMan MicroRNA Assay kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. The
PCR primers used indluded: miR-30a forward, 5′-TGTAAACATCCTCGAC-3′
and reverse, 5′-ACATCCAGTGTAGCATA-3′; U6 forward,
5′-CTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CAGGGGCCATGCTAAATCTTC-3′. The PCR reaction was performed for 35
cycles at 94°C for 30 sec, 50°C for 30 sec and 72°C for 30 min.
To measure the expression of MDR1 mRNA, RT
was carried out with 2 µg extracted total RNA using a First Strand
cDNA Synthesis kit (Toyobo Life Science, Osaka, Japan). For
semi-quantitative RT-PCR, 100 ng cDNA was amplified with
gene-specific primers using Taq DNA polymerase in 10X PCR buffer,
4X dNTP mixture and MgCl2. The following primers
(Shanghai GeneCore BioTechnologies Co., Ltd., Shanghai, China) were
used for the specific amplification of MDR1 forward,
5′-AGACATGACCAGGTATGCCTAT-3′ and reverse,
5′-AGCCTATCTCCTGTCGCATTA-3′. The expression of GAPDH (forward,
5′-GAGGGGCCATCCACAGTCTT-3′ and reverse, 5′-TTCATTGACCTCAACTACAT-3′)
was used as an internal control. The PCR reaction was performed for
35 cycles at 94°C for 30 sec, 56°C for 30 sec and 65°C for 1
min.
The PCR products were separated on 2% agarose gels
containing 0.5 µg/ml ethidium bromide and photographed under a UV
transilluminator, and AlphaEaseFC software was used to analyze the
relative light intensities. Three independent experiments with
triplicate samples were performed.
Detection of cell chemosensitivity to
CDDP
Cell chemosensitivity was detected in the following
four cell groups: SGC7901 (chemosensitive) cells, SGC7901/CDDP
(chemoresistant) control cells, SGC7901-NC miRNA cells (transfected
with NC miRNA), and SGC7901/CDDP-miR30a mimics cells (transfected
with miR30a mimics). The cells of the different groups were seeded
in 96-well plates (5×103 cells/well) and incubated at
37°C in a humidified 5% CO2 atmosphere for 24 h.
Subsequently, CDDP was added at final concentrations of 0.02, 0.1,
0.5, 2.5, 12.5, and 62.5 µg/ml to the culture medium or an equal
volume of vehicle was added as control treatment. At 48 h after
CDDP administration, cell viability was assessed using a CCK-8
assay (Toyobo Life Science) according to the manufacturer's
instructions. The absorbance at 450 nm was measured, from which the
cell growth inhibition rate and half maximal inhibitory
concentration (IC50) of CDDP was calculated.
Additionally, the resistance index of the SGC7901/CDDP cells was
calculated as: IC50 of sensitive cell line/IC50 of resistant cell
line. Three independent experiments were performed in
triplicate.
Cell apoptosis measurement by flow
cytometry
Cell apoptosis was measured using an Annexin
V-propidium iodide (PI) apoptosis detection kit (BD556547™; BD
Pharmingen; BD Biosciences, Franklin Lakes, USA). Cell apoptosis
was analyzed in the following cell groups: SGC7901 sensitive cells,
SGC7901/CDDP resistant control cells, SGC7901-NC miRNA cells and
SGC7901/CDDP-miR30a mimics cells. Following treatment with 5 µg/ml
CDDP for 24 h, the cells in the different groups were collected and
washed twice with precooled PBS, and then re-suspended in 400 µl of
Annexin V binding buffer. Subsequently, the cells were incubated
with fluorescein isothiocyanate (FITC)-Annexin V (5 µl) for 15 min
at room temperature in the dark, and then with PI (5 µl) for 5 min
at 4°C in the dark prior to analysis by flow cytometry.
Western blotting
Total protein was extracted from the cells in each
group and protein concentration was measured using the
bicinchoninic acid method. The protein samples were subjected to
SDS-PAGE and transferred to PVDF membranes (EMD Millipore,
Billerica, MA, USA) for 2 h. After washing, Tris-buffered saline
with Tween solution containing 5% skimmed milk powder was used to
block the membranes for 1 h. Primary antibodies diluted to the
appropriate concentrations were incubated with the membranes
overnight at 4°C: Rabbit anti-LC3A/B (#12741, 1:1,000; Cell
Signaling Technology, Danvers, MA, USA), mouse anti-Mdr-1
(sc-13131, 1:800; Santa Cruz Biotechnology, Inc., Dallas, TX, USA),
and rabbit anti-β-actin (ab8227, 1:5,000; Abcam, Cambridge, UK) as
internal control. The membranes were then washed and incubated with
secondary antibody (goat anti-rabbit, ZB-2301, 1:5,000; goat
anti-mouse, ZDR5307) (both purchased from ZSGB-Bio, Beijing, China)
for 1 h at room temperature. After washing, ECL substrate (EMD
Millipore) was added and the protein bands were analyzed with a gel
electrophoresis image analyzer (FR980; Shanghai Furi Science &
Technology Co., Ltd., Shanghai, China).
Statistical analysis
SPSS 18.0 software (SPSS Inc., Chicago, IL, USA) was
used to analyze the data. All data were expressed as the mean ± SD.
One-way analysis of variance was used for comparisons among three
or more groups, and the post hoc LSD test was used for subsequent
comparisons between two groups. P<0.05 was considered to
indicate statistical significance.
Results
Expression of miR-30a in
chemoresistant cells
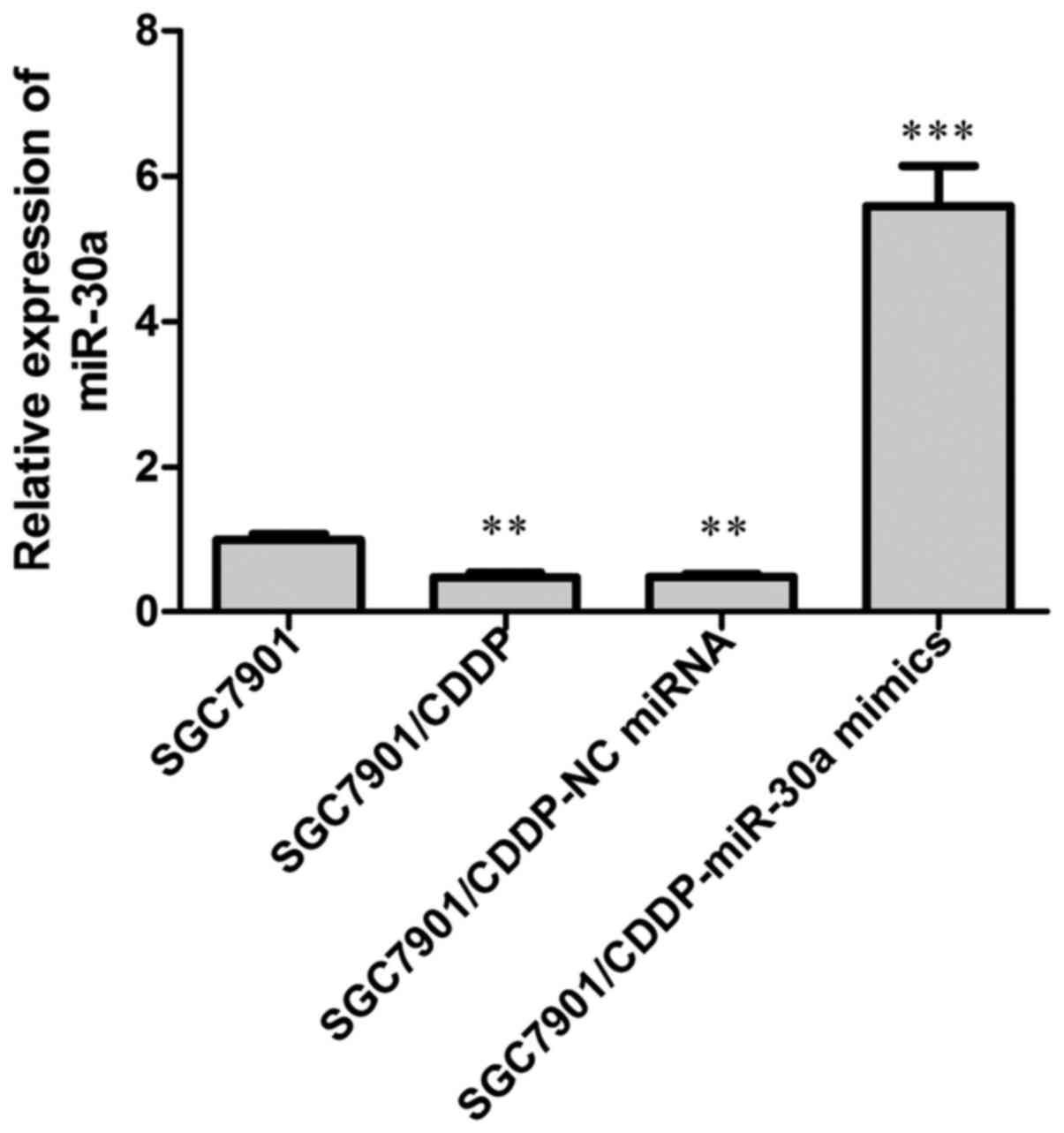
The expression of miR-30a was examined by RT-PCR
analysis. Our results showed that in the chemoresistant
SGC7901/CDDP cells, the expression of miR-30a was significantly
decreased when compared with that in the chemosensitive SGC7901
cells (Fig. 1; P<0.01).
Subsequently, we overexpressed miR-30a endogenous miR-30a
expression in the SGC7901/CDDP cells by transfection with miR30a
mimics, with NC miRNA transfection performed in parallel, and the
transfection efficiency was detected by RT-PCR analysis of miR-30a
expression, our data showed that after miR-30a mimics transfection,
the miR-30a level in SGC7901/CDDP cells was elevated to about
6-fold of the SGC7901/CDDP control cells.
miR-30a expression is related to
gastric cancer cell chemoresistance
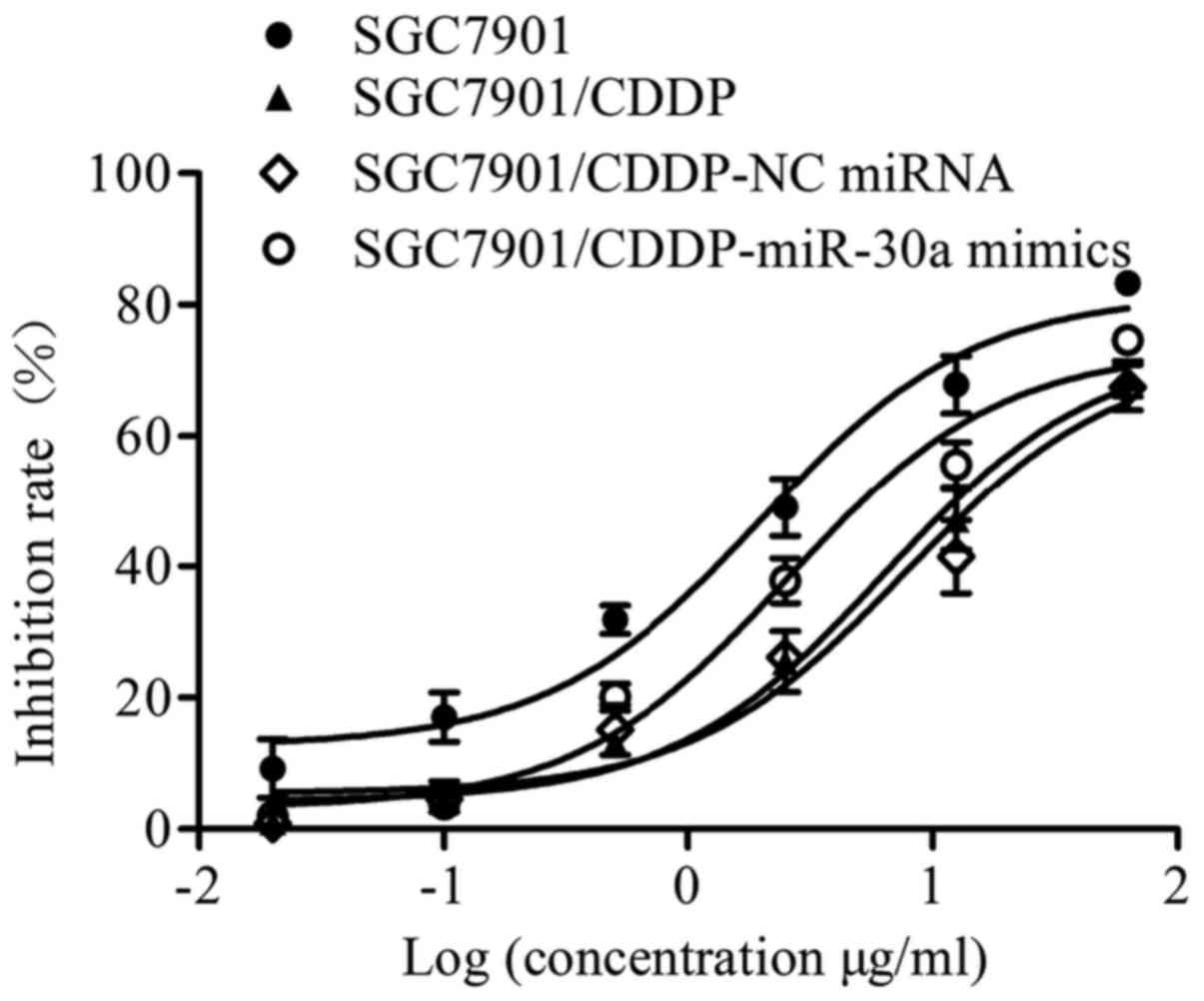
To determine the effect of miR-30a on the
chemosensitivity of gastric cancer cells, a CCK-8 assay was
performed. Our results (Fig. 2 and
Table I) showed that the
chemosensitivity of SGC7901/CDDP cells was significantly decreased
compared with that of SGC7901 cells; notably, the IC50 value of
CDDP was elevated from 6.63 µM in SGC7901 cells to 21.28 µM in
SGC7901/CDDP cells. Furthermore, the resistance index of the
SGC7901/CDDP cells was 3.21. No significant difference in IC50 was
observed between the SGC7901/CDDP (21.28 µM) and SGC7901/CDDP-NC
miRNA (25.77 µM) groups (P>0.05). By contrast, the IC50 of CDDP
in the SGC7901/CDDP-miR-30a mimics group was decreased to 8.56 µM
(P<0.001 vs. SGC7901/CDDP group), indicating increased
chemosensitivity following miR-30a transfection.
 | Table I.Cytotoxicity of CDDP in different
groups. |
Table I.
Cytotoxicity of CDDP in different
groups.
|
|
|
| SGC7901/CDDP |
|---|
|
|
|
|
|
|---|
| Variable | SGC7901 | SGC7901/CDDP | NC miRNA | miR-30a mimics |
|---|
| IC50
(µM) | 1.99 | 6.39a | 7.74 | 2.57b |
| IRmax
(%) | 83.27±2.03 | 68.76±4.68 | 67.42±5.96 | 74.66±3.10 |
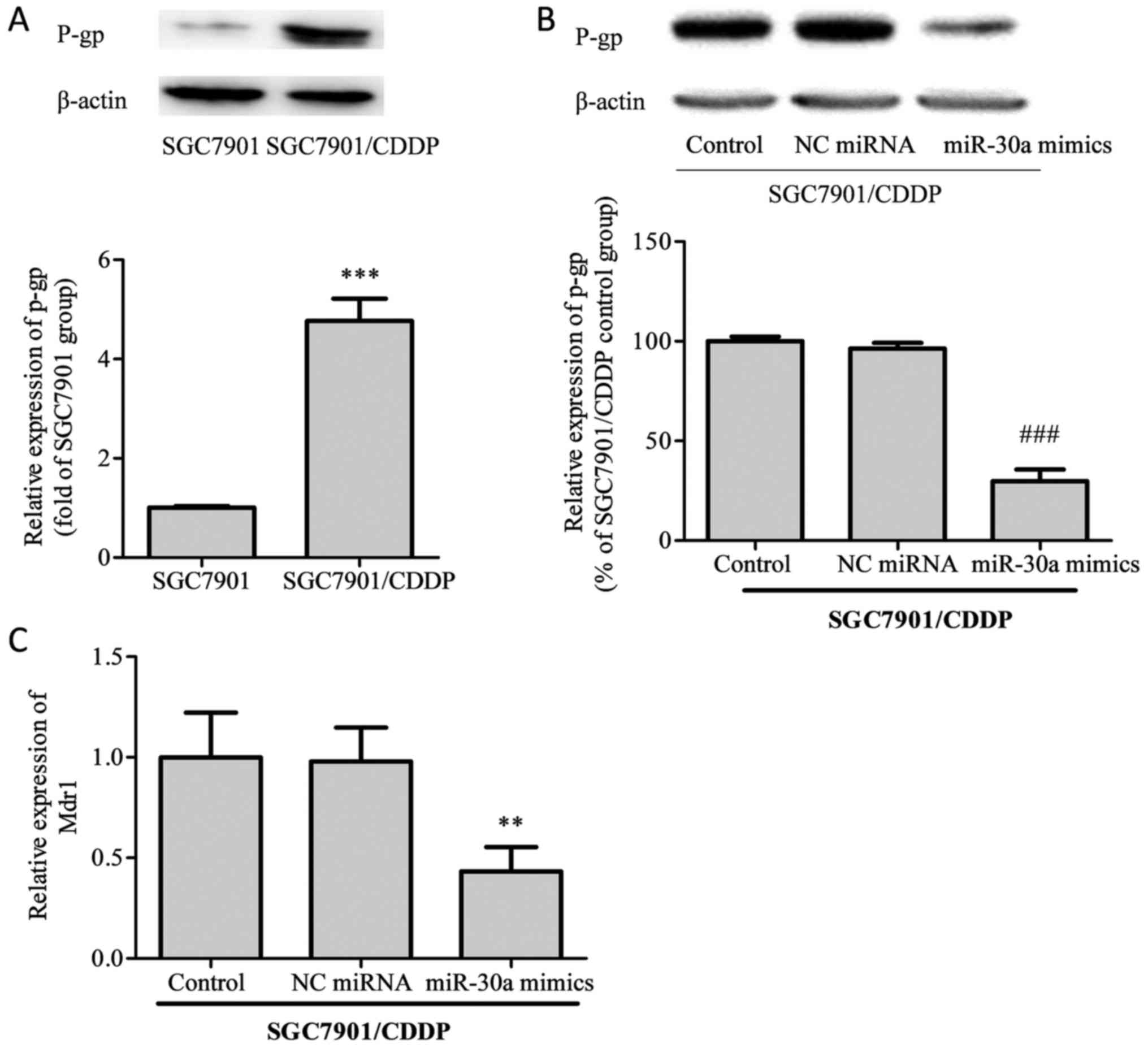
miR-30a regulates MDR-related
factors
Subsequently, we examined the protein levels of
P-glycoprotein (P-gp) and the mRNA expression of its corresponding
gene, MDR1, in SGC7901/CDDP cells. As shown in Fig. 3, the expression of P-gp protein was
notably elevated in SGC7901/CDDP cells compared with SGC7901 cells.
After transfection with miR-30a mimics, P-gp expression in
SGC7901/CDDP cells was significantly decreased. On MDR1 mRNA
analysis, the same trends as for P-gp protein expression were
identified.
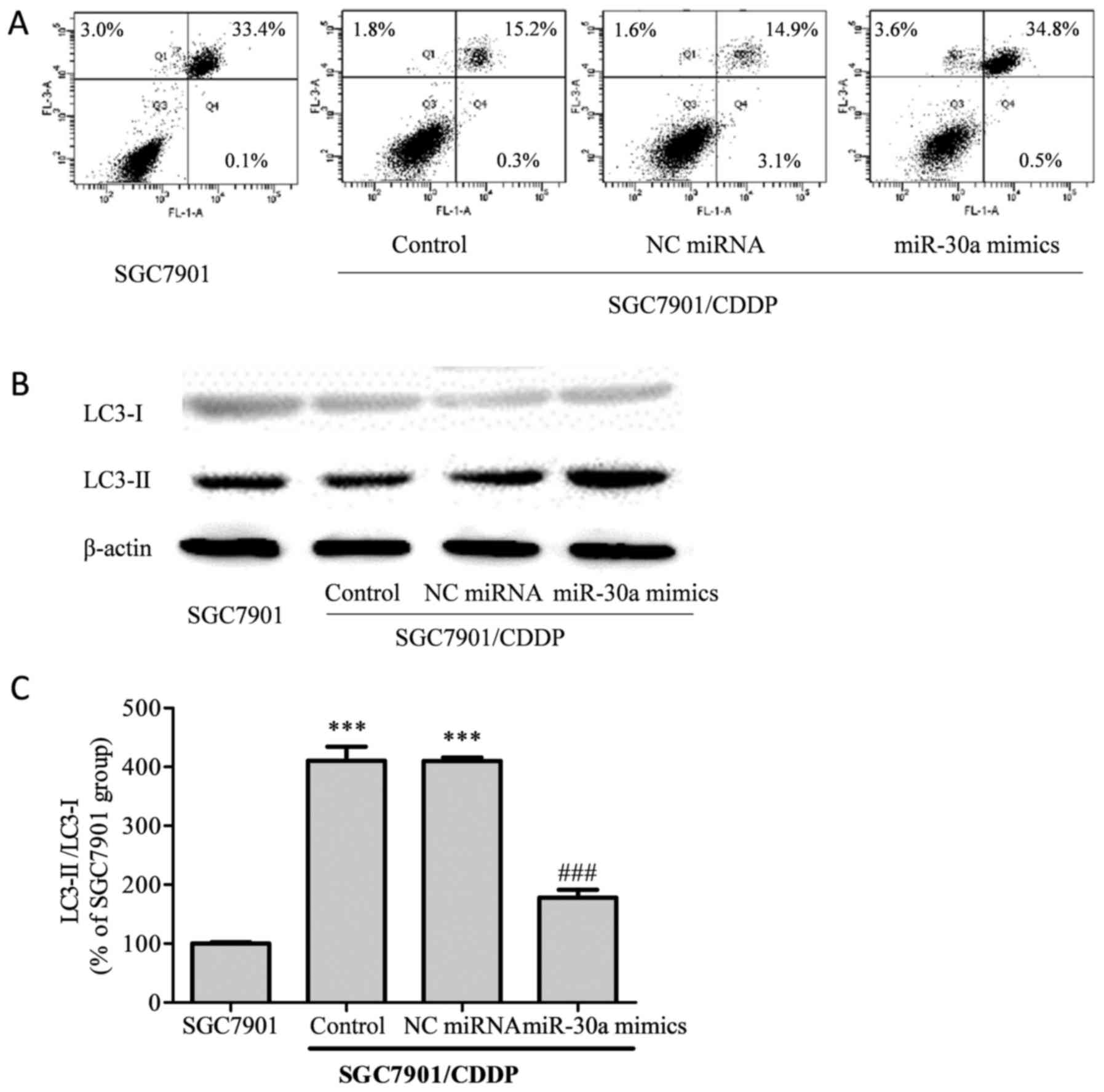
miR-30a modulates cell autophagy and
increases apoptosis in chemoresistant gastric cancer cells
Cell apoptosis induced by CDDP treatment was
analyzed by flow cytometry (Fig.
4A). The total apoptotic rate of SGC7901 cells treated with 5
µg/ml CDDP was 36.5%, while in the SGC7901/CDDP group, the total
apoptotic rate was decreased to 17.3% (P<0.001 vs. SGC7901
group). Compared with the SGC7901/CDDP group, the total apoptotic
rate did not differ significantly in the SGC7901/CDDP-NC miRNA
group (19.6%, P>0.05); however, in the SGC7901/CDDP-miR-30a
mimics group, the total apoptotic rate was elevated to 38.9%
(P<0.001), thus indicating that the upregulation of miR-30a
improved chemosensitivity in the SGC7901/CDDP cells.
Light chain (LC)3, which exists as a cytosolic form
(LC-I) under normal homeostatic conditions, becomes conjugated to
phosphatidylethanolamine (PE) to form the membrane-bound form
LC3-II upon autophagy induction (14). Our results showed that in the
chemoresistant SGC7901/CDDP cells, the LC3-II/LC3-I ratio was
significantly higher than that in the chemosensitive SGC7901 cells
(Fig. 4B, P<0.001). Meanwhile, on
upregulation of miR-30a in the SGC7901/CDDP cells, the elevated
LC3-II/LC3-I ratio was attenuated.
Discussion
Clinical treatment methods for patients with gastric
cancer include surgery, radiation therapy, and chemotherapy. Tumors
that develop drug resistance to chemotherapy are a major obstacle
for successful cancer chemotherapy. Previously reported mechanisms
of chemoresistance include increased drug efflux, mutation of
target genes, inactivation of detoxification enzymes, dysfunction
of pro-apoptotic proteins and enhancement of DNA repair activity,
although the precise mechanisms involved in cancer cell
chemoresistance remain to be fully elucidated (4,15). The
present study identified miR-30a as a novel
chemosensitivity-associated miRNA related to autophagy and
apoptosis.
Increasing evidence has demonstrated that miRNAs are
involved in chemoresistance in many tumor types (8,16,17). In
this research, we confirmed that miR-30a expression was
downregulated in the chemoresistant SGC7901/CDDP cell line.
Additionally, our results showed that upregulating miR-30a by
transfection with miR-30a mimics could attenuate the
chemoresistance of SGC7901/CDDP cells, observed as a decrease in
the IC50 of CDDP from 21.28 to 8.56 µM (P<0.05, SGC7901/CDDP vs.
SGC7901/CDDP-miR-30a mimics group). These results suggest that
miR-30a plays an important role in the chemoresistance of gastric
cancer cells. P-gp, a 170-kDa transmembrane glycoprotein encoded by
the human MDR1 [ATP-binding cassette (ABC) subfamily B
member 1] gene, is a member of the ABC transporters family, and
serves as a drug efflux pump that extrudes a wide spectrum of
chemotherapeutic agents from MDR cancer cells (18). Accordingly, P-gp substrates can
generally induce MDR1/P-gp expression in MDR cancer cells
(19). In this study, we found that
the expression level of P-gp in the SGC7901/CDDP cells was
significantly higher than that in the SGC7901 cells, suggesting
that overexpression of P-gp may be a mechanism by which SGC7901
cells develop drug resistance. Several miRNAs have been identified
as critical regulators of P-gp-mediated drug resistance in many
cancers (13,16,17).
Similarly, we revealed that the expression of P-gp protein and its
cognate gene MDR1 could be affected by miR-30a expression,
indicating that miR-30a may arrest the chemoresistance and growth
of SGC7901/CDDP cells by inhibiting P-gp activity and expression.
This was consistent with previous research (13).
Apoptosis has been widely investigated in the past
few decades and is established as the major mechanism of programmed
cell death (PCD) (20).
Additionally, apoptosis is believed to be the major type of cell
death triggered by chemotherapeutic drugs. However, apoptosis is
not the sole route of PCD; both apoptosis (‘self-killing’) and
autophagy (‘self-eating’) serve a role in self-destructive
processes in cells, and interact to execute gastric cancer cell
death (21,22). Autophagy is a survival strategy
employed by cells experiencing nutrient deprivation or other
stresses, which is a tightly regulated by the lysosomal degradation
pathway, and is considered to be essential for cell growth,
differentiation, development, survival and homeostasis. However,
while autophagy is generally considered to be a survival mechanism
under normal homeostatic conditions, it has both pro-death and
pro-survival roles in cancer (21,23).
Chemotherapeutic drugs can induce both apoptosis and
autophagy in the treatment of cancer, and it was previously
reported that autophagy helps cancer cells evade apoptosis, and
thus contributes to chemoresistance (24). Additionally, cancer cells that
respond to drugs by inducing autophagy are more drug-resistant
(25): For example, in response to
5-fluorouracil (5-FU) and cisplatin, chemosensitive cell lines were
found to undergo apoptosis, whereas chemoresistant populations
underwent autophagy (25–28). It is believed that autophagy can play
both positive and negative roles in promoting apoptosis. Previous
research demonstrated the cytoprotective role of autophagy in
response to 5-FU treatment in gastric cancer cells (27,28),
while a different study showed that 5-FU may suppress miR-30 to
upregulate beclin-1 and thus induce autophagic cell death and cell
proliferation arrest in gastric cancer cells (29). Nevertheless, many studies have shown
that autophagy protects various tumor cells against apoptosis
induced by chemotherapeutic drugs (23,30,31).
LC3 is one of the biomarkers of autophagy, and on
initiation of autophagy, cytosolic LC3-I is converted into
membrane-bound LC3-II, which is an essential step in autophagosome
formation, and thus the abundance of LC3-II is correlated with the
number of autophagosomes (32). In
this research, we confirmed that autophagy was induced while cell
apoptosis was mitigated in the SGC7901/CDDP resistant cells after
treatment with CDDP, relative to CDDP-treated SGC7901 cells. These
findings were consistent with previous reports (30–32).
It has previously been confirmed that the autophagic
protein beclin-1 is a putative target of miR-30a (33). To understand the mechanisms of
miR-30a in gastric cancer chemoresistance, we examined the effect
of miR-30a overexpression on the autophagy status of cells. We
found that low expression of miR-30a was related to chemoresistance
in gastric cancer cells, and in the chemoresistant cell line
SGC7901/CDDP, CDDP-induced apoptosis was weakened. Furthermore, the
LC3-II/LC3-I ratio was elevated in the SGC7901/CDDP cells when
compared with that in the chemosensitive SGC7901 cells (P<0.05),
while transfection with miR-30a mimics decreased the LC3-II/LC3-I
ratio in the SGC7901/CDDP cells (P<0.05 vs. SGC7901/CDDP control
cells). These results suggested that autophagy might contribute to
chemoresistance in gastric cancer cells, and that the reduction of
LC3-II in response to miR-30a upregulation might inhibit
chemoresistance-related autophagy in gastric cancer cells.
In summary, as autophagy plays a dual role in tumor
promotion and suppression, further investigation is required to
determine whether the activation of autophagy leads to the survival
or death of cancer cells during chemotherapy. The present study
demonstrated that miR-30a was associated with chemoresistance in
SGC7901 cells, and that the underlying mechanism involves, at least
in part, the modulation of chemoresistance-related autophagy.
Understanding the novel functions of miRNAs may allow us to develop
promising therapeutic strategies for enhancing the effects of
chemotherapy, which may ultimately improve clinical outcomes in the
treatment of cancer patients.
References
|
1
|
Xu W, Yang Z and Lu N: Molecular targeted
therapy for the treatment of gastric cancer. J Exp Clin Cancer Res.
35:12016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hu Y, Ying M, Huang C, Wei H, Jiang Z,
Peng X, Hu J, Du X, Wang B, Lin F, et al: Oncologic outcomes of
laparoscopy-assisted gastrectomy for advanced gastric cancer: A
large-scale multicenter retrospective cohort study from China. Surg
Endosc. 28:2048–2056. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
O'Connor R: The pharmacology of cancer
resistance. Anticancer Res. 27:1267–1272. 2007.PubMed/NCBI
|
|
5
|
Choi JH, Lim HY, Joo HJ, Kim HS, Yi JW,
Kim HC, Cho YK, Kim MW and Lee KB: Expression of multidrug
resistance-associated protein1, P-glycoprotein and thymidylate
synthase in gastric cancer patients treated with 5-fluorouracil and
doxorubicin-based adjuvant chemotherapy after curative resection.
Br J Cancer. 86:1578–1585. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Morin PJ: Drug resistance and the
microenvironment: Nature and nurture. Drug Resist Updat. 6:169–172.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lippert TH, Ruoff HJ and Volm M: Intrinsic
and acquired drug resistance in malignant tumors. The main reason
for therapeutic failure. Arzneimittelforschung. 58:261–264.
2008.PubMed/NCBI
|
|
8
|
Zhang M and Du X: Noncoding RNAs in
gastric cancer: Research progress and prospects. World J
Gastroenterol. 22:6610–6618. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tan P and Yeoh KG: Genetics and molecular
pathogenesis of gastric adenocarcinoma. Gastroenterology.
149:1153–1162.e3. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Song JH and Meltzer SJ: MicroRNAs in
pathogenesis, diagnosis, and treatment of gastroesophageal cancers.
Gastroenterology. 143:35–47.e2. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang M and Du X: Noncoding RNAs in
gastric cancer: Research progress and prospects. World J
Gastroenterol. 22:6610–6618. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu Z, Chen L, Zhang X, Xu X, Xing H,
Zhang Y, Li W, Yu H, Zeng J, Jia J, et al: RUNX3 regulates vimentin
expression via miR-30a during epithelial-mesenchymal transition in
gastric cancer cells. J Cell Mol Med. 18:610–623. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li C, Zou J, Zheng G and Chu J: miR-30a
decreases multidrug resistance (MDR) of gastric cancer cells. Med
Sci Monit. 0:02016.PubMed/NCBI
|
|
14
|
Lai YC, Chuang YC, Chang CP and Yeh TM:
Macrophage migration inhibitory factor has a permissive role in
concanavalin A-induced cell death of human hepatoma cells through
autophagy. Cell Death Dis. 6:e20082015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shi WJ and Gao JB: Molecular mechanisms of
chemoresistance in gastric cancer. World J Gastrointest Oncol.
8:673–681. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
An Y, Zhang Z, Shang Y, Jiang X, Dong J,
Yu P, Nie Y and Zhao Q: miR-23b-3p regulates the chemoresistance of
gastric cancer cells by targeting ATG12 and HMGB2. Cell Death Dis.
6:e17662015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao L, Wang Y, Jiang L, He M, Bai X, Yu L
and Wei M: MiR-302a/b/c/d cooperatively sensitizes breast cancer
cells to adriamycin via suppressing P-glycoprotein(P-gp) by
targeting MAP/ERK kinase kinase 1 (MEKK1). J Exp Clin Cancer Res.
35:252016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Abraham I, Jain S, Wu CP, Khanfar MA,
Kuang Y, Dai CL, Shi Z, Chen X, Fu L, Ambudkar SV, et al: Marine
sponge-derived sipholane triterpenoids reverse P-glycoprotein
(ABCB1)-mediated multidrug resistance in cancer cells. Biochem
Pharmacol. 80:1497–1506. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fayad W, Fryknäs M, Brnjic S, Olofsson MH,
Larsson R and Linder S: Identification of a novel topoisomerase
inhibitor effective in cells overexpressing drug efflux
transporters. PLoS One. 4:e72382009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu G, Pei F, Yang F, Li L, Amin AD, Liu
S, Buchan JR and Cho WC: Role of autophagy and apoptosis in
non-small-cell lung cancer. Int J Mol Sci. 18:pii: E3672017.
View Article : Google Scholar
|
|
21
|
Qian HR and Yang Y: Functional role of
autophagy in gastric cancer. Oncotarget. 7:17641–17651. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Talaiezadeh A, Jalali F, Galehdari H and
Khodadadi A: Time depended Bcl-2 inhibition might be useful for a
targeted drug therapy. Cancer Cell Int. 15:1052015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cai Y, Xu P, Yang L, Xu K, Zhu J, Wu X,
Jiang C, Yuan Q, Wang B, Li Y and Qiu Y: HMGB1-mediated autophagy
decreases sensitivity to oxymatrine in SW982 human synovial sarcoma
cells. Sci Rep. 6:378452016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
An X, Sarmiento C, Tan T and Zhu H:
Regulation of multidrug resistance by microRNAs in anti-cancer
therapy. Acta Pharm Sin B. 7:38–51. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
O'Donovan TR, O'Sullivan GC and McKenna
SL: Induction of autophagy by drug-resistant esophageal cancer
cells promotes their survival and recovery following treatment with
chemotherapeutics. Autophagy. 7:509–524. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kumar A, Singh UK and Chaudhary A:
Targeting autophagy to overcome drug resistance in cancer therapy.
Future Med Chem. 7:1535–1542. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li LQ, Xie WJ, Pan D, Chen H and Zhang L:
Inhibition of autophagy by bafilomycin A1 promotes chemosensitivity
of gastric cancer cells. Tumor Biol. 37:653–659. 2016. View Article : Google Scholar
|
|
28
|
Bhattacharya B, Low SH, Soh C, Mustapa N
Kamal, Beloueche-Babari M, Koh KX, Loh J and Soong R: Increased
drug resistance is associated with reduced glucose levels and an
enhanced glycolysis phenotype. Br J Pharmacol. 171:3255–3267. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang C and Pan Y: Fluorouracil induces
autophagy-related gastric carcinoma cell death through Beclin-1
upregulation by miR-30 suppression. Tumour Biol. Jul 25–2015.(Epub
ahead of print).
|
|
30
|
Liu J, Zhang Y, Qu J, Xu L, Hou K, Zhang
J, Qu X and Liu Y: β-Elemene-induced autophagy protects human
gastric cancer cells from undergoing apoptosis. BMC Cancer.
11:1832011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mathew R, Karantza-Wadsworth V and White
E: Role of autophagy in cancer. Nat Rev Cancer. 7:961–967. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lee J, Giordano S and Zhang J: Autophagy,
mitochondria and oxidative stress: Cross-talk and redox signalling.
Biochem J. 441:523–540. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhu H, Wu H, Liu X, Li B, Chen Y, Ren X,
Liu CG and Yang JM: Regulation of autophagy by a beclin 1-targeted
microRNA, miR-30a, in cancer cells. Autophagy. 5:816–823. 2009.
View Article : Google Scholar : PubMed/NCBI
|