Introduction
Lumbar disc herniation is a common disease that
initially manifests as lumbar back pain. It subsequently shifts to
the hips and lower limbs and may cause severe dysfunction of the
lower limbs and paralysis. The effects of the disease may therefore
cause a considerable economic burden on society (1,2). It has
been suggested that the gradual degeneration of the intervertebral
disc may be responsible (3,4). This initially occurs in the nucleus
pulposus (NP) and involves the loss of existing cells and changes
of the extracellular matrix (5). The
outer layer of the fibrous ring may change its normal lamellar
arrangement, progressively, cracks may begin to gradually emerge
from the inner part of the fiber ring to the outside, leading to a
change in the overall mechanics of the lumbar disk (6). These changes increase the force that
passes to the end of the vertebral body resulting in minor
fractures and marginal osteophyte formation (7). Certain environmental factors, including
weightlifting, vibration massage, trauma, smoking, diabetes,
cerebrovascular disease and infections may induce mechanical stress
or alter the metabolism of the intervertebral disc (8). Therefore, intervertebral disc
degeneration is a result of long-term effects by multiple damaging
factors and further research is required to fully understand the
potential causes and its molecular mechanisms of action.
Microarrays are high-throughput platforms used to
analyze gene expression, which may be used to examine a broad range
of signaling pathways and have a high degree of reliability
(9–11). In the present study the downregulated
miRNA miR-125b-1-3p was investigated. Follow-up experiments were
performed to assess the target genes of miR3150A.
The microRNA miR-125b-1-3p has diverse functions,
including in development, differentiation, cell proliferation and
apoptosis (12,13). Previous research has implied that it
may serve as a tumor suppressor or an oncogene (14); it is downregulated in malignancies
originating in the ovaries, bladder and breast, but upregulated in
leukemia, prostate cancer and glioma (15). At present the exact influence of
miR-125b-1-3p on NP cells remains unclear.
Homeobox genes serve key roles in the
standardization and patterning development in different parts of
the body (16,17). The human genome contains at ≥178
homeobox sequences, among which 160 genes may be translated into
homeodomains within functional proteins (18). Teashirt zinc finger homeobox 3
(TSHZ3) encodes a zinc-finger transcription factor and is highly
expressed in the developing human neocortex (19). TSHZ3 also serves a primary role in
smooth muscle formation (20) but
its specific functions remain ambiguous. The present study aimed to
determine whether TSHZ3 is a direct target of miR-125b-1-3p.
Furthermore, the potential association between miR-125b-1-3p, TSHZ3
and the degeneration of rat NP cells was investigated.
Materials and methods
Microarray data
In the present study, the gene expression profile
GSE63492 was downloaded from the GEO database. GSE63492 was based
on an Agilent GPL19449 platform which entitled: Exiqon miRCURY LNA
microRNA Array, 7th generation REV-hsa, mmu & rno (miRBase
v18.0), submitted by Liu et al (21). The GSE63492 dataset contained 10
samples in total, 5 of which were samples from patients with IDD
and the other 5 were tissues from individuals with normal discs. A
total of 134 micro (mi)RNAs (DEMs) have been identified as
differentially expressed in intervertebral disc degeneration (IDD)
by gene expression profiling studies using microarray technology
(22). MicroRNA (miR)3150A was
selected from the upregulated miRNAs and miR-125b-1-3p was selected
from the downregulated miRNAs as these were identified as the most
significant. Their target genes were predicted by finding
overlapping genes across different databases (Targetscan,
http://www.targetscan.org/; miRDB,
http://mirdb.org/ and DIANA, http://diana.imis.athena-innovation.gr/). It was
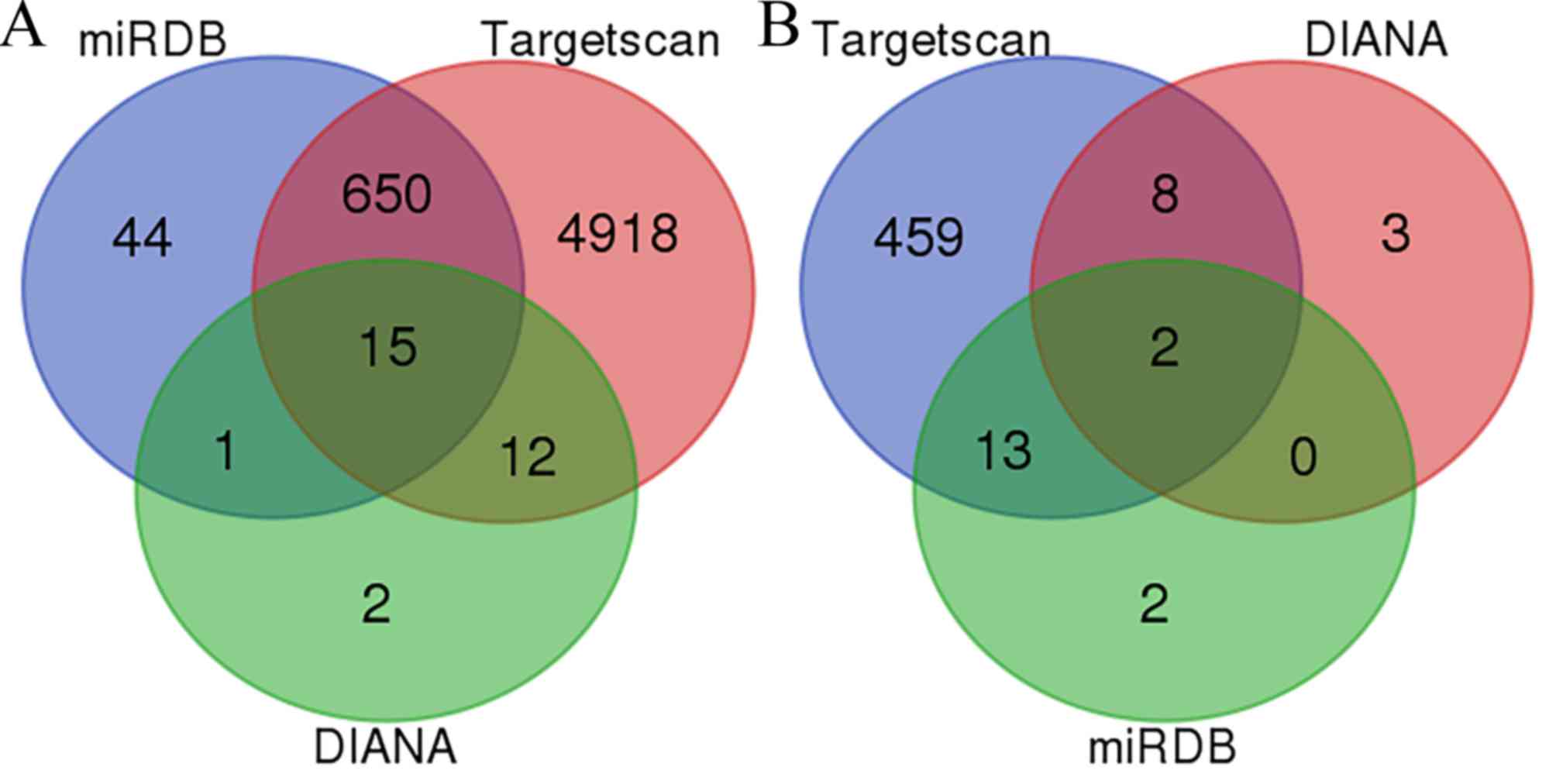
observed that miR3150A had 15 overlapping genes, whereas
miR-125b-1-3p had only two: GLRA2 and TSHZ3 (Fig. 1). The follow-up experiments were
performed to assess the target genes of miR3150A.
Cell culture and reagents
A total of 20 male Sprague Dawley rats (8 weeks old,
150±20 g) were obtained from Tongji University (Shanghai, China).
Rats were maintained in a room with a 12-h light/dark cycle
(temperature, 18–26°C; humidity, 40–70%) and had free access to
food (5 g for 100 g weight/24 h) and drinking water (8–11 ml for
100 g weight/24 h). Rats were euthanized prior to the isolation of
NP on a super-clean bench. NP tissues were minced to 1
mm3 and digested with trypsin for 25 min at 37°C.
Tissues were washed three times using phosphate buffered solution
(PBS) following centrifugation at 240 × g for 10 min at room
temperature and added to Dulbecco's Modified Eagle's medium (DMEM)
F-12 (HyClone, Laboratories; GE Healthcare Life Sciences, Logan,
UT, USA) containing 15% fetal bovine serum (FBS) (Clark Bioscience,
Richmond, VA, USA) and 0.25% collagenase type II (COL2;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 3 h at 37°C. The
tissues digested by collagenase type II were finally isolated in
15%FBS following three washes using phosphate buffered solution
(PBS) and centrifuged at 170 × g for 5 min at room temperature. The
NP cells were adherent and then cultured in DMEM F-12 with 15% FBS
(replaced every 3 days) in 5% CO2 at 37°C. When the
primary cells formed a monolayer, they were digested by trypsin and
passaged to the third and fifth generation for the follow-up
experiment.
The present study was performed according to
international, national and institutional rules concerning animal
experiments, clinical studies and biodiversity rights. The study
protocol was approved by the Laboratory Animal Welfare and Ethical
Committee of Shanghai Tenth People's Hospital affiliated to Tongji
University (Shanghai, China).
MiR-125b-1-3p inhibitor and TSHZ3
small interfering (si)RNA transfection
Cells were cultured in DMEM with F12 supplemented
with 10% FBS in a humidified incubator at 5% CO2 and
37°C. When they reached 70–80% confluence, cells were transfected
with Lipofectamine 2000® (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and either miR-125b-1-3p
inhibitor (Shanghai GeneChem Co., Ltd., Shanghai, China) or TSHZ3
siRNA (Genechem), following the manufacturer's protocol.
Luciferase assay
The wild type or mutant TSHZ3 3′-untranslated region
(UTR; Shanghai GeneChem Co., Ltd.) was cloned into the pmirGLO
luciferase reporter vector (Promega Corporation, Madison, WI, USA)
and co-transfected using Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) with either the miR-590-3p mimic or the
miR-negative control (NC; Shanghai GeneChem Co., Ltd.) into 293T
cells using Lipofectamine 2000®. The pRL-TK
Renilla luciferase reporter vector (Promega Corp., Madison,
WI, USA) was transfected into cells as an internal control. At 24 h
post-transfection, luciferase activity was measured using a
Dual-Luciferase® Reporter assay system (Promega Corp.),
following cell lysis with passive lysis buffer included in the
Reporter assay system. Relative luciferase activity was calculated
as the ratio of firefly luciferase activity to Renilla
luciferase activity.
Western blot analysis
NP cells were treated with different concentrations
of H2O2 (0.05, 0.1 and 0.2 mM/l) to simulate
DNA oxidative damage. Cells were collected at 0, 0.5, 1 and 2 h
following administration of H2O2. Total
proteins were extracted from the NP cells using
radioimmunoprecipitation assay lysis buffer (Beyotime Institute of
Biotechnology, Shanghai, China) for 30 min at 4°C. The supernatant
containing total protein was harvested and proteins were quantified
using the bicinchoninic acid method. Aliquots containing 50 µg
protein were separated by 12% SDS-PAGE and transferred to
polyvinylidene difluoride membranes at 60 V for 2 h at 4°C.
Membranes were then soaked in 5% blocking buffer, containing 25 mg
bovine serum albumin (Beyotime Institute of Biotechnology) in Tris
buffered saline (TBS) buffer to final volume of 0.5 l, at 4°C for 2
h and subsequently incubated with primary antibodies against TSHZ3
(cat. no. ab176117;), COL2 (cat. no. ab34712), cyclin D1 (cat. no.
ab134175), cyclin B1 (cat. no. ab2949; all Abcam, Cambridge, MA,
USA) at 1:5,000 dilution overnight at 4°C. This was followed by
incubation with goat anti-rabbit peroxidase-conjugated secondary
antibodies (cat. no. ab6721; Abcam) at 1:10,000 dilution for 2 h at
room temperature. The DNR imaging system (DNR Bio-Imaging Systems,
Ltd., Jerusalem, Israel) was used to visualize specific bands, and
the optical density of each band was measured using ImageJ software
(version 1.51; NIH, Bethesda, MD, USA). The ratio between the
target proteins and β-actin was calculated and presented
graphically.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) assay
Total RNA was extracted from NP cells using the
E.Z.N.A.® Total RNA Midi kit (Omega Bio-Tek, Inc.,
Norcross, GA, USA) following the manufacturer's protocol and
quantified spectrophotometrically at 260 nm with acceptable
CCLX/280 ratios between 1.8 and 2.0. RNA quality was determined by
1% agarose gel electrophoresis and staining with 1 µg/ml ethidium
bromide (room temperature, 5 min). qPCR was performed on
LightCycler® 480 High-Resolution Melting Master (Roche
Diagnostics, Basel, Switzerland) using SYBR Premix Ex
Taq™ II (Takara Biotechnology Co., Ltd., Dalian, China).
Specific primers for miR-125b-1-3p (forward primer,
5′ACGGGTTAGGCTCTTGG3′; and reverse primer, 5′CAGTGCGTGTCGTGGAGT3′)
and TSHZ3 (forward primer, 5′-GCAGCAGCCTATGTTTCCGATG-3′; and
reverse primer, 5′-GTAGCTAGGGCAGGCTTTGC-3′) were obtained from
Shanghai GeneChem Co., Ltd. Amplifications were performed in a
total reaction volume of 20 µl and cycled 40 times following
initial denaturation (95°C for 30 sec) using the following
parameters: 95°C for 5 sec and 60°C for 30 sec. β-actin (forward
primer, TCCTCCCTGGAGAAGAGCTA, and reverse primer,
TCAGGAGGAGCAATGATCTTG) were used as the internal controls. Analysis
of the melting curve supported the reliability of the results.
RT-qPCR data was quantified using the 2−ΔΔCq method
(23).
Statistical analysis
SPSS 20.0 software (IBM Corp., Armonk, NY, USA) was
used analyze the data. The one sample t-test was used to evaluate
the differences among different generations under the same
treatment factors. One-way analysis of variance which followed by
the student Newman-Keuls test was used to evaluate the differences
among different groups treated with different concentrations of
H2O2 and times. All data are presented as the
mean ± standard error of the mean and a minimum of three
independent repeats were performed for each experiment. P<0.05
was considered to indicate a statistically significant difference.
N-fold values in gene expression ≤0.5 and >2 were taken to be
significant, in accordance with values obtained from control
genes.
Results
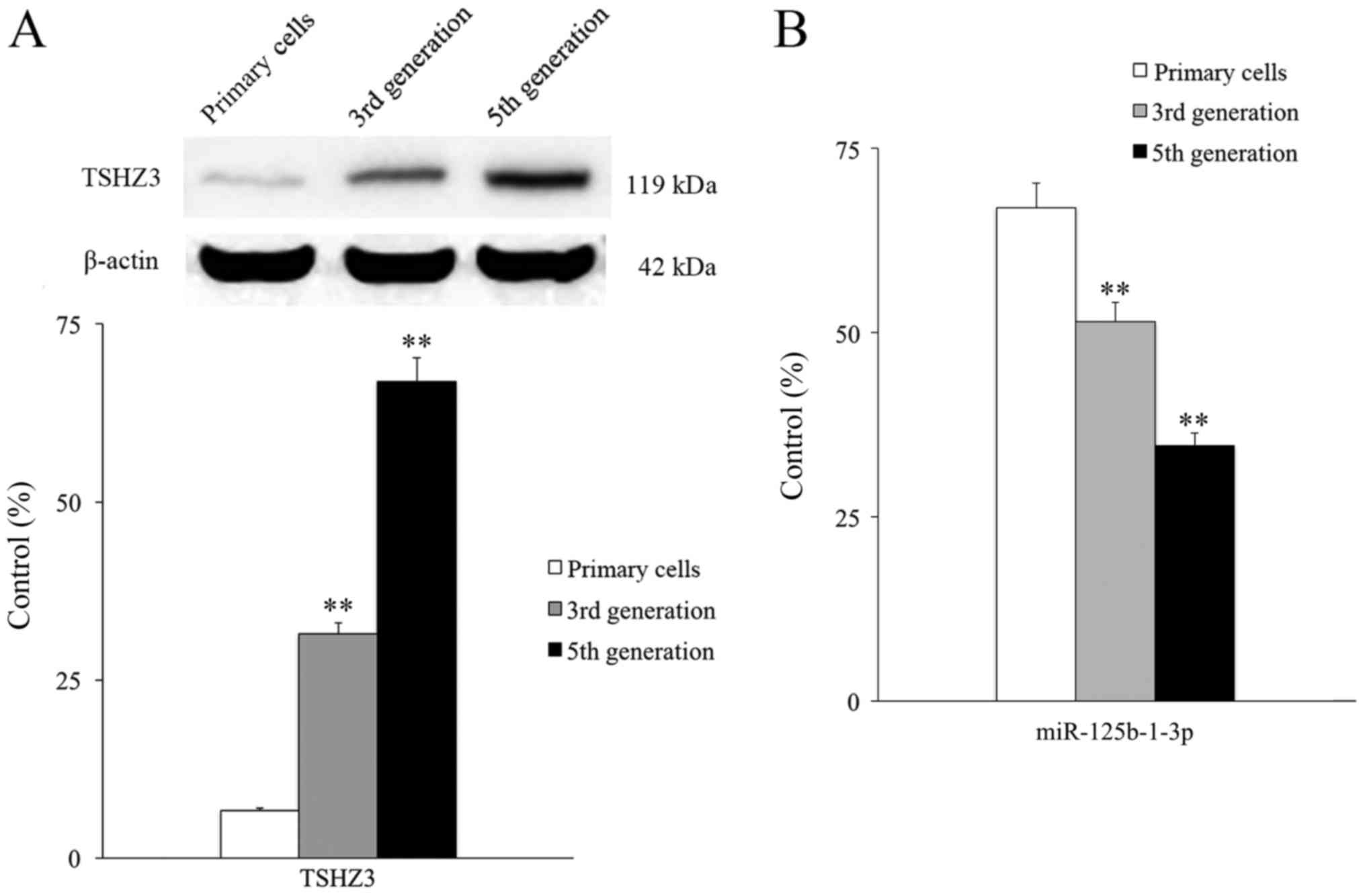
The expression of TSHZ3 was assessed by extracting
membrane proteins from normal rat NP cells at different passages
(primary cells, the third-generation cells and the fifth-generation
cells) and measuring the expression of TSHZ3 by western blotting.
Expression of TSHZ3 was low in primary NP cells, however TSHZ3
expression exhibited a passage-dependent increase; third and fifth
generation cells expressed TSHZ3 at a significantly higher level
than the primary cells (P<0.01; Fig.
2A). Expression levels of miR-125b-1-3p exhibited a
passage-dependent change and there were significant differences
observed between generations (Fig.
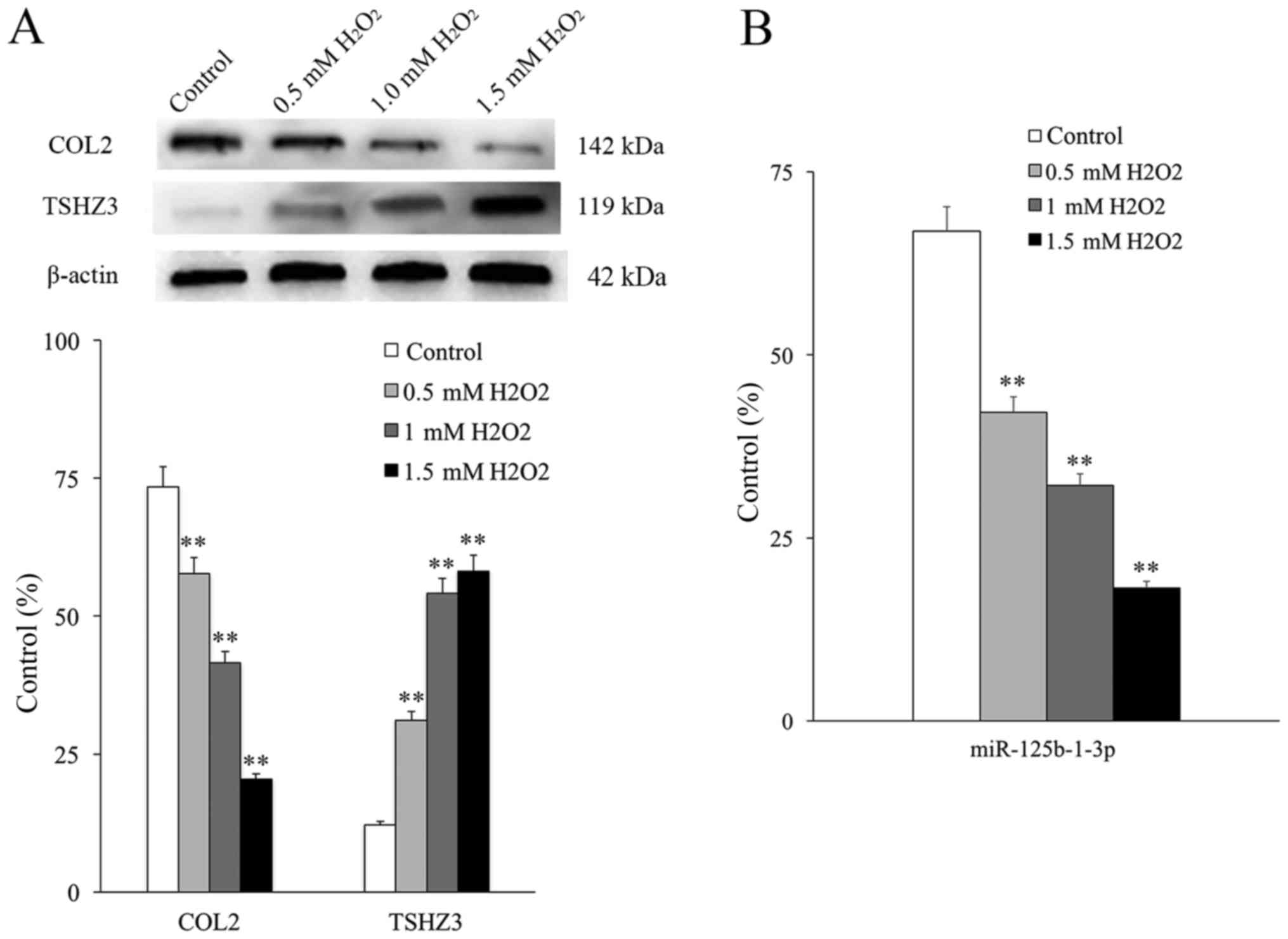
2B). When the NP cells were cultured in vitro to the
third generation, degeneration was clear and mainly characterized
by the markedly decreased expression level of COL2 (Fig. 3A).
It had been demonstrated that
H2O2 may stimulate DNA oxidative damage in
the NP cells of rats (24) and in
the present research a DNA oxidative damage model was established
by applying different concentrations of H2O2
(0.05, 0.1 and 0.2 mM/l). COL2 expression was used to monitor the
degeneration of NP cells, following a previously described protocol
(25). The expression of COL2
decreased as increasing concentrations of
H2O2 were used, whereas the expression of
TSHZ3 increased in an H2O2
concentration-dependent manner (Fig.
3A). The expression of COL2 was significantly decreased in all
groups compared with the control and the expression of TSHZ3 was
significantly increased in all groups compared with the control
(P<0.01). This indicates that TSHZ3 is associated with DNA
damage and degeneration in NP cells. The expression of
miR-125b-1-3p also decreased in an H2O2
concentration-dependent manner and was significantly decreased in
all groups compared with the control (P<0.01; Fig. 3B).
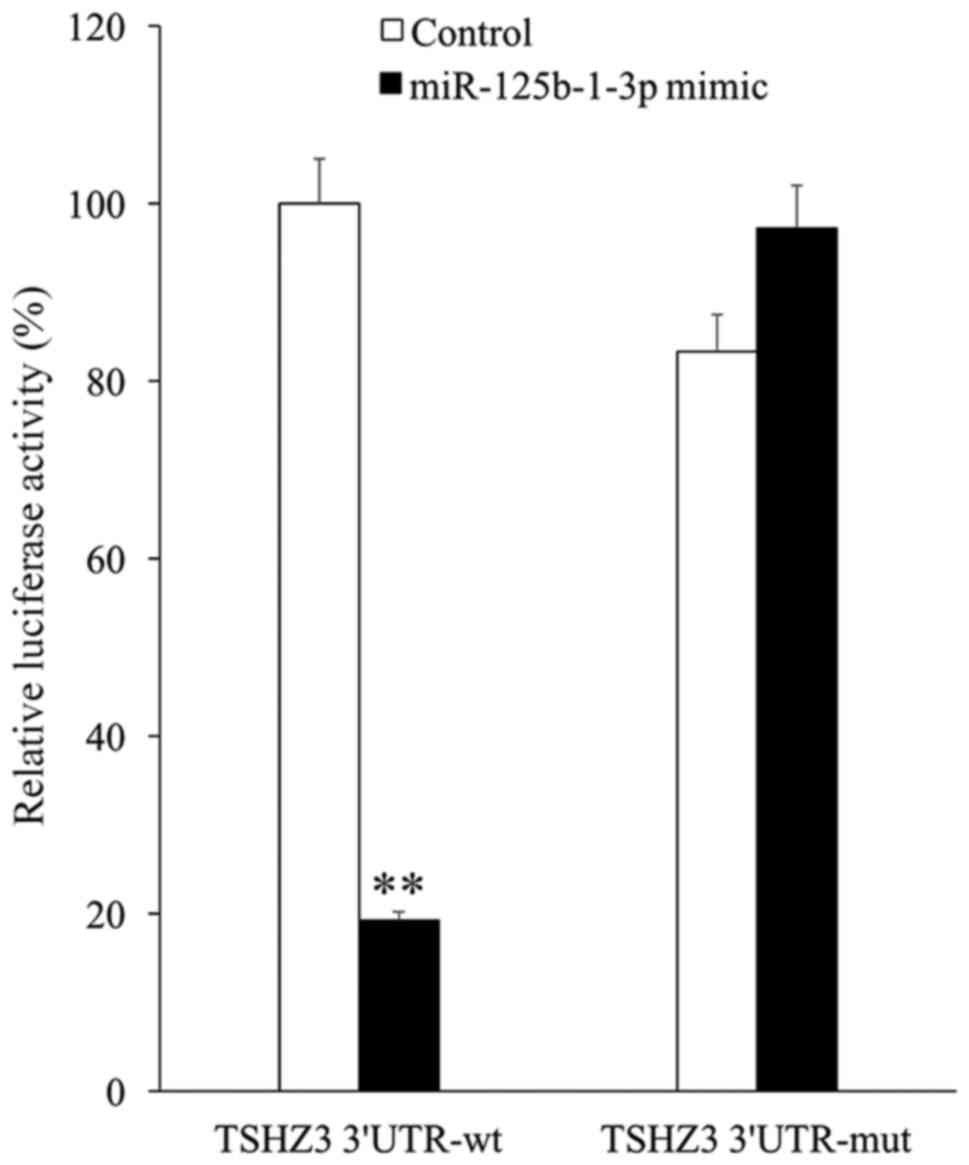
To elucidate the association between TSHZ3 and
miR-125b-1-3p, TSHZ3 3′UTRs containing the wild type or mutant
potential target site of miR-125b-1-3p were constructed and
co-transfected with the miR-125b-1-3p mimic into 293T cells. The
results of the luciferase reporter assay demonstrated that the
miR-125b-1-3p mimic significantly decreased the luciferase activity
of wild type TSHZ3 3′UTR compared with the control (P<0.01),
whereas the luciferase activity of the mutant TSHZ3 3′UTR was
unaffected by the miR-125b-1-3p mimic (Fig. 4).
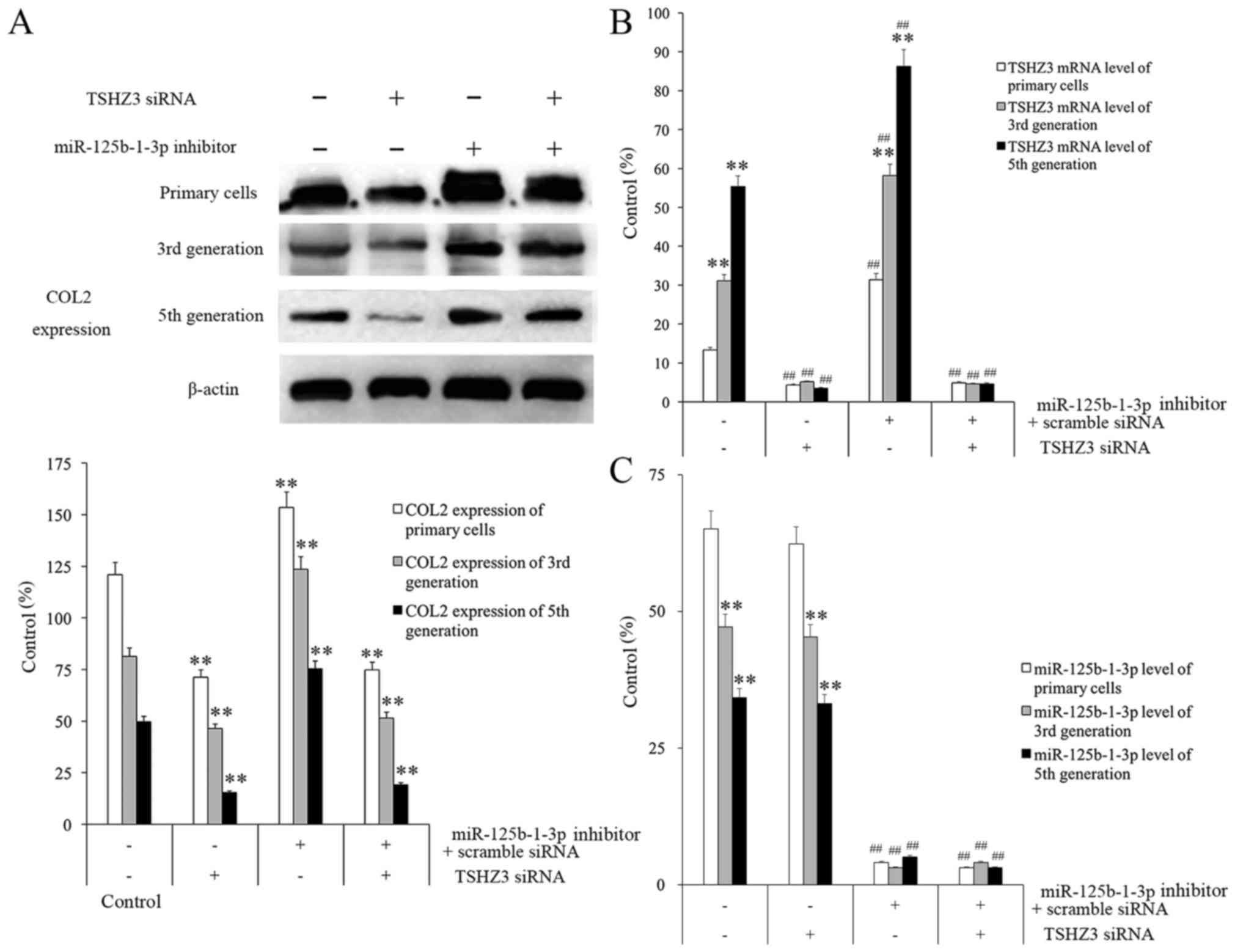
NP cells were treated with different combinations of
TSHZ3 siRNA and miR-125b-1-3p inhibitor and the results
demonstrated that, although COL2 expression decreased in all of the
groups in a passage-dependent manner, the overall expression levels
at each passage differed between each treatment group (Fig. 5A). The group treated with TSHZ3 siRNA
(alone or in combination with miR-125b-1-3p inhibitor) exhibited
the lowest expression level of COL2 in each generation among all of
the groups, whereas the group treated with the miR-125b-1-3p
inhibitor alone exhibited the highest COL2 expression in every
generation among all groups (Fig.
5A).
It was observed that NP cells treated with 60 nmol/l
miR-125b-1-3p inhibitor for 24 h had the highest TSHZ3 mRNA levels
and NP cells treated with only TSHZ3 siRNA had the highest
miR-125b-1-3p mRNA levels compared to negatively treated cells in
each group (Fig. 5B and C). It was
clear that the expression of TSHZ3 in the miR-125b-1-3p inhibiting
group increased significantly compared with the miR-125b-1-3p
inhibitor treated group (P<0.01; Fig.
5B). These results indicate that the expression of TSHZ3 in
normal NP cells may be an important factor protecting them against
degeneration and that miR-125b-1-3p inhibits its expression in
normal circumstances.
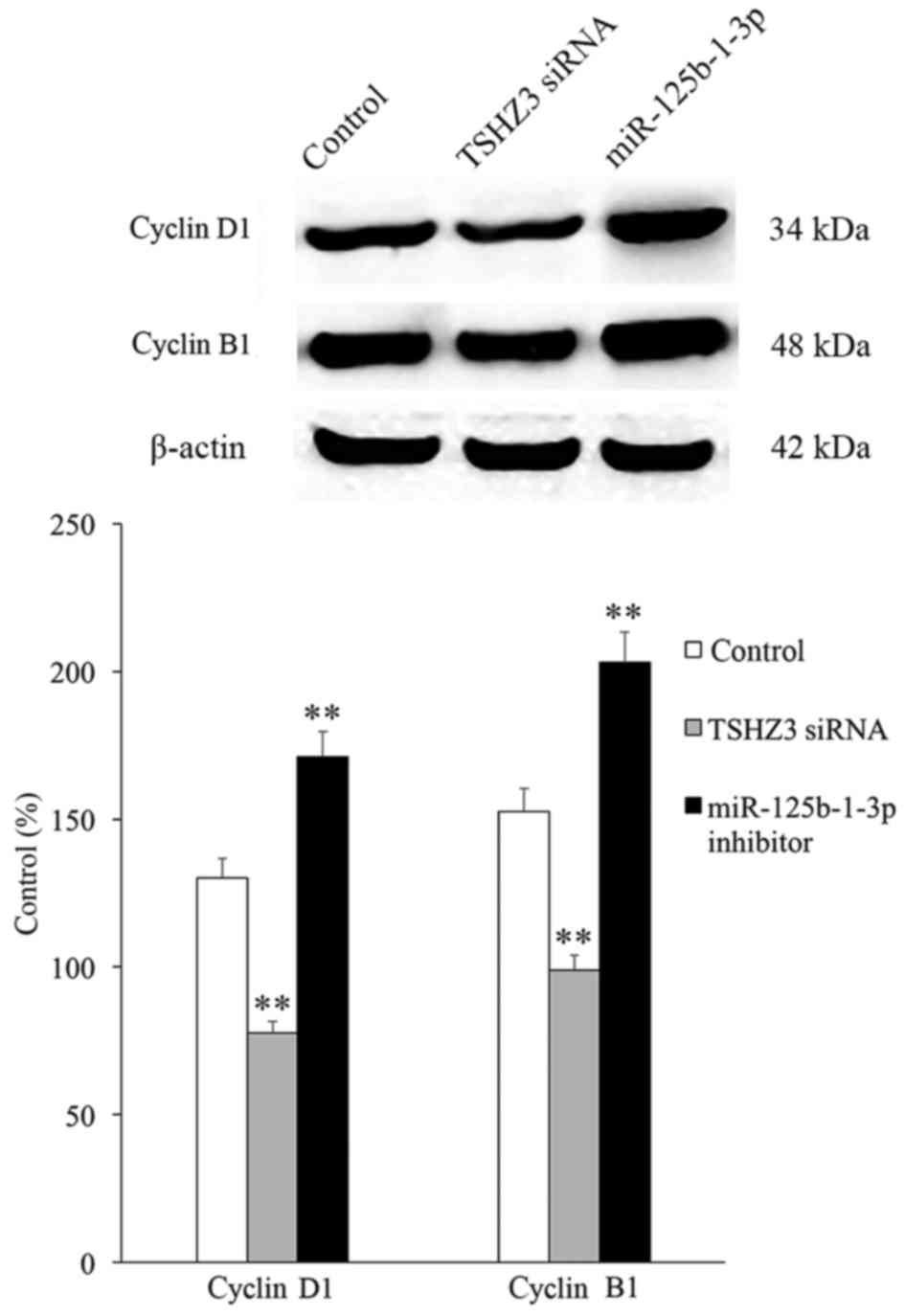
The expression of the cell cycle associated proteins
(cyclin D1 associated with G1 phase and cyclin B1
associated with G2/M phase) was investigated and the
results revealed that in the TSHZ3 siRNA treatment group, the
expression of cyclins D1 and B1 were significantly downregulated
compared with the control group (Fig.
6), which may induce G1 and G2/M phase
arrest and thus inhibit cell proliferation. By contrast, the
expression of cyclins D1 and B1 were significantly increased in the
miR-125b-1-3p treatment group compared with the control (Fig. 6).
Discussion
Degeneration of the intervertebral disc occurs due
to morphological and histological changes to the intervertebral
discs, as well as changes in the molecular biology of the NP, which
are induced by internal and external factors (26). The proliferation and apoptosis of NP
cells and the abnormal metabolism of the extracellular matrix
components are key causes of intervertebral disk degeneration
(27). The regeneration of normal NP
cells is closely associated with their proliferation ability and
maintenance of the extracellular matrix (28–31).
Therefore, maintaining NP cell activity and slowing down the
degeneration progress may be a novel method of preventing and
treating degenerative diseases affecting the intervertebral
discs.
Previous studies have demonstrated that certain
cytokines may activate degeneration by inducing apoptosis,
including by activating the tumor necrosis factor-associated
apoptosis inducing-ligand that belongs to the TNK superfamily
(3,4,32–34). In
addition, certain cytokines promote the proliferation and inhibit
the degeneration of NP cells, including insulin-like growth factor
and thymosin β-4 (35–37).
The primary function of the extracellular matrix of
the NP cells is to provide support and nutrients, whereas a
decrease in the amount of extracellular matrix increases the
susceptibility of NP cells to degeneration (5). Cytokines involved in the generation and
degradation of the extracellular matrix include tumor necrosis
factor ligand superfamily member, 12 transforming growth factor-β1,
matrix metalloproteinase and β-catenin (38–40).
COL2 is widely expressed in the extracellular matrix
of NP cells. It provides the fibrous scaffold to the cell and
therefore greatly influences the structure and function of NP cells
(41). It has been previously
reported that COL2 expression is decreased in degenerative NP cells
and this may be used as an index to evaluate the proliferation
capability and activity of NP cells (42). The results of the present study
demonstrate that, following treatment with different concentrations
of H2O2 for a specified period of time, the
expression of COL2 decreases significantly indicating that
degeneration is occurring.
When the NP cells were cultured in vitro to
the third generation, degeneration was clear and mainly
characterized by slow growth. In the present study, the expression
of COL2 in the NP cells decreased in a passage-dependent manner,
whereas the expression of TSHZ3 increased in a passage-dependent
manner. This indicates that during cellular senescence, TSHZ3 may
be a protective factor against degeneration.
The expression of miR-125b-1-3p exhibited no
significant changes in third and fifth generation cells compared
with the primary cells. It has been reported that miR-125b-1-3p
acts as a tumor suppressor or an oncogene (14) and has diverse functions, including on
cell development, differentiation, proliferation and apoptosis
(12,13). It has been demonstrated that
miR-125b-1-3p is downregulated in malignancies originating in the
ovary, bladder and breast but is upregulated in leukemia, prostate
cancer and glioma (15).
Additionally, the results of the luciferase reporter assay
demonstrated that TSHZ3 was a direct target of miR-125b-1-3p.
NP cells were treated with
H2O2 to stimulate DNA damage, as previously
documented by Zhou et al (24). Following treatment with increasing
concentrations of H2O2, the expression of
COL2 significantly decreased and the expression of TSHZ3 was
significantly increased compared with the control groups.
Therefore, it may be hypothesized that TSHZ3 also serves a role in
DNA repair.
TSHZ3 siRNA and the miR-125b-1-3p inhibitor were
administered alone or in combination to further investigate the
association between miR-125b-1-3p, TSHZ3 and NP cell
degeneration.
The expression of cell cycle associated proteins in
NP cells was determined by western blot analysis following TSHZ3
silencing. The results revealed that the expression of cyclin D1
associated with the G1 phase and cyclin B1 associated
with the G2/M phase of the cell cycle were significantly
decreased, indicating that the inhibition of TSHZ3 may induce
G1 and G2/M phase arrest, thus blocking the
proliferation of NP cells.
In conclusion, the results of the present study
demonstrate that TSHZ3, which is one of the target genes of
miR-125b-1-3p, may serve a protective role in the degeneration and
proliferation of NP cells in rats. This provides a novel molecular
mechanism by which to study and more effectively understand
intervertebral disc degeneration. However, further studies are
required to elucidate its underlying mechanisms of action more
comprehensively.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81472044) and the
Shenyang Science and Technology Program-Population and Health
Special (grant no. 17-230-9-04).
References
|
1
|
Millward-Sadler SJ, Costello PW, Freemont
AJ and Hoyland JA: Regulation of catabolic gene expression in
normal and degenerate human intervertebral disc cells: Implications
for the pathogenesis of intervertebral disc degeneration. Arthritis
Res Ther. 11:R652009. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Maniadakis N and Gray A: The economic
burden of back pain in the UK. Pain. 84:95–103. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Takada T, Nishida K, Doita M and Kurosaka
M: Fas ligand exists on intervertebral disc cells: A potential
molecular mechanism for immune privilege of the disc. Spine (Phila
Pa 1976). 27:1526–1530. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wiley SR, Schooley K, Smolak PJ, Din WS,
Huang CP, Nicholl JK, Sutherland GR, Smith TD, Rauch C, Smith CA,
et al: Identification and characterization of a new member of the
TNF family that induces apoptosis. Immunity. 3:673–682. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bachmeier BE, Nerlich A, Mittermaier N,
Weiler C, Lumenta C, Wuertz K and Boos N: Matrix metalloproteinase
expression levels suggest distinct enzyme roles during lumbar disc
herniation and degeneration. Eur Spine J. 18:1573–1586. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wilkie R and Pransky G: Improving work
participation for adults with musculoskeletal conditions. Best
Pract Res Clin Rheumatol. 26:733–742. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Freemont AJ, Watkins A, Le Maitre C, Baird
P, Jeziorska M, Knight MT, Ross ER, O'Brien JP and Hoyland JA:
Nerve growth factor expression and innervation of the painful
intervertebral disc. J Pathol. 197:286–292. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Roberts S, Evans EH, Kletsas D, Jaffray DC
and Eisenstein SM: Senescence in human intervertebral discs. Eur
Spine J. 15 Suppl 3:S312–S316. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kulasingam V and Diamandis EP: Strategies
for discovering novel cancer biomarkers through utilization of
emerging technologies. Nat Clin Pract Oncol. 5:588–599. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu J, Keisling MP, Samkari A, Halligan G,
Pascasio JM and Katsetos CD: Malignant glioma with primitive
neuroectodermal tumor-like component (MG-PNET): Novel microarray
findings in a pediatric patient. Clin Neuropathol. 35:353–367.
2016. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Decock A, Ongenaert M, Van Criekinge W,
Speleman F and Vandesompele J: DNA methylation profiling of primary
neuroblastoma tumors using methyl-CpG-binding domain sequencing.
Sci Data. 3:1600042016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Inui M, Martello G and Piccolo S: MicroRNA
control of signal transduction. Nat Rev Mol Cell Biol. 11:252–263.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li W, Xie L, He X, Li J, Tu K, Wei L, Wu
J, Guo Y, Ma X, Zhang P, et al: Diagnostic and prognostic
implications of microRNAs in human hepatocellular carcinoma. Int J
Cancer. 123:1616–1622. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee YS and Dutta A: MicroRNAs in cancer.
Annu Rev Pathol. 4:199–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gong J, Zhang JP, Li B, Zeng C, You K,
Chen MX, Yuan Y and Zhuang SM: MicroRNA-125b promotes apoptosis by
regulating the expression of Mcl-1, Bcl-w and IL-6R. Oncogene.
32:3071–3079. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
McGinnis W, Levine MS, Hafen E, Kuroiwa A
and Gehring WJ: A conserved DNA sequence in homoeotic genes of the
Drosophila Antennapedia and bithorax complexes. Nature.
308:428–433. 1984. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Scott MP and Weiner AJ: Structural
relationships among genes that control development: Sequence
homology between the Antennapedia, Ultrabithorax, and fushi tarazu
loci of Drosophila. Proc Natl Acad Sci USA. 81:pp. 4115–4119. 1984;
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Macdonald PM and Struhl G: A molecular
gradient in early Drosophila embryos and its role in specifying the
body pattern. Nature. 324:537–545. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Caubit X, Gubellini P, Andrieux J,
Roubertoux PL, Metwaly M, Jacq B, Fatmi A, Had-Aissouni L, Kwan KY,
Salin P, et al: TSHZ3 deletion causes an autism syndrome and
defects in cortical projection neurons. Nat Genet. 48:1359–1369.
2016. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Faralli H, Martin E, Coré N, Liu QC,
Filippi P, Dilworth FJ, Caubit X and Fasano L: Teashirt-3, a novel
regulator of muscle differentiation, associates with
BRG1-associated factor 57 (BAF57) to inhibit myogenin gene
expression. J Biol Chem. 286:23498–23510. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu X, Che L, Xie YK, Hu QJ, Ma CJ, Pei
YJ, Wu ZG, Liu ZH, Fan LY and Wang HQ: Noncoding RNAs in human
intervertebral disc degeneration: An integrated microarray study.
Genom Data. 5:80–81. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liang B, Li C and Zhao J: Identification
of key pathways and genes in colorectal cancer using bioinformatics
analysis. Med Oncol. 33:1112016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou X, Zhang HL, Gu GF, Ding Y, Jia JB,
Fu QS and He SS: Investigation of the relationship between
chromobox homolog 8 and nucleus pulposus cells degeneration in rat
intervertebral disc. In Vitro Cell Dev Biol Anim. 49:279–286. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kim KS, Yoon ST, Park JS, Li J, Park MS
and Hutton WC: Inhibition of proteoglycan and type II collagen
synthesis of disc nucleus cells by nicotine. J Neurosurg. 99(3
Suppl): S291–S297. 2003.
|
|
26
|
Cao Y, Liao S, Zeng H, Ni S, Tintani F,
Hao Y, Wang L, Wu T, Lu H, Duan C and Hu J: 3D characterization of
morphological changes in the intervertebral disc and endplate
during aging: A propagation phase contrast synchrotron
micro-tomography study. Sci Rep. 7:430942017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang J, Pan H, Li X, Zhang K, Li Z, Wang
H, Zheng Z and Liu H: Hypoxia suppresses serum deprivation-induced
degradation of the nucleus pulposus cell extracellular matrix
through the JNK and NF-κB pathways. J Orthop Res. 35:2059–2066.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Luoma K, Riihimäki H, Luukkonen R,
Raininko R, Viikari-Juntura E and Lamminen A: Low back pain in
relation to lumbar disc degeneration. Spine (Phila Pa 1976).
25:487–492. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sobajima S, Shimer AL, Chadderdan RC,
Kompel JF, Kim JS, Gilbertson LG and Kang JD: Quantitative analysis
of gene expression in a rabbit model of intervertebral disc
degeneration by real-time polymerase chain reaction. Spine J.
5:14–23. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jiricny J: The multifaceted
mismatch-repair system. Nat Rev Mol Cell Biol. 7:335–346. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Urban JP and Roberts S: Degeneration of
the intervertebral disc. Arthritis Res Ther. 5:120–130. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dai X, Zhang J, Arfuso F, Chinnathambi A,
Zayed ME, Alharbi SA, Kumar AP, Ahn KS and Sethi G: Targeting
TNF-related apoptosis-inducing ligand (TRAIL) receptor by natural
products as a potential therapeutic approach for cancer therapy.
Exp Biol Med (Maywood). 240:760–773. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Heyde CE, Tschoeke SK, Hellmuth M,
Hostmann A, Ertel W and Oberholzer A: Trauma induces apoptosis in
human thoracolumbar intervertebral discs. BMC Clin Pathol. 6:52006.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Buckwalter JA: Aging and degeneration of
the human intervertebral disc. Spine (Phila Pa 1976). 20:1307–1314.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pasold J, Zander K, Heskamp B, Grüttner C,
Lüthen F, Tischer T, Jonitz-Heincke A and Bader R: Positive impact
of IGF-1-coupled nanoparticles on the differentiation potential of
human chondrocytes cultured on collagen scaffolds. Int J
Nanomedicine. 10:1131–1143. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Niu M and Nachmias VT: Increased
resistance to apoptosis in cells overexpressing thymosin beta four:
A role for focal adhesion kinase pp125FAK. Cell Adhes Commun.
7:311–320. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tapp H, Deepe R, Ingram JA, Yarmola EG,
Bubb MR, Hanley EN Jr and Gruber HE: Exogenous thymosin beta4
prevents apoptosis in human intervertebral annulus cells in vitro.
Biotech Histochem. 84:287–294. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Huh H, Lee YJ, Kim JH, Kong MH, Song KY
and Choi G: The effects of TWEAK, Fn14, and TGF-beta1 on
degeneration of human intervertebral disc. J Korean Neurosurg Soc.
47:30–35. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Reno F, Sabbatini M, Stella M, Magliacani
G and Cannas M: Effect of in vitro mechanical compression on
Epilysin (matrix metalloproteinase-28) expression in hypertrophic
scars. Wound Repair Regen. 13:255–261. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gruber HE, Ingram JA, Hoelscher GL,
Zinchenko N, Norton HJ and Hanley EN Jr: Matrix metalloproteinase
28, a novel matrix metalloproteinase, is constitutively expressed
in human intervertebral disc tissue and is present in matrix of
more degenerated discs. Arthritis Res Ther. 11:R1842009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yokoyama K, Hiyama A, Arai F, Nukaga T,
Sakai D and Mochida J: C-Fos regulation by the MAPK and PKC
pathways in intervertebral disc cells. PLoS One. 8:e732102013.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lohmander LS, Neame PJ and Sandy JD: The
structure of aggrecan fragments in human synovial fluid. Evidence
that aggrecanase mediates cartilage degradation in inflammatory
joint disease, joint injury, and osteoarthritis. Arthritis Rheum.
36:1214–1222. 1993. View Article : Google Scholar : PubMed/NCBI
|