Introduction
As a kind of anti-apoptosis protein, survivin is
critical to cell apoptosis (1).
Also, survivin is a member in inhibitor of apoptosis protein (IAP)
family, and was separated via hybridization screening of cDNA of
the effector cell protease receptor-1 (EPR-1) with the human genome
library by Chen (2). Survivin is
specifically expressed in the G2/M phase (3), and correlated with the inhibition of
cell apoptosis (2–4), cell cycle regulation (5,6) and drug
resistance of tumors (7–9). Over the years, studies of survivin
mostly focus on the solid tumors (10) and the leukemia of adults, and few
studies reported acute leukemia (AL) of children, especially those
on the polymorphism level. Single nucleotide polymorphism (SNP) can
result in the variation in transcriptional activity of genes, and
some SNPs in coding sequence of protein can affect the amino acid
sequence of the key genes after translation, thereby altering the
protein functions and further affecting the susceptibility of
individuals to the disease (11). In
this study, we investigated the correlation between the SNP of
survivin and the AL of children, so as to provide theoretical
support for prophylaxis of leukemia.
Materials and methods
Subjects
We collected 100 peripheral blood specimens from AL
children (12), and 82 peripheral
blood specimens from the children with acute myelogenous leukemia
(AML) (13). This study was approved
by the Ethics Committee of SunYat-Sen University (Guangdong,
China). Signed informed consents were obtained from the patients'
guardians. In these children, there were 101 males and 81 females
aged between 45 days and 18 years (median age of 7.55 years). In
addition, the peripheral blood samples collected from 200 healthy
blood donators were enrolled as the control group. All blood
samples were treated with heparin sodium and preserved at
−80°C.
Extraction of whole blood genomic
DNA
In accordance with the methods in literature, we
extracted the genomic DNA from 3 ml of blood, and assayed the
concentration and purity of DNA with ultraviolet spectrometer and
through agarose electrophoresis, in which the concentration of DNA
was adjusted to 100 µg/ml. Thereafter, DNA was preserved at −20°C
for later use.
Identification of survivin SNPs
After reviewing the relevant literature, we found
that the family of glutathione S-transferases (GSTs) (14) is involved in antagonizing the
toxicity of mutagens, which is correlated with the distribution of
GSTM1 polymorphism in the population.
Detection of the genotypes of GSTM1
and GSTT1
According to the nucleotide sequence of human GSTM1
and GSTT1 in National Center of Biotechnology Information (NCBI)
database, the primers were designed with primer 5. GSTM1 forward,
5′-GACTCCCTGAAAAGCTAAAGC-3′ and reverse,
5′-GTTGGGCTCAAATATACGGTCG-3′; GSTT1 forward,
5′-TTCCTTACTGGTCCTCACATCTC-3′ reverse, 5′-TCACCGGATCATGGCCAGCA-3′.
Albumin gene was used as the internal reference (forward,
5′-GCCCTCTGCTAACAAGTCCTAC-3′ and reverse,
5′-GCCCTAAAAAGAAAATCGCCAATC-3′). Reaction was carried out in a 50
µl amplification system [25 µl Ex Taq + 19 µl ddH2O + 2
µl forward primer (10 mM) and 2 µl reverse primer (10 mM) + 2 µl
genomic DNA] under the following reaction conditions: 94°C for 5
min followed by a total of 30 cycles (94°C for 30 sec, 56°C for 45
sec and 72°C for 1 min) and extension at 72°C for 10 min, in which
water was added in the negative control group. Five microliters of
amplification product was taken for 1.0 agarose electrophoresis
(300 V, 15 min) and stained with ethidium bromide (EB) followed by
photographing with a gel imaging analyzer and image
preservation.
Statistical analysis
Chi-square test was performed for comparison of data
with the GSTM1/T1 homozygous deletion genotype as control, and we
calculated the odds ratio (OR) and 95% confidence interval (95%
CI).
Identification of the alleles C and G
in SNPs of rs9904341C/G and rs8073069C/G in survivin
In this study, 182 AL children 15 and 200 healthy
volunteers were enrolled as subjects, and in accordance with the
case-control method, we extracted 5 ml peripheral venous blood and
recorded the material of these subjects. Phenol extraction method
was applied in DNA extraction from peripheral blood, and genetic
typing was performed for SNPs, rs9904341C/G and rs8073069C/G
(15–18), through polymerase chain
reaction-ligase detection reaction (PCR-LDR) (19). Primer of rs9904341C/G on survivin
forward, 5′-CGCCTCTACTCCCAGAAGGC-3′ and reverse,
5′-GAGATGCGGTGGTCCTTGAGAA-3′. Primer of rs8073069C/G on survivin
forward, 5′-TCGTGCAGTCAACGATGTACT-3′ and reverse,
5′-ACAGGAGAGCTTTACAGGGTG-3′.
PCR reaction system for PCR-LDR products of
rs9904341 and rs8073069: 2 µl PCR products + 1 µl 10X Taq DNA
ligase buffer + 0.125 µl Taq DNA ligase (40 U/µl) + 10 µl probe (10
bp, 0.01 µl/probe and diluted to 10 µl with water). Reaction
conditions: denaturation at 95°C for 3 min followed by 30 cycles
with two temperatures in each cycle, 94°C for 30 sec and 56°C for 3
min.
Sequencing
Sequencing was performed on ABI3730XL sequencer
(SeqGen; Inc., Torrance, CA, USA). Briefly, 1 µl ligation product
was added into 10 µl loading buffer with the corresponding marker
followed by denaturation at 95°C for 3 min. Thereafter, the product
was placed in the ice bath and used for data analysis with
GeneMapper® Software (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA).
Results
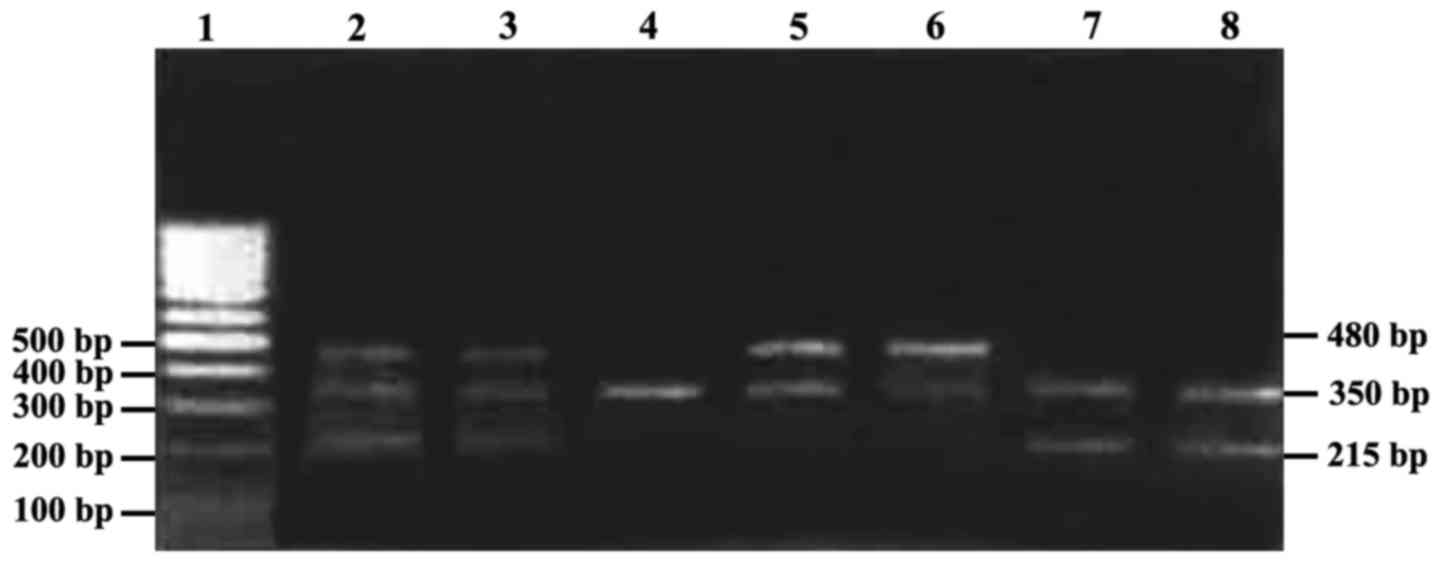
Results of electrophoresis are shown in Fig. 1. The amplification fragments of
GSTM1, GSTT1 (20) and Albumin were
215, 480 and 350 bp, respectively. As for GSTM1, homozygous
deletion was recorded as GSTM/T1-0, and heterozygous deletion as
GSTM-1. In the AL children, homozygous deletion rate of GSTM1/T1
was significantly higher than that in the control group (case
group, 76.12 vs. 52.74%, OR=2.856, 95% CI=1.493–5.465, P=0.001;
control group, 50.74 vs. 24.66%, OR=3.148, 95% CI=1.712–5.789,
P=0.0001); in AML children, homozygous deletion rate of GSTM1 was
significantly higher than that in the control group (71.88 vs.
52.74%, OR=2.290, 95% CI=0.992–5.285, P=0.048); comparisons of the
homozygous deletion rates between the control group (49.32%) and
the AL patients (62.29%) and the AML patients (59.38%) showed no
statistically significant difference (P>0.05).
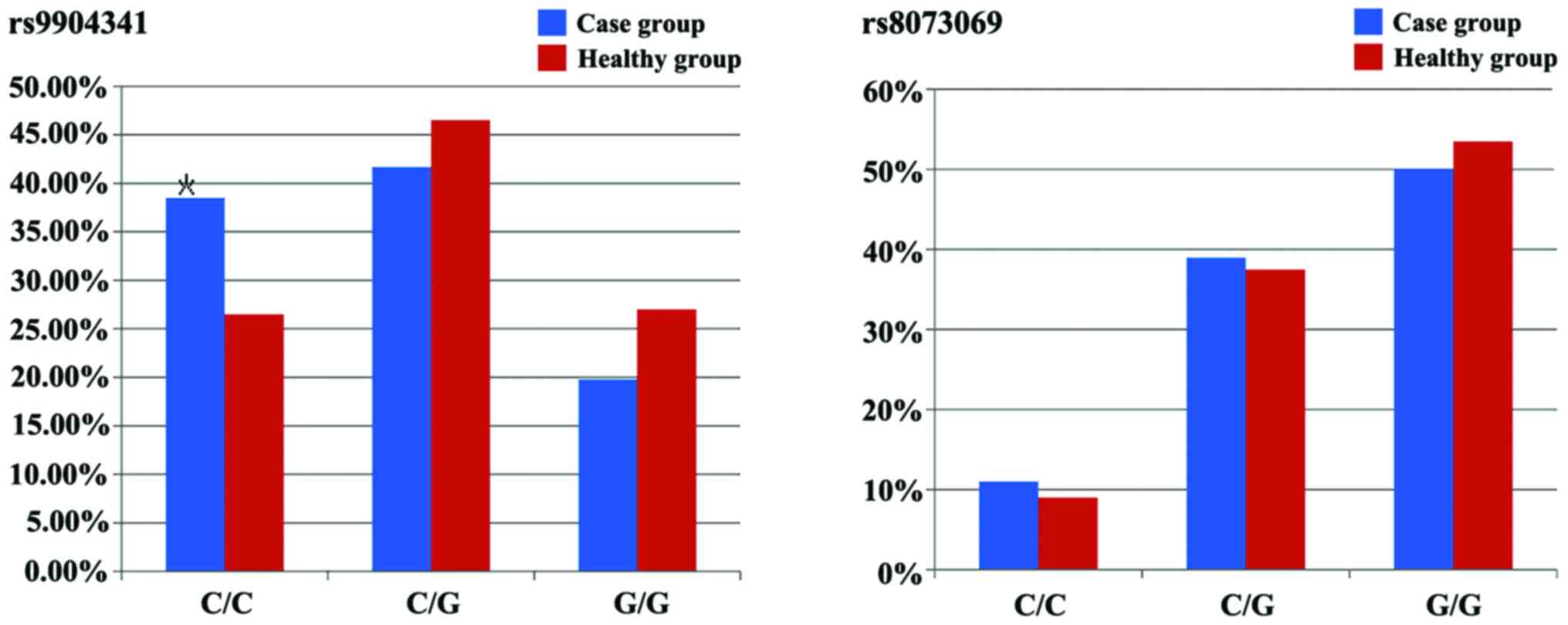
Alleles C and G in rs9904341C/G of survivin. In the
case group and healthy group, the frequencies of C and G alleles
wer e 59.3 and 41.7%, 46.7 and 50.3%, respectively, and the
coherence comparison showed that the difference had statistical
significance (P=0.008). In the case group and healthy group, the
frequencies of C/C, C/G and G/G were 38.5, 41.7, 19.8% and 26.5,
46.5, 27.0%, respectively, and the comparison between the two
groups showed that the differences had statistical significance
(P=0.033). The frequency of C/C genotype (38.5%) in the case group
was significantly higher than that (26.5%) in the healthy group,
and compared with C/C genotype, C/G and G/G genotypes could
decrease the risk coefficient of leukemia.
In the case group and healthy group, the frequencies
of C and G alleles on rs8073069C/G in survivin were 30.5 and 69.5,
27.7 and 72.3%, respectively, and the comparison showed that the
difference was not statistically significant (P=0.404). In the case
group and healthy group, the frequencies of C/C, C/G and G/G were
11, 39.0, 50.0% and 9.0, 37.5, 53.5%, respectively, and the
comparison between the two groups showed that the differences were
not statistically significant (P=0.62). Compared with C/C genotype,
C/G and G/G genotypes showed no influences on the risk of leukemia
(Figs. 2 and 3).
Comparison via ABI3730XL sequencer (SeqGen, Inc.,
Torrance, CA, USA) showed a linkage disequilibrium in the SNPs of
survivin, rs9904341 and rs8073069 (D'=0.628). The most common
haplotype was rs9904341C-8073069G, and its frequency in the healthy
group was 49.8% following the rs9904341G-8073069C (27.5%),
rs9904341G-8073069G (22.5%) and rs9904341C-8073069C (0.2%). In
population, the haplotype of rs9904341C-8073069C may result in an
increase in risk of onset, and compared with its haplotype, the
onset risk of AL could be dramatically decreased in population with
rs9904341G-8073069C (Table I).
 | Table I.Comparison of the haplotypes of
rs9904341 and rs8073069 on survivin between the healthy group and
the case group. |
Table I.
Comparison of the haplotypes of
rs9904341 and rs8073069 on survivin between the healthy group and
the case group.
| Haplotype | Healthy group
(%) | Case group (%) |
|---|
|
rs9904341C-8073069C | 1
(0.2) | 44
(12.1) |
|
rs9904341C-8073069G | 199 (49.8) | 172 (47.3) |
|
rs9904341G-8073069C | 110 (27.5) | 67
(18.4) |
|
Rs9904341G-8073069G | 90
(22.5) | 81
(22.3) |
Discussion
As known, the incidence rate of AL in children is
increasing year by year, and AL has become the prevalent malignant
tumor in children. The development of AL in children is regulated
by multiple genes and affected by a variety of cytokines (21). Survivin, a new member of the
anti-apoptosis protein family, is a protein with the smallest
relative molecular weight in this family, it can directly or
indirectly modulate the activity of asparagine-specific proteinase
through regulating the cell proliferation, thereby controlling cell
apoptosis; survivin is mainly distributed in the malignant tumor
tissues, and scarcely or not expressed in the normal tissues
(22). Survivin gene exhibits
tremendous effect on structural variation of amino acid. For
example, in α-helical structure of survivin protein, the
replacement of lysine (an essential basic amino acid) by glutamic
acid (an acidic amino acid with two carboxyl groups) can result in
the two kinds of variations in biological functions: First, it can
lead to alteration in focal spatial structure; second, it can
induce an increase in the number of phosphorylation sites (23,24).
SNP refers to the polymorphism in DNA sequence
generated by the mutation of a nucleotide in the chromosome
genomes, including the conversion and transversion. Specifically,
it involves the changes between the purines and pyrimidines
(25,26). SNPs have been used as the 3rd
generation of molecular marker with promising development potential
(27). At present, increasing number
of studies have focused on SNPs (28,29). For
example, SNPs in promotor regions of 31C/G, −24T/C, −625G/C,
−644T/C and 1547A/G, can affect the genetic predisposition of
tumors (30). Studies have confirmed
that the abnormal polymorphism of survivin may be closely related
to the metastasis and prognosis of malignant tumors.
In conclusion, the results of this study showed that
SNP of rs9904341C/G on survivin may affect the onset risk of AL,
and patients with C/C genotype may suffer from an increase in onset
of risk. Compared with C/C genotype, C/G and G/G genotype can
significantly reduce the onset risk of AL. The results on the
rs8073069C/G SNP suggested that this SNP is not correlated with the
onset risk of AL. In conclusion, the study on SNPs of survivin can
be used for assessment of disease condition and prognosis of AL in
children, which can serve as the evidence base for clinical
practice.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Deveraux QL, Takahashi R, Salvesen GS and
Reed JC: X-linked IAP is a direct inhibitor of cell-death
proteases. Nature. 388:300–304. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen G: The relationship between the
expression of TAM, survivin and the degree of necrosis of the tumor
after cisplatin treatment in osteosarcoma. Eur Rev Med Pharmacol
Sci. 21:490–497. 2017.PubMed/NCBI
|
|
3
|
Musacchio A: Spindle assembly checkpoint:
The third decade. Philos Trans R Soc Lond B Biol Sci.
366:3595–3604. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kuo ML, Shen SC, Yang CH, Chuang SE, Cheng
AL and Huang TS: Bcl-2 prevents topoisomerase II inhibitor
GL331-induced apoptosis is mediated by down-regulation of
poly(ADP-ribose)polymerase activity. Oncogene. 17:2225–2234. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
O'Connor DS, Grossman D, Plescia J, Li F,
Zhang H, Villa A, Tognin S, Marchisio PC and Altieri DC: Regulation
of apoptosis at cell division by p34cdc2 phosphorylation of
survivin. Proc Natl Acad Sci USA. 97:pp. 13103–13107. 2000;
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shin S, Sung BJ, Cho YS, Kim HJ, Ha NC,
Hwang JI, Chung CW, Jung YK and Oh BH: An anti-apoptotic protein
human survivin is a direct inhibitor of caspase-3 and −7.
Biochemistry. 40:1117–1123. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Suzuki A, Ito T, Kawano H, Hayashida M,
Hayasaki Y, Tsutomi Y, Akahane K, Nakano T, Miura M and Shiraki K:
Survivin initiates procaspase 3/p21 complex formation as a result
of interaction with Cdk4 to resist Fas-mediated cell death.
Oncogene. 19:1346–1353. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Olie RA, Simões-Wüst AP, Baumann B, Leech
SH, Fabbro D, Stahel RA and Zangemeister-Wittke U: A novel
antisense oligonucleotide targeting survivin expression induces
apoptosis and sensitizes lung cancer cells to chemotherapy. Cancer
Res. 60:2805–2809. 2000.PubMed/NCBI
|
|
9
|
Tamm I, Wang Y, Sausville E, Scudiero DA,
Vigna N, Oltersdorf T and Reed JC: IAP-family protein survivin
inhibits caspase activity and apoptosis induced by Fas (CD95), Bax,
caspases, and anticancer drugs. Cancer Res. 58:5315–5320.
1998.PubMed/NCBI
|
|
10
|
Kappler M, Köhler T, Kampf C,
Diestelkötter P, Würl P, Schmitz M, Bartel F, Lautenschläger C,
Rieber EP, Schmidt H, et al: Increased survivin transcript levels:
An independent negative predictor of survival in soft tissue
sarcoma patients. Int J Cancer. 95:360–363. 2001.PubMed/NCBI
|
|
11
|
Pui CH and Evans WE: Treatment of acute
lymphoblastic leukemia. N Engl J Med. 354:166–178. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Campana D, Coustan-Smith E, Manabe A,
Buschle M, Raimondi SC, Behm FG, Ashmun R, Aricò M, Biondi A and
Pui CH: Prolonged survival of B-lineage acute lymphoblastic
leukemia cells is accompanied by overexpression of bcl-2 protein.
Blood. 81:1025–1031. 1993.PubMed/NCBI
|
|
13
|
Flotho C, Coustan-Smith E, Pei D, Iwamoto
S, Song G, Cheng C, Pui CH, Downing JR and Campana D: Genes
contributing to minimal residual disease in childhood acute
lymphoblastic leukemia: Prognostic significance of CASP8AP2. Blood.
108:1050–1057. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jaskoll T, Chen H, Min Zhou Y, Wu D and
Melnick M: Developmental expression of survivin during embryonic
submandibular salivary gland development. BMC Dev Biol. 1:52001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pui CH, Relling MV and Downing JR: Acute
lymphoblastic leukemia. N Engl J Med. 350:1535–1548. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Krajinovic M, Labuda D, Richer C, Karimi S
and Sinnett D: Susceptibility to childhood acute lymphoblastic
leukemia: Influence of CYP1A1, CYP2D6, GSTM1, and GSTT1 genetic
polymorphisms. Blood. 93:1496–1501. 1999.PubMed/NCBI
|
|
17
|
Canalle R, Burim RV, Tone LG and Takahashi
CS: Genetic polymorphisms and susceptibility to childhood acute
lymphoblastic leukemia. Environ Mol Mutagen. 43:100–109. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lanciotti M, Dufour C, Corral L, Di
Michele P, Pigullo S, De Rossi G, Basso G, Leszl A, Luciani M, Lo
Nigro L, et al: Genetic polymorphism of NAD(P)H:quinone
oxidoreductase is associated with an increased risk of infant acute
lymphoblastic leukemia without MLL gene rearrangements. Leukemia.
19:214–216. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bolufer P, Barragan E, Collado M, Cervera
J, López JA and Sanz MA: Influence of genetic polymorphisms on the
risk of developing leukemia and on disease progression. Leuk Res.
30:1471–1491. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Aydin-Sayitoglu M, Hatirnaz O, Erensoy N
and Ozbek U: Role of CYP2D6, CYP1A1, CYP2E1, GSTT1, and GSTM1 genes
in the susceptibility to acute leukemias. Am J Hematol. 81:162–170.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
d'Errico A, Mamo C, Costa G, Filippi M and
Crosignani P: Use of pension records for occupational health
surveillance: Example of record-linkage with hospital discharge
records to study the association between work and the incidence of
leukaemias, lung and bladder cancer, and miscarriage. Med Lav.
96(Suppl): s147–s160. 2005.(In Italian). PubMed/NCBI
|
|
22
|
LaCasse EC, Baird S, Korneluk RG and
MacKenzie AE: The inhibitors of apoptosis (IAPs) and their emerging
role in cancer. Oncogene. 17:3247–3259. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gilbert ME and Shafer TJ: In vitro
exposure to aluminum does not alter long-term potentiation or
glutamate release in rat hippocampal slices. Neurotoxicol Teratol.
18:175–180. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Takashima A, Noguchi K, Michel G, Mercken
M, Hoshi M, Ishiguro K and Imahori K: Exposure of rat hippocampal
neurons to amyloid beta peptide (25–35) induces the inactivation of
phosphatidyl inositol-3 kinase and the activation of tau protein
kinase I/glycogen synthase kinase-3 beta. Neurosci Lett. 203:33–36.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mullikin JC, Hunt SE, Cole CG, Mortimore
BJ, Rice CM, Burton J, Matthews LH, Pavitt R, Plumb RW, Sims SK, et
al: An SNP map of human chromosome 22. Nature. 407:516–520. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Altshuler D, Pollara VJ, Cowles CR, Van
Etten WJ, Baldwin J, Linton L and Lander ES: An SNP map of the
human genome generated by reduced representation shotgun
sequencing. Nature. 407:513–516. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gray IC, Campbell DA and Spurr NK: Single
nucleotide polymorphisms as tools in human genetics. Hum Mol Genet.
9:2403–2408. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Carlson CS, Eberle MA, Rieder MJ, Yi Q,
Kruglyak L and Nickerson DA: Selecting a maximally informative set
of single-nucleotide polymorphisms for association analyses using
linkage disequilibrium. Am J Hum Genet. 74:106–120. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nowotny P, Kwon JM and Goate AM: SNP
analysis to dissect human traits. Curr Opin Neurobiol. 11:637–641.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jang JS, Kim KM, Kang KH, Choi JE, Lee WK,
Kim CH, Kang YM, Kam S, Kim IS, Jun JE, et al: Polymorphisms in the
survivin gene and the risk of lung cancer. Lung Cancer. 60:31–39.
2008. View Article : Google Scholar : PubMed/NCBI
|