Introduction
Pancreatic cancer (PC), with almost as many deaths
(n=432,000) as cases (n=459,000), is a highly lethal malignancy and
the seventh leading cause of cancer-associated mortality in 2018
worldwide (1). Indeed, the incidence
of PC continues to rise, and it is predicted to become the second
leading cause of cancer-associated mortality by the year 2030 in
the United States (2). The poor
prognosis of PC is attributed to its asymptomatic nature until the
late stages and invasive features, with a <20% chance of being
operable at the time of diagnosis (3). Furthermore, >50% of cases of early
PC after surgery encounter recurrences within 12 months (4), with a 5-year survival rate of up to 45%
after curative surgery (5). The poor
prognosis of PC is due to most cases of PC being diagnosed at the
late stage when they have missed the time window for curative
surgery (3) and the fact that its
molecular pathogenesis remains largely elusive. To improve outcomes
for patients with PC, investigation into prognostic biomarkers and
their molecular mechanisms in the early stages of disease is
imperative to prolong patient survival. Using reliable biomarkers,
high-risk patients may be followed up more frequently after surgery
instead of being dictated by the routine schedule.
MicroRNAs (miRNAs/miRs), a class of short RNAs
(6), are promising prognostic
predictors for various types of cancer (7–9),
including PC (7). miRNAs have
crucial roles in transcriptional regulation of gene expression via
several mechanisms (10). Competing
endogenous RNAs (ceRNAs) constitute one of these mechanisms. This
type of RNA crosstalk exists between protein-coding mRNAs and
non-coding RNAs, including miRNAs and long non-coding RNAs
(lncRNAs) (11–13). Various studies have proposed
miRNA-based signatures for prognosis prediction for PC (14–16);
however, few of them focused on early PC and corresponding
potential ceRNA networks.
In the present study, the mature miRNA expression
profiles and clinical information of cases of early PC in The
Cancer Genome Atlas (TCGA) database were comprehensively analyzed
using Bioinformatics. To the best of our knowledge, the present
study is the first to not only propose a novel seven-miRNA
prognostic signature, but also predict a ceRNA network for early
PC.
Materials and methods
Data processing
The pre-processed mature miRNA expression profiles
for PC and pancreatic tissues from healthy individuals in The
Cancer Genome Atlas (TCGA) database (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga/using-tcga/citing-tcga),
displayed as log2-converted reads per million [log2(RPM+1)], were
downloaded from the University of California Santa Cruz Xena
database (17). The mature miRNA
expression profiles covered 2,050 miRNAs. Corresponding clinical
information was downloaded from the TCGA database (accession date,
10/14/2018). The mature miRNA expression profiles contained 182
samples (178 PC tissues and four pancreatic tissues from healthy
individuals) based on the IlluminaHiSeq_miRNASeq platform (Illumina
Inc.). The inclusion criteria for screening differentially
expressed miRNAs were as follows: i) The dataset included miRNA
expression profiles; ii) the samples were from patients with stage
IA-IIB disease according to the American Joint Committee on Cancer
Tumor-Node-Metastasis (TNM) system (https://cancerstaging.org/) (18), and iii) the miRNAs were expressed in
>20% of samples of PC. The data for a total of 167 samples of PC
with stage IA-IIB disease and four matched healthy pancreatic
tissues were extracted for screening differentially expressed
miRNAs. As all of the data were publicly available and open-access,
ethical approval was not required, and the study adhered to the
TCGA publication guidelines and data access policies (https://cancergenome.nih.gov/).
Screening of differentially expressed
miRNAs
The differentially expressed miRNAs between PCs and
pancreatic tissues from healthy individuals were analyzed using the
‘limma’ package (19) in R. The fold
changes (FCs) in the expression of individual miRNAs were
calculated and differentially expressed miRs with |log2FC|>1 and
P<0.05 adjusted by the false discovery rate (FDR) were
considered as significant (8,9,20).
Identification of the miRNA-based
prognostic signature for early PC
The expression values of differentially expressed
miRNAs in each sample of PC were extracted. Patients with a
survival time of <30 days from the day of diagnosis were removed
and 161 patients remained for survival analysis. The method of
least absolute shrinkage and selection operator (LASSO) for
regression may be used for optimal selection of features in
high-dimensional data with a strong predictive value and low
correlation between one another to prevent overfitting (21). This approach may be applied to the
Cox proportional hazard regression model for survival analysis with
high-dimensional data (22).
Therefore, as in a previous high-quality study (23), the LASSO Cox regression model was
used to select the most useful predictive markers among all
differentially expressed miRNAs, and a multi-miRNA-based signature
was constructed for predicting prognosis. The analysis was
performed using the ‘glmnet’ package (https://CRAN.R-project.org/package=glmnet) in R. A
risk score was created using the regression coefficients from the
LASSO analysis to weight the expression value of the selected
miRNAs with the following formula:
Prognostic Index (PI)=exprmiRNA1 ×
Coef1 + exprmiRNA2 × Coef2 +
exprmiRNA3 × Coef3+…
Where the ‘Coef’ value is the estimated regression
coefficient of a certain miRNA and is derived from the LASSO Cox
regression analysis, and ‘expr’ indicates the expression value of
the miRNAs. Patients with early PC were divided into low-risk and
high-risk groups, according to the median PI. Time-dependent
receiver operating characteristic (ROC) analysis was used to assess
the predictive value of the miR-based signature for 2-year-survival
of PC, performed with the ‘survivalROC’ package in R (24).
Prediction of the target genes of the
prognostic miRNA-based signature and pathway enrichment
analysis
The miRNet tool integrates data from 11 different
miRNA databases (25) (http://www.mirnet.ca/). Using miRNet, the targets
(mRNAs and lncRNAs) of the prognostic miRNA-based signature were
predicted and pathway enrichment analysis of the target mRNAs was
performed. A lncRNA-miRNA-gene network was constructed. The network
was further optimized to improve visualization using Cytoscape
software (26).
Gene set enrichment analysis (GSEA)
and construction of ceRNA network of the miRNA-based signature
Given that a single miRNA is able to modulate the
levels of several hundreds of target mRNAs and affect a myriad of
cellular processes, it is promising to explore the ultimate effects
of the interactions of these cellular processes that these seven
miRNAs are implicated in. The normalized fragments per kilobase of
transcript per million mapped reads values from the RNA-sequencing
data of the corresponding 161 patients with early PC were obtained
from TCGA Data Portal (https://portal.gdc.cancer.gov/). GSEA was performed by
the JAVA program (http://www.broadinstitute.org/gsea) using the MSigDB
c2.cp.kegg.v6.2.symbols.gmt gene set collection. Gene sets
with FDR of <0.25 after performing 1,000 permutations were
considered to be significantly enriched (27). The LncRNADisease database (www.cuilab.cn/lncrnadisease) is a resource that
curates the experimentally supported lncRNA-disease association
data (28). The PC-associated
lncRNAs were obtained from the lncRNADisease database. The common
lncRNAs between target lncRNAs predicted by miRNet and the
PC-associated lncRNAs, and the common genes between target genes
predicted by miRNet and the core enrichment genes provided by GSEA
were extracted. The ceRNA network of the miR-based signature in
early PC was constructed using Cytoscape software.
Statistical analysis
The unpaired t-test was used to screen
differentially expressed miRNAs. Univariate/multivariate Cox
proportional hazards analyses and Kaplan-Meier survival analysis
with log-rank test were used to compare survival between the two
groups of patients. The survival analysis was performed using SPSS
statistics software version 22.0 (IBM Corp.). All tests were
two-sided and P<0.05 was considered to indicate statistical
significance.
Results
Differentially expressed miRNAs
between early PC and pancreatic tissues from healthy
individuals
The detailed clinical characteristics of the
patients with PC at the early stage, including sex, age at
diagnosis and TNM stage are listed in Table I. According to the cutoff criteria
(P<0.05 and |log2FC|>1), 62 miRNAs were identified to be
differentially expressed between PC and pancreatic tissues from
healthy individuals. These include eight miRNAs that were
upregulated and 54 miRNAs that were downregulated in PC tissues.
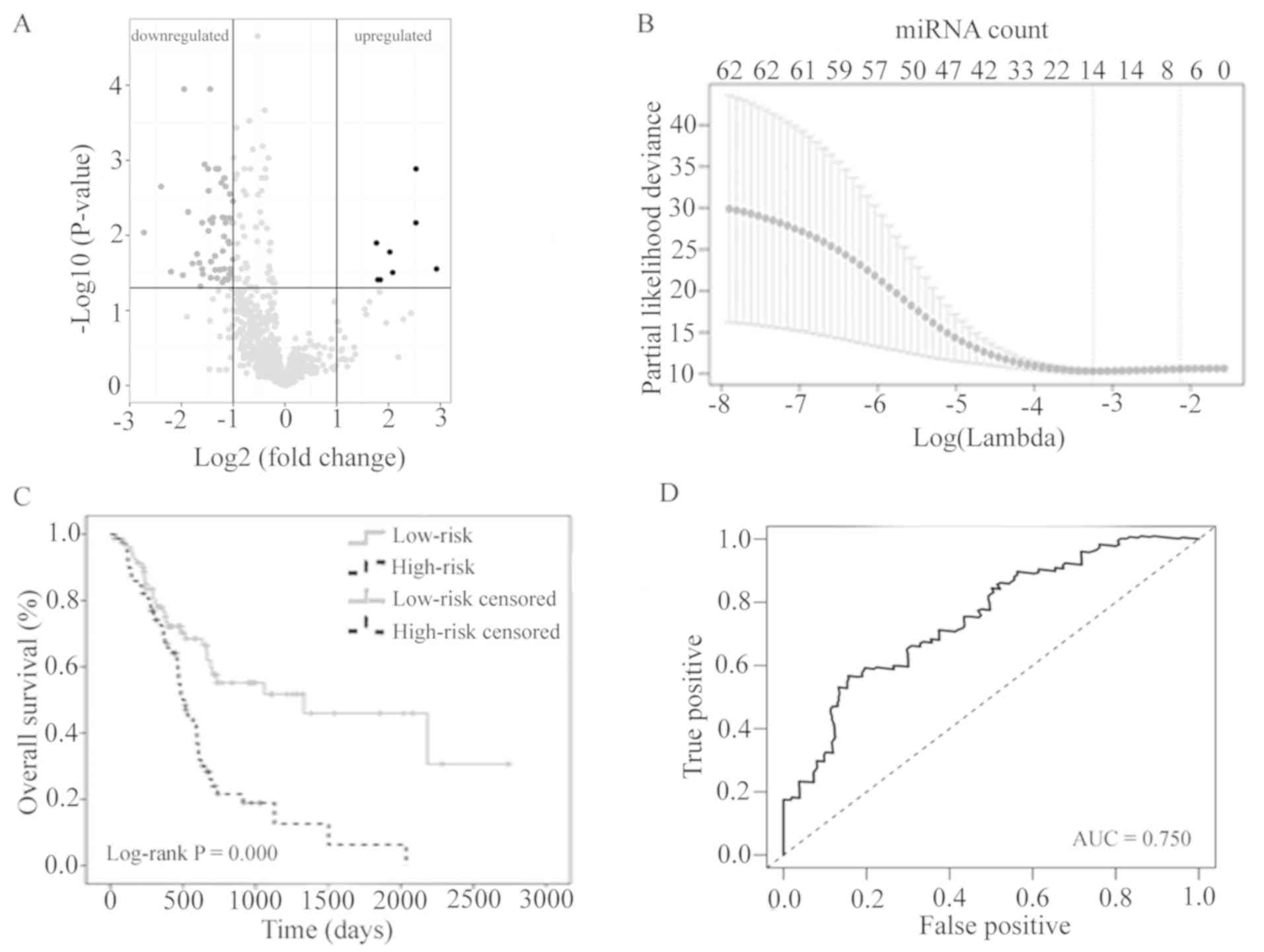
The results of the expression analysis are presented as a volcano
plot (Fig. 1A) to demonstrate that
the distribution of P-values and |log2FC| was reasonable with
respect to each other.
 | Table I.Clinicopathological features of 161
patients with pancreatic at the early stage. |
Table I.
Clinicopathological features of 161
patients with pancreatic at the early stage.
| Characteristic | n (%) |
|---|
| Sex |
|
Male | 89 (55.28) |
|
Female | 72 (44.72) |
| Age (years) |
|
<65 | 75 (46.58) |
|
≥65 | 86 (53.42) |
| T-stage |
|
T1/2 | 27 (16.77) |
|
T3/4 | 134 (83.23) |
| N-stage |
| N1 | 42 (26.09) |
| N0 | 115 (71.43) |
| Nx | 4 (2.48) |
| TNM Stage |
| I | 19 (11.80) |
| II | 142 (88.20) |
| Grade |
|
G1/2 | 111 (68.94) |
|
G3/4 | 48 (29.81) |
|
Unknown | 2 (1.24) |
Identification of seven-miRNA
prognostic signature for early PC
In order to develop a miRNA-based signature for
predicting prognosis in early PC, LASSO Cox regression was
performed using the expression profiles of the 62 differentially
expressed miRNAs. Using the LASSO method and 20-fold
cross-validation, seven miRNAs were identified with non-zero
regression coefficients (Fig. 1B). A
risk score was created by establishing the following formula:
PI=expmiR-424-5p × 0.111 +
expmiR-139-5p × (−0.029) + expmiR-5586-5p ×
(−0.070) + expmiR-126-3p × (−0.026) +
expmiR-3613-5p × (−0.199) + expmiR-454-3p ×
(−0.031) + expmiR-1271-5p × (−0.094).
The 161 patients with early PC were divided into a
low-risk group (<median PI; n=80) and a high-risk group (≥median
PI; n=81). The survival time was compared between the two groups
using Kaplan-Meier analysis with the log-rank test. The overall
survival in the low-risk group was significantly longer than that
in the high-risk group (log-rank P<0.001; Fig. 1C). The seven-miRNA signature PI was
indicated to be a promising biomarker for predicting the
2-year-survival rate of patients with PC at the early stage with an
area under the ROC curve (AUC)=0.750 (Fig. 1D). Furthermore, as opposed to routine
clinicopathological features, this seven-miRNA signature PI was a
powerful independent prognostic factor, as statistical significance
was retained after multivariate logistic regression analysis
[P=0.002, hazard ratio (HR)=2.204, 95% CI: 1.285–3.196; Table II].
 | Table II.Univariate/multivariate analysis of
routine clinicopathological features and the PI of the
seven-microRNA signature. |
Table II.
Univariate/multivariate analysis of
routine clinicopathological features and the PI of the
seven-microRNA signature.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
|
Characteristics | P-value | HR | HR (95% CI) | P-value | HR | HR (95% CI) |
|---|
| Sex
(male/female) | 0.538 | 1.141 | 0.750–1.735 |
|
|
|
| Age (≥65 y/<65
y) | 0.076 | 1.473 | 0.960–2.259 |
|
|
|
| T (T2/T1) | 0.019 | 2.217 | 1.142–4.304 | 0.428 | 1.597 | 0.502–5.082 |
| N (N1/N0) | 0.010 | 2.056 | 1.192–3.547 | 0.062 | 1.948 | 0.966–3.927 |
| Stage (II/I) | 0.036 | 2.305 | 1.056–5.032 | 0.540 | 0.621 | 0.136–2.845 |
| Grade
(G3-4/G1-2) | 0.089 | 1.469 | 0.943–2.289 |
|
|
|
| PI (high risk/low
risk) | 0.000 | 2.420 | 1.548–3.782 | 0.002 | 2.024 | 1.285–3.186 |
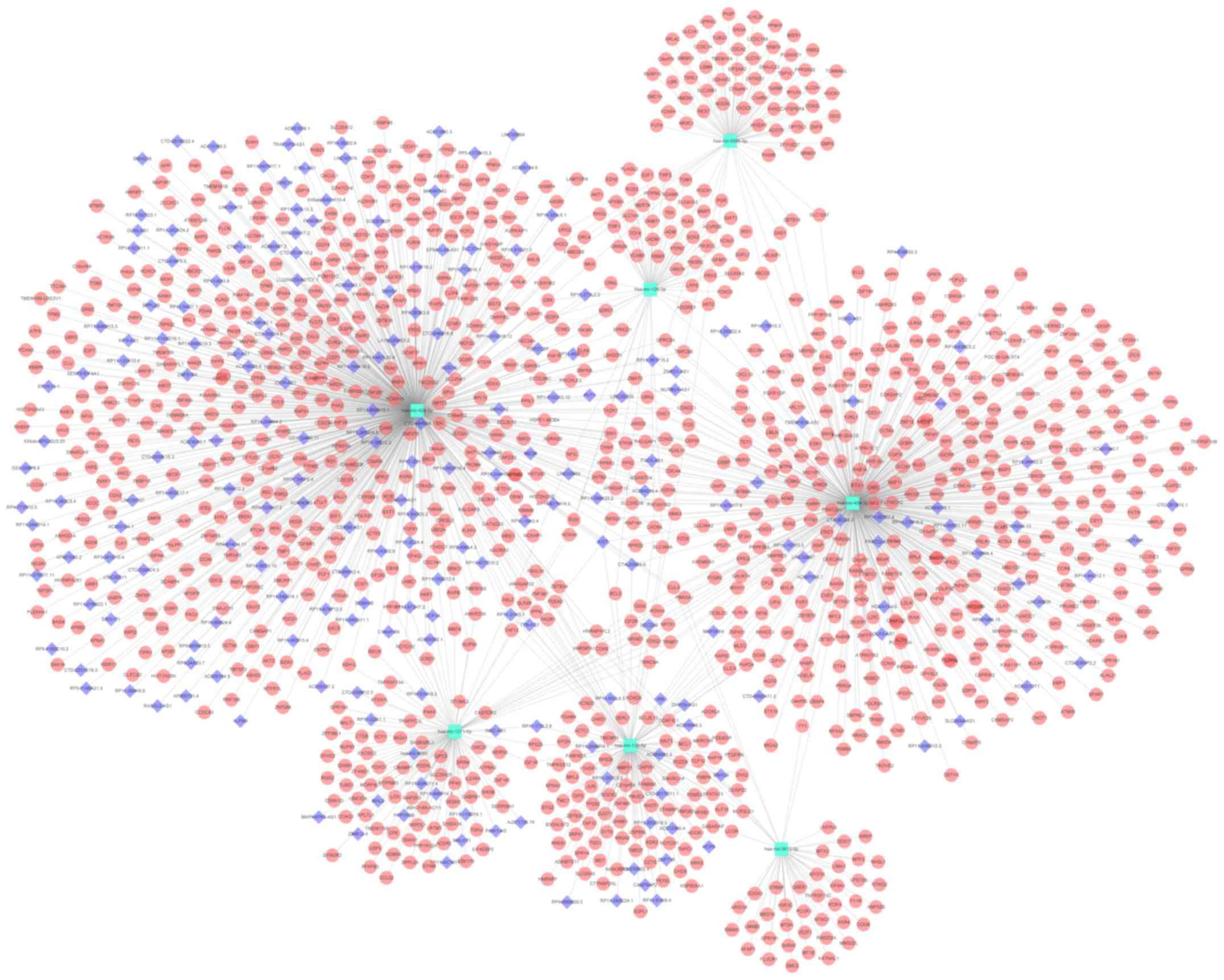
Targets of the seven miRNAs and
pathway enrichment analysis
A total of 1,126 genes and 219 lncRNAs were
predicted by using the miRNet tool based on the seven miRNAs
selected (miR-424-5p, miR-139-5p, miR-5586-5p, miR-126-3p,
miR-3613-5p, miR-454-3p and miR-1271-5p). The miRNA-gene and
lncRNA-miRNA networks were merged together and a lncRNA-miRNA-gene
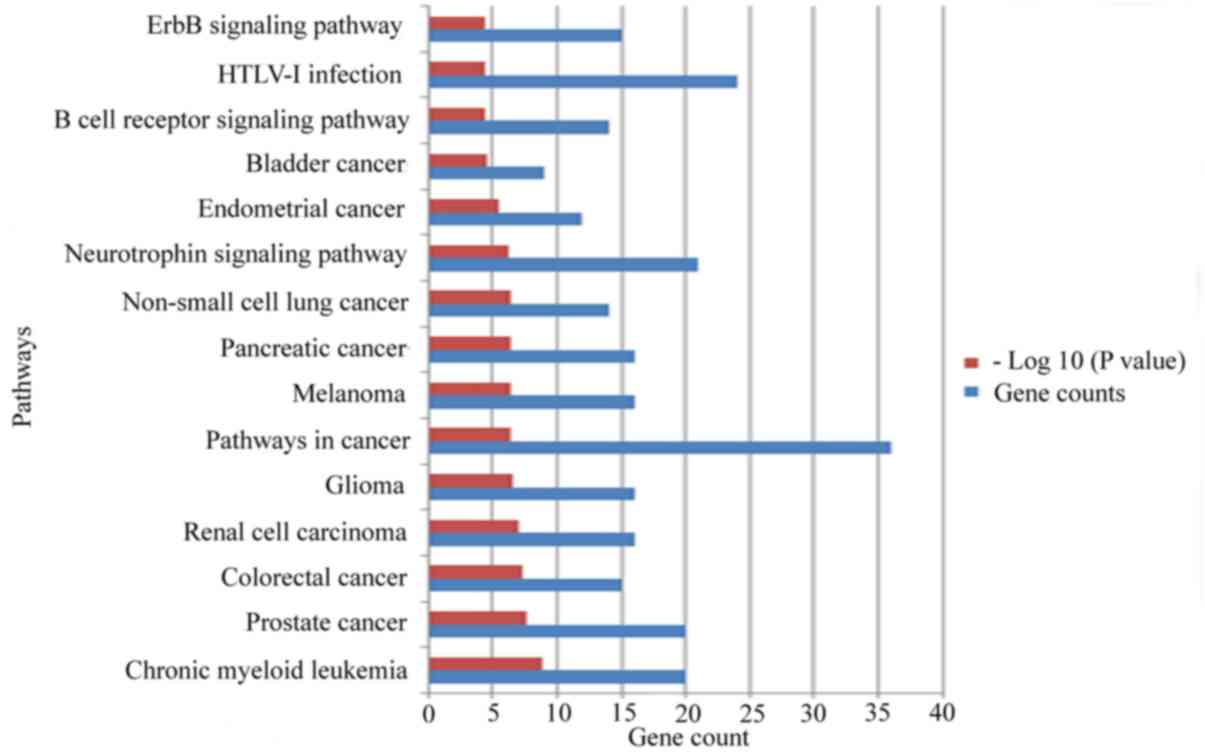
network was constructed using Cytoscape software (Fig. 2). Kyoto Encyclopedia of Genes and
Genomes pathway enrichment analyses of the target genes were
performed in miRNet using the hypergeometric test algorithm, and
the top 15 pathways are presented in Fig. 3. All of the results of the pathway
enrichment analysis are provided in Table SI. The analysis indicated that the
target genes of seven miRNAs are involved in various pathways
associated with cancer, including the ‘pancreatic cancer’
pathway.
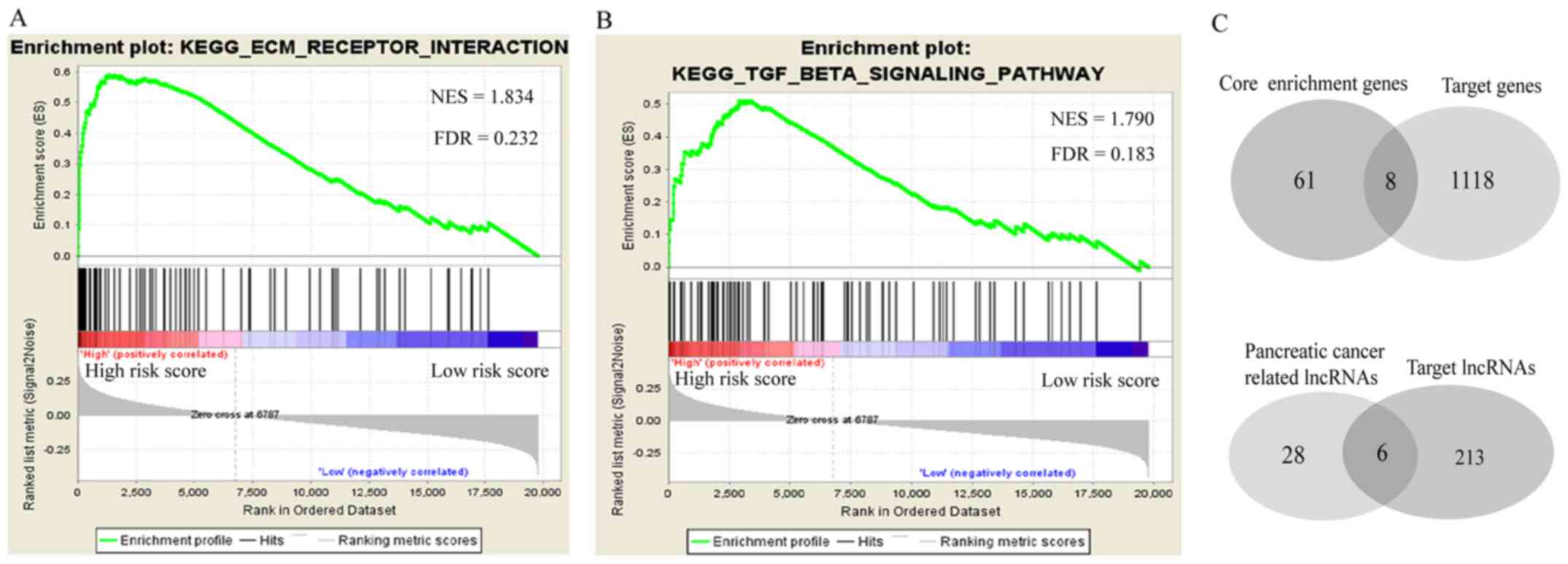
Results of GSEA and the ceRNA network
of the seven-miRNA signature
The GSEA was performed to identify potential
associated biological pathways affected by the seven-miRNA
signature. The extracellular matrix (ECM) receptor interaction
pathway (Fig. 4A) and transforming
growth factor (TGF)-β signaling pathway (Fig. 4B) were significantly enriched in the
high-risk group, suggesting that the seven-miRNA signature may
affect the prognosis by targeting those genes involved in the two
pathways. A total of 69 core enrichment genes were provided by the
GSEA, eight of which were shared among the 1,126 target genes of
these seven miRNAs (Fig. 4C, upper
panel). A total of 34 PC-associated lncRNAs were obtained from the
lncRNADisease database, six of which were shared among the 219
target lncRNAs of these seven miRNAs (Fig. 4C, lower panel). According to miRNet,
there is no record of PC-associated lncRNA targeting by miR-5586-5p
or miR-3613-5p. The ceRNA network contained 16 edges and 19 nodes
(five miRNAs, eight genes and six lncRNAs) (Fig. 5). Gene laminin subunit γ1 (LAMC1) and
integrin subunit α2 (ITGA2) were involved in the ECM receptor
interaction pathway. Rho associated coiled-coil containing protein
kinase (ROCK)2, ROCK1, activin A receptor type 2A (ACVR2A), SMAD
specific E3 ubiquitin protein ligase 1, ACVR1 and TGF-β receptor 2
(TGFBR2) were involved in the TGF-β signaling pathway.
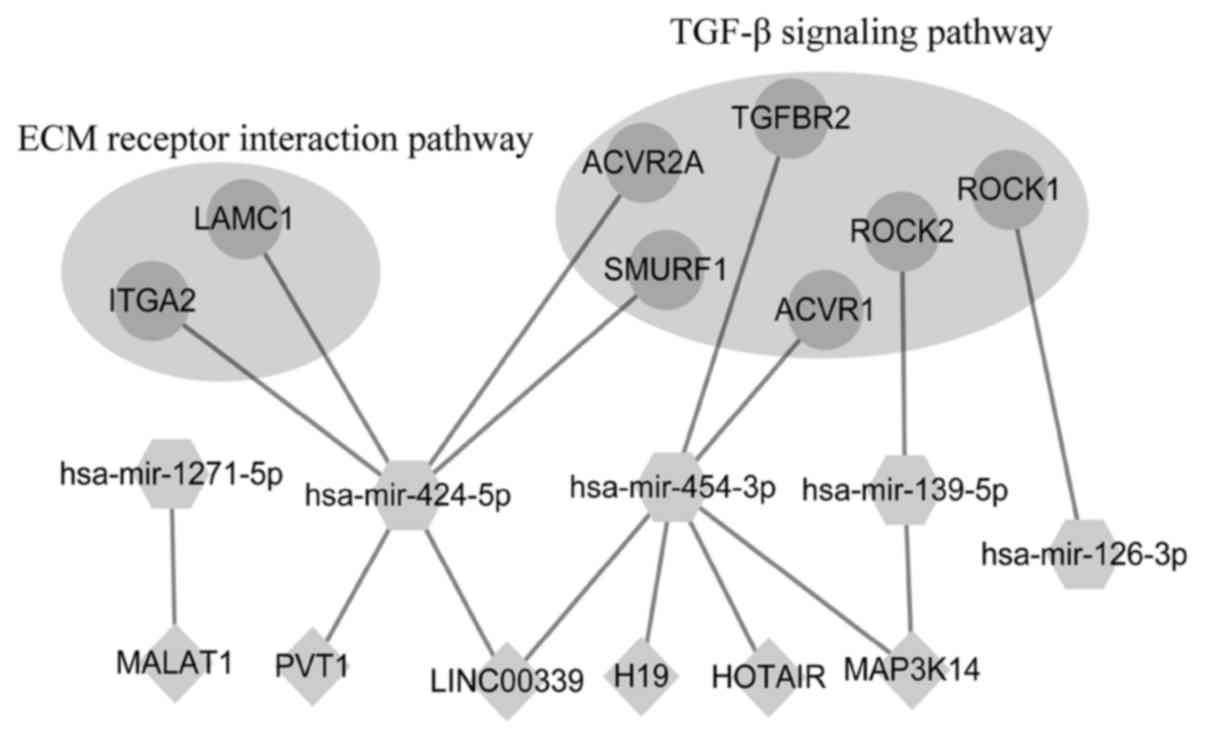 | Figure 5.Predicted competing endogenous RNA
network of the seven-miRNA signature. Dots indicate genes, diamonds
indicate lncRNAs and hexagons indicate miRNAs. ECM, extracellular
matrix; TGF, transforming growth factor; lncRNA, long non-coding
RNA; miRNA/miR, microRNA; hsa, Homo sapiens; MAP3K14,
mitogen-activated protein kinase kinase kinase 14; MALAT1,
Metastasis Associated Lung Adenocarcinoma Transcript 1; PVT1, Pvt1
oncogene; LINC00339, long intergenic non-protein coding RNA 339;
H19, H19 imprinted maternally expressed transcript; HOTAIR, HOX
transcript antisense RNA; ROCK, Rho associated coiled-coil
containing protein kinase; ACVR, activin A receptor; ITGA2,
integrin subunit α2; LAMC1, laminin subunit γ1; SMURF1, SMAD
specific E3 ubiquitin protein ligase 1; TGFBR2, transforming growth
factor-β receptor 2. |
Discussion
Only <20% PCs are diagnosed at the early stages
due to its asymptomatic nature (3),
however, >50% of early PC cases encounter recurrences within 12
months after surgery (4). Thanks to
TCGA, large samples of early PCs can be obtained for this present
study, which attempted to provide novel information on the
mechanism on early PC pathogenesis. Previous studies have indicated
that certain miRNAs are promising diagnostic and prognostic markers
for PC (7,29,30).
Given the development of high-throughput sequencing technologies,
an increasing number of miRNAs has been identified in recent years.
Furthermore, therapies based on miRNAs hold promise to
revolutionize PC treatment due to their increased efficacy compared
to conventional chemoradiation-based therapies (31). Therefore, it may be worthwhile to
further explore prognostic miRNA biomarkers for PC. In the present
study, a novel seven-miRNA prognostic signature for predicting
prognosis of early PC was obtained by comprehensively analyzing the
mature miRNA expression profiles and clinical information in TCGA
database using a Bioinformatics method. The seven miRNAs
(miR-424-5p, miR-139-5p, miR-5586-5p, miR-126-3p, miR-3613-5p,
miR-454-3p and miR-1271-5p) were obtained using LASSO and the Cox
regression method. Based on the seven-miRNA signature, high-risk
individuals were successfully identified among patients with early
PC. In clinical practice, such high-risk individuals may be
followed up more frequently. Furthermore, treatments for patients
with early PC remain limited (32)
and high-risk individuals have a poorer prognosis; therefore,
high-risk individuals are encouraged to receive more aggressive
management than the low-risk individuals, including participation
in a clinical trial.
A number of previous studies have proposed various
pre-miRNA-based signatures for predicting the prognosis of patients
with PC (7,33–36). Dou
et al (36) proposed an
eight-miRNA signature (miR-1301, miR-598, miR-1180, miR-155,
miR-496, miR-203, miR-193b and miR-135b), Liang et al
(33) proposed a five-miRNA
signature (miR-1301, miR-125a, miR-376c, miR-328 and miR-376b) and
Yu et al (34) proposed a
two-miRNA signature (miR-424 and miR-126). It is worth noting that
the data of only several cases of PC with advanced stage (III/IV)
disease are contained in TCGA; however, none of the previous
studies removed these samples to focus on early-stage PC. In the
present study, mature miRNA expression profiles were used instead
of pre-miRNA to establish a novel miRNA-based prognostic signature
for early-stage PC. The seven-miRNA signature PI is a promising
biomarker for predicting 2-year-survival rate of early PC with an
AUC=0.750. Several previous studies did not provide data from ROC
analyzes (33,34,36). In
contrast to routine clinicopathological features, the seven-miRNA
signature provided in the present study is an independent
prognostic factor.
In the present study, GSEA was also performed and
the potential ceRNA network of the seven miRNAs was constructed to
explore their ultimate downstream effects. The potential ceRNA
network comprised a total of 1,126 genes and 219 lncRNAs as
predicted by using the miRNet tool, and this network provided a
large number of interactions between these seven miRNAs and other
genes. This may provide a valuable resource of information, as
miRNA-based therapy is a promising treatment strategy for PC
(37). The TGF-β signaling pathway
has been reported to facilitate the genesis of PC (38), and the present study suggested that
this may be associated with downregulation of miR-454-3p,
miR-139-5p and miR-126-3p. It has been reported that miR-424-5p
(39) and miR-139-5p (40) are associated with PC. TGFBR2 is one
of the most frequently altered genes in patients with PC (41), and the present study suggested that
it may be targeted by miR-454-3p. HOTAIR has been reported to be a
negative prognostic factor with an oncogenic role in PC (42), and the present study indicated that
it may promote the development of PC by upregulating TGFBR2 and
ACVR1 via adsorbing miR-454-3p. The ECM receptor interaction
pathway has been reported to be involved in the development of
various cancer types, including lung cancer (43), bladder cancer (44) and breast cancer (45). The present study suggests that
miR-424-5p is downregulated by adsorption of PVTI and LINC00339,
leading to upregulation of ITGA2 and LAMC1 involved in the ECM
receptor interaction pathway to promote PC. This suggests that the
ceRNA network constructed in the present study warrants further
exploration to expand the understanding of the pathogenesis of
PC.
To the best of our knowledge, the present study was
the first to not only propose a novel seven-miRNA prognostic
signature for early PC, but also to provide their potential ceRNA
network. However, the study has certain limitations. There was a
lack of validation, as well as absence of prospective follow-up
data from other databases or clinical trials. It is essential to
validate and even improve this seven-miRNA signature in a larger
independent cohort. Particularly, due to most cases of PC are
diagnosed at late stage, the findings derived from the present
study would need to be validated on late-stage PC samples. In
addition, although a potential ceRNA network was constructed, the
exact roles of these seven miRNAs in PC require further assessment;
it remains elusive whether these miRNAs are causal or are merely
prognostic markers for early PC.
In conclusion, a seven-miRNA expression-based
prognostic signature for predicting the prognosis of patients with
PC at the early stage was proposed in the present study. Using this
signature, patients with early PC may be divided into high-risk and
low-risk groups. The corresponding potential ceRNA network of the
seven-miRNA expression-based prognostic signature was preliminarily
explored.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the Youth Science
Foundation of Guangxi Medical University (grant no. GXMUYSF
201716).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding authors on reasonable
request.
Authors' contributions
LH and YL conceived and designed the study. XB and
DL performed data analyses and prepared the manuscript. YL
contributed significantly to the data analyses and manuscript
revision. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rahib L, Smith BD, Aizenberg R, Rosenzweig
AB, Fleshman JM and Matrisian LM: Projecting cancer incidence and
deaths to 2030: The unexpected burden of thyroid, liver, and
pancreas cancers in the United States. Cancer Res. 74:2913–2921.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wagner M, Redaelli C, Lietz M, Seiler CA,
Friess H and Buchler MW: Curative resection is the single most
important factor determining outcome in patients with pancreatic
adenocarcinoma. Br J Surg. 91:586–594. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yadav D and Lowenfels AB: The epidemiology
of pancreatitis and pancreatic cancer. Gastroenterology.
144:1252–1261. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Beger HG, Thorab FC, Liu Z, Harada N and
Rau BM: Pathogenesis and treatment of neoplastic diseases of the
papilla of Vater: Kausch-Whipple procedure with lymph node
dissection in cancer of the papilla of Vater. J Hepatobiliary
Pancreat Surg. 11:232–238. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schultz NA, Andersen KK, Roslind A,
Willenbrock H, Wøjdemann M and Johansen JS: Prognostic microRNAs in
cancer tissue from patients operated for pancreatic cancer-five
microRNAs in a prognostic index. World J Surg. 36:2699–2707. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu L, Xiang L, Feng J, Li B, Zhou Z, Li J,
Lin Y, Lv Y, Zou D, Lei Z and Zhang J: miRNA-21 and miRNA-223
expression signature as a predictor for lymph node metastasis,
distant metastasis and survival in kidney renal clear cell
carcinoma. J Cancer. 9:3651–3659. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lin Y, Lv Y, Liang R, Yuan C and Zhang J,
He D, Zheng X and Zhang J: Four-miRNA signature as a prognostic
tool for lung adenocarcinoma. Onco Targets Ther. 11:29–36. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wiemer EA: The role of microRNAs in
cancer: No small matter. Eur J Cancer. 43:1529–1544. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bak RO and Mikkelsen JG: miRNA sponges:
Soaking up miRNAs for regulation of gene expression. Wiley
Interdiscip Rev RNA. 5:317–333. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Duell EJ, Lujan-Barroso L, Sala N, Deitz
McElyea S, Overvad K, Tjonneland A, Olsen A, Weiderpass E, Busund
LT, Moi L, et al: Plasma microRNAs as biomarkers of pancreatic
cancer risk in a prospective cohort study. Int J Cancer.
141:905–915. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Namkung J, Kwon W, Choi Y, Yi SG, Han S,
Kang MJ, Kim SW, Park T and Jang JY: Molecular subtypes of
pancreatic cancer based on miRNA expression profiles have
independent prognostic value. J Gastroenterol Hepatol.
31:1160–1167. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee KH, Lee JK, Choi DW, Do IG, Sohn I,
Jang KT, Jung SH, Heo JS, Choi SH and Lee KT: Postoperative
prognosis prediction of pancreatic cancer with seven microRNAs.
Pancreas. 44:764–768. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Goldman M, Craft B, Hastie M, Repečka K,
Kamath A, McDade F, Rogers D, Brooks AN, Zhu J and Haussler D: The
UCSC Xena platform for public and private cancer genomics data
visualization and interpretation. bioRxiv. 3264702019.
|
|
18
|
Amin MB, Greene FL, Edge SB, Compton CC,
Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR and
Winchester DP: The Eighth Edition AJCC Cancer Staging Manual:
Continuing to build a bridge from a population-based to a more
‘personalized’ approach to cancer staging. CA Cancer J Clin.
67:93–99. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xie M, Lv Y, Liu Z, Zhang J, Liang C, Liao
X, Liang R, Lin Y and Li Y: Identification and validation of a
four-miRNA (miRNA-21-5p, miRNA-9-5p, miR-149-5p, and miRNA-30b-5p)
prognosis signature in clear cell renal cell carcinoma. Cancer
Manag Res. 10:5759–5766. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wu TT, Chen YF, Hastie T, Sobel E and
Lange K: Genome-wide association analysis by lasso penalized
logistic regression. Bioinformatics. 25:714–721. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tibshirani R: The lasso method for
variable selection in the Cox model. Stat Med. 16:385–395. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lin Z, Cai YJ, Chen RC, Chen BC, Zhao L,
Xu SH, Wang XD, Song M, Wu JM, Wang YQ, et al: A microRNA
expression profile for vascular invasion can predict overall
survival in hepatocellular carcinoma. Clin Chim Acta. 469:171–179.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Heagerty PJ, Lumley T and Pepe MS:
Time-dependent ROC curves for censored survival data and a
diagnostic marker. Biometrics. 56:337–344. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fan Y, Siklenka K, Arora SK, Ribeiro P,
Kimmins S and Xia J: miRNet-dissecting miRNA-target interactions
and functional associations through network-based visual analysis.
Nucleic Acids Res. 44:W135–W141. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Demchak B, Hull T, Reich M, Liefeld T,
Smoot M, Ideker T and Mesirov JP: Cytoscape: The network
visualization tool for GenomeSpace workflows. F1000Res. 3:1512014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen
X, Zhang Q, Yan G and Cui Q: LncRNADisease: A database for
long-non-coding RNA-associated diseases. Nucleic Acids Res.
41:D983–D986. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tavano F, di Mola FF, Piepoli A, Panza A,
Copetti M, Burbaci FP, Latiano T, Pellegrini F, Maiello E,
Andriulli A and di Sebastiano P: Changes in miR-143 and miR-21
expression and clinicopathological correlations in pancreatic
cancers. Pancreas. 41:1280–1284. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Giovannetti E, van der Velde A, Funel N,
Vasile E, Perrone V, Leon LG, De Lio N, Avan A, Caponi S, Pollina
LE, et al: High-throughput microRNA (miRNAs) arrays unravel the
prognostic role of MiR-211 in pancreatic cancer. PLoS One.
7:e491452012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gurbuz N and Ozpolat B: MicroRNA-based
targeted therapeutics in pancreatic cancer. Anticancer Res.
39:529–532. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chiaravalli M, Reni M and O'Reilly EM:
Pancreatic ductal adenocarcinoma: State-of-the-art 2017 and new
therapeutic strategies. Cancer Treat Rev. 60:32–43. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liang L, Wei DM, Li JJ, Luo DZ, Chen G,
Dang YW and Cai XY: Prognostic microRNAs and their potential
molecular mechanism in pancreatic cancer: A study based on The
Cancer Genome Atlas and bioinformatics investigation. Mol Med Rep.
17:939–951. 2018.PubMed/NCBI
|
|
34
|
Yu Y, Feng X and Cang S: A two-microRNA
signature as a diagnostic and prognostic marker of pancreatic
adenocarcinoma. Cancer Manag Res. 10:1507–1515. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhou X, Huang Z, Xu L, Zhu M, Zhang L,
Zhang H, Wang X, Li H, Zhu W, Shu Y and Liu P: A panel of 13-miRNA
signature as a potential biomarker for predicting survival in
pancreatic cancer. Oncotarget. 7:69616–69624. 2016.PubMed/NCBI
|
|
36
|
Dou D, Yang S, Lin Y and Zhang J: An
eight-miRNA signature expression-based risk scoring system for
prediction of survival in pancreatic adenocarcinoma. Cancer
Biomark. 23:79–93. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pai P, Rachagani S, Are C and Batra SK:
Prospects of miRNA-based therapy for pancreatic cancer. Curr Drug
Targets. 14:1101–1109. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Principe DR, DeCant B, Mascariñas E, Wayne
EA, Diaz AM, Akagi N, Hwang R, Pasche B, Dawson DW, Fang D, et al:
TGFβ signaling in the pancreatic tumor microenvironment promotes
fibrosis and immune evasion to facilitate tumorigenesis. Cancer
Res. 76:2525–2539. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wu K, Hu G, He X, Zhou P, Li J, He B and
Sun W: MicroRNA-424-5p suppresses the expression of SOCS6 in
pancreatic cancer. Pathol Oncol Res. 19:739–748. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ma J, Zhang J, Weng YC and Wang JC:
EZH2-mediated microRNA-139-5p regulates epithelial-mesenchymal
transition and lymph node metastasis of pancreatic cancer. Mol
Cells. 41:868–880. 2018.PubMed/NCBI
|
|
41
|
Ku JL, Yoon KA, Kim WH, Jang Y, Suh KS,
Kim SW, Park YH and Park JG: Establishment and characterization of
four human pancreatic carcinoma cell lines. Genetic alterations in
the TGFBR2 gene but not in the MADH4 gene. Cell Tissue Res.
308:205–214. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kim K, Jutooru I, Chadalapaka G, Johnson
G, Frank J, Burghardt R, Kim S and Safe S: HOTAIR is a negative
prognostic factor and exhibits pro-oncogenic activity in pancreatic
cancer. Oncogene. 32:1616–1625. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li D, Yang W, Zhang Y, Yang JY, Guan R, Xu
D and Yang MQ: Genomic analyses based on pulmonary adenocarcinoma
in situ reveal early lung cancer signature. BMC Med Genomics. 11
(Suppl 5):S1062018. View Article : Google Scholar
|
|
44
|
Gao X, Chen Y, Chen M, Wang S, Wen X and
Zhang S: Identification of key candidate genes and biological
pathways in bladder cancer. PeerJ. 6:e60362018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yeh MH, Tzeng YJ, Fu TY, You JJ, Chang HT,
Ger LP and Tsai KW: Extracellular matrix-receptor interaction
signaling genes associated with inferior breast cancer survival.
Anticancer Res. 38:4593–4605. 2018. View Article : Google Scholar : PubMed/NCBI
|