Introduction
The ubiquitin-proteasome system (UPS) is a major
pathway for intracellular protein degradation and regulates a
number of key cellular processes. Its target proteins include a
broad array of regulatory proteins that play important roles in
cell cycle progression, cell development and differentiation, DNA
damage response and tumorgenesis. This system allows the cells to
modulate their protein expression patterns in response to changing
physiological conditions and plays a critical role in health and
disease (1,2). The UPS has therefore been
extensively studied as a novel molecular target for the development
of novel drugs in an attempt to restore protein homeostasis, as the
ultimate therapeutic strategy (3,4).
The proteasome is a massive multicatalytic protease responsible for
degrading a large number of cellular proteins. In order to be
degraded by the proteasome, these target proteins are first tagged
with ubiquitin (Ub), which can then target the substrate protein to
the 26S proteasome for destruction. The 20S proteasome, the core of
the 26S proteasome complex, has at least three distinct catalytic
activities, including chymotrypsin-like activity (cleavage after
hydrophobic residues by the β5 subunit). Several studies have shown
that the inhibition of the proteasomal chymotrypsin-like activity
results in the accumulation of several target proteins and the
induction of apoptosis in various types of tumor cells (5,6).
Zinc (Zn) was recognized as a trace element with
important roles in various metabolic processes in living organisms
almost a century ago. Zinc is the second most abundant transition
metal ion in the human body and an essential element for the proper
function of many different enzymes and the tight control of gene
expression (7–9). Cobalt (Co) is also needed in the
body and is an essential trace element found in small amounts in
different organs and bones. It is an integral part of vitamin B12,
which is vital to the formation of red blood cells (10,11). Additionally, cadmium (Cd) has been
shown to affect cell proliferation, differentiation and apoptosis
(12). The interest in
metal-based anticancer drugs has increased since the development of
cisplatin (13–16); however, due to the fact that there
are many pitfalls in the use of metal-based anticancer drugs, the
search for other metals and ligands that may produce more specific
antitumor effects is an ongoing process, in an effort to synthesize
and characterize novel potential metal-based antitumor drugs that
have less toxicity and higher clinical effectiveness (18–20). Our laboratory has studied a number
of novel metal-based drugs, including organic copper-, zinc- and
cadmium-based complexes, capable of inhibiting the tumor cell
proteasome and thus, proliferation, thereby inducing cancer cell
death (3,17,21–23). It has also been reported that
cobalt-based complexes effectively inhibit chymotrypsin-like
activity in the purified proteasome and PC-3 prostate cancer cells
(24).
2,3-Indolinedione (isatin; formula,
C8H5O2N), an endogenous indole in
marine and mammalian organisms, possesses a wide range of
biological activities, including anxiogenic, sedative and
anticonvulsant activities, and is a potent antagonist of atrial
natriuretic peptide receptors. Studies have shown that
2,3-indolinedione and its derivatives have pro-apoptotic functions
in human cancer and mouse neuroblastoma cells (25,26).
Considering the importance of the UPS and the
properties of 2,3-indolinedione, we aimed to investigate whether
2,3-indolinedione derivatives have the ability to inhibit
proteasome activity, and whether structure is an essential factor
affecting antitumor activity. To investigate our hypothesis, we
synthesized six novel metal compounds (Table I) with 2,3-indolinedione,
2-amino-5-methoxyphenol (N1), 2-amino-5-methylphenol (N2),
3-hydroxy-4-aminobenzoic acid (N3), L-tryptophane (N4) and
L-phenylalanine (N5) with Cd (M1), Zn (M2) and Co (M3),
respectively (Table I). The
compounds were then tested in human breast cancer metal-based
complexes in human breast (MDA-MB-231) cells to determine whether
compound structure affects proteasome-inhibitory and
apoptosis-inducing abilities.
 | Table IChemical structures of L, N1-N5 and
compounds C1-C6. |
Table I
Chemical structures of L, N1-N5 and
compounds C1-C6.
Materials and methods
Materials
Compounds C1-C6 were synthesized by the laboratory
at the Ocean University of China, Qingdao, China.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT),
dimethyl sulfoxide (DMSO) and other chemicals were purchased from
Sigma-Aldrich (St. Louis, MO, USA). All compounds were dissolved in
DMSO at stock concentrations of 80 mM and stored at 4°C. Fetal
bovine serum (FBS) was purchased from Aleken Biologicals (Nash, TX,
USA). Dulbecco’s modified Eagle’s medium/F12 medium and
penicillin/streptomycin were purchased from Invitrogen (Carlsbad,
CA, USA). Rabbit polyclonal antibody against human poly(ADP-ribose)
polymerase (PARP; H-250), mouse monoclonal antibodies against Ub
(P4D1), Bax (B-9), goat polyclonal antibody against β-actin (C-11)
and donkey anti-goat secondary antibody were from Santa Cruz
Biotechnology, Inc. (Santa Cruz, CA, USA). Goat anti-rabbit and
goat anti-mouse secondary antibodies were from Bio-Rad (Hercules,
CA, USA).
Metal complex syntheses
C1, C2, C3, C4, C5 and C6: these compounds were
synthesized by the laboratory at the Ocean University of China. The
ligand (2 mM) was dissolved in 15 ml of ethanol. M
(CH3COO)2·2H2O (2 mM) dissolved in
10 ml of anhydrous ethanol was added dropwise to the above solution
with stirring and the mixture was reacted for 4 h at 50°C to yield
a precipitate, which was filtered off, to produce the final
complexes.
C1: yield, 81%; Anal. Calc. for C1 {%, [Cd
(C15H11O3N2)
(CH3COO)], FW=438.71 g·mol-1}; C, 46.54; H,
3.22; N, 6.39. Found (%): C, 47.02; H, 3.19; N, 6.56. λmax (nm):
232, 285. IR data (KBr, cm-1): 3267.68, υ (-NH-);
1651.35, υ (-C=O); 1618.32, υ (-C=N-); 1590.95, υas
(COO-); 1319.99, υs (COO-); 1197.76, υ
(-OCH3); 1108.84, υ (-Ph-OH); 478.89, υ (Cd-O). 1H NMR
(DMSO, 600 MHz; s, singlet; d, doublet; t, triplet): δ (ppm) 10.498
(s, 1H, -NH-); 7.497 (s, 1H, -Ph-H); 7.340 (d, 1H, -Ph-H); 7.066
(s, 1H, -Ph-H); 6.726 (s, 1H, -Ph-H); 6.462 (s, 1H, -Ph-H); 6.438
(d, 1H, -Ph-H); 6.306 (s, 1H, -Ph-H); 3.323 (d, 3H, -OCH3); 2.614
(t, 3H, -CH3). Thermogravimetric (TG) analysis, residue 30.22%
(calculated 29.27%, CdO). Molar conductivity, Λm
(S·cm2·mol-1): 19.88.
C2: yield, 85%; Anal. Calc. for C2 {%, [Cd
(C15H11O2N2)
(CH3COO)], FW=422.72 g·mol-1}; C, 48.30; H,
3.34; N, 6.63. Found (%): C, 48.41; H, 3.31; N, 7.01. λmax (nm):
236, 655. IR data (KBr, cm-1): 3189.68, υ (-NH-);
1648.35, υ (-C=O); 1610.52, υ (-C=N-); 1580.95, υas
(COO-); 1327.89, υs (COO-); 1199.56, υ (-Ph-OH); 470.89,
υ (Cd-O). 1H NMR (DMSO, 600 MHz; s, singlet; d, doublet; t,
triplet): δ (ppm) 10.498 (s, 1H, -NH-); 7.497 (s, 1H, -Ph-H); 7.340
(d, 1H, -Ph-H); 7.066 (s, 1H, -Ph-H); 6.726 (s, 1H, -Ph-H); 6.462
(s, 1H, -Ph-H); 6.438 (d, 1H, -Ph-H); 6.306 (s, 1H, -Ph-H); 2.614
(t, 3H, -CH3); 2.541 (d, 3H, -CH3). TG analysis: residue 31.32%
(calculated 30.74%, CdO). Molar conductivity, Λm
(S·cm2·mol-1): 19.60.
C3: yield, 79%; Anal. Calc. for C3 {%, [Co
(C15H9O4N2)
(CH3COO)], FW=399.22 g·mol-1}; C, 51.15; H,
3.03; N, 7.02. Found (%): C, 52.25; H, 2.96; N, 7.21. λmax (nm):
238, 564. IR data (KBr, cm-1): 3215.46, υ (-NH-);
1678.66, υ (-C=O); 1602.19, υ (-C=N-); 1540.99, υas
(COO-); 1321.62, υs (COO-); 1212.36, υ (-Ph-OH); 477.02,
υ (Co-O). 1H NMR (DMSO, 600 MHz; s, singlet; d, doublet; t,
triplet): δ (ppm) 11.308 (s, 1H, -COOH); 10.964 (s, 1H, -NH-);
7.584 (d, 1H, -Ph-H); 7.497 (s, 1H, -Ph-H); 7.340 (d, 1H, -Ph-H);
7.065 (s, 1H, -Ph-H); 6.724 (s, 1H, -Ph-H); 6.630 (d, 1H, -Ph-H);
6.335 (s, 1H, -Ph-H); 2.613 (t, 3H, -CH3); TG analysis: residue
19.65% (calculated 18.77%, CoO). Molar conductivity, Λm
(S·cm2·mol-1): 16.91.
C4: yield, 85%; Anal. Calc. for C4 {%, [Co
(C15H11O2N2)
(CH3COO)], FW=369.24 g·mol-1}; C, 55.30; H, 3.82; N,
7.59. Found (%): C, 55.01; H, 3.79; N, 7.20. λmax (nm): 237, 286.
IR data (KBr, cm-1): 3289.67, υ (-NH-); 1672.52, υ
(-C=O); 1612.31, υ (-C=N-); 1526.21 υas (COO-); 1320.54,
υs (COO-); 1226.34, υ (-Ph-OH); 473.04, υ (Co-O). 1H NMR
(DMSO, 600 MHz; s, singlet; d, doublet; t, triplet): δ (ppm) 10.496
(s, 1H, -NH-); 7.497 (s, 1H, -Ph-H); 7.340 (d, 1H, -Ph-H); 7.066
(s, 1H, -Ph-H); 6.726 (s, 1H, -Ph-H); 6.462 (s, 1H, -Ph-H); 6.438
(d, 1H, -Ph-H); 6.306 (s, 1H, -Ph-H); 2.614 (t, 3H, -CH3); 2.541
(d, 3H, -CH3). TG analysis: residue 20.95% (calculated 20.29%,
CoO). Molar conductivity, Λm (S·cm2·mol-1):
18.12.
C5: yield, 80%; Anal. Calc. for C5 {%, [Zn
(C19H14O3N3)
(CH3COO)], FW=456.77 g·mol-1}; C, 55.22; H,
3.75; N, 9.20. Found (%): C, 55.15; H, 3.71; N, 9.52. λmax (nm):
231, 296. IR data (KBr, cm-1): 3399.68, υ (-NH-);
1711.36, υ (-C=O); 1611.65, υ (-C=N-); 1608.21, υas
(COO-); 1316.82, υs (COO-); 452.54, υ (Zn-O). 1H NMR
(DMSO, 600 MHz; s, singlet; d, doublet; t, triplet; m, multiplet):
δ (ppm) 10.749 (s, 1H, -NH-); 9.762 (s, 1H, -NH-); 7.919 (m, 4H,
-Ph-H); 7.497 (s, 1H, -Ph-H); 7.340 (d, 1H, -Ph-H); 7.067 (s, 1H,
-Ph-H); 6.724 (s, 1H, -Ph-H); 3.779 (s, 1H, -C-H-); 3.16 (d, 1H,
-C-H-); 3.021 (t, 2H, -CH2-); 2.613 (t, 3H, -CH3). TG analysis:
residue 19.06% (calculated 18.21%, ZnO). Molar conductivity, Λm
(S·cm2·mol-1): 23.56.
C6: yield, 85%; Anal. Calc. for C6 {%, [Zn
(C17H13O3N2)
(CH3COO)], FW=417.73 g·mol-1}; C, 54.63; H, 3.86; N,
6.71. Found (%): C, 54.51; H, 3.81; N, 7.01. λmax (nm): 232, 652.
IR data (KBr, cm-1): 3267.68, υ (-NH-); 1686.32, υ
(-C=O); 1612.55, υ (-C=N-); 1590.95, υas (COO-);
1339.93, υs (COO-); 445.30, υ (Zn-O). 1H NMR (DMSO, 600
MHz; s, singlet; d, doublet; t, triplet): δ (ppm) 10.760 (1H, s,
-NH-); 7.498 (s, 1H, -Ph-H); 7.342 (d, 1H, -Ph-H); 7.140 (t, 2H,
-Ph-H); 7.067 (s, 1H, -Ph-H); 7.050 (t, 2H, -Ph-H); 7.024 (s, 1H,
-Ph-H); 6.724 (s, 1H, -Ph-H); 3.779 (s, 1H, -C-H-); 3.020 (t, 2H,
-CH2-); 2.614 (t, 3H, -CH3). TG analysis: residue 19.65%
(calculated 19.30%, ZnO). Molar conductivity, Λm
(S·cm2·mol-1): 21.42.
Elemental analysis and NMR
spectroscopy
Elemental analysis (C, H and N) was performed using
a 2400 PerkinElmer analyzer (Perkin-Elmer, Inc., Wellesley, MA,
USA). Infrared spectrum was recorded as KBr pellets on the Nicolet
170SX spectrophotometer (Thermo Fisher Scientific Inc., Waltham,
MA, USA) in the 4,000–400 cm-1 region. 1H NMR spectrum
was recorded on an Avance III (600 MHz) spectrometer (Bruker
Biospin Group, Zurich, Switzerland).
Cell culture and whole-cell extract
preparation
MDA-MB-231, LNCaP and PC-3 cell lines were obtained
from the American Type Culture Collection (ATCC; Manassas, VA,
USA). The MDA-MB-231 human breast cancer cells were cultured in
DMEM/F-12 (1:1) and LNCaP and the PC-3 human prostate cancer cells
were cultured in RPMI-1640 medium. All media were supplemented with
10% FBS, 100 μg/ml streptomycin and 100 U/ml penicillin (Life
Technologies, Carlsbad, CA USA). All cells were maintained in a
humidified atmosphere containing 5% CO2 at 37°C. The
cells were harvested, washed with phosphate-buffered saline (PBS),
lysed in lysis buffer [50 mM tris(hydroxymethyl)aminomethane
Tris-HCl, pH 8.0, 150 mM NaCl, 0.5% NP40], vortexed at 4°C for 30
min, and centrifuged at 13,000 × g for 14 min (27). The supernatants were collected as
whole-cell extracts and used for the measurement of
chymotrypsin-like activity and western blot analysis, as previously
described (28).
Cell proliferation assays
The effects of the complexes the growth of cancer
cells were determined by MTT assay. The MDA-MB-231 (breast cancer),
and the LNCaP and PC-3 (prostate cancer) cells were seeded in
triplicate in 96-well plates and cultured until 70–80% confluency
at 37°C, followed by treatment with the indicated concentrations of
each compound for 24 h. The medium was then removed and MTT
solution (1 mg/ml) was added. After 2 h of incubation at 37°C, MTT
was removed, and 100 μl DMSO were added to dissolve the metabolized
MTT product. The inhibition of cell proliferation was measured at
an absorbance of 560 nm on a Wallac Victor3 multi-label plate
reader (Perkin-Elmer, Inc.).
Proteasomal chymotrypsin-like activity in
MDA-MB-231 breast cancer cells
The MDA-MB-231 cells were treated with C1-C6 at the
indicated concentrations (5, 10, 20, 30, 40 μM) or for different
periods of time (0, 2, 4, 8, 16, 24 h; C=30 μM), lysed, and the
protein concentrations were measured using a Bio-Rad protein assay
(Bio-Rad). Whole-cell lysates (10 μg) were incubated for 2 h at
37°C in 100 μl assay buffer (20 mM Tris-HCl, pH 7.5) with 20 μM
fluorogenic peptide substrate Suc-LLVY-AMC (AnaSpec, Fremont, CA,
USA). Proteasomal CT-like activity was measured using the Wallac
Victor3 multi-label counter with an excitation filter of 365 nm and
an emission filter of 460 nm, as previously described (3).
In vitro proteasomal activity assay in
MDA-MB-231 breast cancer cell extracts
The MDA-MB-231 cell extracts (10 μg total protein)
were incubated in 100 μl assay buffer (20 mM Tris-HCl, pH 7.5) and
20 μM chymotrypsin-like substrate Suc-LLVY-AMC, with various
concentrations of C1-C6 or DMSO as the vehicle control for 2 h at
37°C. Following incubation, proteasome CT-like activity was
measured using the Wallac Victor3 multi-label counter with an
excitation filter of 365 nm and an emission filter of 460 nm, as
previously described (3).
Western blot analysis
Proteins (30 μg) from whole-cell lysates were
separated by sodium dodecyl sulfate polyacrylamide gel
electrophoresis (SDS-PAGE) and then transferred onto nitrocellulose
membranes. Western blot analysis was performed using specific
antibodies against Ub, Bax, PARP and β-actin, followed by
visualization with enhanced chemiluminescence reagent (Denville
Scientific, Inc., Metuchen, NJ, USA), as previously described
(29).
Analysis of cellular morphology
Cellular morphological changes were observed using
an Axiovert 25 phase contrast microscope (Carl Zeiss Inc.,
Thornwood, NY, USA), as previously described (29).
Results
IR spectral studies
The structural explanation of the complexes is
supported by infrared spectroscopy (IR spectra). There are medium
strong bands at 3,189–3,399 cm-1 in the spectra of
compounds C1-C6 which are assigned to υ (-NH-) vibration. The IR
spectra also shows sharp bands at 1,622–1,636 cm-1
corresponding to υ (-C=N-) in ligands that are not shown in this
article, which shifted to 1,602–1,618 cm-1 in compounds
1-C6, indicating the coordination complex between nitrogen and
metal. Further evidence of the complexation of nitrogen is obtained
from the appearance of new bands at 445–478 cm-1 which
is assignable to υ (M–N) for the complexes. The loss of OH proton
is indicated by the absence of bands at ~3,500 cm-1 in
the complexes C1-C6. The difference between the value of
υas (COO-) and υs (COO-) is greater than 200
cm-1, thus confirming that carboxylic radical is in the
form of monodentate in the coordination complex.
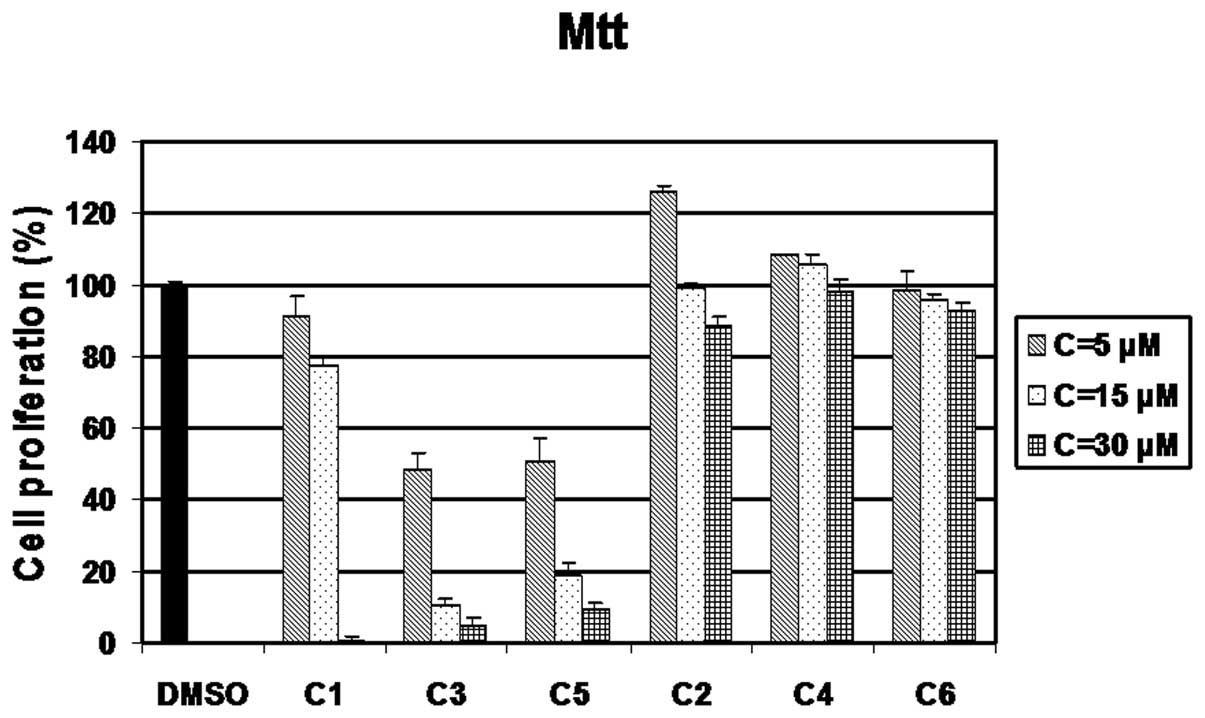
Growth inhibitory effect of compounds
C1-C6 in MDA-MB-231 breast cancer cells
First, to investigate whether compounds C1-C6 had
anti-proliferative ability, the MDA-MB-231 breast cancer cells were
treated with 5, 15 or 30 μM of C1 to C6 for 24 h, followed by
analysis by MTT assay. Cells treated with DMSO were used as a
control. C3 and C5 had similar growth-inhibitory activities,
resulting in approximately 50% inhibition at 5 μM, 90 and 82%
inhibition at 15 μM, respectively and at >92% inhibition at 30
μM (Fig. 1). C1 was also a potent
inhibitor, displaying 5, 12 and 99% in inhibition at 5, 15 and 30
μM, respectively (Fig. 1).
However, C2, C4 and C6 showed a slight inhibitory effect at 5, 15,
30 μM after 24 h of treatment (Fig.
1).
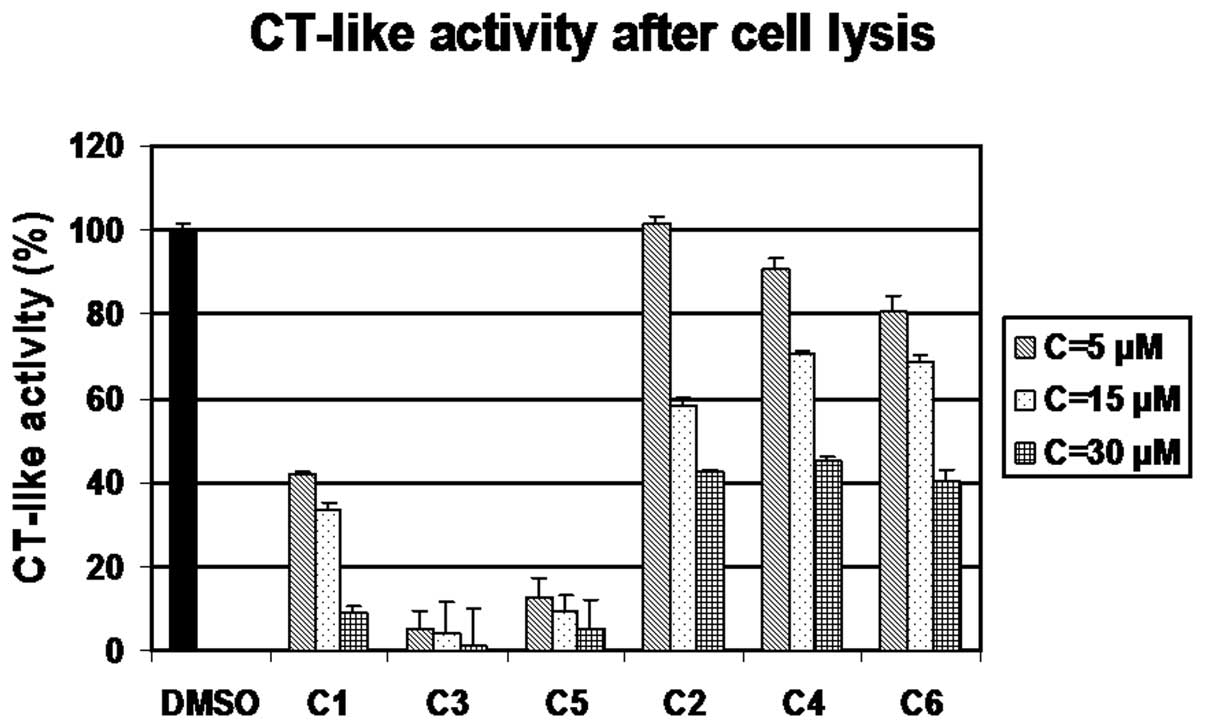
Inhibition of intact 26S proteasome
activity by C1-C6 in vitro
To investigate whether these complexes were capable
of inhibiting proteasome activity with effects similar to those
observed on cell proliferation, the MDA-MB-231 cell extracts were
treated with various concentrations of C1-C6 (5, 15, 30 μM) for 2
h, with DMSO treatment as a control, followed by the measurement of
proteasomal CT-like activity (using a fluorogenic substrate
specific for the CT-like subunit). The results clearly indicated
that the compounds C1, C3 and C5 were the most potent against
proteasomal chymotrypsin-like activity (Fig. 2), similar to their effects on cell
proliferation, while C2, C4 and C6 were much less potent (Fig. 2), similar to their effects on cell
proliferation as measured by MTT assay.
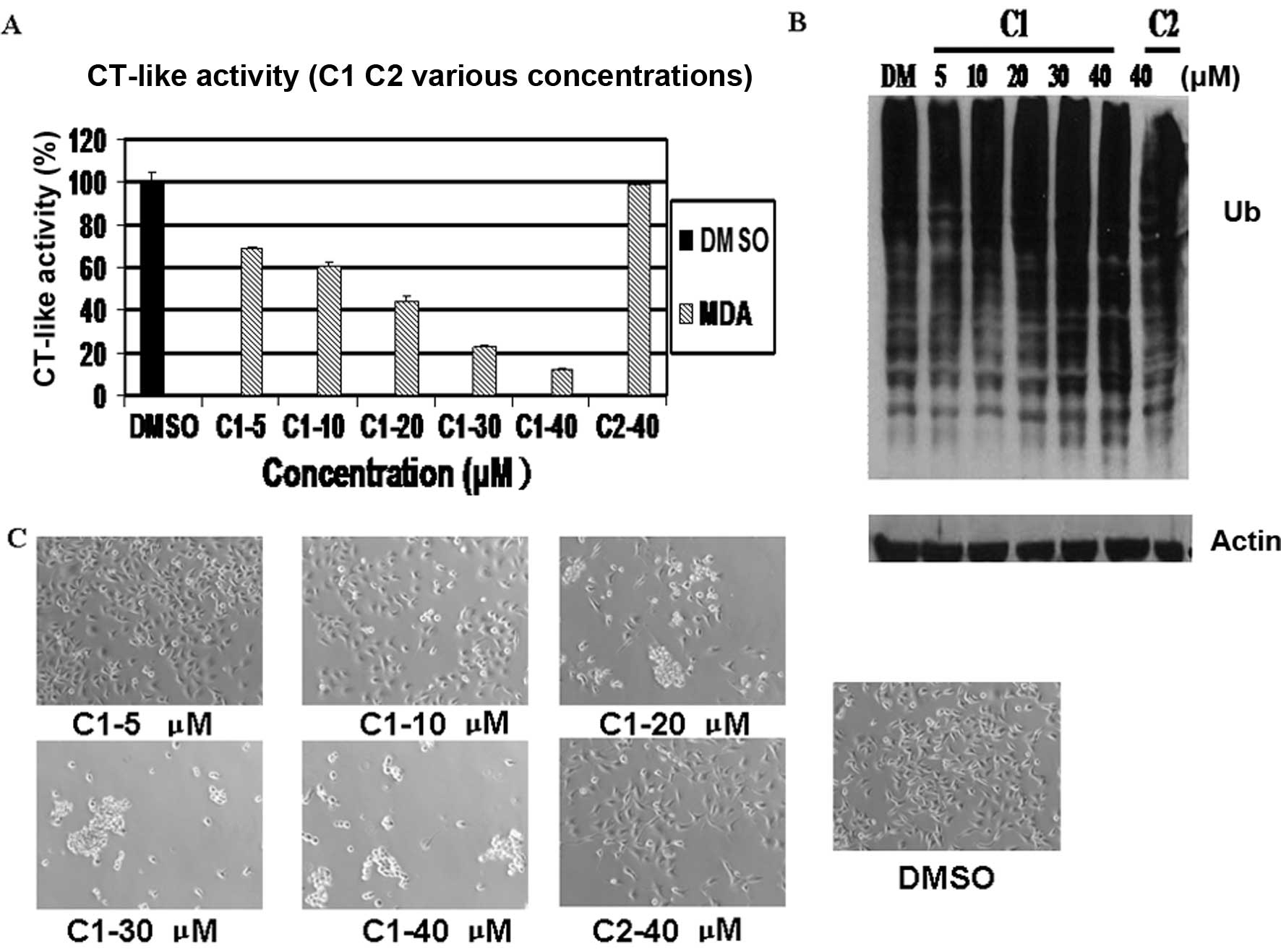
Concentration-dependent proteasome
inhibition and induction of apoptosis by C1, but not C2 in
MDA-MB-231 cells
To determine whether chemical structure is important
for these compounds to inhibit tumor cellular proteasome activity
and induce apoptosis, the MDA-MB-231 cells were treated with
various concentrations of the cadmium-based compounds, C1 and C2,
which are similar in structure, or DMSO as a control, for 24 h. C1
inhibited proteasomal chymotrypsin-like activity in a
concentration-dependent manner, inhibiting >90% activity at 40
μM, while C2 showed no inhibitory effect even at 40 μM (Fig. 3A).
The accumulation of target proteins has been shown
to be associated with the inhibition of proteasome activity
(29). Western blot analysis
revealed a dose-dependent accumulation of ubiquitinated proteins
induced by treatment with C1 (Fig.
3B).
It has been reported that the inhibition of tumor
cellular proteasome activity is also associated with the induction
of apoptosis. In the same experiment, apoptosis associated with
changes in cellular morphology were also observed in the cells
treated with C1 at concentrations as low as 10 μM. These changes
did not occur in the cells treated with C2, even at the highest
concentration (40 μM) (Fig. 3C).
These results suggested that C1, but not C2, inhibited cellular
proteasome activity and induced apoptosis in the intact MDA-MB-231
cells.
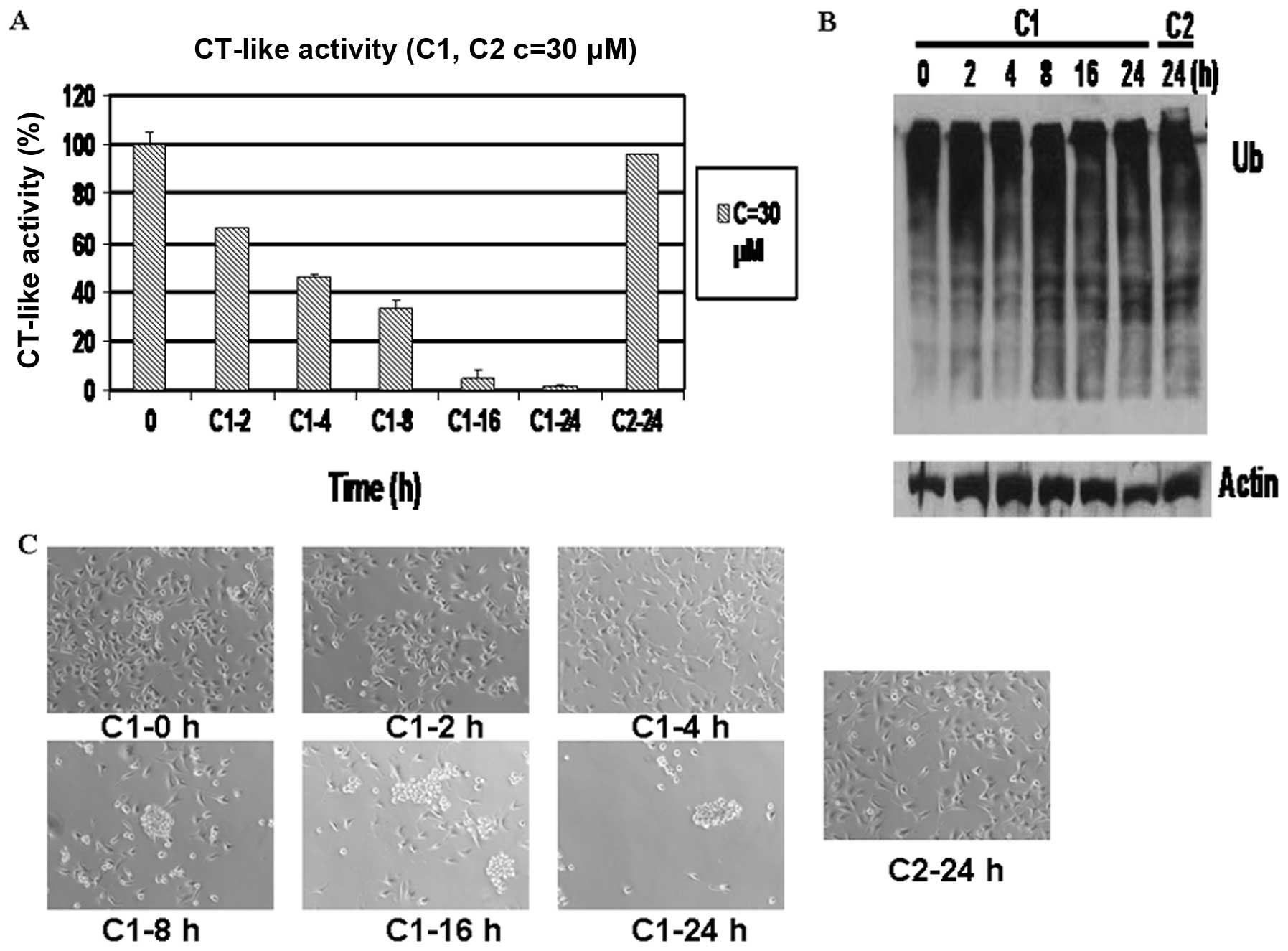
Time-dependent proteasome inhibition and
induction of cell death by C1, but not C2 in MDA-MB-231 cells
To verify that the observed apoptotic changes were a
result of proteasome inhibition, the MDA-MB-231 cells were treated
with 30 μM C1 for 0, 2, 4, 8, 16 and 24 h. Treatment with C2 for 24
h served as a control. The results revealed that proteasomal
chymotrypsin-like activity was inhibited by 36% in these breast
cancer cells after only 2 h of treatment with C1 (Fig. 4A) and further decreased in a
time-dependent manner. After 24 h of treatment with C1, proteasomal
chymotrypsin-like activity was inhibited by 98%, whereas C2
treatment resulted in only 5% inhibition (Fig. 4A).
Western blot analysis revealed that the accumulation
of ubiquitinated proteins appeared at 8 h of treatment with C1
(Fig. 4B). Morphological changes,
indicative of cellular apoptosis, were observed after 8 h of
treatment with C1 and almost increased to 100% after 24 h (Fig. 4C). By contrast, apoptosis was
observed in the cells treated with C2 for 24 h (Fig. 4B and C).
C3 and C5, but not C4 or C6, has
proteasome-inhibitory and apoptosis-inducing activities in
MDA-MB-231 cells
To confirm our findings further, we compared the
biological activities of another two pairs of complexes [C3 vs. C4
and C5 vs. C6 (Table I)] in the
MDA-MB-231 breast cancer cells.
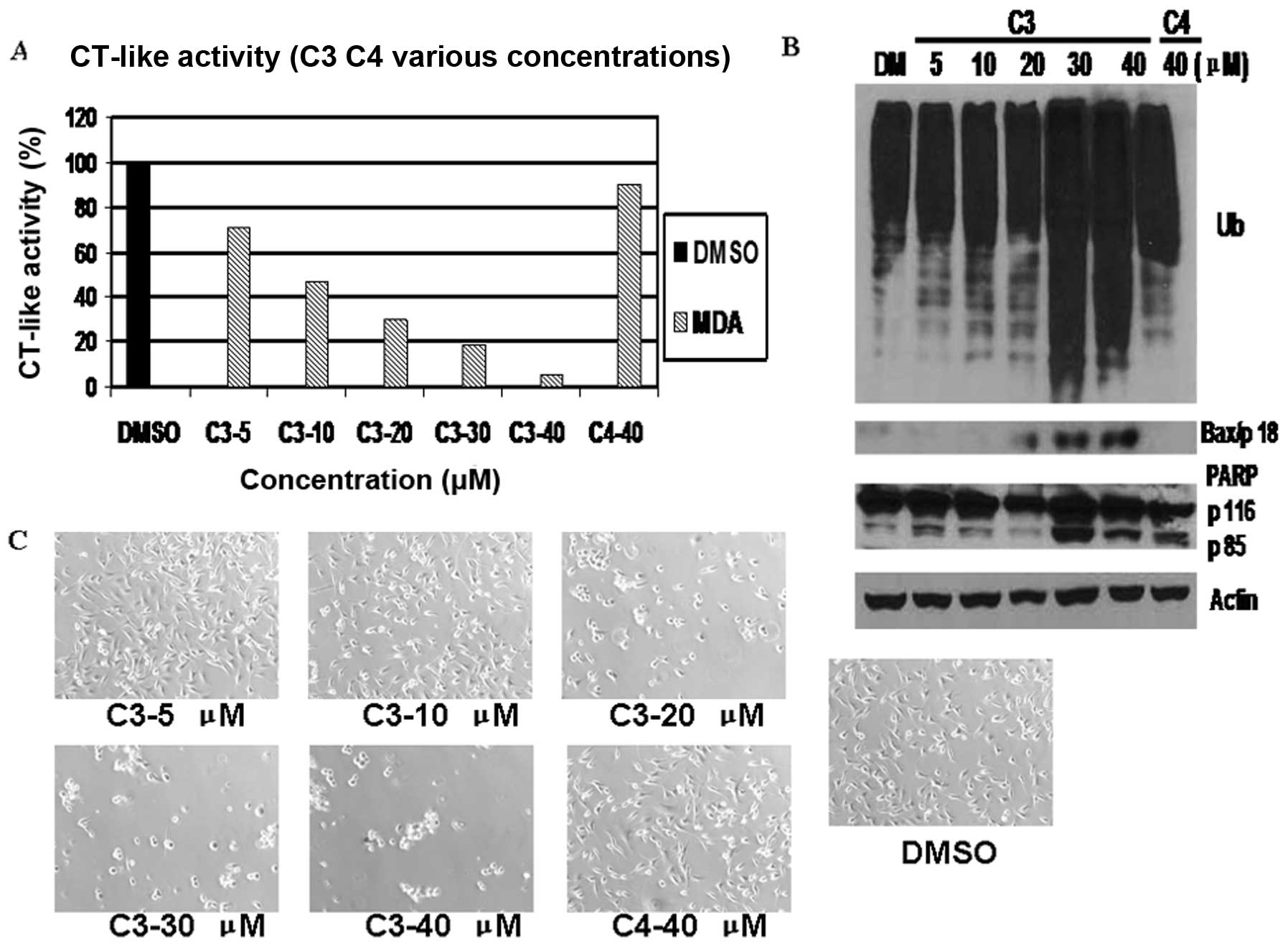
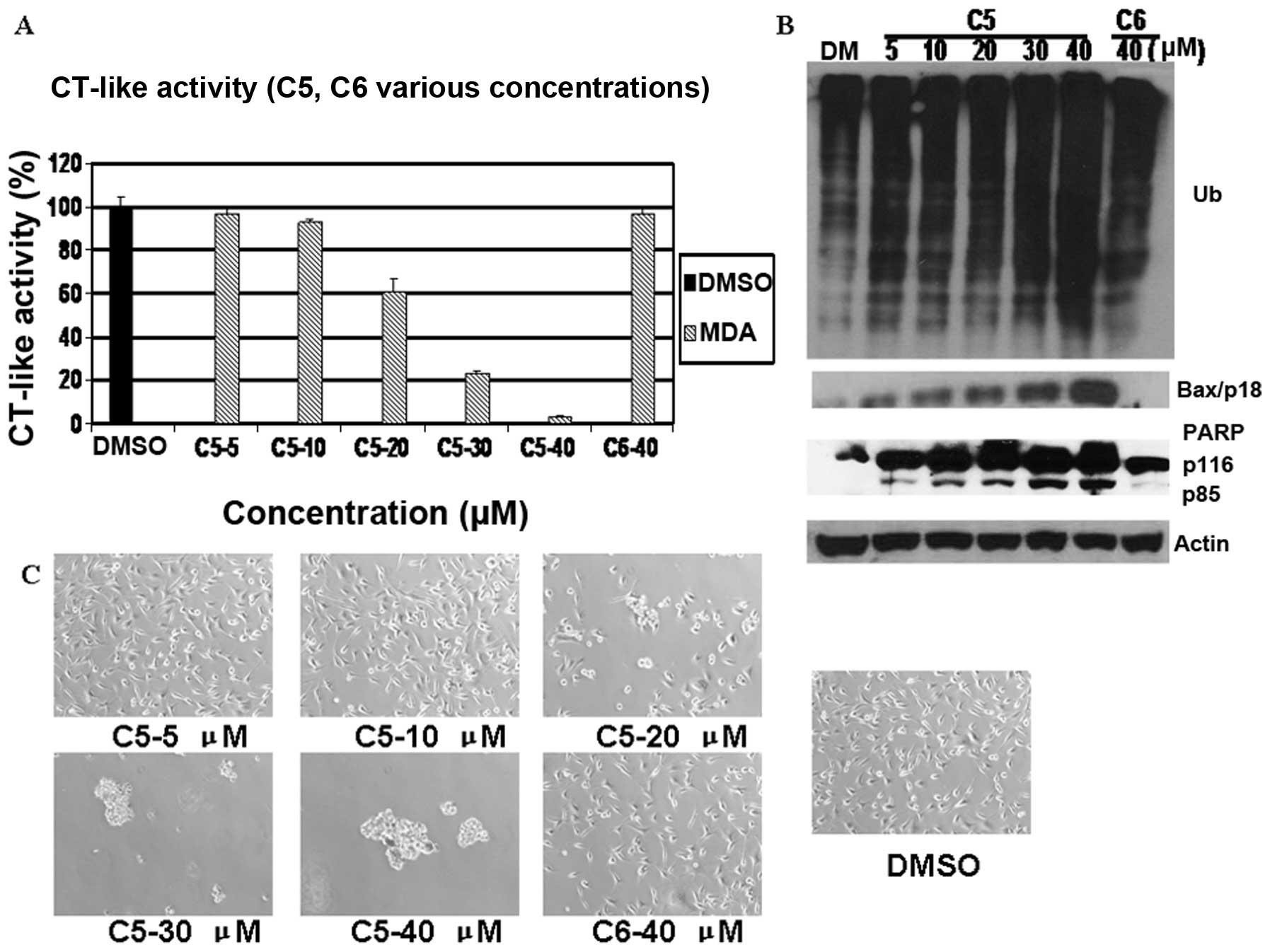
The results indicated that C3 and C5, but not C4 or
C6, inhibited the tumor cell proteasome in a dose-dependent manner,
as shown by CT-like activity assay (Figs. 5A and 7A). C3 inhibited cell proliferation by
30% at 5 μM and by approximately 50, 75, 81 and 95% at 10, 20, 30
and 40 μM, respectively (Fig.
5A). As shown in Fig. 7A, C5
at 40 μM exhibited marked effects on cellular proteasome activity,
causing 96% inhibition. By contrast, C4 and C6 had very little
inhibitory effect even at 40 μM (Figs. 5A and 7A). The levels of Bax increased at 20 μM
and, more clearly, at 40 μM in the MDA-MB-231 cells treated with C3
(Fig. 5B). They were observed at
5 μM, with the highest levels at 40 μM following treatment with C5
(Fig. 7B). The 85 kDa PARP
cleaved fragment appeared at DMSO treatment and accumulated in a
dose-dependent manner, while it appeared at 5 μM C5 treatment and
accumulated as the C5 dose increased (Figs. 5B and 7B). However, slight accumulation was
observed when 40 μM of C4 and C6 was used in the MDA-MB-231 cells
(Figs. 5B and 7B). The MDA-MB-231 cells began to show
morphological signs of apoptosis at 20 μM C3 and C5 treatment, with
100% cell death at 40 μM, whereas almost no morphological changes
were observed in the cells treated with C4 and C6 at 40 μM
(Figs. 5C and 7C).
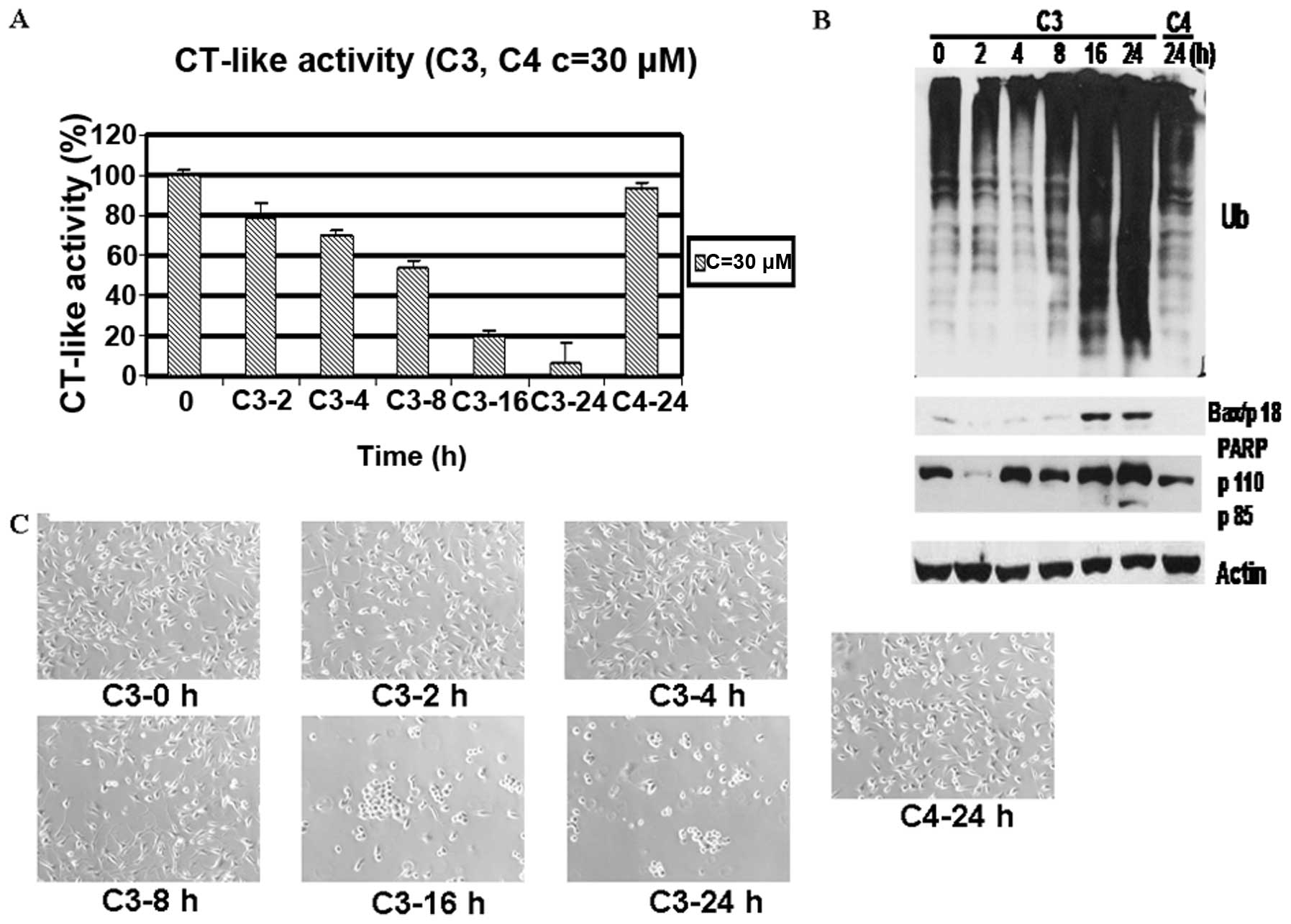
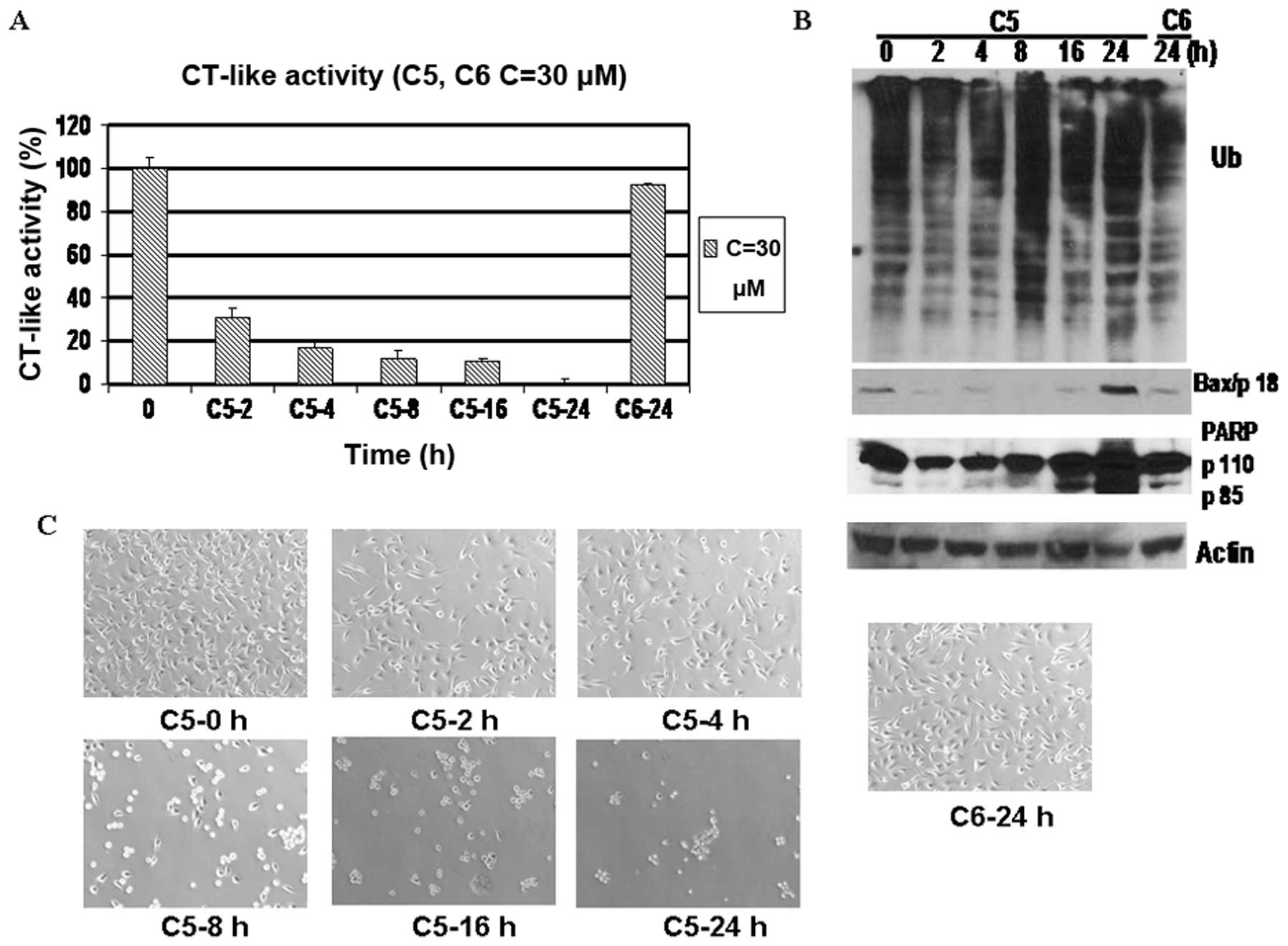
A kinetic experiment also indicated that the
MDA-MB-231 cells treated with C3 and C5 demonstrated time-dependent
proteasome inhibition from 2 to 24 h (Figs. 6A and 8A), which was associated with the
accumulation of ubiquitinated proteins, the increased levels of
Bax, increased PARP cleavage and increased morphological changes
(Figs. 6B and C; 8B and C). On the contrary, treatment
with C4 and C6 for 24 h failed to generate any of the above effects
except for a modest decrease in CT-like activity (Figs. 6 and 8).
Discussion
We have previously reported that metal-based
proteasome inhibitors potently induce apoptosis (30,31). However, the structure-activity
relationship between the metal-based complexes and mechanisms of
inhibition remains undefined. One overarching hypothesis in our
study is that the structure of the complexes affects the delivery
of the metal to the proteasome, causing proteasome inhibition
through direct interaction and tumor cell death. In order to
investigate whether specific structures have the structure-activity
relationship, we synthesized six novel Schiff base complexes that
contained different metals and tested their biological
activity.
First, we measured the anti-proliferative activity
of these complexes by MTT assay (Fig.
1) and found that C1, C3 and C5 possessed a strong ability to
inhibit cell proliferation in breast cancer cells in a
concentration-dependent manner, whereas C2, C4 and C6 did not. In
addition, C1, C3 and C5 at 30 μM inhibited the proliferation of the
MDA-MB-231 cells by 99, 95 and 90%, respectively, after 24 h of
treatment (Fig. 1). Secondly, we
investigated whether these compounds were capable of proteasome
inhibition using MDA-MB-231 breast cancer cell extracts (Fig. 2). C1, C3 and C5 inhibited the
proteasomal CT-like activity (Fig.
2), and proteasome inhibition was confirmed by the increased
levels of the proteasome target protein, Bax, as shown by western
blot analysis, in dose- and time-dependent experiments. By
contrast, C2, C4 and C6 had little to no proteasome-inhibitory and
cell death-inducing activities in the MDA-MB-231 cells.
In this study, we compared the structures and
proteasome-inhibitory potential of various metal-containing
complexes (Figs. 1 and 2; Table
I). While C2 has a very similar structure to C1, their
activities differ greatly (Figs.
3 and 4). The structural
difference between C1 and C2 is that C1 has a functional methoxyl
group, while C2 has only a methyl group. It is well established
that the CT-like activity of the 26S proteasome, which is primarily
associated with the β5 subunit, depends on the presence of the
N-terminal threonine residue that is responsible for catalyzing the
cleavage of peptides by nucleophilic attack (32). It is also known that methoxyl
attracts with electron groups which easily occurs during
nucleophilic attack, particularly with aromatic compounds (33,34). Thus, we hypothesized that the
aromatic compounds with electron-attracting cabailities, such as
methoxyl, can change the electron density and make metal complexes
highly susceptible to nucleophilic attack and therefore likely to
inhibit proteasomal CT-like function.
In order to test the aforementioned hypothesis, we
investigated the structure and activity of three paired complexes,
i.e., C2-C4, C3-C4 and C5-C6 (Figs.
1–8 and Table I). The result indicated that the
compounds C2 and C4 had almost the same structure, but not the same
metal base. We changed the metal and found that there was no change
in the activity (Figs. 3–6). Additionally, we found that the
cobalt-based C3 complex had very a similar structure to, but more
activity than C4 (Figs. 5 and
6). This inhibitory activity was
strongly associated with the abrogation (>90% at 40 μM) of the
CT-like activity of the proteasome (Fig. 5A), the accumulation of
ubiquitinated proteins, and the aggregation of a prime proteasome
target protein, Bax (Figs. 5B and
6B), indicating the occurrence of
apoptosis, which was also associated with phenotypic morphological
changes (Figs. 5C and 7C). This may be due to the fact that C3
has the electron-attracting carboxyl group, whereas C4 does not
(Table I). Similar results were
observed in the cobalt-based complexes C5 and C6 (Figs. 7 and 8). The only difference between C5 and C6
was that C5 had an indole ring that is able to attract electron
groups.
To confirm our hypothesis further, we investigated
the effect of each complex in human prostate cancer cells (LNCaP
and PC-3 cells) with the same concentration treatments. The results
showed that the complexes C1, C3 and C5 were potent inhibitors of
cell proliferation. These results were similar to those observed in
the MDA-MB-231 breast cancer cells (data not shown).
All the experiments showed that C1, C3 and C5 had
inhibitory activity. The preliminary mechanism of the activity is
possibly the special structure, the aromatic ring with
electron-attracting cabailities, can transport metal into cancer
cells more easily by changing the electron density and nucleophilic
attack.
We investigated the growth-inhibitory activity of
six metal-based complexes along with their simple mechanism of
action. C1, C3 and C5 are potent proteasome inhibitors and
apoptosis inducers in some human cancer cells. In conclusion, our
study suggests that the metal-based complexes, including aromatic
compounds with electron-attracting groups, may be promising in the
development of novel anti-cancer drugs.
Acknowledgements
The authors thank Ms. Sara Schmitt for her critical
reading of the manuscript. This study was supported by grants from
the National Science Foundation of China (no. 21371161 and 21071134
to C.B.; and no. 20971115 to Y.F.), the National Cancer Institute
(1R01CA20009, 3R01CA120009-04S1 and 5R01CA127258-05 to P.D.), the
Special Foundation for Young Teachers of Ocean University of China
(no. 201113025 to X.Z.), and a scholarship from the Chinese
Scholarship Council (to P.Z.).
References
|
1
|
Du W and Mei QB: Ubiquitin-proteasome
system, a new anti-tumor target. Acta Pharmacol Sin. 34:187–188.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Campello L, Esteve-Rudd J, Cuenca N and
Martín-Nieto J: The ubiquitin-proteasome system in retinal health
and disease. J Mol Neurobiol. 47:790–810. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang Z, Bi C, Buac D, et al: Organic
cadmium complexes as proteasome inhibitors and apoptosis inducers
in human breast cancer cells. J Inorg Biochem. 123:1–10. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Adams J: The proteasome: a suitable
antineoplastic target. Nat Rev Cancer. 4:349–360. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang H, Chen D, Cui QC, Yuan X and Dou QP:
Celastrol, a triterpene extracted from the chinese ‘Thunder of God
Vine,’ is a potent proteasome inhibitor and suppresses human
prostate cancer growth in nude mice. Cancer Res. 66:4758–4765.
2006.
|
|
6
|
Seemüller E, Lupas A, Stock D, Löwe J,
Huber R and Baumeister W: Proteasome from Thermoplasma
acidophilum: a threonine protease. Science. 268:579–582.
1995.
|
|
7
|
Andreini C, Banci L, Bertini I and Rosato
A: Zinc through the three domains of life. J Proteome Res.
5:3173–3178. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cvek B, Milacic V, Taraba J and Dou QP:
Ni(II), Cu(II), and Zn(II) diethyldithiocarbamate complexes show
various activities against the proteasome in breast cancer cells. J
Med Chem. 51:6256–6258. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Maverakis E, Fung MA, Lynch PJ, Draznin M,
Michael DJ, Ruben B and Fazel N: Acrodermatitis enteropathica and
an overview of zinc metabolism. J Am Acad Dermatol. 56:116–124.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang Y, Rodionov DA, Gelfand MS and
Gladyshev VN: Comparative genomic analyses of nickel, cobalt and
vitamin B12 utilization. BMC Genomics. 10:78–103. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Banerjee R and Ragsdale SW: The many faces
of vitamin B12: catalysis by cobalamin-dependent enzymes. Annu Rev
Biochem. 72:209–247. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Casano C, Agnello M, Sirchia R and
Luparello C: Cadmium effects on p38/MAPK isoforms in MDA-MB-231
breast cancer cells. Biometals. 23:83–92. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Abrams MJ and Murrer BA: Metal compounds
in therapy and diagnosis. Science. 261:725–730. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rosenberg B, Vancamp L and Krigas T:
Inhibition of cell division in Escherichia coli by
electrolysis products from a platinum electrode. Nature.
205:698–699. 1965.PubMed/NCBI
|
|
15
|
Wong E and Giandomenico CM: Current status
of platinum-based antitumor drugs. Chem Rev. 99:2451–2466. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Thompson KH and Orvig C: Boon and bane of
metal ions in medicine. Science. 300:936–939. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xiao Y, Bi CF, Fan YH, Cui QC, Zhang X and
Dou QP: L-glutamine Schiff base copper complex as a proteasome
inhibitor and an apoptosis inducer in human cancer cells. Int J
Oncol. 33:1073–1079. 2008.PubMed/NCBI
|
|
18
|
Hartinger CG, Zorbas-Seifried S, Jakupec
MA, Kynast B, Zorbas H and Keppler BK: From bench to
bedside-preclinical and early clinical development of the
anticancer agent indazolium
trans-[tetrachlorobis(1H-indazole)ruthenate(III)] (KP1019 or
FFC14A). J Inorg Biochem. 100:891–904. 2006.PubMed/NCBI
|
|
19
|
Ang WH and Dyson PJ: Classical and
non-classical ruthenium-based anticancer drugs: towards targeted
chemotherapy. Eur J Inorg Chem. 20:4003–4018. 2006.
|
|
20
|
Alderden RA, Hall MD and Hambley TW: The
discovery and development of cisplatin. J Chem Educ. 83:728–734.
2006. View Article : Google Scholar
|
|
21
|
Zhang X, Bi CF, Fan YH, Cui QC, Chen D,
Xiao Y and Dou QP: Induction of tumor cell apoptosis by taurine
Schiff base copper complex is associated the with inhibition of
proteasomal activity. Int J Mol Med. 22:677–682. 2008.PubMed/NCBI
|
|
22
|
Milacic V, Chen D, Giovagnini L, Diez A,
Fregona D and Dou QP: Pyrrolidine dithiocarbamate-zinc(II) and
-copper(II) complexes induce apoptosis in tumor cells by inhibiting
the proteasomal activity. Toxicol Appl Pharmacol. 231:24–33. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zuo J, Bi C, Fan Y, Buac D, Nardon C,
Daniel KG and Dou QP: Cellular and computational studies of
proteasome inhibition and apoptosis induction in human cancer cells
by amino acid Schiff base-copper complexes. J Inorg Biochem.
118:83–93. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tomco D, Schmitt S, Ksebati B, Heeg MJ,
Dou QP and Verani CN: Effects of tethered ligands and of metal
oxidation state on the interactions of cobalt complexes with the
26S proteasome. J Inorg Biochem. 105:1759–1766. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pandeya SN, Smitha S, Jyoti M and Sridhar
SK: Biological activities of isatin and its derivatives. Acta
Pharm. 55:27–46. 2005.PubMed/NCBI
|
|
26
|
Song J, Hou L, Ju C, Zhang J, Ge Y and Yue
W: Isatin inhibits proliferation and induces apoptosis of SH-SY5Y
neuroblastoma cells in vitro and in vivo. Eur J Pharmacol.
702:235–241. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Motaghed M, Al-Hassan FM and Hamid SS:
Thymoquinone regulates gene expression levels in the estrogen
metabolic and interferon pathways in MCF7 breast cancer cells. Int
J Mol Med. 33:8–16. 2014.PubMed/NCBI
|
|
28
|
Daniel KG, Gupta P, Harbach RH, Guida WC
and Dou QP: Organic copper complexes as a new class of proteasome
inhibitors and apoptosis inducers in human cancer cells. Biochem
Pharmacol. 67:1139–1151. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen D, Daniel KG, Chen MS, Kuhn DJ,
Landis-Piwowar KR and Dou QP: Dietary flavonoids as proteasome
inhibitors and apoptosis inducers in human leukemia cells. Biochem
Pharmacol. 69:1421–1432. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Adsule S, Barve V, Chen D, Ahmed F, Dou
QP, Padhye S and Sarkar FH: Novel Schiff base copper complexes of
quinoline-2 carboxaldehyde as proteasome inhibitors in human
prostate cancer cells. J Med Chem. 49:7242–7246. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Padhye S, Yang H, Jamadar A, Cui QC,
Chavan D, Dominiak K, McKinney J, Banerjee S, Dou QP and Sarkar FH:
New difluoro Knoevenagel condensates of curcumin, their Schiff
bases and copper complexes as proteasome inhibitors and apoptosis
inducers in cancer cells. Pharm Res. 26:1874–1880. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Groll M, Ditzel L, Lowe J, Stock D,
Bochtler M, Bartunik HD and Huber R: Structure of 20S proteasome
from yeast at 2.4 Å resolution. Nature. 386:463–471.
1997.PubMed/NCBI
|
|
33
|
Yuko M and Yasuo K: Synthesis of
6-methoxyindoles and indolines. Regioselective C-6 bromination of
indolines and subsequent nucleophilic substitution with a methoxyl
group. J Heterocyclic Chem. 20:349–352. 1983. View Article : Google Scholar
|
|
34
|
Daniel S, Rigobert P and Uli K: Chelated
ester enolates as versatile nucleophiles for direct nucleophilic
attack on aromatic nitro groups. Synlett. 10:1616–1618. 2006.
|