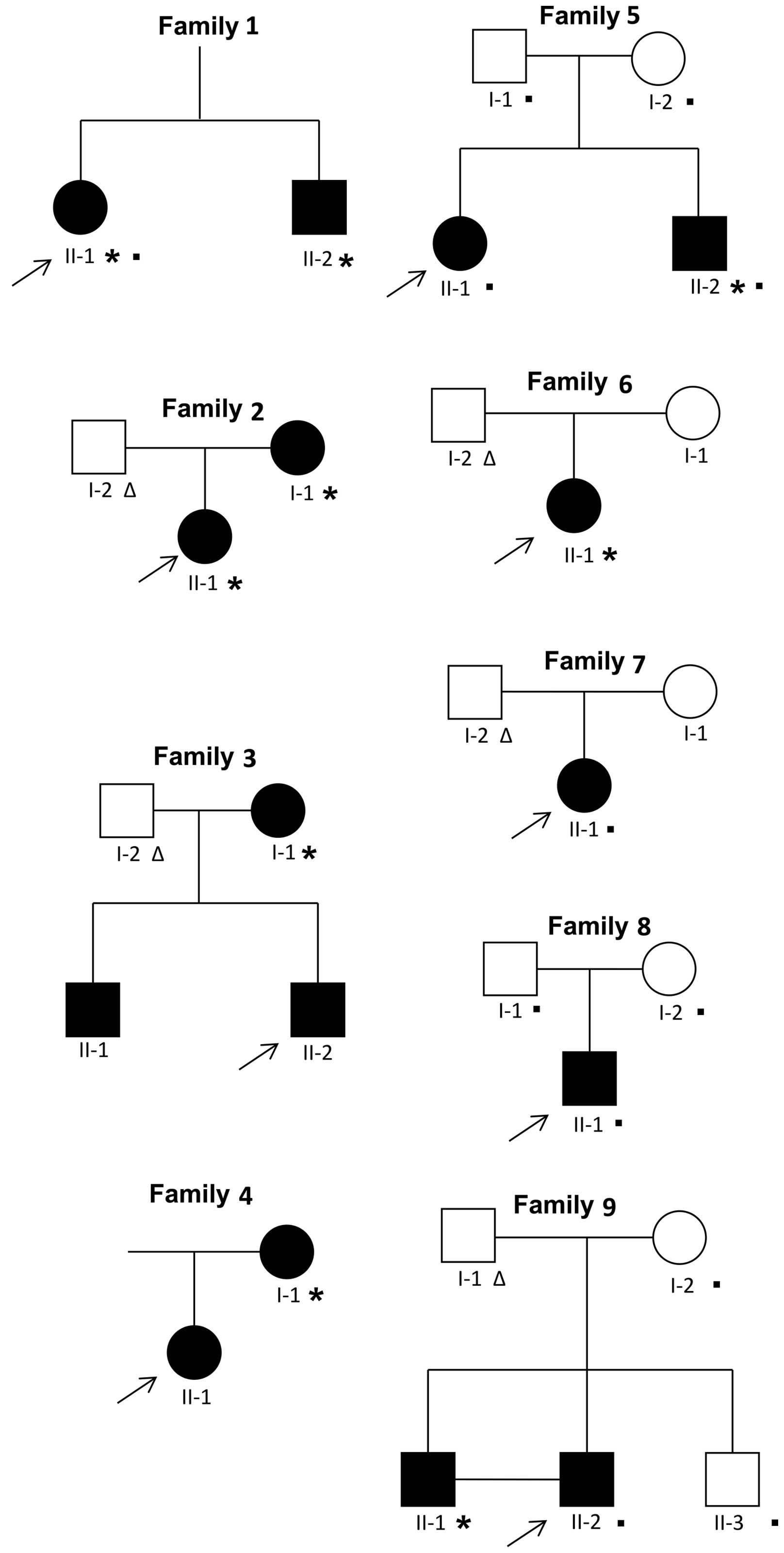
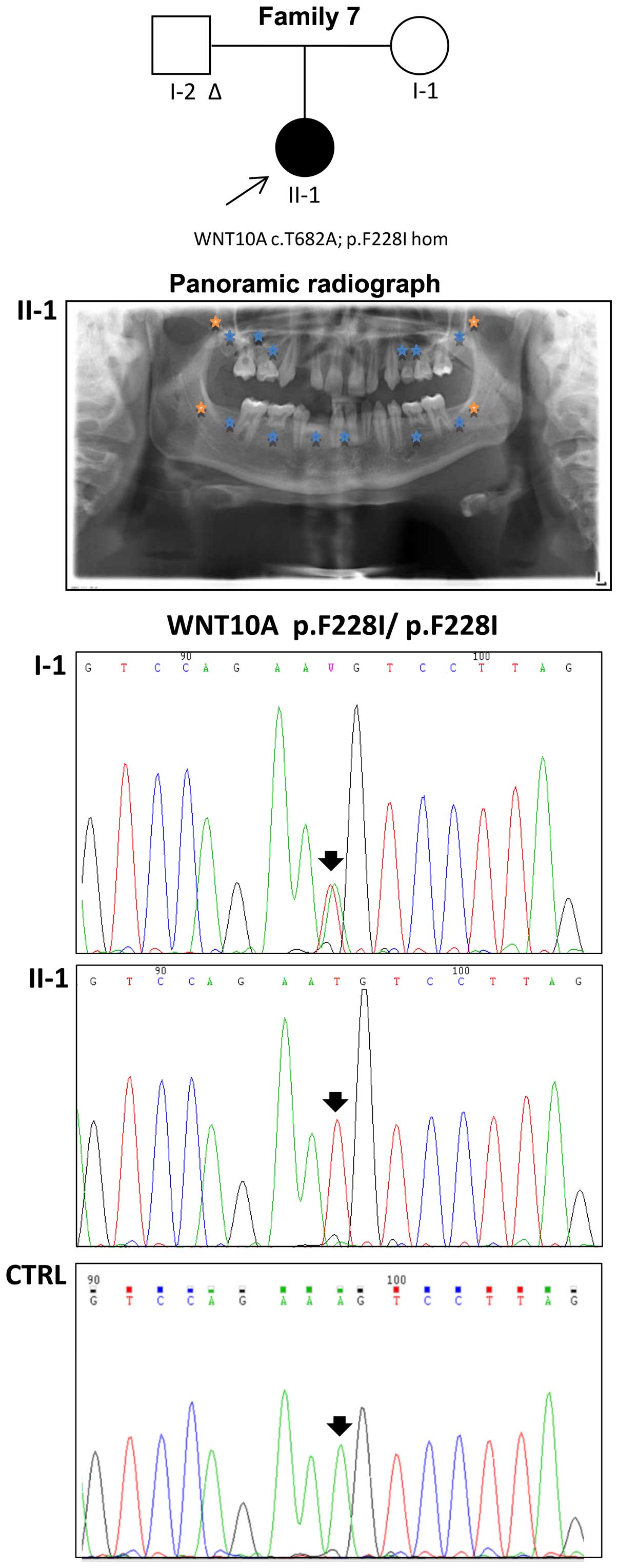
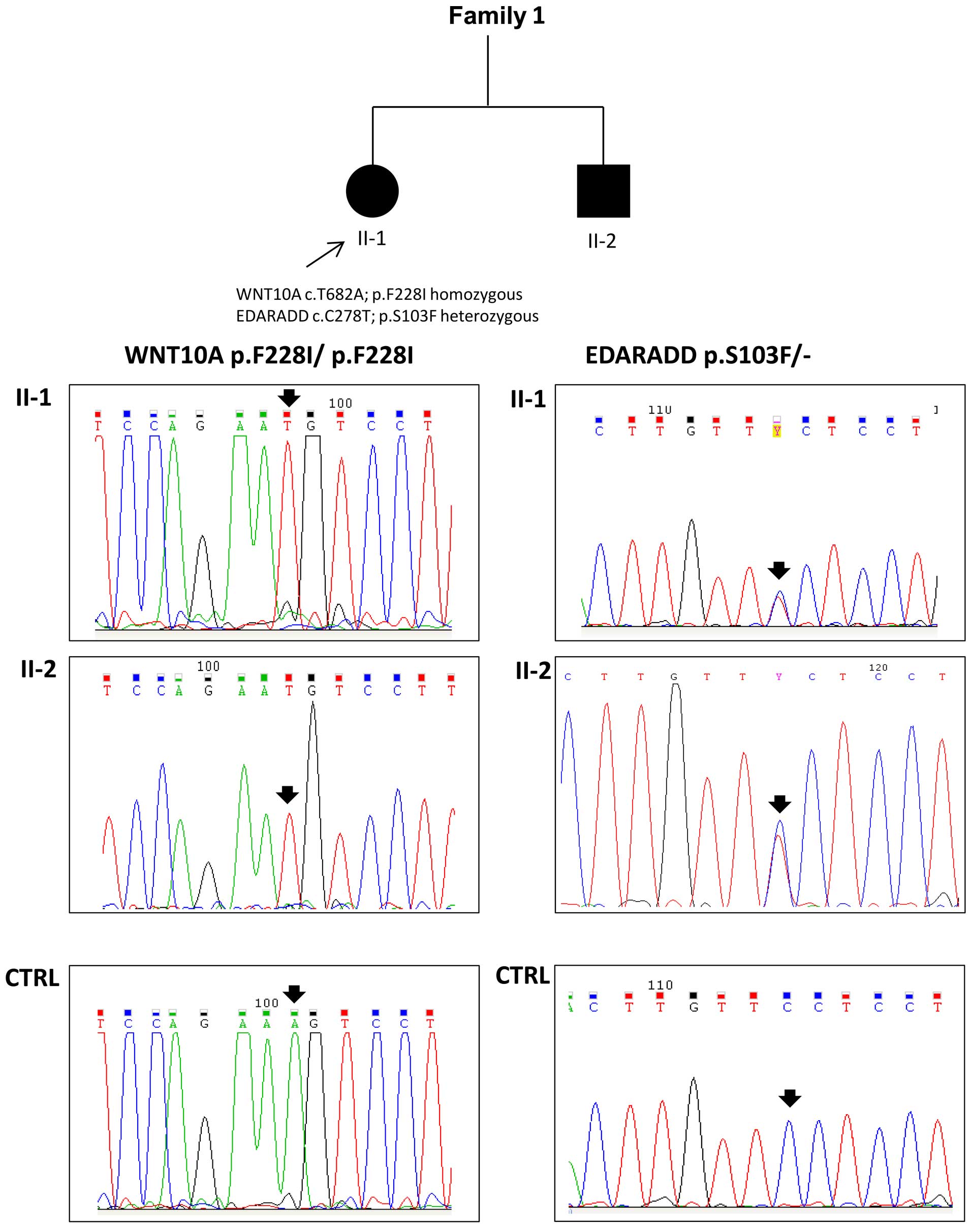
|
1
|
Bozga A, Stanciu RP and Mănuc D: A study
of prevalence and distribution of tooth agenesis. J Med Life.
7:551–554. 2014.
|
|
2
|
Brook AH, Jernvall J, Smith RN, Hughes TE
and Townsend GC: The dentition: The outcomes of morphogenesis
leading to variations of tooth number, size and shape. Aust Dent J.
59(Suppl 1): 131–142. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brook AH: Multilevel complex interactions
between genetic, epigenetic and environmental factors in the
aetiology of anomalies of dental development. Arch Oral Biol.
54(Suppl 1): S3–S17. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cobourne MT: Familial human hypodontia -
is it all in the genes? Br Dent J. 203:203–208. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yin W and Bian Z: The Gene Network
Underlying Hypodontia. J Dent Res. 94:878–885. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang YD, Chen Z, Song YQ, Liu C and Chen
YP: Making a tooth: Growth factors, transcription factors, and stem
cells. Cell Res. 15:301–316. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Feng C, Xu Z, Li Z, Zhang D, Liu Q and Lu
L: Down-regulation of Wnt10a by RNA interference inhibits
proliferation and promotes apoptosis in mouse embryonic palatal
mesenchymal cells through Wnt/β-catenin signaling pathway. J
Physiol Biochem. 69:855–863. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nassif A, Senussi I, Meary F, Loiodice S,
Hotton D, Robert B, Bensidhoum M, Berdal A and Babajko S: Msx1 role
in cranio-facial bone morphogenesis. Bone. 66:96–104. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ogawa T, Kapadia H, Feng JQ, Raghow R,
Peters H and D'Souza RN: Functional consequences of interactions
between Pax9 and Msx1 genes in normal and abnormal tooth
development. J Biol Chem. 281:18363–18369. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Suryadeva S and Khan MB: Role of homeobox
genes in tooth morphogenesis: A review. J Clin Diagn Res.
9:ZE09–ZE12. 2015.PubMed/NCBI
|
|
11
|
Lammi L, Arte S, Somer M, Jarvinen H,
Lahermo P, Thesleff I, Pirinen S and Nieminen P: Mutations in AXIN2
cause familial tooth agenesis and predispose to colorectal cancer.
Am J Hum Genet. 74:1043–1050. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Galluccio G, Castellano M and La Monaca C:
Genetic basis of non-syndromic anomalies of human tooth number.
Arch Oral Biol. 57:918–930. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Song S, Zhao R, He H, Zhang J, Feng H and
Lin L: WNT10A variants are associated with non-syndromic tooth
agenesis in the general population. Hum Genet. 133:117–124. 2014.
View Article : Google Scholar
|
|
14
|
Arzoo PS, Klar J, Bergendal B, Norderyd J
and Dahl N: WNT10A mutations account for ¼ of population-based
isolated oligodontia and show phenotypic correlations. Am J Med
Genet A. 164A:353–359. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nikopensius T, Annilo T, Jagomägi T,
Gilissen C, Kals M, Krjutškov K, Mägi R, Eelmets M, Gerst-Talas U,
Remm M, et al: Non-syndromic tooth agenesis associated with a
nonsense mutation in ectodysplasin-A (EDA). J Dent Res. 92:507–511.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Klein ML, Nieminen P, Lammi L, Niebuhr E
and Kreiborg S: Novel mutation of the initiation codon of PAX9
causes oligodontia. J Dent Res. 84:43–47. 2005. View Article : Google Scholar
|
|
17
|
De Muynck S, Schollen E, Matthijs G,
Verdonck A, Devriendt K and Carels C: A novel MSX1 mutation in
hypodontia. Am J Med Genet A. 128A:401–403. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim JW, Simmer JP, Lin BP and Hu JC: Novel
MSX1 frameshift causes autosomal-dominant oligodontia. J Dent Res.
85:267–271. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Arte S, Parmanen S, Pirinen S, Alaluusua S
and Nieminen P: Candidate gene analysis of tooth agenesis
identifies novel mutations in six genes and suggests significant
role for WNT and EDA signaling and allele combinations. PLoS One.
8:e737052013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li H and Durbin R: Fast and accurate
long-read alignment with Burrows-Wheeler transform. Bioinformatics.
26:589–595. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Van der Auwera GA, Carneiro MO, Hartl C,
Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen
D, Thibault J, et al: From FastQ data to high confidence variant
calls: the Genome Analysis Toolkit best practices pipeline. Curr
Protoc Bioinformatics. 43:111011–33. 2013.
|
|
22
|
Wang K, Li M and Hakonarson H: ANNOVAR:
Functional annotation of genetic variants from high-throughput
sequencing data. Nucleic Acids Res. 38:e1642010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Landrum MJ, Lee JM, Benson M, Brown G,
Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, et al:
ClinVar: Public archive of interpretations of clinically relevant
variants. Nucleic Acids Res. 44:D862–D868. 2016. View Article : Google Scholar :
|
|
24
|
Petrovski S, Wang Q, Heinzen EL, Allen AS
and Goldstein DB: Genic intolerance to functional variation and the
interpretation of personal genomes. PLoS Genet. 9:e10037092013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Adzhubei IA, Schmidt S, Peshkin L,
Ramensky VE, Gerasimova A, Bork P, Kondrashov AS and Sunyaev SR: A
method and server for predicting damaging missense mutations. Nat
Methods. 7:248–249. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shihab HA, Gough J, Cooper DN, Stenson PD,
Barker GL, Edwards KJ, Day IN and Gaunt TR: Predicting the
functional, molecular, and phenotypic consequences of amino acid
substitutions using hidden Markov models. Hum Mutat. 34:57–65.
2013. View Article : Google Scholar :
|
|
27
|
Vieira AR, Meira R, Modesto A and Murray
JC: MSX1, PAX9, and TGFA contribute to tooth agenesis in humans. J
Dent Res. 83:723–727. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tallón-Walton V, Manzanares-Céspedes MC,
Carvalho-Lobato P, Valdivia-Gandur I, Arte S and Nieminen P:
Exclusion of PAX9 and MSX1 mutation in six families affected by
tooth agenesis. A genetic study and literature review. Med Oral
Patol Oral Cir Bucal. 19:e248–e254. 2014. View Article : Google Scholar :
|
|
29
|
van den Boogaard MJ, Créton M, Bronkhorst
Y, van der Hout A, Hennekam E, Lindhout D, Cune M and Ploos van
Amstel HK: Mutations in WNT10A are present in more than half of
isolated hypodontia cases. J Med Genet. 49:327–331. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
MacArthur DG, Manolio TA, Dimmock DP, Rehm
HL, Shendure J, Abecasis GR, Adams DR, Altman RB, Antonarakis SE,
Ashley EA, et al: Guidelines for investigating causality of
sequence variants in human disease. Nature. 508:469–476. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lubitz SA, Sinner MF, Lunetta KL, Makino
S, Pfeufer A, Rahman R, Veltman CE, Barnard J, Bis JC, Danik SP, et
al: Independent susceptibility markers for atrial fibrillation on
chromosome 4q25. Circulation. 122:976–984. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Antunes LS, Küchler EC, Tannure PN, Lotsch
PF, Costa MC, Gouvêa CV, Olej B and Granjeiro JM: TGFB3 and BMP4
polymorphism are associated with isolated tooth agenesis. Acta
Odontol Scand. 70:202–206. 2012. View Article : Google Scholar
|
|
33
|
Mu Y, Xu Z, Contreras CI, McDaniel JS,
Donly KJ and Chen S: Phenotype characterization and sequence
analysis of BMP2 and BMP4 variants in two Mexican families with
oligodontia. Genet Mol Res. 11:4110–4120. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Capasso M, Ayala F, Russo R, Avvisati RA,
Asci R and Iolascon A: A predicted functional single-nucleotide
polymorphism of bone morphogenetic protein-4 gene affects mRNA
expression and shows a significant association with cutaneous
melanoma in Southern Italian population. J Cancer Res Clin Oncol.
135:1799–1807. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang L, Sun HY, Chen DZ, Lu MD, Tang Y and
Xiao JP: Explore the dynamic alternation of gene PLAC4 mRNA
expression levels in maternal plasma in second trimester for
nonivasive detection of trisomy 21. Obstet Gynecol Sci. 58:261–267.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cabral RM, Kurban M, Wajid M, Shimomura Y,
Petukhova L and Christiano AM: Whole-exome sequencing in a single
proband reveals a mutation in the CHST8 gene in autosomal recessive
peeling skin syndrome. Genomics. 99:202–208. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Parker MJ, Fryer AE, Shears DJ, Lachlan
KL, McKee SA, Magee AC, Mohammed S, Vasudevan PC, Park SM, Benoit
V, et al: De novo, heterozygous, loss-of-function mutations in
SYNGAP1 cause a syndromic form of intellectual disability. Am J Med
Genet A. 167A:2231–2237. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Nyboe D, Kreiborg S, Kirchhoff M and Hove
HB: Familial craniosynostosis associated with a microdeletion
involving the NFIA gene. Clin Dysmorphol. 24:109–112. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Esposito E, Napolitano G, Pescatore A,
Calculli G, Incoronato MR, Leonardi A and Ursini MV: COMMD7 as a
novel NEMO interacting protein involved in the termination of NF-κB
signaling. J Cell Physiol. 231:152–161. 2016. View Article : Google Scholar
|