Introduction
Small vesicles released from cells have recently
emerged as important mediators of inter-cellular communication.
These vesicles that have been termed extracellular vesicles (EVs)
are inclusive of exosomes released from the endosomal cell-membrane
compartment and of microvesicles released from the cell surface by
plasma membrane budding. The EV content of proteins, lipids and
nucleic acids varies with the cell of origin and after
incorporation into recipient cells, they may transfer information
that may change the phenotype and function of recipient cells
(1–3). Previous studies have addressed the
role of EVs in physiological and pathological conditions based on
their biological activity and molecular constituents (1–3).
Additionally, since EVs retain the signature of the cell of origin
and are present in all body fluids, their potential use as
diagnostics in different pathological conditions has been
suggested. A fundamental issue remains on how to isolate EVs from
cultured cells in order to study their biological functions or from
biological fluids for diagnostic purposes. Since fetal bovine serum
frequently used for cell culture is enriched in EVs, in
vitro experiments require the use of serum depletion of EVs
(4). By contrast, the isolation
of EVs from body fluids leads to the management of the complexity
due to the concomitant presence of EVs of different cell origin.
Therefore, in order to identify a potential biomarker it is
critical to discriminate cell origin on the base of EV molecular
expression or content by proteomic or genomic analysis.
Following removal of cell debris by centrifugation,
the three main methods used for isolation of EVs include
differential ultracentrifugation in the absence or presence of
sucrose gradient, size exclusion chromatography, and immune
affinity. These methods have some advantages mainly associated with
the possibility to discriminate between different EV populations
and concerns related to the risk to damage vesicles during
purification with loss of biological activity, the need of a
sufficiently large sample and the efficiency of isolation (reviewed
in ref. 5). In addition,
polymeric precipitation has been suggested as an alternative method
mainly focused on the evaluation of RNA and protein content
(6). The methods of polymeric
precipitation are based on the formation of a mesh-like net that
embeds EVs with a size ranging from 60 to 180 nm. These methods may
be applied to culture media or to body fluids. In particular,
polymeric precipitation methods may have the advantage in the
detection of biomarkers in vesicles derived from small biological
samples.
The aim of the present study was to investigate the
possibility to implement polymeric precipitation with a
charge-based precipitation of EVs. For this purpose, we first
measured the charge of EVs from different biological sources.
Taking into account the EV-negative charge, we precipitated EVs in
the presence of positively charged protamine in a polymeric matrix
and compared the efficiency with ultracentrifugation in terms of
yield of recovered vesicles, efficiency of RNA extraction, exosomal
protein expression and biological activity.
Materials and methods
Biological samples
Saliva was obtained from adult normal volunteers
(n=5). The study of exosomes/microvesicles in saliva and serum of
healthy human volunteers was approved by the Internal Ethics
Committee of the Molecular Biotechnology Center. Human serum from
healthy blood donors (n=5) was provided by the Blood Bank of Città
della Salute e della Scienza di Torino, after informed consent and
approval by the internal Review Board of Blood Bank were
obtained.
Adult human liver stem cells (HLSCs)
HLSCs were isolated from human cryopreserved normal
adult hepatocytes (Lonza, Basel, Switzerland), cultured and
characterized as previously described (7). Briefly, hepatocytes were first
cultivated for 2 weeks in Hepatozyme-SFM medium (Gibco, Grand
Island, NY, USA), then in α-MEM/EBM-1 (3:1) (Invitrogen, Carlsbad,
CA, USA) media added with HEPES (12 mM, pH 7.4), L-glutamine (5 mM)
penicillin (50 IU/ml), streptomycin (50 µg/ml) (all from
Sigma, St. Louis, MO, USA), and fetal calf serum (FCS) (10%)
(Invitrogen). The cells were expanded and characterized. The
characterization of HLSCs by cytofluorimetric analysis demonstrated
the expression of the mesenchymal stem cell markers but not of the
endothelial and hematopoietic markers as previously described
(7). HLSCs also expressed
α-fetoprotein, human albumin, vimentin and nestin resident stem
cell markers, but not CD34, CD117 and cytokeratin 19 oval cell
markers (7). In addition, HLSCs
were positive for the Nanog, Sox2, Oct4 and SSEA4 embryonic stem
cell markers (8). HLSCs under
appropriate culture conditions underwent endothelial, osteogenic
and hepatic differentiation (7).
Keratinocytes
Keratinocytes (HaCaT) were purchased and cultured
with KBM-gold basal medium (Lonza, Basel, Switzerland) at 37°C with
5% CO2. The cells were seeded at density
3.5×102 cell/cm2, using 1 ml of
medium/cm2 and subcultured when cell confluence was
70–80%. Briefly, flasks were washed with HEPES buffer saline
solution, incubated with trypsin solution for 6 min and then
trypsin was neutralized with medium containing 10% FCS. If the
cells were not completely detached within 7 min, incubation with
trypsin was repeated.
Renal tubular epithelial cells (TEC)
TEC line immortalized by infection with a hybrid
Adeno5/SV40 virus was previously developed by Cantaluppi et
al (9). Cells were grown
using Dulbecco's modified Eagle's medium (DMEM) (Lonza) containing
10% FCS (Gibco) and 2 mM glutamine (Life Technologies, Carlsbad,
CA, USA). TEC showed negative staining for von Willebrand factor,
minimal staining for desmin and vimentin, and marked staining with
antibodies directed to cytokeratins and actin. TEC was also
positive for markers of fully differentiated proximal TEC such as
alkaline phosphatase, amino peptidase A and megalin.
Isolation of EVs
EVs were purified from the HLSC culture media, human
serum and saliva. EVs isolated from the supernatants of HLSCs
(2×106 cells/T75) were obtained after 24 h culture in
RPMI-1640 deprived of FCS. At the time of EV isolation, 97–99% of
cells was viable by trypan blue exclusion assay, although the TUNEL
assay did not detect apoptotic cells.
Saliva samples (5 ml) were collected in sterile
tubes and kept in ice during harvest. Serum samples were collected
from healthy donors using serum separating tubes (BD) and
centrifuged at 1,500 × g for 15 min.
Prior to the isolation procedures, HLSC supernatant,
saliva and serum samples were submitted to two centrifugations at
3,000 × g for 20 min to remove cell debris and other contaminants.
The saliva samples were diluted 1:1 with phosphate-buffered saline
(PBS) and filtered with 0.22 µm filters.
Differential ultracentrifugation
Following the removal of cell debris and apoptotic
bodies by two centrifugations at 3,000 × g for 20 min, EVs were
purified as previously described by Théry et al (10) by a first ultracentrifugation at
10,000 × g followed by ultracentrifugation at 100,000 × g for 1 h
at 4°C (Beckman Coulter Optima L-90K; Beckman Coulter, Fullerton,
CA, USA).
Charge-based precipitation
In preliminary experiments, samples were incubated
with various doses of protamine (1.0, 0.5, 0.25 and 0.1 mg/ml) to
determine the optimal protamine concentration. The biological
samples ready for the precipitation procedure were transferred in
sterile vials and added with the protamine (P) (Sigma)/Polyethylene
glycol (PEG 35,000; Merck KGaA, Darmstadt, Germany) precipitation
solution (P/PEG) (1 volume precipitation solution:4 volume sample).
Control P or PEG 35,000 alone (PEG) served as the controls. The
composition of precipitation solution was 0.2 g PEG 35,000 (Merck
KGaA) and 1 mg protamine chloride/ml (Sigma) of distilled
water.
After overnight incubation at 4°C, the mixture was
centrifuged at 1,500 × g for 30 min at 22°C and the supernatant was
discarded. The pellet was re-suspended in the appropriate buffer to
study biological activities or in lysis buffer for RNA extraction
and western blot analysis.
To remove the lipoproteins, Sephadex G-100 (GE
Healthcare Bio-Sciences AB, Uppsala, Sweden) spin columns were
prepared and samples were centrifuged at 1,000 × g for 1 min. EVs
were recovered in the void volumes.
Measure of EV charge
The analysis was performed by Zeta-sizer
nanoinstrument (size range, 0.3 nm-10 µm; Malvern
Instruments SA, Vénissieux, France). The ζ potential (slipping
plane) was generated at × distance from the particle indicating the
degree of electrostatic repulsion between adjacent, similarly
charged particles in a dispersion. Negative ζ-potential indicated a
high grade of dispersion across the particles.
Nanoparticle tracking analysis (NTA)
NanoSight LM10 (Malvern Instruments, Malvern, UK)
was used to analyze the concentration and size distribution of EVs
by means of the NTA software (Malvern Instruments SA). The Brownian
movements of EVs present in the sample subjected to a laser light
source were recorded by a camera and converted into size and
concentration parameters by NTA through the Stokes-Einstein
equation [https://en.wikipedia.org/wiki/Einstein_relation_(kinetic_theory)].
Transmission electron microscopy
Transmission electron microscopy was performed on
EVs isolated by ultracentrifugation or charge-based precipitation
resuspended in PBS, placed on 200 mesh nickel formvar carbon-coated
grids (Electron Microscopy Science, Hatfield, PA, USA) and left to
adhere for 20 min. The grids were then incubated with 2.5%
glutaraldehyde containing 2% sucrose and after washings in
distilled water the EVs were negatively stained with NanoVan
(Nanoprobes, Yaphank, NK, USA) and observed using a Jeol JEM 1010
electron microscope (Jeol, Tokyo, Japan).
Western blot analysis
Protein content of the EV preparations was
quantified using the Bradford method (Bio-Rad, Hercules, CA, USA).
Protein samples were separated by 4–15% gradient sodium dodecyl
sulfate-polyacrylamide gel electrophoresis and subjected to
immunoblotting with rabbit polyclonal antibodies (1:1000 dilutions)
anti-CD9 (Cat. no. ab155825), CD63 (Cat. no. ab199921), CD81 (Cat.
no. ab109201), anti-apolipoprotein B100 (ApoB100; 1:5,000 dilution;
Cat. no. ab20737) and goat polyclonal antibody anti-apolipoprotein
A1 (ApoA1; 1:5,000 dilution; Cat. no. ab7613) (Abcam, Cambridge,
UK). The protein bands were visualized using an enhanced
chemiluminescence (ECL) detection kit and ChemiDoc™ XRS + System
(Bio-Rad). Cell and EV lysates were loaded at concentrations of 30
µg/well.
RNA extraction
The mirVana RNA isolation kit (Thermo Fisher
Scientific, Waltham, MA, USA) was used to extract total RNA from
EVs following the manufacturer's protocol. RNA was
spectrophotometrically quantified (NanoDrop ND-1000; NanoDrop,
Wilmington, DE, USA).
miRNA and mRNA profiling by quantitative
PCR
Quantitative PCR was carried out as previously
described (11) using a 48-well
StepOne™ Real-Time system (Applied Biosystems, Waltham, MA, USA).
In brief, 0.2 mg RNA was first reverse-transcribed using a miScript
reverse transcription kit (Qiagen, Valencia, CA, USA).
Subsequently, 3 ng of cDNA in triplicate were employed to identify
and measure significant miRNAs performing RT-qPCR using a miScript
SYBR-Green PCR kit (Qiagen, Valencia, CA, USA). miRNA-specific
primers to hsa-miR-16 (5′-TAG CAG CAC GTA AAT ATT GGC G-3′), 29a
(5′-TAG CAC CAT CTG AAA TCG GTT A-3′), 99b (5′-CCC GTA GAA CCG ACC
TTG C-3′), 191 (5′-CAA CGG AAT CCC AAA AGC AG-3′), 223 (5′-TGT CAG
TTT GTC AAA TAC CCC A-3′) were used in separate reactions. The
RNU44 (purchased by Qiagen) and RNU48 (5′-AAC TCT GAG TGT GTC GCT
GAT G-3′) snoRNAs served as the positive controls and 10 µl
of water were used as the negative control in place of the RNA.
RT-qPCR analysis was also performed for the presence
of mRNA of ID1 (F, 5′-GGC GGC ATG CGT TCC-3′ and R, 5′-TTG TTC TCC
CTC AGA TCC GG-3′) in serum, Annexin A1 (F, 5′-CGG AAC GCT TTG CTT
TCT CTT and R, 5′-CAA GGC CCT GGC ATC TGA-3′) in saliva and DCR1
(F, 5′-CGT TAT CAT TCC AAG ATA TCG CAA-3′ and R, 5′-GGG TAA GAT CAG
TGT ACA CAT CAG CT-3′) in HLSC EVs.
Cell proliferation assays
Immortalized TEC was seeded at a density of
3×103 cells/well in 96-well plates in DMEM supplemented
with 10% FCS. After 12 h, TEC was starved with medium without FCS
for 2 h, stimulated with HLSC EVs and then 10 µM BrdU was
added overnight. The plates were analyzed by BrdU kit (BrdU; Roche
Diagnostics, Mannheim, German) and the absorption values were
determined at a 405 nm wavelength.
In vitro scratch wound-healing assay
HaCaT cells were seeded at a density of
~50×103 cells/well in 24-well plates in DMEM
supplemented with 10% FCS. When the cells reached complete
confluence, they were starved with medium without FCS overnight.
The following day, scratch wounds were created with a sterile tip.
Prior to stimulation (t=0), micrographs of the well were obtained
using a Leica microscope (Leica, Wetzlar, Germany). The cells were
then stimulated with EVs (50,000 EVs/target cells) isolated from
the saliva of three different donors. The 'wound closure'
phenomenon was monitored for 36 h using the Leica microscope and
images were analyzed by ImageJ software (Bethesda, MD, USA)
observing the decrease of the wound area in cells stimulated with
saliva EVs in comparison to cells not stimulated with EVs.
Statistical analysis
Data were presented as mean ± SD. Statistical
analysis was performed using ANOVA with Dunnet's multicomparison
tests when appropriate. P<0.05 was considered significant.
Results
The analysis of the ζ potential was performed on
different biological samples showing that EVs have a negative
charge (Table I). In preliminary
experiments, serum was incubated with different doses of protamine
(1.0, 0.5, 0.25 and 0.1 mg/ml) overnight at 4°C and precipitated
EVs were recovered by centrifugation at 3,000 × g for 30 min
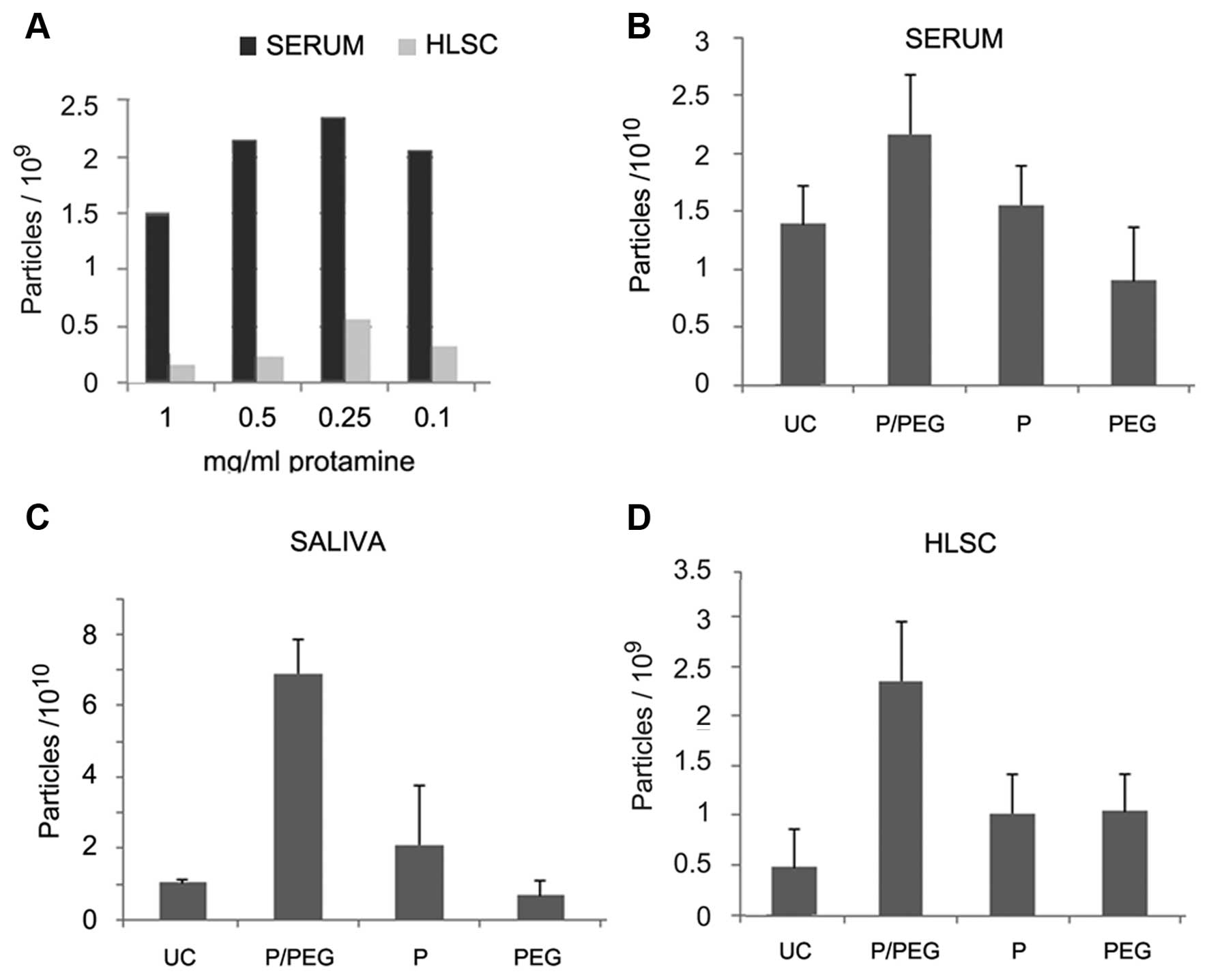
(Fig. 1A). However, the EV pellet
was easily re-suspended with the dose of 0.25 mg protamine/ml serum
whereas higher concentrations generated pellets that were more
difficult to re-suspend. We observed that the addition of PEG
35,000 Da to protamine favored resuspension. On this basis, a
precipitation strategy was established to favor precipitation of
negatively charged EVs into a polymeric matrix that would allow the
recovery of EVs following centrifugation without the need of an
ultracentrifugation step.
 | Table IAnalysis of ζ potential on biological
samples. |
Table I
Analysis of ζ potential on biological
samples.
| ζ potential of
EVs | mV |
|---|
| HLSC | −13.800 |
| Serum | −7.825 |
| Saliva | −8.54 |
Fig. 1 shows the
comparison by NTA of EV recovery from serum, saliva and cell free
supernatant of HLSCs after ultra centrifugation (UC) or
precipitation with P/PEG, PEG alone and protamine alone. The
results indicated that P/PEG precipitation was more efficient than
other conditions in terms of number of EVs detected by NTA. The
comparison between serum and saliva of P/PEG-precipitated EVs
indicated an enrichment of vesicles in saliva in respect to serum.
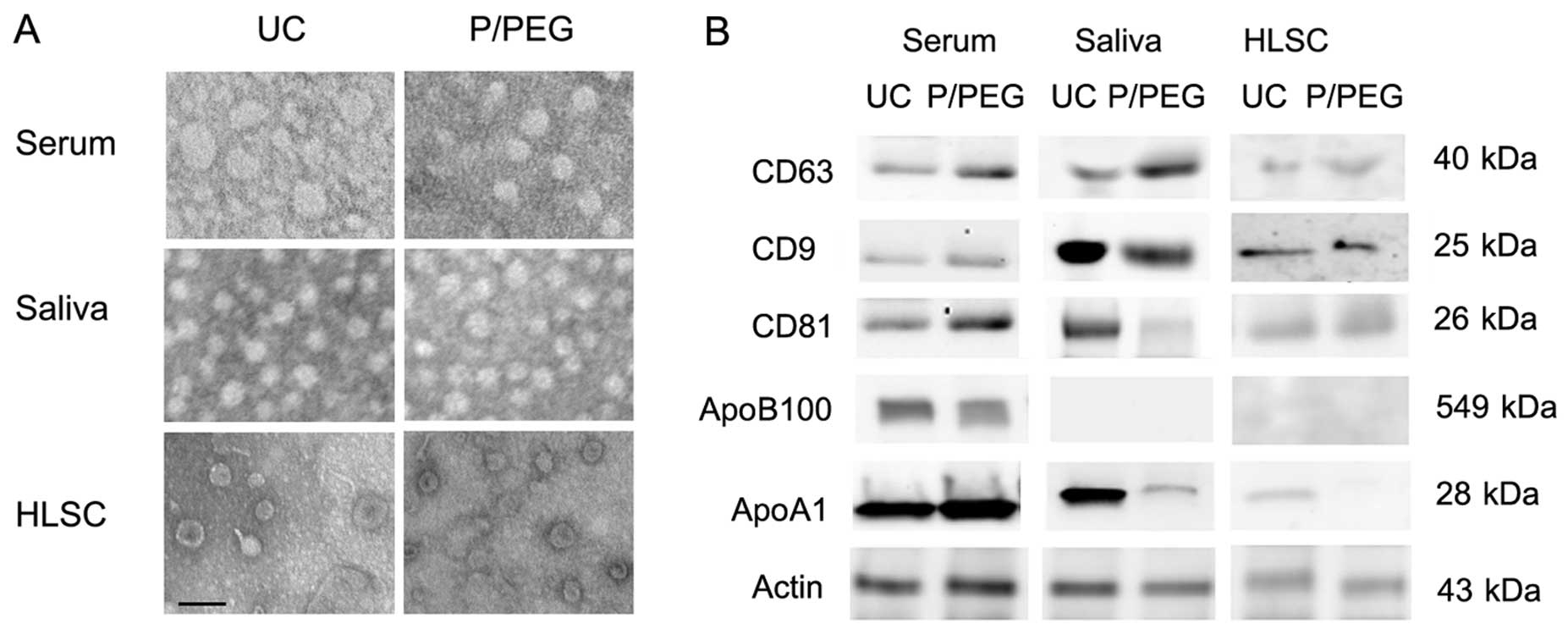
The size of EVs isolated in the different conditions was similar as
observed by transmission electron microscopy. Serum-derived EVs
ranged from 35 to 95 nm, whereas those derived from saliva were a
more homogeneous population with a size ranging from 45 to 65 nm.
EVs derived from HLSCs ranged from 45 to 75 nm (Fig. 2A).
As for EVs obtained by ultracentrifugation, the
western blot analysis of EVs precipitated from serum, saliva and
HLSCs by P/PEG showed the expression of CD63, CD9 and CD81 exosomal
markers (Fig. 2B). Since it has
been suggested that precipitation techniques co-isolate contaminant
lipoproteins (12,13), using western blot analysis, we
evaluated the presence of ApoB100 and ApoA1 in EVs obtained by
ultracentrifugation and P/PEG precipitation. As shown in Fig. 2B, ApoB100 and ApoA1 were detected
in serum EVs obtained by ultracentrifugation and precipitation. In
saliva EVs, ApoB100 was absent. ApoA1 was detectable in EVs
obtained from saliva following ultracentrifugation, whereas ApoA1
was barely detectable in P/PEG precipitation samples. ApoB100 was
absent in EVs purified with HLSC culture media by
ultracentrifugation and precipitation, whereas ApoA1 was detectable
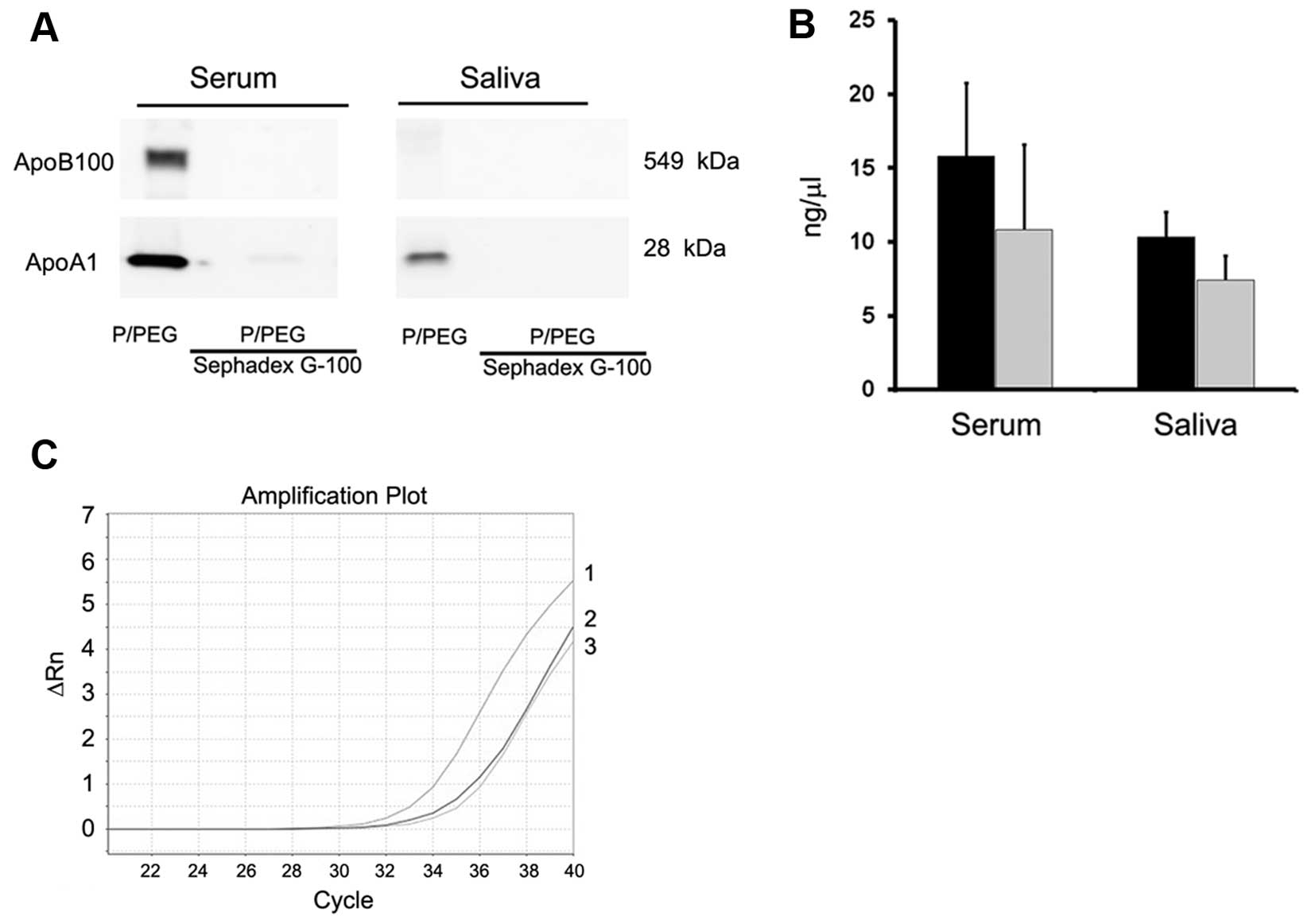
only in EVs purified with ultracentrifugation. To remove
lipoproteins, Sephadex G-100 spin columns were used and the EVs
were recovered in the void volumes whereas apolipoproteins were
retained. As shown in Fig. 3A,
gel-filtration with Sephadex G-100 reduced ApoB100 and ApoA1 in
serum EVs and ApoA1 in saliva EVs.
Detection of RNAs in EVs
As shown in Fig.
3B, the amount of RNA extracted after Sephadex G-100
pre-absorption was reduced in serum and saliva but the difference
was not statistically significant (P>0.05). PCR analysis showed
also a reduction of ~2 cycles of a representative mRNA present in
serum EVs (ID1 mRNA) (Fig. 3C).
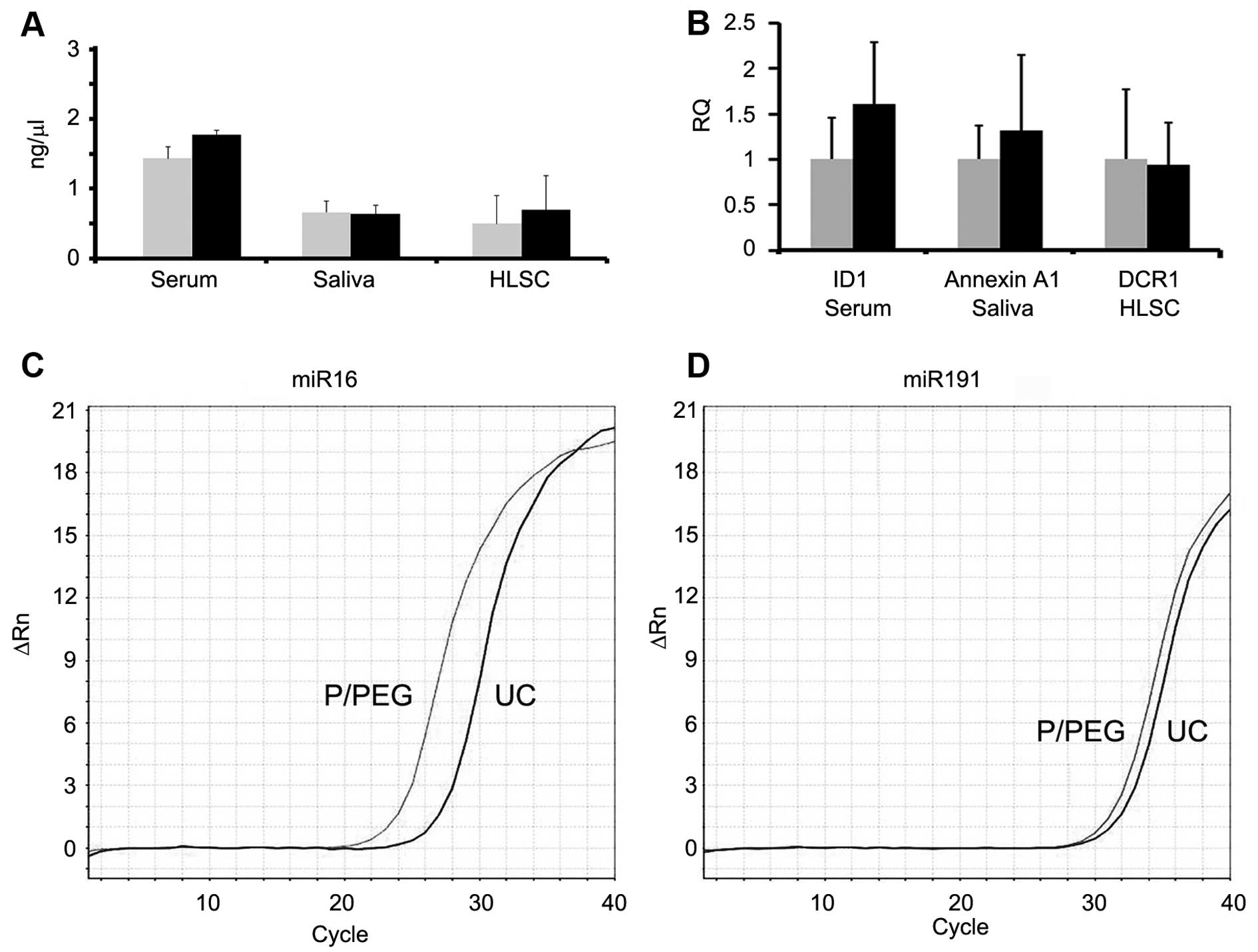
Fig. 4 shows a comparison between
RNA extracted from EVs prepared by P/PEG precipitation and
ultracentrifugation. No significant difference of RNA content was
observed between EVs isolated using the two methods. To evaluate
whether RNA extracted from EVs prepared by P/PEG precipitation was
suitable for the detection of miRNAs or mRNA, RT-qPCR analysis was
performed. RT-qPCR analysis revealed the presence of comparable
amounts of miR-16, -29a, -99b, -191 and -223, in EVs isolated from
normal subjects using the two techniques. By contrast, miR-500,
-142-3p, -127-3p and -155 were undetectable or detectable at very
low levels (not shown). Fig. 4B
shows a representative amplification plot for miR-191 in EVs
obtained by ultracentrifugation and precipitation. We also
performed RT-qPCR analysis for the detection of mRNA. As shown in
Fig. 4C, comparable amounts of
selected mRNA were detected in EVs derived from serum, saliva and
HLSCs, whether purified by ultracentrifugation or P/PEG
precipitation.
Evaluation of the ability of EVs isolated
by charge-based precipitation to retain biological activities
The biological activity of EVs obtained by
ultracentrifugation and by P/PEG precipitation was evaluated for
saliva and HLSC EVs.
To examine the biological activity of saliva EVs we
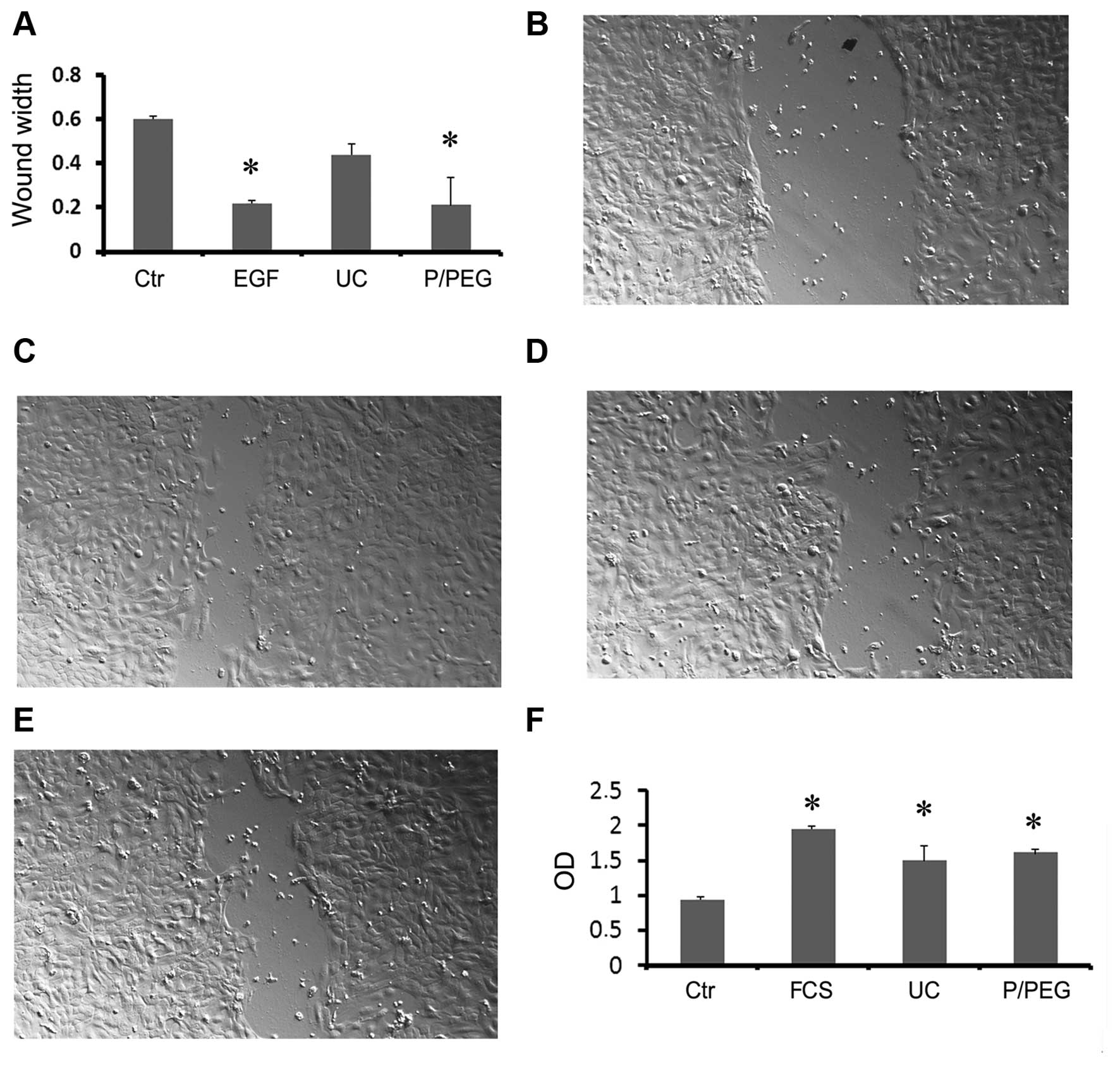
performed an in vitro wound-closure assay using human HaCaT
keratinocytes. Saliva EVs obtained by P/PEG induced a significant
wound closure comparable to that of EGF (Fig. 5A–E). In particular, precipitated
EVs were more effective than EVs obtained by ultracentrifugation
(Fig. 5E).
To examine the biological activity of HLSC EVs we
performed in vitro proliferation of TEC. Both precipitated
and ultra-centrifuged EVs were able to significantly increase cell
proliferation (Fig. 5F).
Discussion
EVs have recently emerged as an important vehicle of
information exchange among cells in the body involved in many
physiological and pathological processes. Since they retain several
molecular markers of the originator cell, EVs isolated from
biological fluids may be exploited as a diagnostic tool (3,6).
Nevertheless, techniques of EV purification and consequent analysis
of EVs remain a challenge. In the present study, we suggest a
charge-based precipitation of EVs from biological fluids and cell
supernatants. The analysis of ζ potential revealed that EVs have a
negative charge that allows the interaction with a positively
charged molecule such as protamine. Protamine was shown to induce
EV precipitation from the biological fluids and cell culture media
avoiding the ultracentrifugation. When protamine-induced
precipitation was performed in a polymeric matrix such as PEG
35,000 Da, the EV recovery was enhanced and pellets were easily
re-suspended.
The 'gold standard' methods of EV purification are
the differential ultracentrifugation or density gradient
ultracentrifugation. These methods, however, are influenced by
several parameters that are difficult to standardize, such as
viscosity of solutions, rotor type, centrifugal radius and g force.
In addition, the integrity of EVs after prolonged high-speed
ultracentrifugation may be damaged. Specifically, membrane debris
were observed by electron microscopy and difficulty in recovering
RNA and exosomal proteins has been reported (13–16). Several other approaches to EV
purification have been investigated. Size exclusion chromatography
may have an advantage on ultracentrifugation in maintaining EV
integrity since they are not subjected to shear stress (17–19). Filtration with membranes with
appropriate pores is also an alternative, but does not guarantee
removal of several small contaminants and loss of EVs by binding to
membranes (20). The
immunoaffinity purification may isolate specific exosome subtypes
maintaining integrity of their cargo (16,20–22). A limitation of most of these
techniques is the efficiency in the recovery of sufficient amounts
of EVs starting from small biological samples. The polymeric
precipitation technique, based on the ability of PEG to entrap EVs,
has been shown to be a rapid approach to EV isolation from
biological samples (5,23–28). This technique has been developed
on the observation that PEG allows virus precipitation (23) and several products based on the
use of PEG with 8,000 Da molecular weight are commercially
available. A recent study suggested heparin affinity chromatography
purification of EVs based on the presence of a putative receptor
for heparin (29). This technique
was shown to allow the purification of EVs with low protein
contamination and detection of mRNA from plasma samples in amounts
comparable to ultracentrifugation.
In the present study, we combined the charge-based
and polymeric precipitation using protamine and PEG 35,000 Da and
we compared this technique with differential ultracentrifugation.
P/PEG was more efficient for the recovery of EVs from small volumes
of serum and saliva as well as from the conditioned medium of
cultured cells than ultracentrifugation as judged by NTA. The size
of vesicles seen by electron microscopy was similar but the
membrane debris present in the ultra-centrifuged EVs were absent in
the P/PEG EV preparations. In particular, EVs precipitated from
saliva were very homogeneous in size and shape. The expression of
exosomal markers in EVs obtained by P/PEG precipitation as well as
the nano-size of vesicles detected by electron microscopy suggest
that this method is more suitable for the isolation of small
exosomes than of larger shed microvesicles. Since one of the main
concerns for EVs obtained by precipitation methods is the presence
of contaminants of non-vesicular origin such as lipoproteins
(12,13,30), we evaluated the presence of
ApoB100 and ApoA1 in the different preparations. The results
obtained indicate that in serum EVs, ApoB100 and ApoA1 were present
in EV precipitates as well as in EVs purified by differential
ultracentrifugation. This may be a limitation for the use of serum
EVs for diagnostic purposes if the intent is to discriminate exRNA
associated with vesicles from those associated with lipoproteins.
However, the detection of exRNA in the biological sample may be
exploited for liquid biopsy independently from their vehicle. In
this case the precipitation techniques may be suitable for this
purpose. ApoB100 was absent in saliva EVs. ApoA1 was present in EV
saliva but more expressed in EVs obtained by ultracentrifugation
than in P/PEG precipitated EVs. ApoB100 and ApoA1 were barely or
absent in EVs purified from culture media by P/PEG preparations,
suggesting that lipoprotein contamination is less relevant for
these biological samples.
The most diffuse precipitation method used,
ExoQuick, developed by System Biosciences (Mountain View, CA, USA)
has solved this problem by a pre-clinical approach to remove
lipoproteins. As an alternative, the use of Sephadex G-25 spin
columns to remove PEG 8,000 Da containing lipoproteins from
precipitated EVs has been suggested (5). Since we precipitated EVs with
protamine in association with PEG 35,000 Da, Sephadex G-100 spin
columns were used to show the effective reduction of apolipoprotein
contaminants. Following absorption, the total RNA was reduced but
was suitable for the detection of miRNA and mRNA content of
EVs.
EVs obtained by P/PEG precipitation retained in
vitro the biological activity as seen by the induction of wound
closure by keratinocytes stimulated with EVs from saliva and of
proliferation of TEC challenged with EVs released by HLSCs.
The methods currently available for EV purification
have both advantages and disadvantages and possibly none are ideal
for each application (5,6,31).
The methods described in the present study have the merit of
simplicity and avoid requirement of expensive equipments. In
addition, the isolated EVs retained the biological activities.
In conclusion, we have shown that charge-based
precipitation of EVs may be used for an efficient isolation of EVs
from biological samples and may be exploited for the search of new
biomarkers.
Abbreviations:
|
EVs
|
extracellular vesicles
|
|
HLSCs
|
human liver stem cells
|
Acknowledgments
The present study was supported by a grant from
Unicyte, SW. M.C.D., F.F., M.F.B., C.T. and G.C. are named as
inventors in EV-related patents. C.T. is a full-time employee of
Fresenius Medical Care and contributed to the study as
researcher.
References
|
1
|
Ratajczak J, Wysoczynski M, Hayek F,
Janowska-Wieczorek A and Ratajczak MZ: Membrane-derived
microvesicles: Important and underappreciated mediators of
cell-to-cell communication. Leukemia. 20:1487–1495. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cocucci E, Racchetti G and Meldolesi J:
Shedding microvesicles: Artefacts no more. Trends Cell Biol.
19:43–51. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Quesenberry PJ, Aliotta J, Deregibus MC
and Camussi G: Role of extracellular RNA-carrying vesicles in cell
differentiation and reprogramming. Stem Cell Res Ther. 6:1532015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shelke GV, Lässer C, Gho YS and Lötvall J:
Importance of exosome depletion protocols to eliminate functional
and RNA-containing extracellular vesicles from fetal bovine serum.
J Extracell Vesicles. Sep 30–2014.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Szatanek R, Baran J, Siedlar M and
Baj-Krzyworzeka M: Isolation of extracellular vesicles: Determining
the correct approach (Review). Int J Mol Med. 36:11–17.
2015.PubMed/NCBI
|
|
6
|
Taylor DD, Zacharias W and Gercel-Taylor
C: Exosome isolation for proteomic analyses and RNA profiling.
Methods Mol Biol. 728:235–246. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Herrera MB, Bruno S, Buttiglieri S, Tetta
C, Gatti S, Deregibus MC, Bussolati B and Camussi G: Isolation and
characterization of a stem cell population from adult human liver.
Stem Cells. 24:2840–2850. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Herrera MB, Fonsato V, Gatti S, Deregibus
MC, Sordi A, Cantarella D, Calogero R, Bussolati B, Tetta C and
Camussi G: Human liver stem cell-derived microvesicles accelerate
hepatic regeneration in hepatectomized rats. J Cell Mol Med.
14:1605–1618. 2010. View Article : Google Scholar
|
|
9
|
Cantaluppi V, Biancone L, Romanazzi GM,
Figliolini F, Beltramo S, Galimi F, Camboni MG, Deriu E, Conaldi P,
Bottelli A, et al: Macrophage stimulating protein may promote
tubular regeneration after acute injury. J Am Soc Nephrol.
19:1904–1918. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Théry C, Amigorena S, Raposo G and Clayton
A: Isolation and characterization of exosomes from cell culture
supernatants and biological fluids. Curr Protoc Cell Biol. Apr
1–2006.Epub ahead of print. View Article : Google Scholar
|
|
11
|
Iavello A, Frech VS, Gai C, Deregibus MC,
Quesenberry PJ and Camussi G: Role of Alix in miRNA packaging
during extracellular vesicle biogenesis. Int J Mol Med. 37:958–966.
2016.PubMed/NCBI
|
|
12
|
Witwer KW, Buzás EI, Bemis LT, Bora A,
Lässer C, Lötvall J, Nolte't Hoen EN, Piper MG, Sivaraman S, Skog
J, et al: Standardization of sample collection, isolation and
analysis methods in extracellular vesicle research. J Extracell
Vesicles. 2:203602013.
|
|
13
|
Momen-Heravi F, Balaj L, Alian S,
Trachtenberg AJ, Hochberg FH, Skog J and Kuo WP: Impact of biofluid
viscosity on size and sedimentation efficiency of the isolated
microvesicles. Front Physiol. 3:1622012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jeppesen DK, Hvam ML, Primdahl-Bengtson B,
Boysen AT, Whitehead B, Dyrskjøt L, Orntoft TF, Howard KA and
Ostenfeld MS: Comparative analysis of discrete exosome fractions
obtained by differential centrifugation. J Extracell Vesicles.
3:250112014.PubMed/NCBI
|
|
15
|
Cvjetkovic A, Lötvall J and Lässer C: The
influence of rotor type and centrifugation time on the yield and
purity of extracellular vesicles. J Extracell Vesicles.
3:32014.
|
|
16
|
Tauro BJ, Greening DW, Mathias RA, Ji H,
Mathivanan S, Scott AM and Simpson RJ: Comparison of
ultracentrifugation, density gradient separation, and
immunoaffinity capture methods for isolating human colon cancer
cell line LIM1863-derived exosomes. Methods. 56:293–304. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Müller G: Novel tools study cell
type-specific exosomes microvesicles. J Bioanal Biomed. 4:46–60.
2012.
|
|
18
|
Taylor DD, Lyons KS and Gerçel-Taylor C:
Shed membrane fragment-associated markers for endometrial and
ovarian cancers. Gynecol Oncol. 84:443–448. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Böing AN, van der Pol E, Grootemaat AE,
Coumans FA, Sturk A and Nieuwland R: Single-step isolation of
extracellular vesicles by size-exclusion chromatography. J
Extracell Vesicles. 3:32014.
|
|
20
|
Taylor DD and Shah S: Methods of isolating
extracellular vesicles impact down-stream analyses of their
cargoes. Methods. 87:3–10. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Caby MP, Lankar D, Vincendeau-Scherrer C,
Raposo G and Bonnerot C: Exosomal-like vesicles are present in
human blood plasma. Int Immunol. 17:879–887. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zarovni N, Corrado A, Guazzi P, Zocco D,
Lari E, Radano G, Muhhina J, Fondelli C, Gavrilova J and Chiesi A:
Integrated isolation and quantitative analysis of exosome shuttled
proteins and nucleic acids using immunocapture approaches. Methods.
87:46–58. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hebert TT: Precipitation of plant viruses
by polyethylene glycol. Phytopathology. 53:3621963.
|
|
24
|
Alvarez ML, Khosroheidari M, Kanchi Ravi R
and DiStefano JK: Comparison of protein, microRNA, and mRNA yields
using different methods of urinary exosome isolation for the
discovery of kidney disease biomarkers. Kidney Int. 82:1024–1032.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Alvarez ML: Isolation of urinary exosomes
for RNA biomarker discovery using a simple, fast, and highly
scalable method. Methods Mol Biol. 1182:145–170. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rekker K, Saare M, Roost AM, Kubo AL,
Zarovni N, Chiesi A, Salumets A and Peters M: Comparison of serum
exosome isolation methods for microRNA profiling. Clin Biochem.
47:135–138. 2014. View Article : Google Scholar
|
|
27
|
Zlotogorski-Hurvitz A, Dayan D, Chaushu G,
Korvala J, Salo T, Sormunen R and Vered M: Human saliva-derived
exosomes: Comparing methods of isolation. J Histochem Cytochem.
63:181–189. 2015. View Article : Google Scholar :
|
|
28
|
Kanchi Ravi R, Khosroheidari M and
DiStefano JK: A modified precipitation method to isolate urinary
exosomes. J Vis Exp. 95:511582015.PubMed/NCBI
|
|
29
|
Balaj L, Atai NA, Chen W, Mu D, Tannous
BA, Breakefield XO, Skog J and Maguire CA: Heparin affinity
purification of extracellular vesicles. Sci Rep. 5:102662015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yuana Y, Levels J, Grootemaat A, Sturk A
and Nieuwland R: Co-isolation of extracellular vesicles and
high-density lipoproteins using density gradient
ultracentrifugation. J Extracell Vesicles. 3:32014.
|
|
31
|
Taylor DD: Isolation and molecular
characterization of extracellular vesicles. Methods. 87:1–2. 2015.
View Article : Google Scholar : PubMed/NCBI
|