Introduction
Esophageal cancer (EC) is one of the six most common
malignancies worldwide, with a higher incidence in males than in
females (1). The lifetime risk of
developing this type of cancer is 0.8% for men and 0.3% for women.
In addition, the risk increases with age, with a mean age at
diagnosis of 67 years (2,3). EC is the seventh leading cause of
cancer-related mortality among American males, particularly African
American males, who have a higher incidence of this disease (13
cases per 100,000 individuals) than do males of other racial or
ethnic origin (2,4). Elucidating molecular alterations in
EC will help us to find new targets for effective therapies.
Cytoplasmic collapsin response mediator protein-1
(CRMP-1), also known as dihydropyrimidinase related protein-1
(DRP-1) is a brain-specific protein that belongs to the
Unc-33-related protein family (5–7).
The dysregulation of cytoplasmic collapsin response mediator
protein 1 (CRMP1) has been reported in brain, lung and pituitary
tumors, and in EC (8–11). In prolactin-secreting pituitary
adenoma, CRMP1 has been shown to be associated with tumor
progression (12). The
downregulation of CRMP1 has been shown to be significantly
associated with advanced disease, metastasis and shorter survival
in non-small cell lung cancer (NSCLC), suggesting that CRMP1 acts
as a novel tumor suppressor gene (13,14). Functional studies have
demonstrated that the depletion of CRMP1 promotes tumor invasion,
whereas its increased expression has an opposite effect in
glioblastoma (9). The expression
level of CRMP1 has been shown to significantly correlate with the
depth of invasion and lymph node metastasis in EC (11). However, its role in EC has not yet
been fully elucidated.
MicroRNAs (miRNAs or miRs) are a class of
conservative single-stranded non-coding RNAs, composed of 17–25
ribonucleotides (15). The
deregulation of miRNA expression was previously detected in EC
(16). It was reported that the
most markedly upregulated miRNAs were hsa-miR-15a, hsa-miR-28-3p,
hsa-miR-31, hsa-miR-99b, hsa-miR-101, hsa-miR-130a, hsa-miR-143,
hsa-miR-196b, hsa-miR-200a-3p, hsa-miR-210, hsa-miR-452 and
hsa-miR-27a, whereas the most markedly downregulated miRNAs
included hsa-miR-30b, hsa-miR-223, hsa-miR-454, hsa-miR-486,
hsa-miR-574-3p and hsa-miR-126 in EC (16). miRNAs are also involved in
numerous physiological processes of cell regulation, including
differentiation, proliferation, migration, invasion, apoptosis and
metastasis (17–20).
In this study, we aimed to elucidate the role and
regulatory mechanisms of CRMP1 in EC. We examined the effects of
miR-200a-3p on EC. As we found that CRMP1 was a direct target of
miR-200a-3p, we also examined the effects of CRMP1 on EC. Our data
provide evidence that miR-200a-3p promotes the proliferation of EC
cells by post-transcriptionally regulating CRMP1.
Materials and methods
Patients and samples
Human tissue samples (tumor tissue and adjacent
normal tissues; n=6) were collected from patients who underwent
surgical resection at People's Hospital of Henan Province,
Zhengzhou, China between 2013 and 2014. All experiments were
performed following the approval of the Medical Ethics Committee of
the People's Hospital of Henan Province (approval no. HN141026) and
after obtaining written informed consent from all patient
donors.
Cell culture, plasmids and
pre-miR-200a-3p/control miR, and cell transfection
The EC cell lines, TE1, TE2, TE3, TE4, TE5 and TE6,
were purchased from the American Type Culture Collection (ATCC,
Manassas, VA, USA and cultured in Dulbecco's modified Eagle's
medium (DMEM) supplemented with 10% fetal bovine serum and 0.01%
penicillin/streptomycine in a 5% CO2 environment at
37°C. The CRMP1-expressing plasmid/empty vector (pcDNA3.1) and the
shCRMP1 plasmid/scramble plasmid were purchased from R&D
Systems (Abingdon, UK). Pre-miR-200a-3p and control-miR were
purchased from Ambion (Austin, TX, USA). For transfection
experiments, the cells were cultured in serum-free medium without
antibiotics at 60% confluence for 24 h. The TE2 cells were
transfected with the CRMP1-expressing plasmid or empty vector, or
with the shCRMP1 plasmid or scramble control plasmid for 48 h using
Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to
manufacturer's instructions. In addition, the cells were
transfected with pre-miR-200a-3p or control-miR. Following
incubation for 6 h, the medium was removed and replaced with normal
culture medium for 48 h, unless otherwise specified. The
transfection efficiency was determined by western blot
analysis.
Western blot analysis
Western blot analysis of different samples was
performed to validate proteomic quantification. Protein was
isolated from the 6 pairs of EC tissues and adjacent normal tissues
(patient nos. 1-6) and from the 6 EC cell lines. Total protein
lysate was prepared using RIPA buffer containing protease
inhibitors and phosphatase inhibitors. Protein lysates were
resolved on a sodium dodecyl sulfate (SDS)-polyacrylamide gel and
electrotransferred onhto PVDF membranes. After blocking, the
membranes were immunoblotted with the following primary antibodies:
anti-CRMP1 (ab199722), anti-p21 (ab109520), anti-CDK4 (ab108357)
and anti-p53 (ab1431) (Abcam, Cambridge, MA, USA) overnight at 4°C
and then immunoblotted with secondary antibody [goat anti-rabbit
IgG H&L (HRP); ab6721, Abcam] at room temperature. The signals
were detected by enhanced chemiluminescence.
Reverse transcription-polymerase chain
reaction (RT-PCR) and quantitative PCR (qPCR) for CRMP1
Total RNA was extracted from the cells using RNeasy
Plus mini kit (Qiagen, Valencia, CA, USA) according to the
manufacturer's instructions. cDNA was synthesized using the
QuantiTect reverse transcription kit (Qiagen). qPCR was carried out
using the 7900HT Fast Real-Time PCR System according to the
manufacturer's instructions (Applied Biosystems, Foster City, CA,
USA). The primer sequences for CRMP1 were as follows: forward,
5′-CCCCAAAAGCGTGTGACAGTA-3′ and reverse, 5′-GGTAGAAGGGATTTGTGCG-3′.
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an
internal control. The primers for GAPDH were as follows: forward,
5′-GAAGGTCGGAGTCAACGGATTTG-3′; and reverse,
5′-ATGGCATGGACTGTGGTCATGAG-3′. PCR amplification was carried out
under the following conditions: denaturation at 95°C for 10 min,
followed by 40 cycles of denaturation at 95°C for 15 sec, annealing
at 60°C for 30 sec, and extension at 72°C for 1 min. The relative
expression level for target genes was normalized by the Ct value of
GAPDH using the 2−ΔΔCt relative quantification
method.
qPCR for miRNA expression
The mirVana miRNA isolation kit (Ambion) was used to
extract the total RNA from the cells according to manufacturer's
instructions. Total RNA was used to generate cDNA using the
specific RT-primer from the TaqMan gene expression assay kit
(Applied Biosystems). qPCR was performed to determine the
expression levels of miRNA. U6 was used as an internal control. The
primer sequences were as follows: miR-200a-3p,
5′-UAACACUGUCUGGUAACGAUGU-3′; and U6,
5-′CGCTTCACGAATTTGCGTGTCAT-3′.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay
The effects on cell proliferation were assessed by
MTT assay (Sigma, St. Louis, MO, USA) which was performed as
previously described (21). In
brief, the cells were plated in 96-well plates in DMEM containing
10% fetal bovine serum at a density of 8×103 cells per
well at 37°C in a 5% CO2 incubator for 12 h. The cells
were transfected with the plasmids. MTT (5 mg/ml) was then added to
the wells (20 µl per well). The plates were then incubated
in a cell incubator for 4 h, and the supernatant was then removed
and 150 µl of dimethyl sulfoxide was added to each well.
Following incubation for 10 min, the absorbance of each well was
measured using a Synergy™ 4 (BioTek Instruments, Winooski, VT, USA)
at a wavelength of 570 nm, with the reference wavelength set at 630
nm. The absorbance was directly proportional to the number of
surviving cells.
In vitro proliferation assay
Cell proliferation was also assessed using a
colorimetric BrdU proliferation kit according to the manufacturer's
instructions (cat no. 74299; Roche, Indianapolis, IN, USA). The
cells were incubated with BrdU for 3 h and wased with
phosphate-buffered saline (PBS). The genomic DNA was fixed in 4%
paraformaldehyde, immunolabeled for BrdU, and counterstained with
the fluorescent nuclear marker, DAPI (Sigma).
Cell cycle analysis
The cells (8.0×105 cells) were seeded
into a 100-mm culture plate and allowed to attach overnight. The
cells were transfected with the plasmids for 24 h, washed twice
with NaCl/Pi, and then centrifuged at 200 × g at room temperature.
The pellet was resuspended in 1 ml cold NaCl/Pi and fixed in 70%
ethanol for at least 12 h at 4°C. The fixed cells were incubated
with 100 µl DNase-free RNase A (200 µg/ml) for 30 min
at 37°C, and then 1 mg/ml propidium iodide was added. The stained
cells were analyzed using a fluorescence-activated cell sorter (BD
Accuri C6; BD Biosciences, Ann Arbor, MI, USA). The percentages of
cells in the G1, S and G2/M phases of the cell cycle were
determined using CellQuest Pro software (FlowJo, Ashland, OR,
USA).
Bioinformatics analysis
The analysis of potential miRNA target sites was
carried out using the commonly used prediction algorithm,
TargetScan (http://www.targetscan.org).
Immunofluorescence staining
The cells were fixed with paraformaldehyde before
blocking with BSA. The cells were then incubated with the rabbit
antibody against CRMP1 (1:200 dilution; Abcam) overnight. After
washing 3 times with PBS, the cells were incubated with the
secondary antibody [goat anti-rabbit IgG H&L (HRP); ab6721,
Abcam]. The labeled cells were detected under a laser scanning
confocal microscope (Olympus, Tokyo, Japan).
Statistics analysis
Data are presented as the means ± SE, and are the
product of 3 independent experiments. Significance among the groups
was determined using a Student's t-test. Statistical significance
was set at P<0.05.
Results
CRMP1 is downregulated in EC and CRMP1
expression differs between different EC cell lines
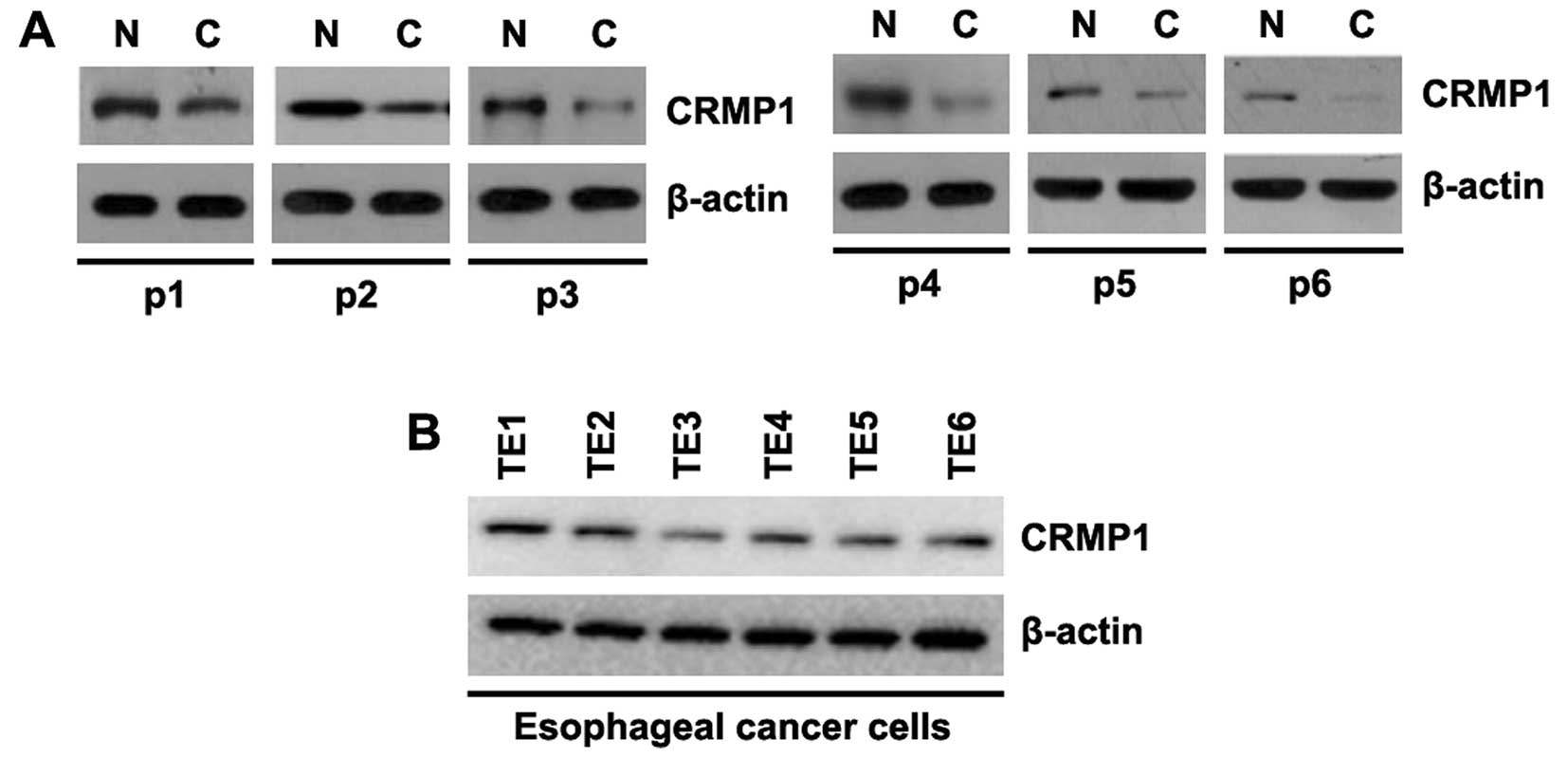
In an aim to identify differences in CRMP1
expression between EC tissues and adjacent normal tissues, we
performed western blot analysis. Protein was isolated from 6 pairs
of EC tissues and normal tissues (patient nos. 1-6). We found that
CRMP1 protein expression was significantly decreased in the tumor
tissues, compared with the adjacent normal tissues (Fig. 1A). This suggested that CRMP1 may
be a tumor suppressor gene in EC. In order to examine CRMP1 protein
expression among different EC cell lines, we performed western blot
analysis using 6 EC cell lines (TE1, TE2, TE3, TE4, TE5 and TE6
cells). Protein isolated from the 6 EC cell lines was examined and
the results revealed that CRMP1 expression differed betwee the
different EC cells (Fig. 1B).
Specifically, CRMP1 expression was lowest in the TE3 cells and
highest in the TE1 cells. However, for our further experiments, we
selected the TE2 cells.
CRMP1 inhibits the proliferation of TE2
EC cells
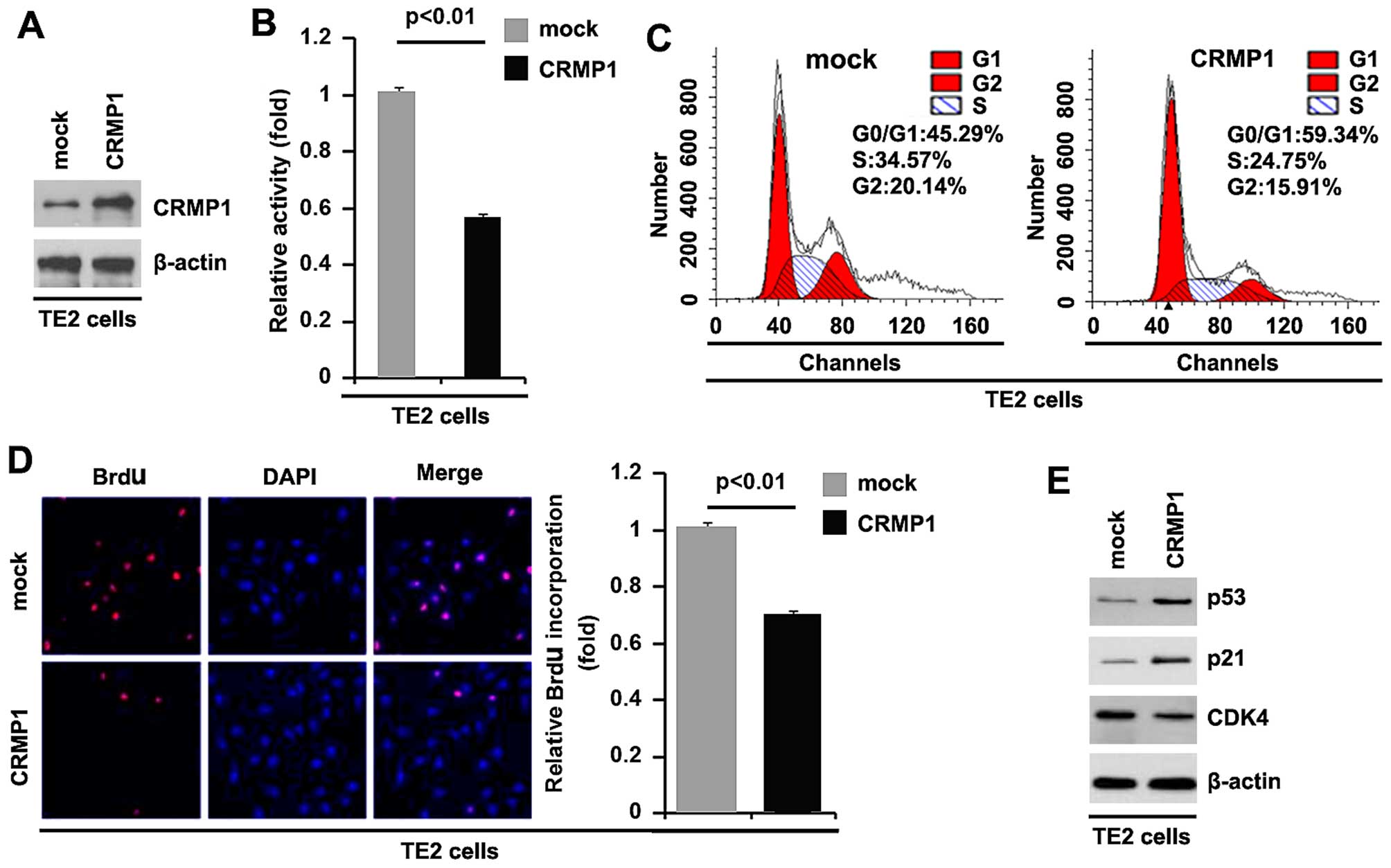
To examine whether CRMP1 affects the proliferation
of EC cells, firstly, by using western blot analysis, we examined
whether a CRMP1-expressing plasmid can be used to stably express
CRMP1 protein in TE2 cells. The results revealed that CRMP1 protein
expression was significantly increased by transfection of the cells
with the CRMP1-expressing plasmid (Fig. 2A). In addition, we performed MTT
assay to examine the proliferation of TE2 cells transfected with
the CRMP1-expressing plasmid. The results revealed that the
overexpression of CRMP1 inhibited the proliferation of TE2 cells
after 48 h of transfection (Fig.
2B). To examine the effects of CRMP1 on cell proliferation, we
also performed cell cycle analysis to examine its effects on the
cell cycle. The results revealed lower S phase and G2 phase
fractions in the TE2 cells transfected with the
CRMP1-overexpressing plamid than in the cells transfected with the
empty vector (Fig. 2C). To
identify whether DNA synthesis inhibition contributes to the lower
S phase fractions in the TE2 cells transfected with the
CRMP1-overexpressing plamid, we performed a BrdU incorporation
assay to detect DNA synthesis in the cells. The results confirmed
that CRMP1 significantly inhibited DNA synthesis in the cells
(Fig. 2D). In addition, we also
performed western blot analysis to further confirm that CRMP1
affects proliferation markers. The results of western blot analysis
demonstrated that CDK4 expression was downregulated and the
expression of p53 and p21 was upregulated by the overexpression of
CRMP1 (Fig. 2E).
Silencing of CRMP1 promotes the
proliferation in of TE2 EC cells
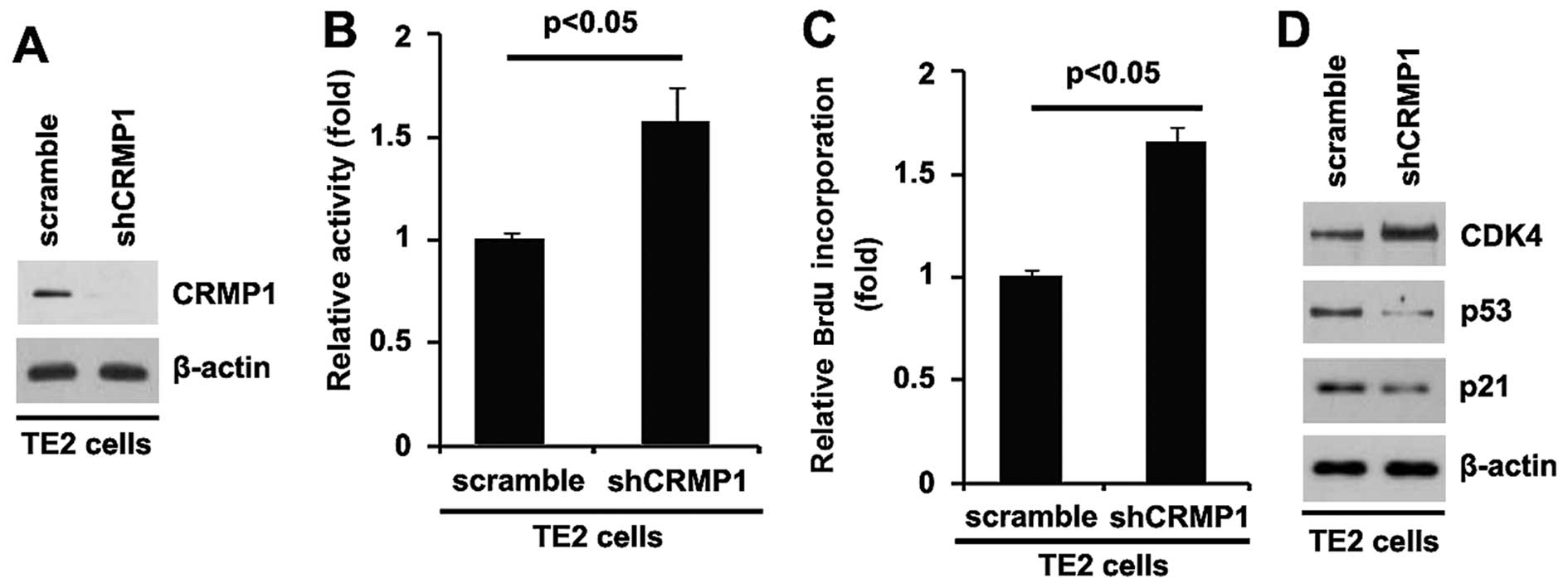
In an attempt to further identify the role of CRMP1
in regulating proliferation of TE2 cells, the cells were
transfected with a shCRMP1 plasmid. Following stable transfection,
CRMP1 protein expression was detected by western blot analysis. The
results revealed that transfection with the shCRMP1 plasmid
evidently suppressed CRMP1 protein expression in the TE2 cells
(Fig. 3A). Moreover, the
proliferation rates of the TE2 cells were examined by MTT assay.
The results revealed that the silencing of CRMP1 promoted the
proliferation of the TE2 cells (Fig.
3B). This was confirmed by BrdU incorporation assay, which
indicated that transfection with the shCRMP1 plasmid resulted in
increased DNA synthesis activity per viable cell in the cells
(Fig. 3C). To further confirm
that CRMP1 regulates the proliferation of TE2 cells, we performed
western blot analysis to detect the expression of proliferation
markers (CDK4, p53 and p21). The results demonstrated that CDK4
expression was upregulated, and that of p53 and p21 was
downregulated in the cells transfected with the shCRMP1 plasmid
(Fig. 3D). The above-mentioned
findings demonstrated that the silencing of CRMP1 promoted the
proliferation of TE2 cells.
miR-200a-3p suppresses CRMP1 protein
expression in TE2 EC cells
miRNAs/miRs are a class of non-coding RNAs able to
regulate gene expression at the post-transcriptional level, by
binding to the 3′UTR of target messenger RNAs (mRNAs) through
partial sequence homology, and causing a block of translation
and/or mRNA degradation (22–24). The upregulation of specific miRNAs
can contribute to the downregulation of tumor suppressor genes
(25–27). Thus, we hypothesized that CRMP1 is
downregulated by miRNAs in EC.
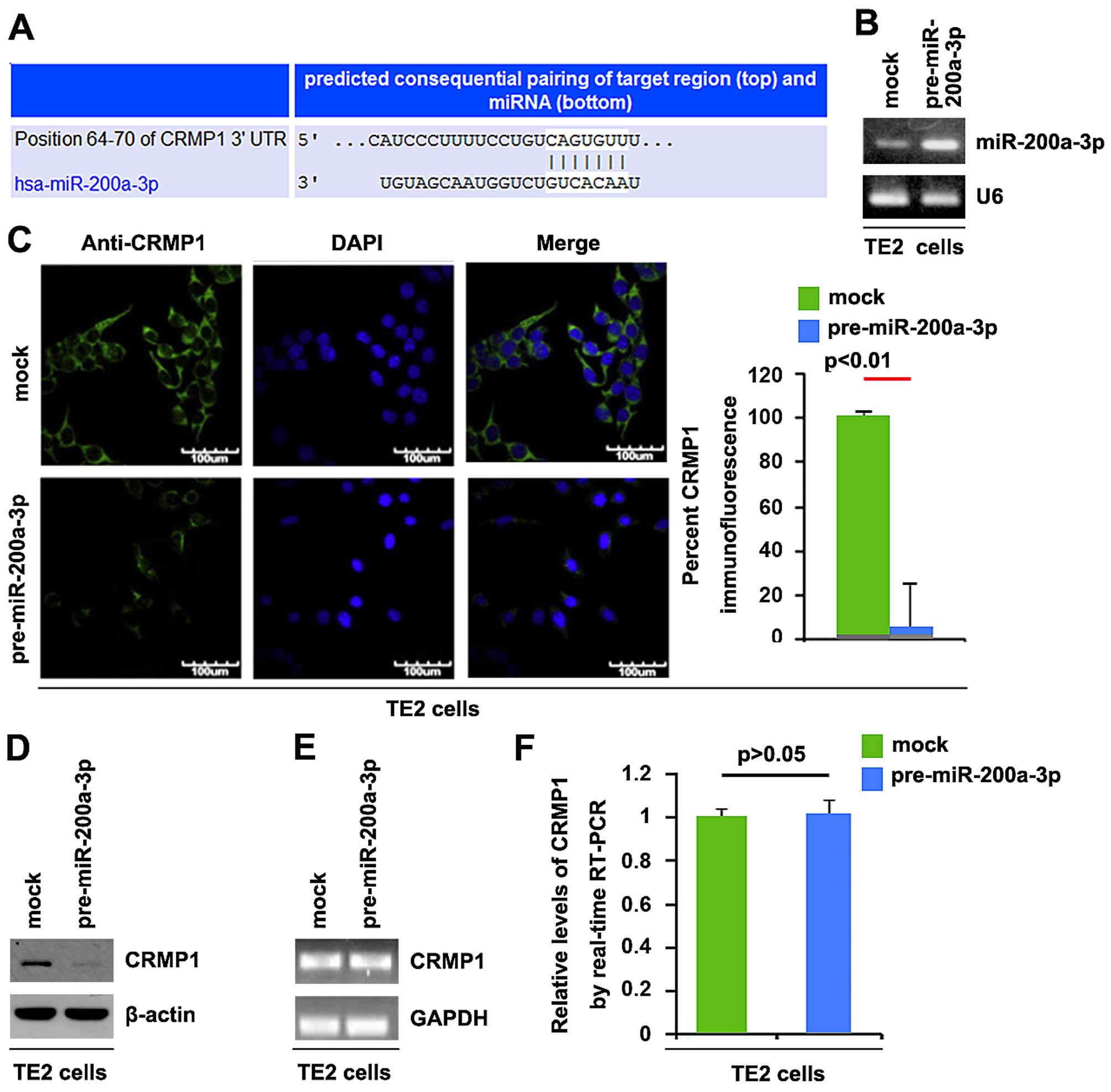
We screened for miRNAs targeting CRMP1 using
TargetScan (http://www.targetscan.org/), and a large number of
target miRNAs were found. However, we were interested in
miR-200a-3p, as it has been previously shown to be upregulated in
EC (16). The target sites on the
3′UTR of CRMP1 are shown in Fig.
4A. We hypothesized that miR-200a-3p downregulates CRMP1
expression by targeting its 3′UTR in EC cells.
In order to determine whether CRMP1 is downregulated
by miR-200a-3p in EC, we transfected the TE2 EC cells with
pre-miR-200a-3p and qPCR was then performed to detect miR-200a-3p
expression in the cells. The results revealed that transfection
with pre-miR-200a-3p significantly upregulated miR-200a-3p
expression (Fig. 4B). To
determine whether CRMP1 protein expression is affected by
miR-200a-3p, we performed immunofluorescence analyses. The results
of immunofluorescence analyses revealed that CRMP1 protein
expression was significantly downregulated by transfection with
pre-miR-200a-3p in the TE2 cells (Fig. 4C). Moreover, western blot analysis
was performed to detect CRMP1 protein expression in TE2 cells
transfected with pre-miR-200a-3p. Consistent with the results of
immunofluorescence analyses, we found that CRMP1 protein expression
was significantly downregulated by transfection with
pre-miR-200a-3p in TE2 cells (Fig.
4D). To determine whether CRMP1 mRNA expression was decreased
by miR-200a-3p, we then performed RT-PCR and qPCR to detect CRMP1
mRNA expression in TE2 cells transfected with pre-miR-200a-3p or
control miR. The results of RT-PCR and qPCR demonstrated that
miR-200a-3p did not affect CRMP1 mRNA expression in TE2 cells
(Fig. 4E and F).
miR-200a-3p promotes the proliferation of
TE2 EC cells
In an attempt to identify the role of miR-200a-3p in
regulating the proliferation of TE2 cells, the cells were
transfected with pre-miR-200a-3p. Following transfection, the
proliferation rates of TE2 cells were examined by MTT assay. The
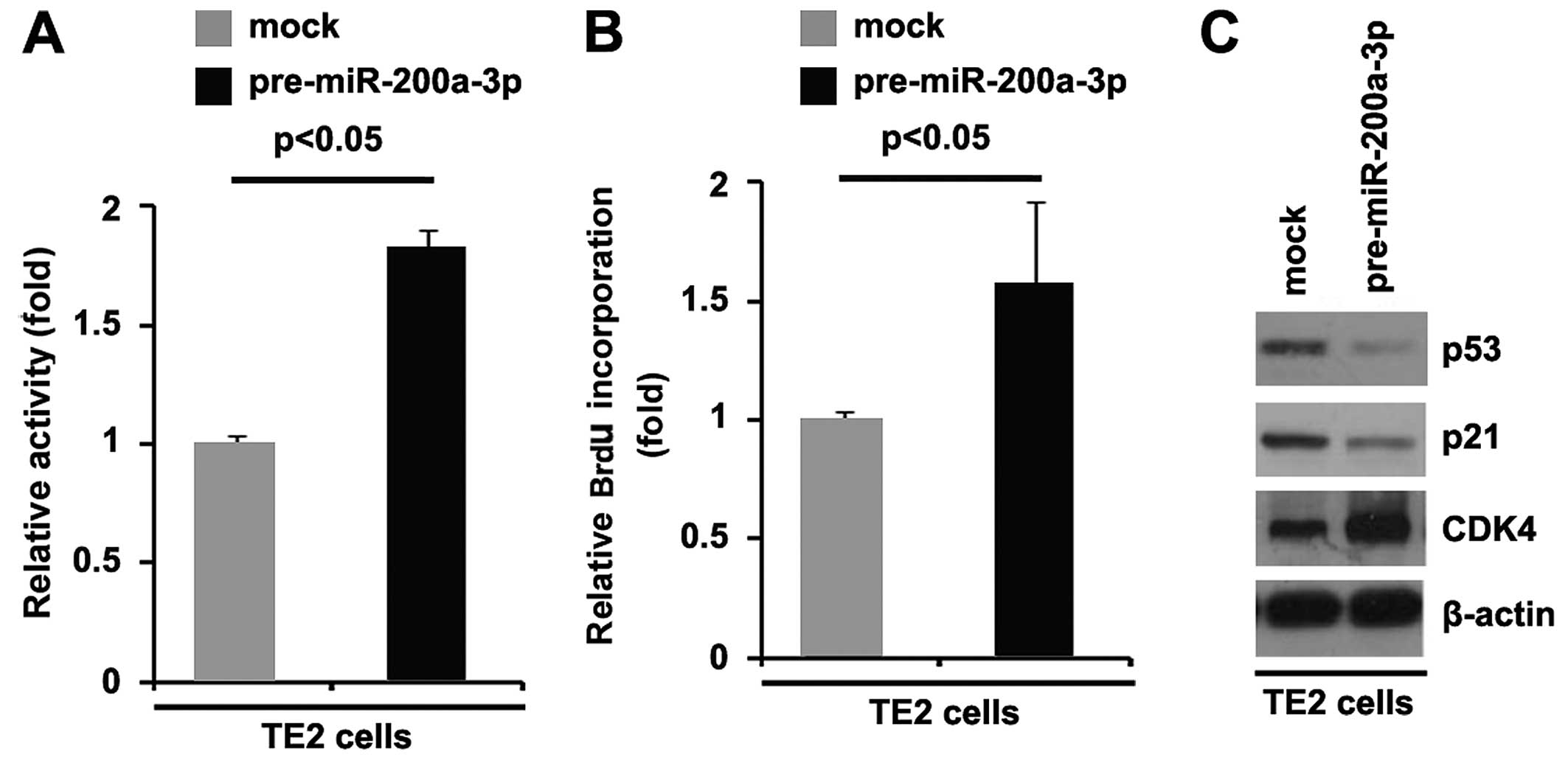
results revealed that transfection with miR-200a-3p promoted the
proliferation of TE2 cells (Fig.
5A). This was further revealed by BrdU incorporation assay
which indicated that transfection with pre-miR-200a-3p resulted in
increased DNA synthesis activity per viable cell (Fig. 5B). To further confirm that
miR-200a-3p can regulate EC cell proliferation, we performed
western blot analysis to detect the expression of proliferation
markers (CDK4, p53 and p21). The results revealed that CDK4
expression was upregulated, and that of p53 and p21 was
downregulated in the cells transfected with pre-miR-200a-3p
(Fig. 5C). The above-mentioned
findings demonstrated that miR-200a-3p promoted the proliferation
of TE2 cells.
Discussion
EC is one of the six most common malignancies
worldwide and is associated with a 5-year survival rate >25%
(1). Understanding the molecular
biology of the disease is a prerequisite to predicting prognosis
and to selecting effective treatment options. The development of EC
is a multistep phenomenon involving genetic events that result in
key abnormalities of cell cycle regulation, growth factor activity
and intercellular adhesion mechanisms (28).
Consistent with previous findings demonstrating that
CRMP1 significantly inhibited the proliferation of medulloblastoma
cells (29), we found that its
overexpression also inhibited the proliferation of EC cells. It has
been demonstrated that CRMP1 significantly inhibits the migration,
invasion of and the formation of filopodia and intense stress
fibers in medulloblastoma cells (29). In addition, the expression level
of CRMP1 has been shown to significantly correlate with the depth
of invasion and lymph node metastasis in EC (11). In the future, we aim to determine
whether CRMP1 affects the migration and invasion of TE2 EC
cells.
The p53 tumor suppressor lies at a nexus of cellular
pathways that sense DNA damage, cellular stress and improper
mitogenic stimulation (30). p53
integrates such signals and, in response, induces growth arrest,
promotes apoptosis, blocks angiogenesis, or mediates DNA repair in
a context-dependent manner (31).
The importance of p53 in preventing tumor formation is indicated by
the presence of mutations in the p53 pathway in almost all types of
cancer (32). In this study, we
found that CRMP1 overexpression significantly upregulated p53
protein expression and the silencing of CRMP1 downregulated p53
protein expression in TE2 EC cells. p21-mediated growth inhibition
has been attributed to two main activities that depend on two
non-overlapping structural domains: the carboxy-terminal
PCNA-binding domain and the amino-terminal CDK-cyclin inhibitory
domain (33,34). By binding to PCNA, p21 competes
for PCNA binding with DNA polymerase-δ and several other proteins
involved in DNA synthesis, thus directly inhibiting DNA synthesis
(35). We demonstrated that CRMP1
overexpression significantly upregulated p21 protein expression and
the silencing of CRMP1 downregulated p21 protein expression in TE2
EC cells. Our results suggeswt that CRMP1 functions as a tumor
suppressor gene by regulating p21 and p53 in EC.
As recently demonstrated through miRNA micro-array
analysis, miR-200 is upregulated in nasopharyngeal carcinoma (NPC).
It was also shown that the endogenous miR-200a-3p expression level
increases with the degree of differentiation in a panel of NPC cell
lines, and that the overexpression of miR-200a-3p inhibits C666-1
cell growth, migration and invasion, whereas its knockdown
stimulates these processes in CNE-1 cells (36). However, the role of miR-200a-3p
remains unclear in EC. In this study, we demonstrated that
miR-200a-3p post-transcriptionally regulates CRMP1 and stimulates
the proliferation of human EC cells.
The recognition of the differential regulation and
function of CRMP1 in EC will ultimately provide a better
understanding of the signaling pathways that can be therapeutically
modulated. We have merely just begun to explore the role of
miR-200a-3p in EC.
References
|
1
|
Enzinger PC and Mayer RJ: Esophageal
cancer. N Engl J Med. 349:2241–2252. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ries LAG, Eisner MP, Kosary C, Hankey BF,
Miller BA, Clegg LX and Edwards BK: SEER Cancer Statistics Review,
1973–1999. National Cancer Institute; Bethesda, MD: 2002
|
|
3
|
Daly JM, Fry WA, Little AG, Winchester DP,
McKee RF, Stewart AK and Fremgen AM: Esophageal cancer: Results of
an American College of Surgeons Patient Care Evaluation Study. J Am
Coll Surg. 190:562–573. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pisani P, Parkin DM, Bray F and Ferlay J:
Erratum: Estimates of the worldwide mortality from 25 cancers in
1990. Int J Cancer. 83:18–29. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Goshima Y, Nakamura F, Strittmatter P and
Strittmatter SM: Collapsin-induced growth cone collapse mediated by
an intracellular protein related to UNC-33. Nature. 376:509–514.
1995. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li W, Herman RK and Shaw JE: Analysis of
the Caenorhabditis elegans axonal guidance and outgrowth gene
unc-33. Genetics. 132:675–689. 1992.PubMed/NCBI
|
|
7
|
Hamajima N, Matsuda K, Sakata S, Tamaki N,
Sasaki M and Nonaka M: A novel gene family defined by human
dihydropyrimidinase and three related proteins with differential
tissue distribution. Gene. 180:157–163. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gao M, Yeh PY, Lu YS, Chang WC, Kuo ML and
Cheng AL: NF-kappaB p50 promotes tumor cell invasion through
negative regulation of invasion suppressor gene CRMP-1 in human
lung adenocarcinoma cells. Biochem Biophys Res Commun. 376:283–287.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mukherjee J, DeSouza LV, Micallef J, Karim
Z, Croul S, Siu KW and Guha A: Loss of collapsin response mediator
Protein1, as detected by iTRAQ analysis, promotes invasion of human
gliomas expressing mutant EGFRvIII. Cancer Res. 69:8545–8554. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wierinckx A, Auger C, Devauchelle P,
Reynaud A, Chevallier P, Jan M, Perrin G, Fèvre-Montange M, Rey C,
Figarella-Branger D, et al: A diagnostic marker set for invasion,
proliferation, and aggressiveness of prolactin pituitary tumors.
Endocr Relat Cancer. 14:887–900. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhai J, Wang Y, Yang F, Hu J, Qi Q and
Zhang Y: DRP-1, ezrin and E-cadherin expression and the association
with esophageal squamous cell carcinoma. Oncol Lett. 8:133–138.
2014.PubMed/NCBI
|
|
12
|
Raverot G, Wierinckx A, Dantony E, Auger
C, Chapas G, Villeneuve L, Brue T, Figarella-Branger D, Roy P,
Jouanneau E, et al: HYPOPRONOS: Prognostic factors in prolactin
pituitary tumors: Clinical, histological, and molecular data from a
series of 94 patients with a long postoperative follow-up. J Clin
Endocrinol Metab. 95:1708–1716. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shih JY, Yang SC, Hong TM, Yuan A, Chen
JJ, Yu CJ, Chang YL, Lee YC, Peck K, Wu CW, et al: Collapsin
response mediator protein-1 and the invasion and metastasis of
cancer cells. J Natl Cancer Inst. 93:1392–1400. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shih JY, Lee YC, Yang SC, Hong TM, Huang
CY and Yang PC: Collapsin response mediator protein-1: A novel
invasion-suppressor gene. Clin Exp Metastasis. 20:69–76. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cho WC: OncomiRs: The discovery and
progress of microRNAs in cancers. Mol Cancer. 6:602007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu SG, Qin XG, Zhao BS, Qi B, Yao WJ,
Wang TY, Li HC and Wu XN: Differential expression of miRNAs in
esophageal cancer tissue. Oncol Lett. 5:1639–1642. 2013.PubMed/NCBI
|
|
17
|
Lee KH, Goan YG, Hsiao M, Lee CH, Jian SH,
Lin JT, Chen YL and Lu PJ: MicroRNA-373 (miR-373)
post-transcriptionally regulates large tumor suppressor, homolog 2
(LATS2) and stimulates proliferation in human esophageal cancer.
Exp Cell Res. 315:2529–2538. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tian Y, Luo A, Cai Y, Su Q, Ding F, Chen H
and Liu Z: MicroRNA-10b promotes migration and invasion through
KLF4 in human esophageal cancer cell lines. J Biol Chem.
285:7986–7994. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kong KL, Kwong DL, Chan TH, Law SY, Chen
L, Li Y, Qin YR and Guan XY: MicroRNA-375 inhibits tumour growth
and metastasis in oesophageal squamous cell carcinoma through
repressing insulin-like growth factor 1 receptor. Gut. 61:33–42.
2012. View Article : Google Scholar
|
|
20
|
Imanaka Y, Tsuchiya S, Sato F, Shimada Y,
Shimizu K and Tsujimoto G: MicroRNA-141 confers resistance to
cisplatin-induced apoptosis by targeting YAP1 in human esophageal
squamous cell carcinoma. J Hum Genet. 56:270–276. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xiang Y, Lu DL, Li JP, Yu CX, Zheng DL,
Huang X, Wang ZY, Hu P, Liao XH and Zhang TC: Myocardin inhibits
estrogen receptor alpha-mediated proliferation of human breast
cancer MCF-7 cells via regulating MicroRNA expression. IUBMB Life.
68:477–487. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pasquinelli AE, Reinhart BJ, Slack F,
Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B,
Müller P, et al: Conservation of the sequence and temporal
expression of let-7 heterochronic regulatory RNA. Nature.
408:86–89. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Reinhart BJ, Slack FJ, Basson M,
Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR and Ruvkun G:
The 21-nucleotide let-7 RNA regulates developmental timing in
Caenorhabditis elegans. Nature. 403:901–906. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhu S, Wu H, Wu F, Nie D, Sheng S and Mo
YY: MicroRNA-21 targets tumor suppressor genes in invasion and
metastasis. Cell Res. 18:350–359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhu S, Si ML, Wu H and Mo YY: MicroRNA-21
targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol
Chem. 282:14328–14336. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tselepis C, Perry I and Jankowski J:
Barrett's esophagus: Disregulation of cell cycling and
intercellular adhesion in the metaplasia-dysplasia-carcinoma
sequence. Digestion. 61:1–5. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li KK, Qi Y, Xia T, Yao Y, Zhou L, Lau KM
and Ng HK: CRMP1 inhibits proliferation of medulloblastoma and is
regulated by HMGA1. PLoS One. 10:e01279102015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Levine AJ, Hu W and Feng Z: The P53
pathway: What questions remain to be explored? Cell Death Differ.
13:1027–1036. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ko LJ and Prives C: p53: puzzle and
paradigm. Genes Dev. 10:1054–1072. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hollstein M, Sidransky D, Vogelstein B and
Harris CC: p53 mutations in human cancers. Science. 253:49–53.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen J, Jackson PK, Kirschner MW and Dutta
A: Separate domains of p21 involved in the inhibition of Cdk kinase
and PCNA. Nature. 374:386–388. 1995. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Luo Y, Hurwitz J and Massagué J:
Cell-cycle inhibition by independent CDK and PCNA binding domains
in p21Cip1. Nature. 375:159–161. 1995. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Moldovan GL, Pfander B and Jentsch S:
PCNA, the maestro of the replication fork. Cell. 129:665–679. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xia H, Ng SS, Jiang S, Cheung WK, Sze J,
Bian XW, Kung HF and Lin MC: miR-200a-mediated downregulation of
ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma
cell growth, migration and invasion. Biochem Biophys Res Commun.
391:535–541. 2010. View Article : Google Scholar
|