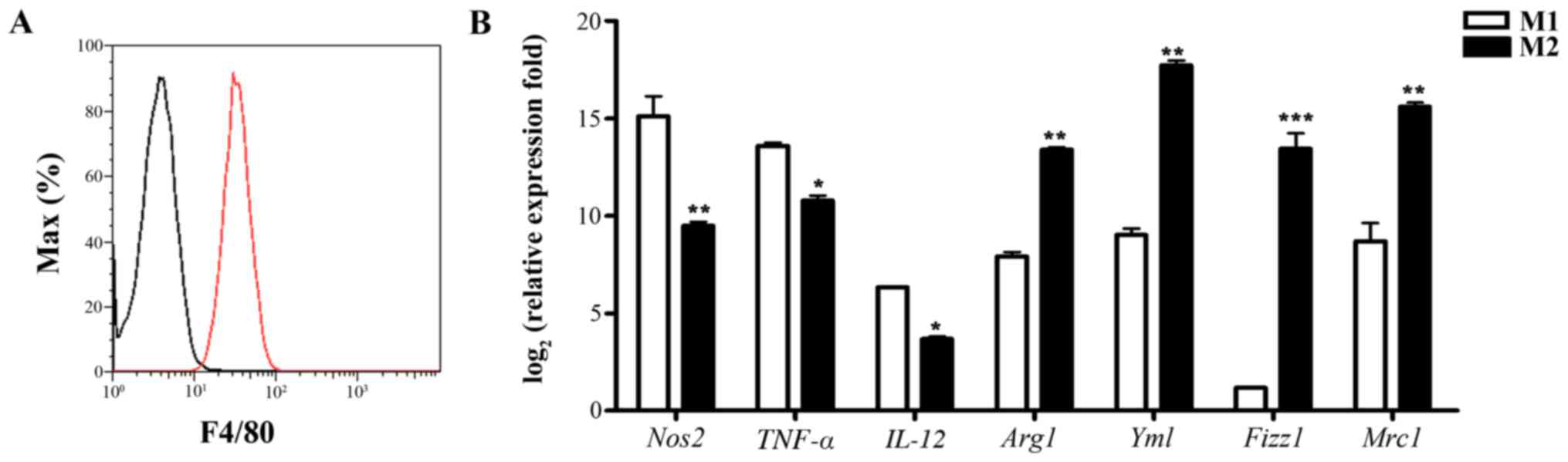
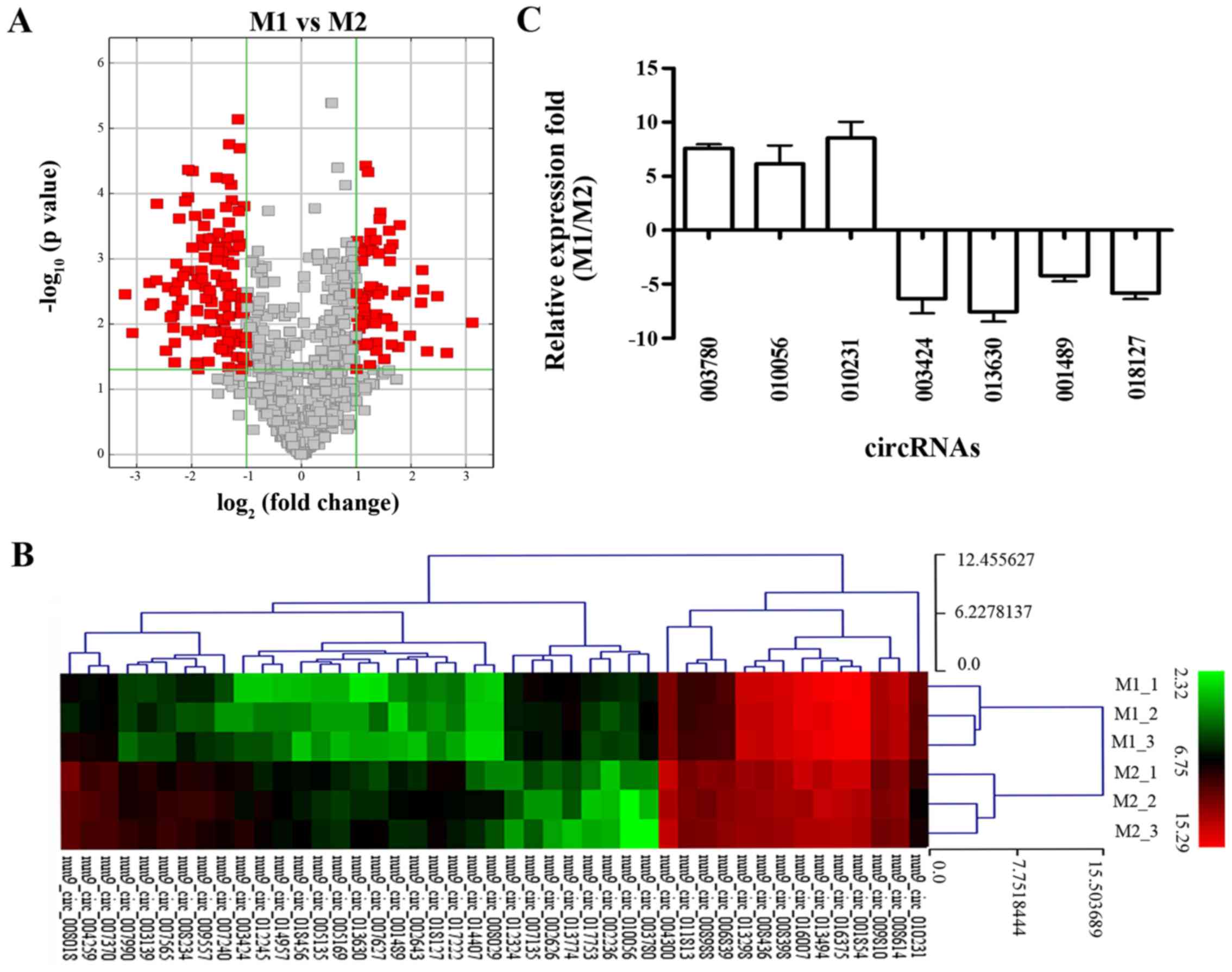
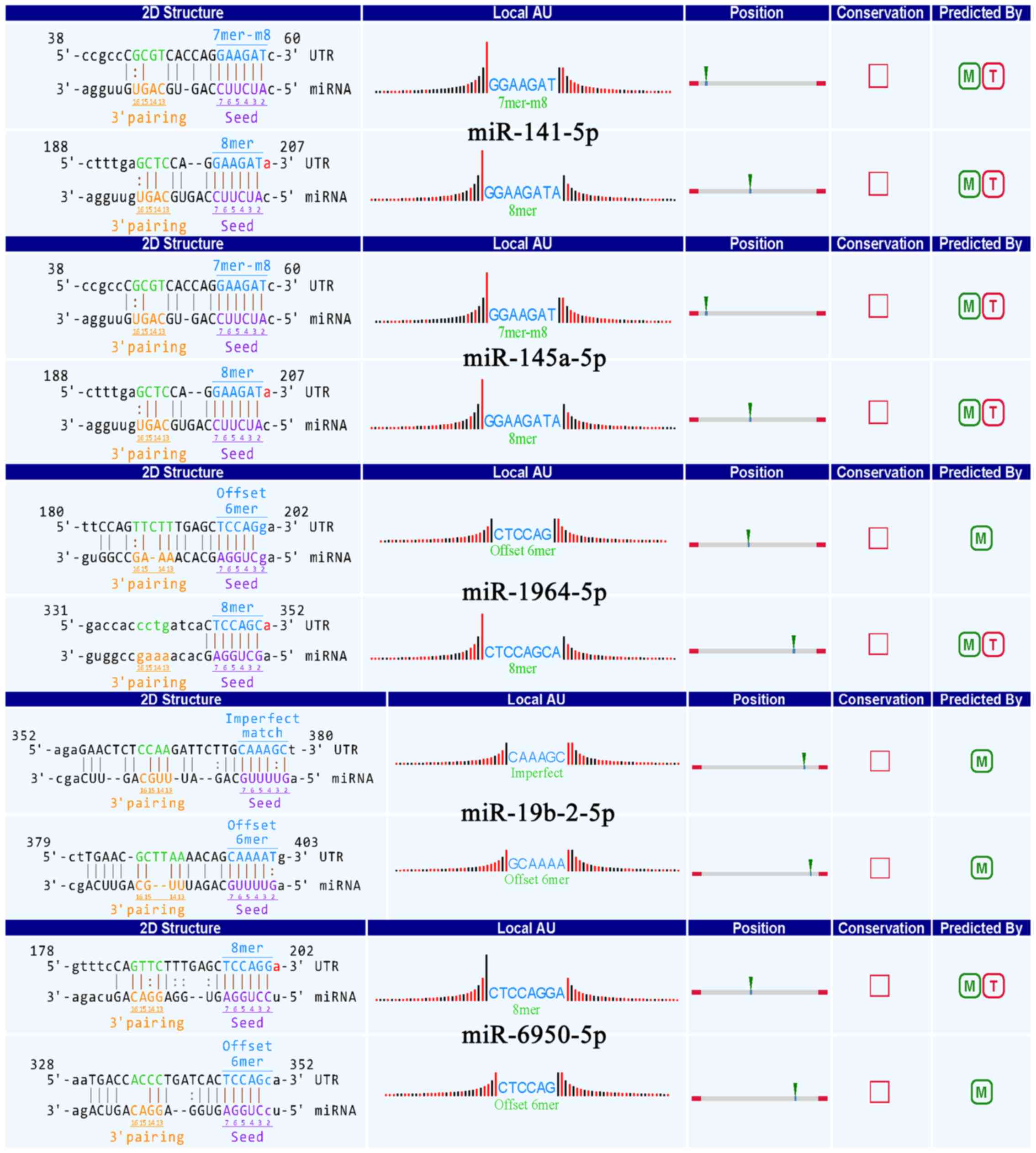
|
1
|
Gordon S and Martinez FO: Alternative
activation of macrophages: Mechanism and functions. Immunity.
32:593–604. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mosser DM: The many faces of macrophage
activation. J Leukoc Biol. 73:209–212. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Biswas SK and Mantovani A: Macrophage
plasticity and interaction with lymphocyte subsets: Cancer as a
paradigm. Nat Immunol. 11:889–896. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mantovani A, Sozzani S, Locati M, Allavena
P and Sica A: Macrophage polarization: Tumor-associated macrophages
as a paradigm for polarized M2 mononuclear phagocytes. Trends
Immunol. 23:549–555. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen T, Huang Z, Wang L, Wang Y, Wu F,
Meng S and Wang C: MicroRNA-125a-5p partly regulates the
inflammatory response, lipid uptake, and ORP9 expression in
oxLDL-stimulated monocyte/macrophages. Cardiovasc Res. 83:131–139.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cheng Y, Kuang W, Hao Y, Zhang D, Lei M,
Du L, Jiao H, Zhang X and Wang F: Downregulation of
miR-27a* and miR-532-5p and upregulation of miR-146a and
miR-155 in LPS-induced RAW264.7 macrophage cells. Inflammation.
35:1308–1313. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Coley W, Van Duyne R, Carpio L, Guendel I,
Kehn-Hall K, Chevalier S, Narayanan A, Luu T, Lee N, Klase Z, et
al: Absence of DICER in monocytes and its regulation by HIV-1. J
Biol Chem. 285:31930–31943. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dalton DK, Pitts-Meek S, Keshav S, Figari
IS, Bradley A and Stewart TA: Multiple defects of immune cell
function in mice with disrupted interferon-gamma genes. Science.
259:1739–1742. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Forrest AR, Kanamori-Katayama M, Tomaru Y,
Lassmann T, Ninomiya N, Takahashi Y, de Hoon MJ, Kubosaki A, Kaiho
A, Suzuki M, et al: Induction of microRNAs, mir-155, mir-222,
mir-424 and mir-503, promotes monocytic differentiation through
combinatorial regulation. Leukemia. 24:460–466. 2010. View Article : Google Scholar
|
|
10
|
Goerdt S and Orfanos CE: Other functions,
other genes: Alternative activation of antigen-presenting cells.
Immunity. 10:137–142. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gordon S: Alternative activation of
macrophages. Nat Rev Immunol. 3:23–35. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cai X, Yin Y, Li N, Zhu D, Zhang J, Zhang
CY and Zen K: Re-polarization of tumor-associated macrophages to
pro-inflammatory M1 macrophages by microRNA-155. J Mol Cell Biol.
4:341–343. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ebert MS, Neilson JR and Sharp PA:
MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian
cells. Nat Methods. 4:721–726. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar :
|
|
16
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lasda E and Parker R: Circular RNAs:
Diversity of form and function. RNA. 20:1829–1842. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Salzman J, Chen RE, Olsen MN, Wang PL and
Brown PO: Cell-type specific features of circular RNA expression.
PLoS Genet. 9:e10037772013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Enright AJ, John B, Gaul U, Tuschl T,
Sander C and Marks DS: MicroRNA targets in Drosophila. Genome Biol.
5:R12003. View Article : Google Scholar
|
|
22
|
Pasquinelli AE: MicroRNAs and their
targets: Recognition, regulation and an emerging reciprocal
relationship. Nat Rev Genet. 13:271–282. 2012.PubMed/NCBI
|
|
23
|
Zhang Y, Zhang M, Zhong M, Suo Q and Lv K:
Expression profiles of miRNAs in polarized macrophages. Int J Mol
Med. 31:797–802. 2013.PubMed/NCBI
|
|
24
|
Rückerl D, Jenkins SJ, Laqtom NN,
Gallagher IJ, Sutherland TE, Duncan S, Buck AH and Allen JE:
Induction of IL-4Rα-dependent microRNAs identifies PI3K/Akt
signaling as essential for IL-4-driven murine macrophage
proliferation in vivo. Blood. 120:2307–2316. 2012. View Article : Google Scholar
|
|
25
|
Chaudhuri AA, So AY, Sinha N, Gibson WS,
Taganov KD, O'Connell RM and Baltimore D: MicroRNA-125b potentiates
macrophage activation. J Immunol. 187:5062–5068. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ghosal S, Das S, Sen R, Basak P and
Chakrabarti J: Circ2Traits: A comprehensive database for circular
RNA potentially associated with disease and traits. Front Genet.
4:2832013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lukiw WJ: Circular RNA (circRNA) in
Alzheimer's disease (AD). Front Genet. 4:3072013. View Article : Google Scholar
|