Introduction
Ovarian cancer is a significant cause of death in
females globally (1). This cancer
is hard to detect at an early stage because of the non-specific
symptoms and misdiagnosis as other disease. The high mortality
associated ovarian cancer is due to delayed diagnosis after
metastasis to other organs (2).
Therefore, it is important to find new biomarkers for detecting and
monitoring of ovarian cancer at early stage (3).
Early stage detection of ovarian cancer has a
survival rate of over 90% (4).
Presently, the cancer antigen CA-125 is increased in more than 80%
of patients with fairly advanced ovarian cancer (5–7). The
level of CA-125 has become established as a useful biomarker for
the assessment of ovarian cancer (8–10).
The serum level of CA-125 and ultrasonography are used to
standardize diagnosis for advanced ovarian cancer determination.
Tumor serum markers could provide reliable and reproducible
information for evaluation of disease. It has been reported that
CA-125 is the most useful target for prognosis of ovarian cancer
(11–14). However, CA-125 monitoring at an
early stage of ovarian cancer is difficult (15). CA-125 lacks sensitivity and
specificity for screening of ovarian cancer. The combined use of
haptoglobin-α (Hp-α) and CA-125 has 91% of sensitivity and 96% of
specificity in the serum of ovarian cancer patients (16). Therefore, many studies have sought
to find biomarkers that can overcome the deficiencies of CA-125
(17). Biomarkers capable of early
detection would improve ovarian cancer patient survival rate.
More recently, biological tools including
microarrays and proteomics have been explored in identifying new
biomarkers for ovarian cancer. In this study, a combined approach
based on the Experion automated electrophoresis system (Bio-Rad,
Hercules, CA) and matrix-assisted laser desorption/ionization
time-of-flight mass spectrometry (MALDI-TOF-MS) was used to
identify highly sensitive and specific serum biomarkers in patient
serum from 14 healthy women and 84 ovarian cancer patients at
stages I–IV. The Experion system is able to quantify protein
expression levels through high throughput screening. These
candidate markers were identified by MALDI-TOF-MS. The distinctive
polypeptides were identified as α-2-macroglobulin (173.7 kDa),
ceruloplasmin (147 kDa), inter-α-trypsin inhibitor family heavy
chain-related protein (IHRP; 126 kDa), C-1 inhibitor (115.2 kDa and
hemoglobin α/β (14.4 kDa).
Materials and methods
Ovarian cancer patients and
specimens
Our analysis included 10 healthy women and 84
ovarian cancer patients. The average age of the ovarian cancer
patients was 46 years (Table I).
The stages of tumors from the ovarian cancer patients were assigned
according to the guidelines provided the International Federation
of Gynecology and Obstetrics. Each serum sample was provided by
Kang Nam St. Mary’s Hospital of Catholic Medical School according
to the procedures approved by the Institutional Review Board of the
Catholic University of Korea (IRB no. KCM07MI020).
 | Table IClinical characteristics and ages of
the patients. |
Table I
Clinical characteristics and ages of
the patients.
| Diagnostic groups and
FIGO stages
| Age
| Histologic subtypes
|
|---|
| a | b | c | n | Mean ± SD
(median) | Epithelial ovarian
tumor | Stromal cell | Other |
|---|
| Control | | | | 10 | 39±5 (40) | | | |
| Stage I | 11 | 1 | 7 | 19 | 45±13 (49) | 11 | 3 | 5 |
| Stage II | 2 | 2 | 6 | 10 | 46±13 (52) | 8 | | 2 |
| Stage III | | 1 | 40 | 41 | 57±13 (62) | 37 | | 4 |
| Stage IV | | | | 14 | 54±11 (56) | 8 | | 6 |
Experion™ system of automated
electrophoresis
The Experion automated electrophoresis system
(Bio-Rad) integrates protein quantitation into a single process in
which protein separation, staining, band detection and quantitation
are automatically executed (18).
All procedures followed the manufacturer’s protocol.
In-gel digestion with trypsin and
extraction of peptides
The procedure for in-gel digestion of protein spots
from silver stained gels were done as previously described
(19). Pieces of stained gel were
washed in 25 mM ammonium bicarbonate buffer (pH 7.8) containing 50%
(v/v) acetonitrile (ACN) for 1 h at room temperature. The gels were
dehydrated by speed vacuum for 10 min and then rehydrated in
trypsin solution (Promega, Madison, WI) at 37°C overnight. The
tryptic peptides were incubated with 5 μl of 0.5%
trifluoroacetic acid (TFA) containing 50% (v/v) ACN for 40 min with
mild sonication. The eluted peptides were enriched up to 1
μl volume using vacuum centrifugation (20). To perform mass spectrometric
analysis, each peptide solution was applied to a desalting column
(GE loader tip; Eppendorf, Hamburg, Germany) (21,22).
Eluted samples from desalting column were dropped onto a MALDI
plate (96×2; Applied Biosystems, Foster City, CA) for analysis as
described below.
Analysis of peptides using MALDI-TOF MS
for identification of proteins
MALDI-TOF mass spectrometry was performed using a
Voyager-DE STR mass spectrometer (Applied Biosystems) in the
reflectron positive ion mode (19). The proteins were matched by peptide
mass fingerprinting searching against the Swiss-Prot and NCBI
databases, using the search program MS-Fit (http://prospector.ucsf.edu).
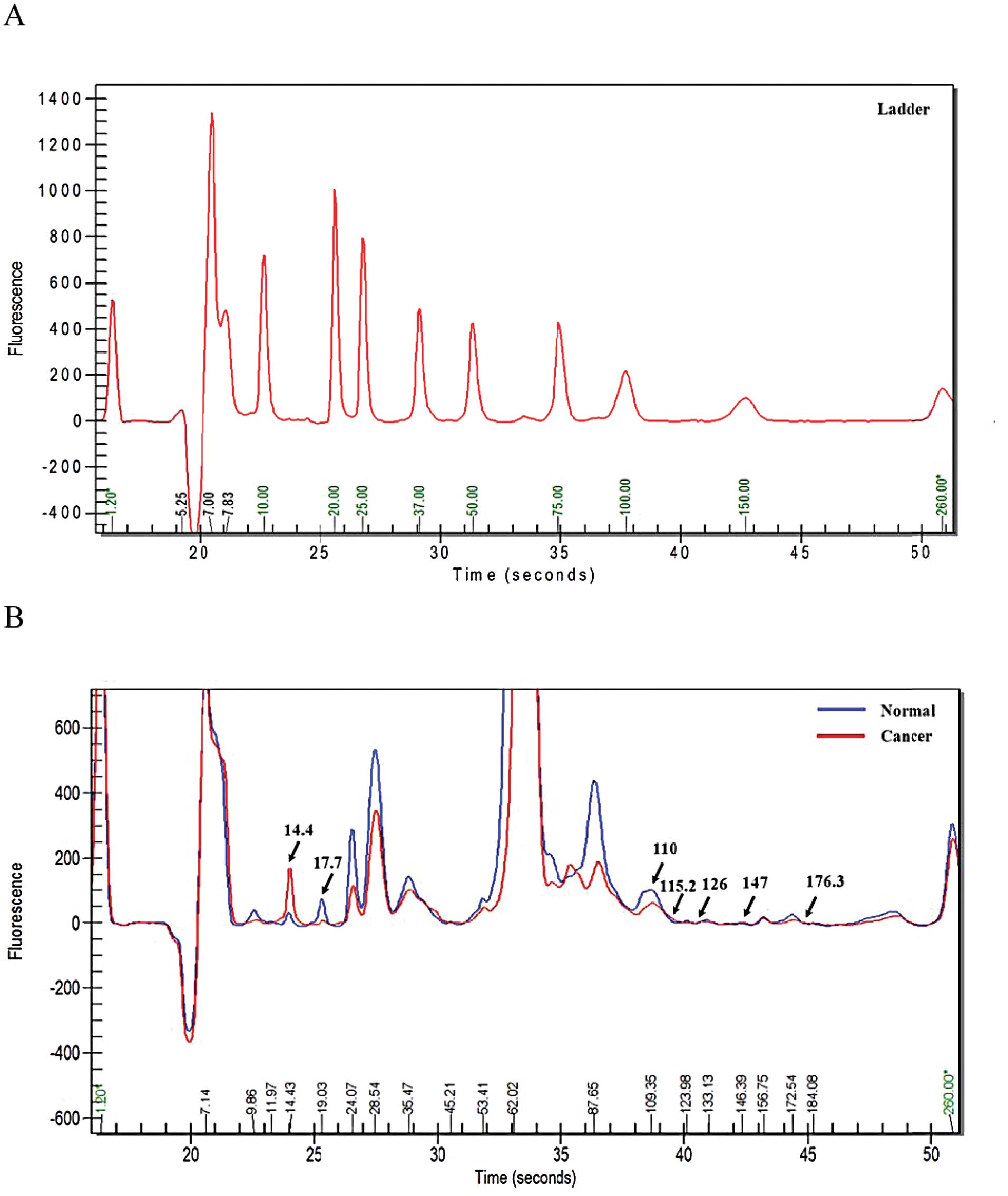
Results
Quantification of protein expression in
ovarian cancer sera
To ascertain the protein expression patterns in the
ovarian cancer patient sera, we used an Experion™ Pro260Chip and
analyzed the distinction between ovarian cancer patient sera and
sera from normal females. The protein quantification profile of the
serum samples revealed higher protein concentrations in the ovarian
cancer sera than normal sera for proteins migrating with an
electrophoretic mobility of 14.4, 115.2, 126, 147 and 176.3 kDa
(Table II). On the other hand,
normal control samples were higher than ovarian cancer serum in
17.1- and 110-kDa proteins. These proteins all displayed
concentration differences exceeding 1.5-fold between normal and
ovarian cancer serum. Although the spectra profiles of the serum of
ovarian cancer and normal were comparable, seven different peak
patterns were expressed (Fig. 1).
These results enabled the detection of potential biomarkers of
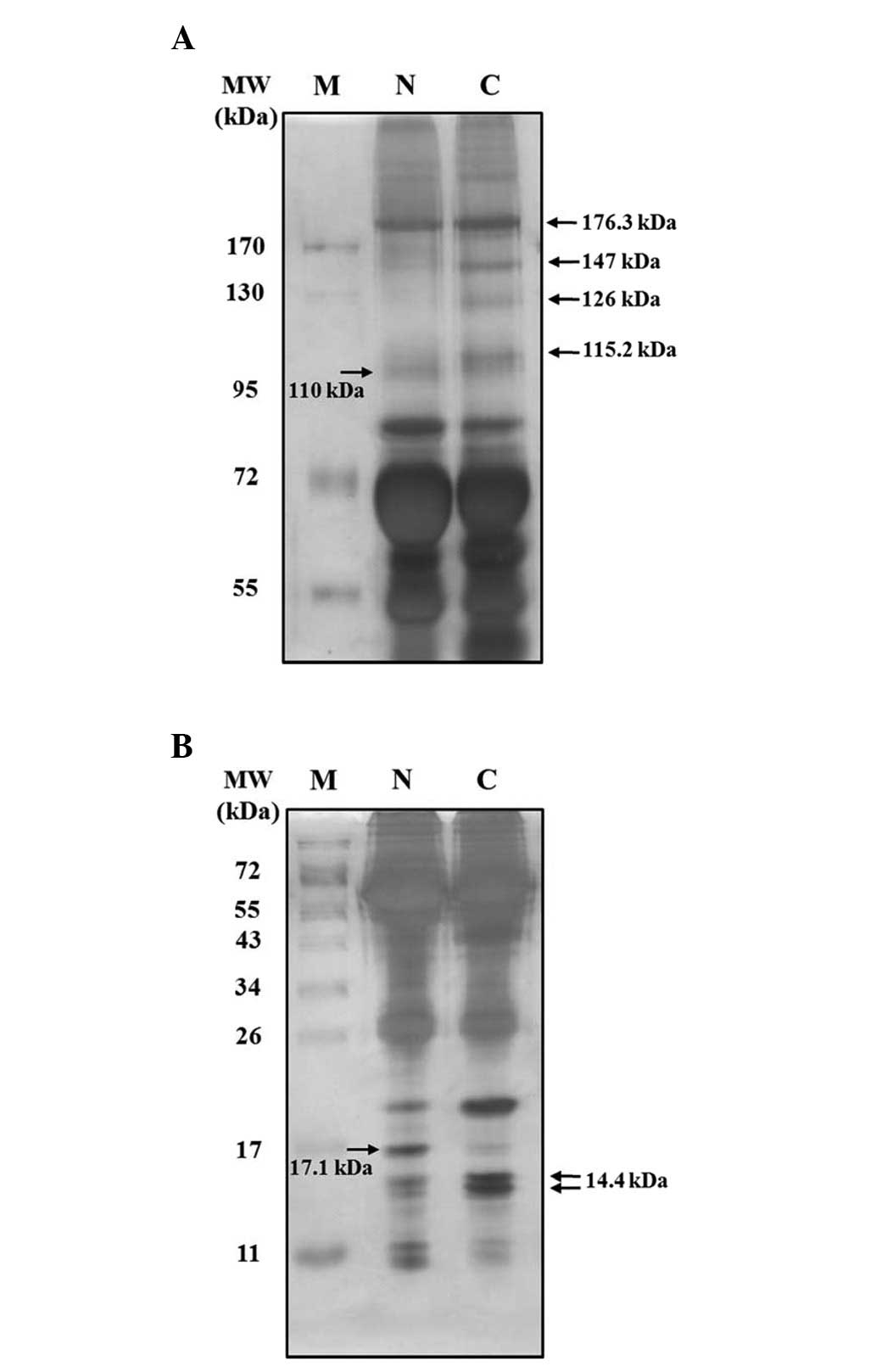
ovarian cancer. To confirm the elevated protein expression level in
ovarian cancer serum, the serum samples were examined using 10 and
15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE) and visualized by silver staining. The silver stained
10% gel images show that proteins of 115.2, 126, 147 and 176.3 kDa
were elevated in ovarian cancer sera, whereas a 110-kDa species was
increased in normal sera (Fig.
2A). In 15% gels, a 14.4- kDa species was increased and a
17.1-kDa protein was decreased in ovarian cancer sera (Fig. 2B).
 | Table IIProtein peaks identified by the
Experion system. |
Table II
Protein peaks identified by the
Experion system.
Healthy women
| Stage I Con
(μg/μl) | Ovarian cancer
patients
|
|---|
| MW (kDa) | Con
(μg/μl) | Stage II Con
(μg/μl) | Stage III Con
(μg/μl) | Stage IV Con
(μg/μl) | Average (stages
I–IV) Con (μg/μl) |
|---|
| 9.9 | 13.2 | 9.4 | 6.3 | 6.3 | 6.1 | 7.0 |
| 12.5 | 1.2 | 5.3 | 0.0 | 0.0 | 0.0 | 1.3 |
| 14.4 | 2.7 | 29.7 | 38.8 | 18.6 | 62.8 | 37.5 |
| 17.1 | 5.4 | 2.1 | 3.2 | 1.1 | 2.2 | 2.2 |
| 19.1 | 11.6 | 12.4 | 10.2 | 12.2 | 17.6 | 13.1 |
| 24.1 | 88.2 | 75.7 | 56.1 | 65.3 | 70.7 | 67.0 |
| 28.8 | 253.3 | 225.4 | 258.2 | 228.1 | 290.6 | 250.6 |
| 36.0 | 125.9 | 99.8 | 104.4 | 112.1 | 135.4 | 112.9 |
| 38.4 | 0.0 | 31.9 | 124.4 | 23.7 | 40.2 | 55.1 |
| 41.4 | 15.7 | 15.7 | 0.0 | 0.0 | 0.0 | 3.9 |
| 47.8 | 0.0 | 1.0 | 3.6 | 6.5 | 2.3 | 3.4 |
| 54.3 | 21.9 | 19.3 | 16.9 | 18.5 | 16.4 | 17.8 |
| 56.5 | 0.0 | 0.0 | 35.2 | 9.8 | 9.4 | 13.6 |
| 64.4 | 2,160.4 | 2,093.9 | 2,419.2 | 1,862.6 | 2,295.0 | 2,167.7 |
| 78.6 | 62.6 | 99.8 | 0.0 | 33.0 | 0.0 | 33.2 |
| 81.9 | 53.7 | 57.8 | 134.0 | 65.9 | 57.0 | 78.7 |
| 90.4 | 208.1 | 189.0 | 180.3 | 123.4 | 161.6 | 163.6 |
| 94.6 | 0.0 | 0.0 | 203.7 | 38.7 | 40.3 | 70.7 |
| 110 | 54.5 | 28.2 | 36.9 | 24.5 | 26.2 | 29.0 |
| 115.2 | 0.0 | 40.2 | 54.4 | 52.0 | 63.7 | 52.6 |
| 126 | 1.4 | 7.5 | 9.5 | 13.3 | 12.4 | 10.7 |
| 147 | 1.1 | 2.3 | 4.5 | 6.7 | 7.9 | 5.4 |
| 176.3 | 6.0 | 11.2 | 9.0 | 14.6 | 15.3 | 12.5 |
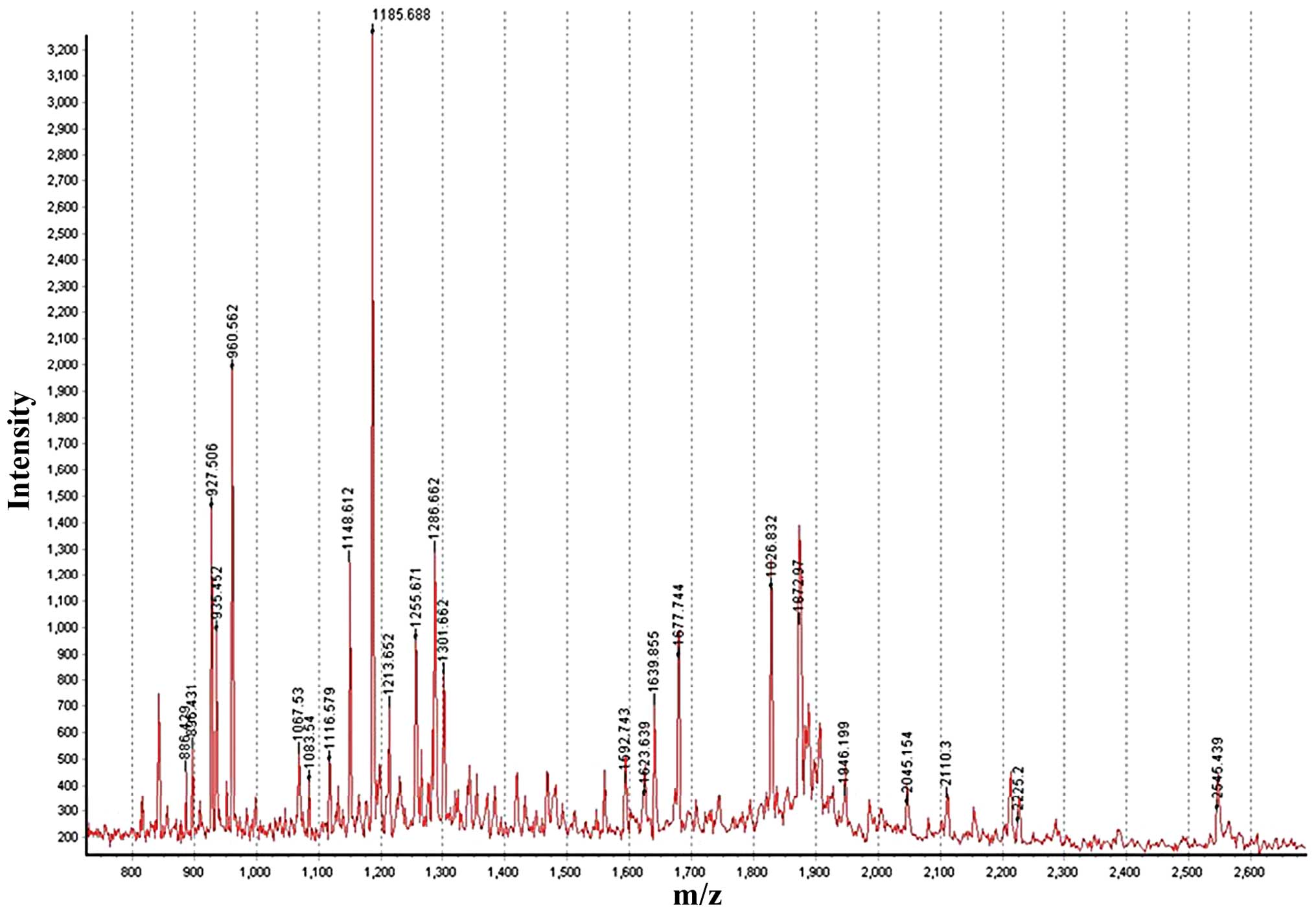
Purification and identification of
biomarker candidates
To further characterize the candidate biomarkers,
the fractions eluted from the gel were analyzed by MALDI-TOF-MS,
which confirmed the purification of the polypeptide peaks (Fig. 3). Analysis focused on regions
showing reproducible differences in increased intensity between
sera of ovarian cancer patients and healthy individuals. The
differential expression of the protein content between the two
groups was determined using MALDI-TOF-MS analysis. Table III lists the identified proteins,
theoretical pI value, molecular weight, Z score and number of
peptides used for identification and protein coverage.
 | Table IIIProteins identified by peptide mass
fingerprinting using MALDI-TOF-MS. |
Table III
Proteins identified by peptide mass
fingerprinting using MALDI-TOF-MS.
| Experion Data (kDa)
up | MALDI-TOF-MS result
|
|---|
| Identified
protein | MW | pI | Est’d Z
(95%≥1.65) | Coverage (%) |
|---|
| 176.3 | C |
α-2-macroglobulin | 164.72 | 6.0 | 2.37 | 21 |
| 147 | C | Ceruloplasmin | 120.87 | 5.4 | 2.24 | 22 |
| 126 | C | IHRP | 103.60 | 6.5 | 1.57 | 18 |
| 115.2 | C | C-1 inhibitor | 32.75 | 8.8 | 2.30 | 24 |
| 110 | N | P130 | 129.78 | 7.4 | 0.89 | 8 |
| 17.1 | N | TTR | 12.83 | 5.3 | 1.42 | 67 |
| 14.4 | C | Hemoglobin β | 15.98 | 6.8 | 2.00 | 45 |
| | Hemoglobin α | 10.69 | 7.1 | 1.34 | 40 |
Seven proteins were determined to be
α-2-macroglobulin (176.3 kDa), ceruloplasmin (147 kDa), IHRP (126
kDa), C-1 inhibitor (115.2 kDa), P130 (110kDa), transthyretin (TTR;
17.1 kDa) and hemoglobin β/hemoglobin α (14.4 kDa) (Table III). Expression of P130 (110 kDa)
and TTR (17.1 kDa) were lower in serum from ovarian cancer patients
than in healthy women, whereas expression of α-2-macroglobulin
(176.3 kDa), ceruloplasmin (147 kDa), IHRP (126 kDa), C-1 inhibitor
(115.2 kDa) and hemoglobin β/hemoglobin α (14.4 kDa) were
increased.
Discussion
In this study, we used the Experion protein
quantification system to detect biomarkers for diagnosis in ovarian
cancer patient. The Experion automated electrophoresis system
easily enables protein quantitation and performs high-throughput
screening for detecting candidate biomarkers in ovarian cancer. The
present findings are hopeful, given that ovarian cancer is one of
the detrimental causes of death in females in the world (1), yet no apparent clinical prognoses or
characteristics have been demonstrated at the initial stage of
ovarian cancer.
We detected hemoglobin β chain, hemoglobin α chain
together with α-2-macroglobulin, ceruloplasmin, IHRP, C-1
inhibitor, P130 and TTR. Of these proteins, P130 and TTR were
decreased in cancer serum samples. Our analysis included 10 healthy
individuals and 84 ovarian cancer patients.
α-2-macroglobulin (A2M) is a protease inhibitor in
mammals (23). It is reported that
A2M is secreted in serum of women with inflammatory and neoplastic
ovarian lesions. A2M has also been semi-quantitatively identified
in ovarian cancer-related proteins (24).
Ceruloplasmin is a member of a family of copper
transport metalloproteins. It has important roles in iron
metabolism and antioxidant defense (25). Ceruloplasmin blocks the copper
ion-activated production of toxic oxygen compounds and protects
cells from oxidative stress (26).
Ceruloplasmin protein is expressed in pancreatic, nasopharyngeal
and germ-line ovarian cancers (27). Ceruloplasmin promoter activation is
specifically and efficiently enhanced in ovarian cancer (28).
IHRP is an acute phase protein and glycoprotein in
mammals, which is cleaved to different length fragments (29). IHRP inhibits polymerization through
binding to actin and protects cells from phagocytosis. IHRP
concentrations are elevated in patient serum of inflammatory
disease. So, IHRP has been implicated as an anti-inflammatory
protein (30).
C-1 inhibitor is a protease inhibitor that regulates
vascular permeability and suppression of inflammation (31). C-1 inhibitor is an acute phase
protein that inhibits complement system protease. Also, C-1
inhibitor is proposed to play a role in inhibition of alternative
complement activation and inflammation.
Hemoglobin is an iron-containing oxygen transporter
in red blood cells. Hemoglobin is overexpressed in ovarian cancer
(16,32). Previously, our group also reported
that the potential value of the hemoglobin-α and -β subunits as
serum biomarkers for the early diagnosis and prognosis of ovarian
cancer (33).
In contrast, our results show that P130 and TTR were
decreased in ovarian cancer patient serum. Commonly, it has been
reported that these protein expressions were decreased in cancer
patient serum.
P130 (also known as pRb2), is a member of a family
of retinoblastoma proteins. Proteins of the pRB family regulate
transcription and progression of the cell cycle (34). P130 may operate as a tumor
suppressor in small-cell lung carcinoma (35). The decline of P130 causes
tumorigenesis in mouse model of human lung adenocarcinoma (36).
TTR is a carrier of the thyroid hormone thyroxine
and retinol in serum and cerebrospinal fluid. TTR is reduced in
ovarian cancer as well as cervical and endometrial carcinomas
(37,38).
The present study demonstrates the potential of the
Experion quantitation method that covers high sensitivity and
specificity biomarker discovery in ovarian cancer. Our group has
previously found that hemoglobin β/α and ceruloplasmin are
increased in ovarian cancer serum using the Experion assay system
(39). Hemoglobin β/α has been
identified as an ovarian cancer biomarker by using the surface
enhanced laser desorption/ionization time-of-flight mass
spectrometry mass method (33).
Thus, hemoglobin β/α must be regarded as a strong candidate ovarian
cancer biomarker. α-2-macroglobulin, IHRP, C-1 inhibitor, P130 and
TTR were all also presently altered in expression (overexpressed or
decreased) in ovarian cancer. Further studies are needed to
determine their relevance as ovarian cancer biomarkers. In
addition, C-1 inhibitor and IHRP were increased in stage I ovarian
cancer serum compared with normal serum (Table II), implicating the two proteins as
strong candidate biomarkers for the early detection of ovarian
cancer.
Our study shows that the Experion system is able to
identify new biomarkers selectively and correctly. Also, the
Experion system could be applied to find other disease
biomarkers.
In conclusion, α-2-macroglobulin, ceruloplasmin,
IHRP, C-1 inhibitor, P130, TTR and hemoglobin β/α were identified
in ovarian cancer using the Experion assay system. The findings
provide evidence for the use of these proteins as new potential
biomarkers for ovarian cancer diagnosis. Identification of
potential biomarkers provides opportunities to develop noninvasive
diagnosis and further improves the understanding of ovarian cancer
development.
Acknowledgements
This research was supported by Basic
Science Research Program through the National Research Foundation
of Korea (NRF) funded by the Ministry of Education, Science and
Technology (2012-000-8766).
References
|
1
|
Jemal A, Siegel R, Ward E, et al: Cancer
statistics, 2008. CA Cancer J Clin. 58:71–96. 2008. View Article : Google Scholar
|
|
2
|
Visintin I, Feng Z, Longton G, et al:
Diagnostic markers for early detection of ovarian cancer. Clin
Cancer Res. 14:1065–1072. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Suh KS, Park SW, Castro A, et al: Ovarian
cancer biomarkers for molecular biosensors and translational
medicine. Expert Rev Mol Diagn. 10:1069–1083. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bast RC Jr, Urban N, Shridhar V, et al:
Early detection of ovarian cancer: promise and reality. Cancer
Treat Res. 107:61–97. 2002.PubMed/NCBI
|
|
5
|
Jemal A, Thomas A, Murray T and Thun M:
Cancer statistics, 2002. CA Cancer J Clin. 52:23–47. 2002.
View Article : Google Scholar
|
|
6
|
Keyes K, Cox K, Treadway P, et al: An in
vitro tumor model: analysis of angiogenic factor expression after
chemotherapy. Cancer Res. 62:5597–5602. 2002.PubMed/NCBI
|
|
7
|
Keyes KA, Mann L, Cox K, et al:
Circulating angiogenic growth factor levels in mice bearing human
tumors using Luminex Multiplex technology. Cancer Chemother
Pharmacol. 51:321–327. 2003.PubMed/NCBI
|
|
8
|
Sasaroli D, Coukos G and Scholler N:
Beyond CA125: the coming of age of ovarian cancer biomarkers. Are
we there yet? Biomark Med. 3:275–288. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Szekanecz E, Sandor Z, Antal-Szalmas P, et
al: Increased production of the soluble tumor-associated antigens
CA19-9, CA125, and CA15-3 in rheumatoid arthritis: potential
adhesion molecules in synovial inflammation? Ann NY Acad Sci.
1108:359–371. 2007. View Article : Google Scholar
|
|
10
|
Gupta D and Lis CG: Role of CA125 in
predicting ovarian cancer survival - a review of the
epidemiological literature. J Ovarian Res. 2:132009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Scholler N and Urban N: CA125 in ovarian
cancer. Biomark Med. 1:513–523. 2007. View Article : Google Scholar
|
|
12
|
Rice LW, Lage JM, Berkowitz RS, et al:
Preoperative serum CA-125 levels in borderline tumors of the ovary.
Gynecol Oncol. 46:226–229. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pearl ML, Yashar CM, Johnston CM, Reynolds
RK and Roberts JA: Exponential regression of CA 125 during salvage
treatment of ovarian cancer with taxol. Gynecol Oncol. 53:339–343.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bast RC Jr, Badgwell D, Lu Z, et al: New
tumor markers: CA125 and beyond. Int J Gynecol Cancer. 15(Suppl 3):
274–281. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kitawaki J, Ishihara H, Koshiba H, et al:
Usefulness and limits of CA-125 in diagnosis of endometriosis
without associated ovarian endometriomas. Hum Reprod. 20:1999–2003.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ye B, Cramer DW, Skates SJ, et al:
Haptoglobin-alpha subunit as potential serum biomarker in ovarian
cancer: identification and characterization using proteomic
profiling and mass spectrometry. Clin Cancer Res. 9:2904–2911.
2003.
|
|
17
|
Raja FA, Hook JM and Ledermann JA:
Biomarkers in the development of anti-angiogenic therapies for
ovarian cancer. Cancer Treat Rev. 38:662–672. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fleige S and Pfaffl MW: RNA integrity and
the effect on the real-time qRT-PCR performance. Mol Aspects Med.
27:126–139. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Park YD, Kim SY, Jang HS, et al: Towards a
proteomic analysis of atopic dermatitis: a
two-dimensional-polyacrylamide gel electrophoresis/mass
spectrometric analysis of cultured patient-derived fibroblasts.
Proteomics. 4:3446–3455. 2004. View Article : Google Scholar
|
|
20
|
Bahk YY, Kim SA, Kim JS, et al: Antigens
secreted from Mycobacterium tuberculosis: identification by
proteomics approach and test for diagnostic marker. Proteomics.
4:3299–3307. 2004.PubMed/NCBI
|
|
21
|
Gobom J, Nordhoff E, Mirgorodskaya E,
Ekman R and Roepstorff P: Sample purification and preparation
technique based on nano-scale reversed-phase columns for the
sensitive analysis of complex peptide mixtures by matrix-assisted
laser desorption/ionization mass spectrometry. J Mass Spectrom.
34:105–116. 1999. View Article : Google Scholar
|
|
22
|
Kim SY, Kim YS and Bahk YY: Proteome
changes induced by expression of tumor suppressor PTEN. Mol Cells.
15:396–405. 2003.PubMed/NCBI
|
|
23
|
Armstrong PB: Proteases and protease
inhibitors: a balance of activities in host-pathogen interaction.
Immunobiology. 211:263–281. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shield-Artin KL, Bailey MJ, Oliva K, et
al: Identification of ovarian cancer-associated proteins in
symptomatic women: A novel method for semi-quantitative plasma
proteomics. Proteomics Clin Appl. 6:170–181. 2012. View Article : Google Scholar
|
|
25
|
Samokyszyn VM, Miller DM, Reif DW and Aust
SD: Inhibition of superoxide and ferritin-dependent lipid
peroxidation by ceruloplasmin. J Biol Chem. 264:21–26.
1989.PubMed/NCBI
|
|
26
|
Healy J and Tipton K: Ceruloplasmin and
what it might do. J Neural Transm. 114:777–781. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pang WW, Abdul-Rahman PS, Wan-Ibrahim WI
and Hashim OH: Can the acute-phase reactant proteins be used as
cancer biomarkers? Int J Biol Markers. 25:1–11. 2010.PubMed/NCBI
|
|
28
|
Lee CM, Lo HW, Shao RP, et al: Selective
activation of ceruloplasmin promoter in ovarian tumors: potential
use for gene therapy. Cancer Res. 64:1788–1793. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Negishi A, Ono M, Handa Y, et al:
Large-scale quantitative clinical proteomics by label-free liquid
chromatography and mass spectrometry. Cancer Sci. 100:514–519.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Choi-Miura NH: Novel human plasma
proteins, IHRP (acute phase protein) and PHBP (serine protease),
which bind to glycosaminoglycans. Curr Med Chem Cardiovasc Hematol
Agents. 2:239–248. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Davis AE III, Mejia P and Lu F: Biological
activities of C1 inhibitor. Mol Immunol. 45:4057–4063. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Maccio A, Madeddu C, Massa D, et al:
Hemoglobin levels correlate with interleukin-6 levels in patients
with advanced untreated epithelial ovarian cancer: role of
inflammation in cancer-related anemia. Blood. 106:362–367. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Woong-Shick A, Sung-Pil P, Su-Mi B, et al:
Identification of hemoglobin-alpha and -beta subunits as potential
serum biomarkers for the diagnosis and prognosis of ovarian cancer.
Cancer Sci. 96:197–201. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
De Falco G and Giordano A: pRb2/p130: a
new candidate for retinoblastoma tumor formation. Oncogene.
25:5333–5340. 2006.PubMed/NCBI
|
|
35
|
Schaffer BE, Park KS, Yiu G, et al: Loss
of p130 accelerates tumor development in a mouse model for human
small-cell lung carcinoma. Cancer Res. 70:3877–3883. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ho VM, Schaffer BE, Karnezis AN, Park KS
and Sage J: The retinoblastoma gene Rb and its family member p130
suppress lung adenocarcinoma induced by oncogenic K-Ras. Oncogene.
28:1393–1399. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Schweigert FJ and Sehouli J:
Transthyretin, a biomarker for nutritional status and ovarian
cancer. Cancer Res. 65:1114author reply 1114,. 2005.PubMed/NCBI
|
|
38
|
Kozak KR, Su F, Whitelegge JP, Faull K,
Reddy S and Farias-Eisner R: Characterization of serum biomarkers
for detection of early stage ovarian cancer. Proteomics.
5:4589–4596. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Min HJ, Bae SM, Kwak SY, et al:
Application of Experion™ assay system for discovery of ovarian
cancer serum biomarkers. Korean J Obstet Gynecol. 50:751–759.
2007.
|