Introduction
Breast cancer is the most frequent cancer and the
most common cause of cancer-related death in women (1). In Brazil alone, ∼50,000 new cases of
breast cancer are expected per year, with an estimated risk of 52
cases for every 100,000 women (2).
Several studies have suggested that different genetic alterations
are linked to different histological types of breast cancer, such
as estrogen receptor expression/lobular type, c-ErbB2
expression/invasive type, among others (3,4).
Recently, molecular studies have shown that breast cancer
development involves not only alterations in oncogenes and tumor
suppressor genes, but also epigenetic alterations in genes related
to cellular growth, survival, motility and differentiation
(5). Disturbance of epigenetic
regulation of gene expression through methylation might contribute
to tumorigenesis. In this context, altered histone modifying
enzymes such as methyltransferases may play important roles in the
process of carcinogenesis.
The mixed lineage leukemia (MLL) family is
characterized by a conserved catalytic SET domain, responsible for
the methyltransferase activity and comprises five genes [MLL
(ALL-1), MLL2, MLL3, MLL4 and MLL5].
The mixed lineage leukemia gene (MLL or
ALL-1, HRX and Htrx1) (Gene ID: 4297) is located in
human chromosome 11q23 and is frequently involved in chromosomal
translocations in leukemia (6,7).
MLL seems to function as a mammalian counterpart of
Drosophila trx, which maintains the expression of multiple
Hox genes during development. Mll-deficiency in mice
results in failure to express Hox genes and in embryonic
lethality. Heterozygosity for disrupted MLL, by homologous
recombination in mouse embryonic stem cells, leads to retarded
growth, hematopoietic abnormalities and skeleton malformations
(8).
The MLL2 gene (Gene ID: 8085) is located on
human chromosome 12q13.12 and encodes a protein with domains
similar to MLL. MLL2 was shown to be associated with Pax7
(paired-box transcription factor), forming the Wdr5-Ash2L-MLL2
histone methyltransferase (HMT) complex that directs
tri-methylation of histone H3 lysine 4, when bound to Myf5. These
chromatin modifications stimulate transcriptional activation of
target genes to regulate entry into the myogenic developmental
program (9). Another study, based
on muscle-specific myogenin gene, suggested that
transcriptional activators Mef2d and Six4 mediate recruitment of
trithorax group proteins Ash2L/MLL2 and UTX to MyoD-bound promoters
to overcome the polycomb-mediated repression of muscle genes,
allowing transcriptional activation of muscle-specific genes
(10). MLL and MLL2
genes show highly similar exon/intron structures. The proteins
encoded by these genes are ubiquitously expressed among adult human
tissues and although they share similar domains, they seem to act
in a different manner. MLL2 could not compensate for the
heterozygosity of the disrupted MLL, by homologous
recombination in mice (8), which
indicates that their functions seem to be at least partially
non-overlapping.
MLL3 (Gene ID: 58508) maps to chromosome
7q36.1, a region usually deleted in hematological malignancies
(11). MLL4 (Gene ID: 9757)
is located on chromosome 19q13.1. MLL3 and MLL4 are
found as part of a complex named ASCOM (for ASC-2 COMplex),
that acts as a p53 co-activator. This complex is required for the
expression of endogenous p53-target genes in response to the DNA
damage (12). Deletions or
truncations in Mll, Mll2 and Mll3 in mice show
different phenotypes, suggesting that MLL protein functions are not
redundant (8,13).
MLL5 (Gene ID: 55904) is located in human
chromosome 7q22.1. The nuclear protein encoded shows single PHD and
SET domains and lacks other conserved sequence elements, like its
Drosophila homolog CG9007. MLL5 shows ubiquitous
tissue expression and is highly conserved. It is the latest
addition to the mammalian Trithorax/MLL gene family and
recent studies show that the MLL5 may function in a different
manner, without methyltranferase activity, which is consistent with
its different sequence of the SET domain compared to the rest of
the MLL family of H3K4 methyltransferases (14). It was suggested that MLL5 would
have an indirect mechanism regulating expression of
histone-modifying enzymes, such as LSD1 and SET7/9 (15). MLL5 was shown to promote myogenic
differentiation through positive regulation of the pro-myogenic
genes Pax7, Myf5 and myogenin, which are
impaired in MLL5 knockdown, leading to serious
differentiation defects (15).
Mll5 deficiency in mice leads to growth retardation, male
infertility and defects in multiple hematopoietic lineages
(16). Therefore, the defects
found could be associated with deregulation of cell cycle
control.
Some studies have shown that MLLs are critical
co-activators in estrogen (E2)-dependent regulation of
E2-responsive genes, through LXXLL motifs (NR boxes) (17,18).
The suppression of MLL2 decreased expression of estrogen
receptor (ER) target genes (18).
MLL3 and MLL4 coordinate with ERs and act in transcriptional
regulation of HOXC10 in the presence of estrogen (19). MLL3 and MLL4, as members of the
ASCOM complex, play essential roles in gene activation mediated
through nuclear receptors, which bind hormone responsive elements
(12). The activity of the MLL
family in hormone-mediated gene activation and signaling could be
helpful to understand its involvement in breast carcinogenesis.
Here we present an analysis of the patterns of
MLL family gene expression in seven breast cancer cells
lines, twelve human normal tissues and eight breast tumors and
their adjacent normal breast tissues. Our results may help to
provide a better understanding of the role of the MLL genes
in breast carcinogenesis.
Materials and methods
Cell lines and culture
Seven breast cancer lines were used on this study:
HCC-1954, MCF-7, CAMA-1, SKBR-3, MDA-MB-231, MDA-MB-436 and
MDA-MB-468. The cell lines HCC-1954, MCF-7 and MDA-MB-231, obtained
from the ATCC, were cultured in our laboratory in recommended media
and checked periodically for mycoplasma contamination. The main
characteristics of the cell lines are summarized in Table I (20). Cells were grown in Dulbecco’s
modified Eagle’s medium (DMEM, Sigma-Aldrich, St. Louis, MO, USA)
supplemented with 10% fetal bovine serum (Sigma-Aldrich) and
penicillin/streptomycin (100 U/ml; Life Technologies, Carlsbad, CA,
USA) at 37°C and 5% CO2. Total RNA from these cells was
extracted and used for expression analysis as described below.
Additionally, total RNA from the remainder cells were kindly
provided by the Ludwig Institute for Cancer Research, São Paulo,
and were included in the gene expression analysis.
 | Table I.Characteristics of the breast cancer
cell lines. |
Table I.
Characteristics of the breast cancer
cell lines.
| Cell lines | Origin | ER | PR | Cerb-B2 | Tumor type |
Characteristics |
|---|
| HCC-1954 | Primary breast
cancer | − | − | + | DC | Ductal carcinoma
with no lymph node metastases |
| MCF-7 | Pleural
effusion | + | + | * | IDC | Invasive ductal
carcinoma; weakly invasive |
| CAMA-1 | Pleural
effusion | + | − | * | AC | Metastatic
adenocarcinoma |
| SKBR-3/AU565 | Pleural
effusion | − | − | + | AC | Metastatic
adenocarcinoma |
| MDA-MB-231 | Pleural
effusion | − | − | * | AC | Metastatic
adenocarcinoma; highly invasive |
| MDA-MB-436 | Pleural
effusion | − | − | * | IDC | Metastatic
adenocarcinoma; highly pleomorphic |
| MDA-MB-468 | Pleural
effusion | − | − | * | AC | Metastatic
adenocarcinoma |
Normal tissues
A commercial panel (OriGene Technologies, Rockville,
MD, USA) of total RNA from twelve human normal tissues from
different organs (lung, small intestine (SI), brain, colon, kidney,
muscle, liver, spleen, heart, testis, stomach and placenta) was
used in this study. Total RNA of each normal tissue was used to
verify the expression levels of the genes studied.
Tumor specimens
The sample collection protocol was approved by the
Research Ethics Committee of the Faculty of Health Sciences,
University of Brasília, Brazil, based on resolution 196/96 of the
National Health Council/Brazilian Ministry of Health, project no.
025/09. Written informed consent was obtained from all
participants. The samples were collected from surgically removed
breast tissue from female patients diagnosed with breast cancer at
the Hospital of the University of Brasilia. A total of 14 samples
(6 normal and 8 tumor samples) was used. The identification of
tumor tissue from the removed samples was based on the
histopathological examination and all types of breast cancer were
included in our analyses. Samples were selected based on tumor
content (minimally 80% tumor) as determined by microscopic
pathological analysis. A sample of normal tissue was collected from
each patient whenever possible. The clinical and histopathological
characteristics of the patients are summarized in Table IV.
 | Table IV.Clinical and histopathological
characteristics of the 8 breast cancer patients. |
Table IV.
Clinical and histopathological
characteristics of the 8 breast cancer patients.
| Samples | Breast
affected | Age | Histological
type | Histological
grade | Stage | Neoadjuvant
chemotherapy | C-erb-B2 | ER | PR |
|---|
| 1 | Left | 58 | DCI | High | T3N2AM0 | Yes | + | − | − |
| 2 | Left | 54 | DCI | Intermediate | T4DN3M0 | Yes | − | + | + |
| 3 | Left | 64 | DCI | Intermediate | YPT4PN1M0 | Yes | − | − | − |
| 4 | Right | 43 | DCI | High | T2N1M0 | Yes | * | * | * |
| 5 | Left | 56 | DCI | High | T2N1AM0 | Yes | − | − | − |
| 6 | Right | 60 | DCI | High | T2N3M0 | Yes | − | + | + |
| 7 | Right | 61 | LCI | Intermediate | T2N0M0 | No | + | + | + |
| 8 | Right | 32 | DCI | Low | T2N0M0 | Yes | + | + | + |
RNA extraction and cDNA synthesis
Total RNA was isolated using TRIzol LS reagent,
according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA,
USA). Single-stranded complementary DNA was generated from total
RNA obtained from cell lines, normal tissues and clinical samples
with reverse transcriptase and random primers, using the Applied
Biosystems High Capacity cDNA Reverse Transcription kit (Applied
Biosystems, Carlsbad, CA, USA).
Quantitative PCR (qPCR)
Reactions of qPCR were performed on a StepOnePlus™
Real-Time PCR System (Applied Biosystems) using TaqMan Universal
PCR MasterMix and TaqMan Gene Expression Assays (Hs01061596_m1 for
MLL, Hs00231606_m1 for MLL2, Hs00419011_m1 for
MLL3, Hs00207065_m1 for MLL4 and Hs00218773_m1 for
MLL5, Applied Biosystems), according to the manufacturer’s
instructions. Briefly, 2 μl of each sample template was
added to a 10 μl final reaction volume per well.
Amplification conditions were as follows: 2 min at 50°C and 10 min
at 95°C on holding stage and then 40 cycles of 15 sec at 95°C and 1
min at 60°C. Gene expression values are reported as ratios between
the amplification levels of genes of interest and that of
endogenous control gene (Hs99999903_m1 for β-actin, Applied
Biosystems), which provides a normalization factor for the amount
of RNA isolated from a specimen. This was subsequently normalized
with the ratio obtained in control samples (relative expression
level). Each assay was performed in triplicate for each sample.
qPCR data analysis
To determine the relative quantification (RQ) of
gene expression, the data were analyzed using the comparative
quantification Ct method (ΔΔCt) (Applied Biosystems). Briefly, the
mean of Ct (cycle threshold) values of the replicates were
calculated and normalized by subtracting the Ct value of the
co-amplified endogenous control gene to yield a ΔCt value. The ΔCt
of a random control calibrator sample (1X sample) was subtracted
from the ΔCt of the other samples to yield a ΔΔCt value. The amount
of target gene, normalized to an endogenous reference and relative
to a calibrator was converted into relative quantification by the
formula: 2−ΔΔCT. For the cell lines, normal tissues and
clinical samples used, the MLL genes expression was
normalized using the β-actin as an endogenous control. Expression
level was considered altered when augmented or diminished ≥2X
compared to the control calibrator sample, chosen for each
analysis. To check the statistical significance of the relative
quantification of the genes, the non-parametric Mann-Whitney U test
was performed for the pools of normal (n=6) and tumor (n=8)
samples. The one-sample Wilcoxon signed-rank test was used for the
expression results of the MLL genes family in seven cancer
cell lines, to test if the samples means significantly differed
from the hypothesized value (sample calibrator relative
quantification: 1) The statistical tests were considered
significant when the P-value was <0.05 (CI 95%). Calculations
were performed using the software SPSS version 20 (SPSS Inc.,
Chicago, IL, USA). Additionally, we searched for MLL gene
expression profiles in breast cancer datasets, available in the
database Oncomine, to compare with our most significant
findings.
Results
MLL genes are downregulated in breast
cancer cell lines
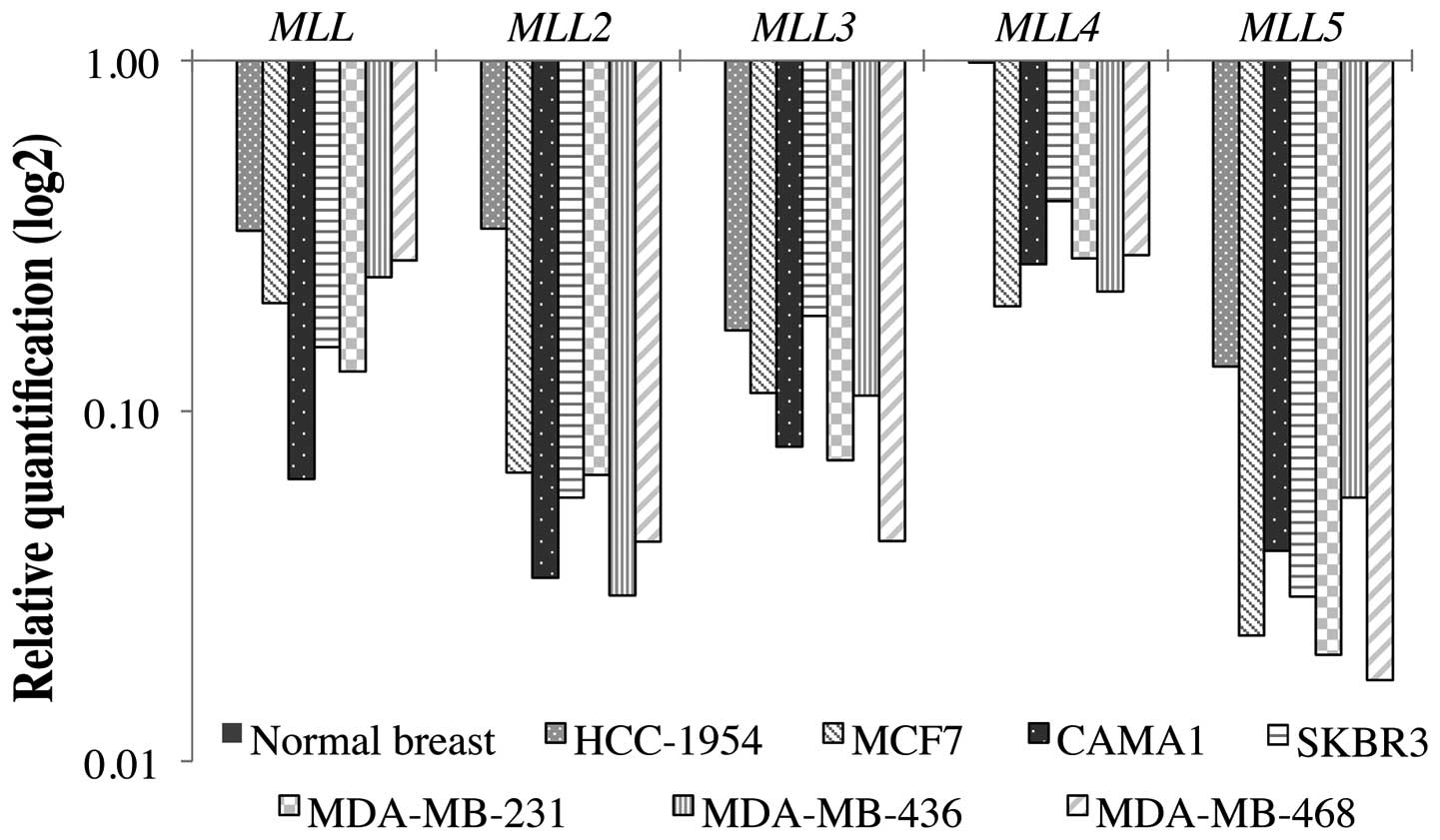
The results of the MLL gene expression
analysis in seven cancer cell lines using the comparative
quantification method ΔΔCT are shown in Fig. 1. All genes showed expression levels
diminished in comparison to the normal breast sample calibrator (1X
sample) (P<0.05). The MLL2 and MLL5 were the genes
with the lowest expression level in all cell lines, with the
exception of HCC-1954. The HCC-1954 lineage is the least aggressive
among the breast cancer cell lines studied, with no lymph node
metastases (Table I). Although all
MLL genes were also downregulated in HCC-1954 cells in
relation to the normal breast calibrator control, this cell line
showed the least reduced expression among the breast cancer cell
lines.
MLL genes are differentially expressed
among normal tissues
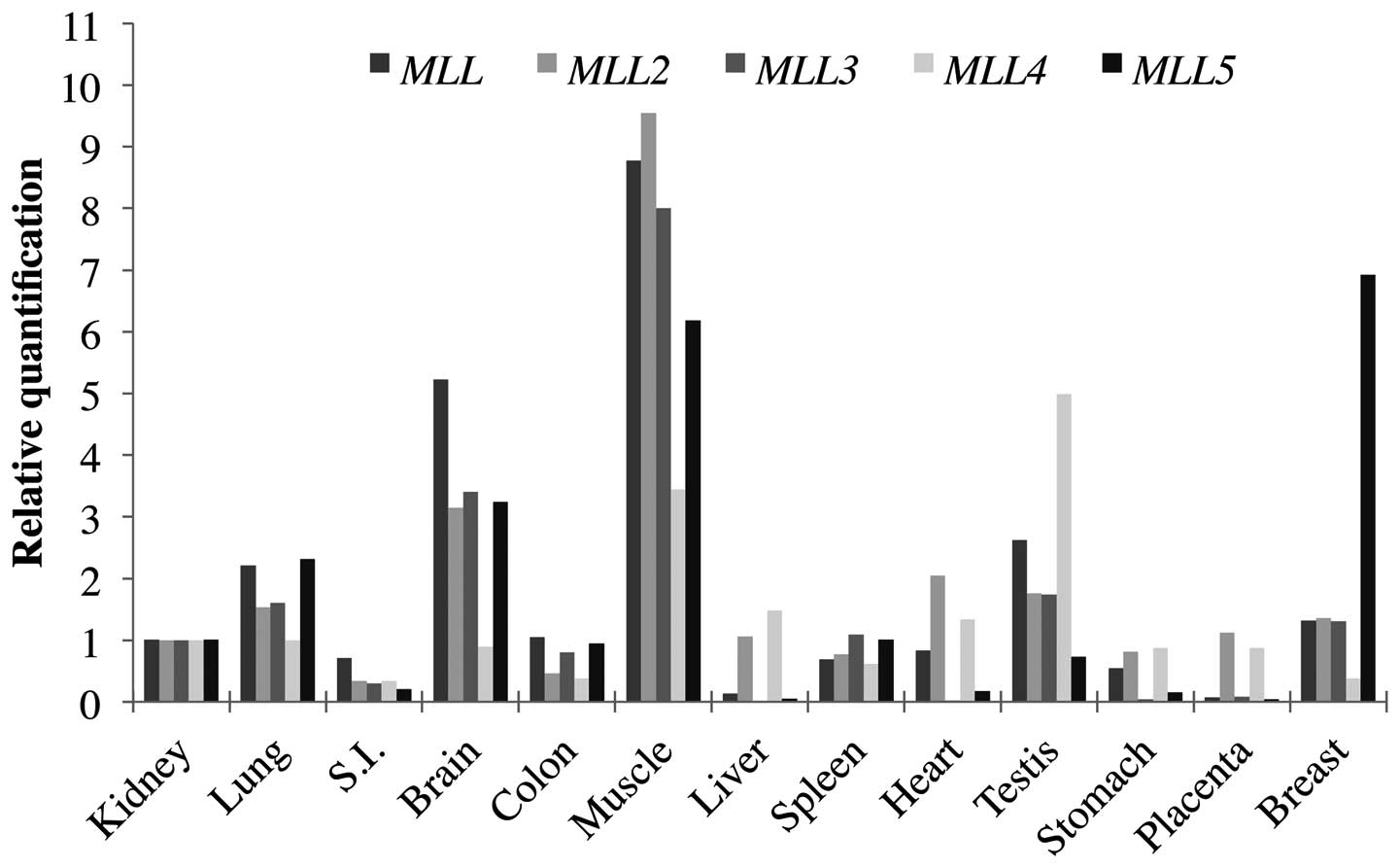
In order to determine the MLL genes
expression pattern in normal human tissues, quantitative real-time
polymerase chain reaction (qPCR) was performed using a commercial
panel of total RNA from normal tissues obtained from
OriGene®. Expression analysis using the comparative
quantification method ΔΔCT was performed. The kidney tissue, which
showed a median expression level, was chosen as a sample calibrator
(1X sample) and a normal breast clinical sample was included for
the relative quantification. Relative quantification for the normal
tissues can be seen in Fig. 2 and
in Table II. These results
evidence a heterogeneous tissue expression for the MLL
family genes, suggesting that MLL proteins functions are not
redundant, but acting in a tissue-specific manner.
 | Table II.Relative quantification of MLL
genes in normal human tissues. |
Table II.
Relative quantification of MLL
genes in normal human tissues.
| MLL | MLL2 | MLL3 | MLL4 | MLL5 |
|---|
| Kidney | 1 | 1 | 1 | 1 | 1 |
| Lung | 2 | 2 | 2 | 1 | 2 |
| Small
intestine | 1 | 3 | 3 | 3 | 5 |
| Brain | 5 | 3 | 3 | 1 | 3 |
| Colon | 1 | 2 | 1 | 3 | 1 |
| Muscle | 9 | 10 | 8 | 3 | 6 |
| Liver | 8 | 1 | 163 | 1 | 20 |
| Spleen | 1 | 1 | 1 | 2 | 1 |
| Heart | 1 | 2 | 50 | 1 | 6 |
| Testis | 3 | 2 | 2 | 5 | 1 |
| Stomach | 2 | 1 | 23 | 1 | 6 |
| Placenta | 14 | 1 | 12 | 1 | 28 |
| Breast | 1 | 1 | 1 | 3 | 7 |
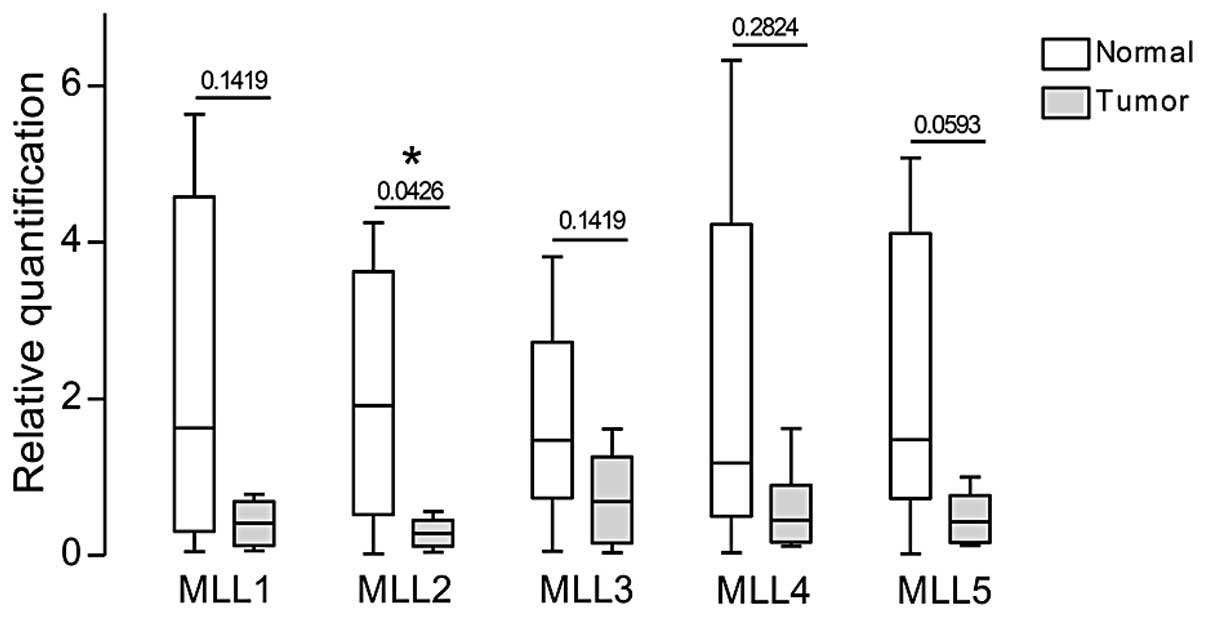
MLL2 is significantly downregulated in
clinical samples
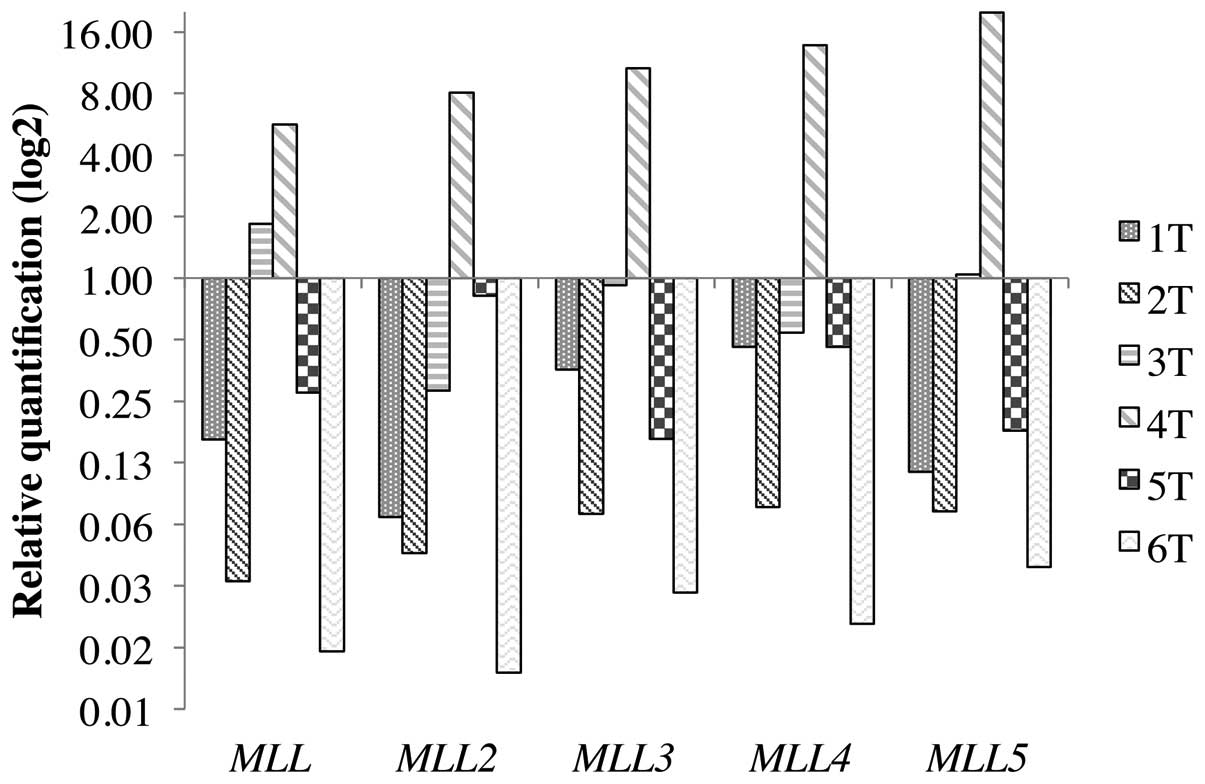
The paired expression analysis between normal and
tumor samples from the same patient can be seen in Fig. 3 and the relative quantification
values in Table III. The
quantification analysis for the pools of normal (n=6) and tumor
(n=8) clinical samples showed a decreased expression level of all
genes in the tumors when compared to the pool of normal samples
(Fig. 4). The expression of
MLL was reduced in 5 of the 8 (62.5%) tumor samples and
remained the same in the 3 other (37.5%) samples. The expression of
MLL2 was reduced in all the 8 tumor samples (100%). The
expression of MLL3 was reduced in 4 of the 8 (50%) tumor
samples, remained the same in the 3 other (37.5%) samples and was
increased in 1 sample (12.5%). The expression of MLL4 was
reduced in 5 of the 8 (62.5%) tumor samples, remained the same in
the other 2 (25%) samples and was increased in 1 sample (12.5%).
For the MLL5, the expression was reduced in 6 of the 8
samples (75%) and remained the same in 2 (25%). Although this
observation reveals a significantly suppressed expression of
MLL2 and MLL5 in the majority of tumors, the small
number of cases does not allow strong associations with the
clinical and pathological characteristics of the patients (Table IV).
 | Table III.Relative quantification of MLL
genes in tumor samples compared to their normal counterparts. |
Table III.
Relative quantification of MLL
genes in tumor samples compared to their normal counterparts.
| MLL | MLL2 | MLL3 | MLL4 | MLL5 |
|---|
| 1T | 6 | 15 | 3 | 2 | 9 |
| 2T | 30 | 22 | 14 | 13 | 14 |
| 3T | 2 | 4 | 1 | 2 | 1 |
| 4T | 6 | 8 | 11 | 14 | 20 |
| 5T | 4 | 1 | 6 | 2 | 6 |
| 6T | 67 | 85 | 34 | 49 | 26 |
To overcome our limited clinical breast cancer
cohort, we searched for MLL genes expression profile
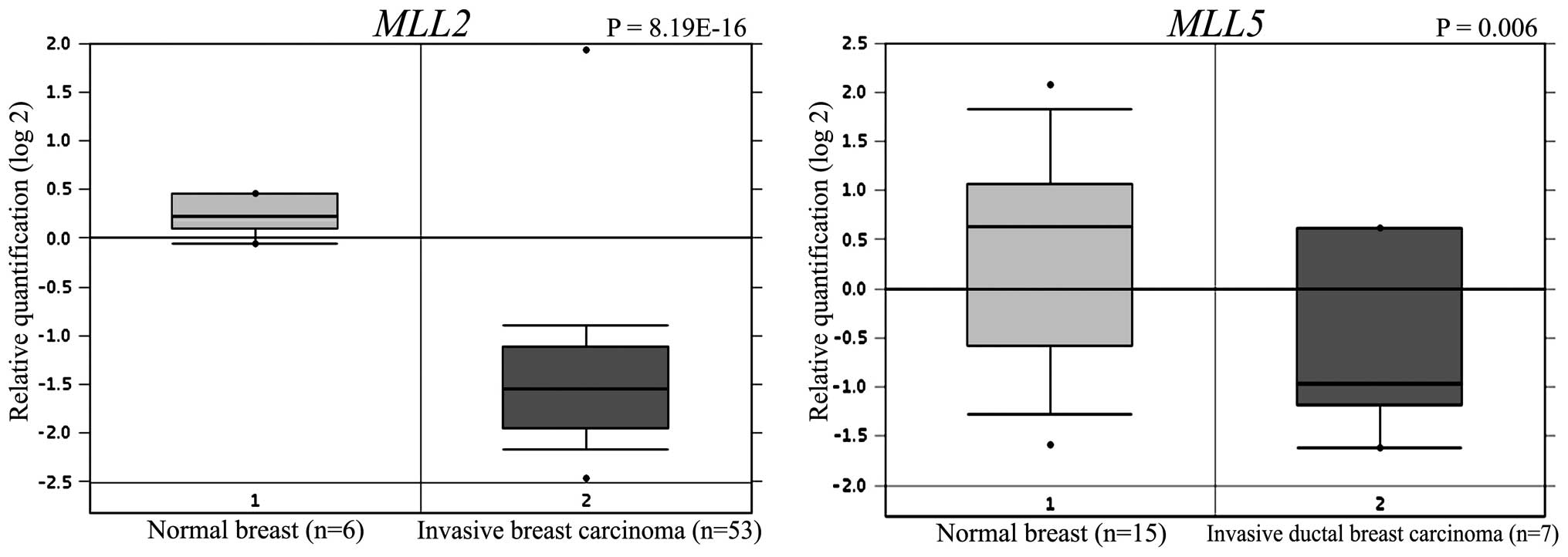
available in the Oncomine microarray database. We found that
MLL2 was significantly downregulated in a set of 53 invasive
breast carcinomas compared to 6 normal breast tissues (P=8.19E-16)
(21). MLL5 was found to be
2-fold downregulated in a set of 7 invasive breast carcinomas
compared to 15 normal breast tissues (P<0.005) (22). These data corroborate our findings
indicating that the levels of MLL2 and MLL5 are
widely downregulated in breast carcinomas (Fig. 5).
Discussion
Despite the advances that have been made in
uncovering potential roles for protein methyltransferases in human
cancers, it is still largely unknown how they contribute to
carcinogenesis. Deregulated methylation mechanisms in cancer cells
may be due to enzyme mutations or altered expression (23,24).
Recent studies have shown a relationship between SMYD3, a lysine
methyltransferase and breast carcinogenesis (25). SMYD3 modulates the chromatin
structure through specific methylation of lysine 4 of histone 3
(H3K4), an epigenetic alteration that leads to uncontrolled cell
proliferation (25). Several other
lysine methyltransferases have already been shown to be involved in
cancer. For example, the gene EZH2 was overexpressed in
different cancer tissues, including breast and prostate (26). The SETD2, a potential tumor
suppressor gene, was downregulated in breast cancer samples and in
high-grade tumors (27).
Deregulation of PRDM14 gene has been associated to several
human tumors, including an overexpression state in breast cancer
cells (28). Although little is
known about the exact roles of MLL family genes in breast
carcinogenesis, some recent studies have linked alterations in
individual MLL genes with cancer progression. The present
study provides the first comparative expression analysis of the
five MLL family genes in breast cancers. In this study, we
investigated the differential expression of MLL genes in
breast tumor samples and cell lines, as well as in normal tissues.
All MLL genes had a significantly decreased expression in
the seven breast cancer cell lines analyzed. It is noteworthy that
although these cell lines represent different stages of the
disease, presenting different receptors and characteristics of
aggressiveness (Table I), they all
showed a similar expression profile for all five MLL genes.
In the HCC-1954 cell line, which represents a less aggressive
breast cancer, with no lymph node metastases, a subtle higher
expression for the MLL genes when compared to the other cell
lines was found (Fig. 1). This
observation suggests that expression of MLL family genes may
decrease as disease progresses. Cell lines exhibit substantial
genomic, transcriptional and biological heterogeneity found in
primary tumors and represents a good system to study primary breast
tumor features (20). Indeed, the
results obtained in cell lines were in accordance to those obtained
in clinical samples.
The analysis of the MLL genes in normal
tissues showed highly heterogeneous expression among the different
tissues studied. This finding indicates a possible tissue-specific
function for each individual gene. The only consensus seems to be
the high expression of all MLL genes in skeletal muscle and
brain. For instance, MLL2 gene showed a markedly increased
expression in skeletal muscle (>10-fold) and heart (>2-fold),
which is in agreement with other studies showing the involvement of
this gene with myogenic developmental program (9,10).
It is of interest that in normal breast tissue, MLL5 has a
markedly higher expression among all MLL family genes. The
analysis of the primary breast clinical samples clearly pointed to
decreased expression of the genes MLL2 and MLL5 in
breast cancer. The other MLL genes did not show a
significant difference in expression between tumor and normal
samples.
The MLL gene had a slightly decreased
expression in the tumor samples, compared to the normal ones (P=
0.142). In all the cancer cell lines MLL was downregulated
in comparison to the pool of normal samples (P<0.05). Mammalian
MLL and MLL2 have a role in long-term maintenance of
Hox and other gene expression patterns during development
(8,13). Several rearrangements of the
MLL gene underlie the majority of infant acute leukemias, as
well as of therapy-related leukemias occurring in cancer patients
treated with inhibitors of topoisomerase II (29). Higher transcript levels of
MLL were associated with patients who remained disease-free
compared to those with bone metastasis. (30). Liu et al identified
MLL as a key constituent of the DNA damage response pathway,
showing that deregulation of the S-phase checkpoint caused by
MLL translocations may contribute to the pathogenesis of
human MLL leukemias (31).
The comparative analysis for MLL2, revealed a
significantly decreased expression of this gene in tumor samples
when compared to normal ones (P= 0.043). In the analysis using
breast cancer cell lines, MLL2 was the second gene with the
most prominent downregulation, after MLL5 (P<0.05). The
Oncomine database results corroborated our data, showing that
MLL2 was found markedly downregulated in a set of 53
invasive breast carcinomas compared to 6 normal breast tissues
(P=8.19E-16) (21). These results
are consistent with several other studies on this gene, in
different types of cancer. Curiously, Huntsman et al
(32) showed that MLL2 is
amplified in some solid tumor cell lines (pancreatic and
glioblastoma cell lines), with the exception of breast cancer cell
lines. Morin et al (33)
detected, in lymphoma patients, several somatic mutations
distributed across MLL2, with some mutations affecting both
MLL2 alleles or leading to loss of heterozygosity (LOH),
which is consistent with the complete, or almost complete, loss of
MLL2 in the tumor cells studied. They suggested that since
the majority of the somatic mutations found in MLL2 were
inactivating, MLL2 probably acts as a tumor suppressor of
significance in lymphomas. Pasqualucci et al (34) found that the MLL2 gene was
the most frequently mutated gene in diffuse large B-cell lymphoma
(DLBCL) with most cases of inactivating mutations, generating
truncated proteins lacking the entire or part of the C-terminal
cluster of conserved domains (including the SET domain). Since most
of the cases affected a single allele, they suggested a role for
MLL2 as a haploinsufficient tumor suppressor. Dalgliesh
et al (35) demonstrated
that the MLL2 is frequently mutated in clear cell renal cell
carcinoma (ccRCC), with silence, missense and truncating mutations.
It was identified as a likely ccRCC cancer gene in statistical
analyses. Finally, MLL2 was shown to be involved in several
cellular signaling pathways, such as p53 and cAMP, regulating
different sets of genes. The link of MLL2 to the p53 pathway
corroborates a possible role of MLL2 as tumor suppressor
gene. Also, among the categories of genes regulated by MLL2,
were those that respond to nuclear hormones, including genes from
the retinoic acid signaling (36).
MLL3 did not show a statistical significant
differential expression between the tumor and normal samples used
in this study (P=0.142), although in Fig. 4, this tendency can be noted. In the
seven breast cancer cell lines analyzed (Fig. 1), there is a clear decreased
expression of MLL3 (P<0.05). The MLL3 seems to be
frequently mutated in many types of human tumors and a recent study
detected that 40% of the primary breast tumor samples showed
reduced expression of this gene, suggesting its possible role in
the development of breast cancer as a tumor suppressor (37). Parsons et al (38) found, that MLL3, as well as
MLL2, harbors several inactivating mutations in
medulloblastoma that seem to be driver mutations. In the case of
MLL2, 67% of the mutations were predicted to truncate the
encoded proteins. This observation led to the conclusion that both
genes act as tumor suppressors inactivated by mutation in
medulloblastoma. MLL3 also showed inactivating mutations in
colorectal cancer samples (5,39)
and reduced expression in primary breast tumor samples, suggesting
a role in cancer development (37).
We found no significant difference between normal
and tumor sample expression of MLL4 (P=0.282). In the seven
breast cancer cell lines (Fig. 1),
MLL4 showed decreased expression, although it was the less
downregulated gene among the other members of the family
(P<0.05). MLL4 is mostly reported as a member of the
complex ASCOM (for ASC-2 COMplex), that acts as a p53
co-activator. Nonetheless, it is suggested that MLL3 and MLL4 are
found in distinct ASC-2-containing complexes rather than in a
common ASC-2 complex, with only partially redundant functions
(12).
For the MLL5 gene, our results revealed a
clear tendency of diminished expression in the clinical tumor
samples (P=0.059). It also showed a significant decreased
expression in the cancer cell lines, representing the most
downregulated gene among the MLL family for the seven
lineages (P<0.05). The Oncomine database results for MLL5
expression in breast cancer also corroborated the tendency of our
data, with a 2-fold downregulation of MLL5 in a set of 7
invasive breast carcinomas compared to 15 normal breast tissues
(P<0.005) (22). MLL5
gene is the latest addition to the mammalian Trithorax/MLL
gene family and recent studies show that the MLL5 may function in a
different manner, without histone methyltransferase activity
(16). MLL5 lacks DNA-binding
motifs of A-T hooks and methyltransferase homology motifs, both
common to other MLL protein members. Instead of binding directly to
DNA, MLL5 may modulate transcription indirectly via protein-protein
interactions through its PHD (plant homeodomain, a nuclear
protein-interaction domain) and SET domains (14). Overexpression of MLL5
prevented cell cycle progression pointing to a role of this gene in
the cell cycle regulatory network (6). MLL5 seems to form intranuclear
protein complexes, similar to other tumor suppressors, that may
play a role in chromatin remodeling and cellular growth suppression
(6). In cell line transfection
studies, overexpression of MLL5 transcripts inhibits cell
cycle progression, suggesting a role for MLL5 in tumor
suppression (6,16). Moreover, MLL5 was identified
as a candidate suppressor gene in 7q22, a chromosomal site
frequently deleted in patients with aggressive acute myeloid
leukemia and high MLL5 transcript levels seem to be
associated with a favorable outcome in this disease (40). Therefore, it is possible that in
cancer, the truncated or absent MLL5 protein is unable to form the
intranuclear protein complexes to interact with chromatin, allowing
the cell cycle to progress.
Although the complete genome screening is essential
to unveil the genetic profile of complex diseases, the comparative
analysis of a specific group of genes can be valuable to better
understand the complexity of cancer mechanisms. In the present
study, we analyzed the expression profiles of the MLL family
in different breast cancer cell lines and clinical samples. Our
results demonstrate a significant decreased expression of the
MLL2 and a clear tendency of MLL5 down-regulation in
tumor samples, which could indicate a possible role as tumor
suppressor genes. Since MLL2 has a role in maintenance of
Hox genes expression during development and MLL5
seems to have a role in cell cycle regulation, they may both be
involved in the regulation of other genes and pathways (8,13).
Furthermore, MLLs seem to have other roles in regulating gene
activation beyond their methyltransferase activities, including
responses under a hormonal environment (12,17–19).
Our results are consistent with several studies on these genes in
different cancer types, as discussed above, confirming that our
high-throughput approach was reliable enough for validation of the
genes expression profiles. Although these results point to an
involvement of MLL2 and MLL5 in breast cancer, the
number of cases does not enable robust statistical analyses neither
associations with the clinical characteristics. Therefore, further
studies using a larger clinical breast cancer cohort are required
to determine the exact role of these methyltransferases as
epigenetic regulators in cancer development.
The correlation between various methyltransferases
and breast cancer highlights the importance of this protein family
in the progression of this disease. Further study is required to
reinforce the importance of the MLL family as a target for
breast cancer therapy and will help elucidate the mechanisms
involved in the regulation of its activity. Our findings reveal the
importance of these genes in breast carcinogenesis and may
contribute to the identification of novel strategies to treat
breast tumors.
Acknowledgements
This study was supported by grants
from CAPES, FAPDF and CNPq. The authors thank Dr Anamaria Camargo
Aranha from the Ludwig Institute for Cancer Research, São Paulo,
for providing total RNA from several breast cancer cell lines. The
authors received financial support from the University of Brasilia
UnB-DPP (Decanato de Pesquisa e Pós-graduação).
References
|
1.
|
IARC: World Cancer Report. IARC; Lyon:
2008
|
|
2.
|
INCA: Estimativa 2012: Incidência de
câncer no Brasil. INCA Rio de Janeiro; 2011, (In Portuguese).
|
|
3.
|
Castro N, Osório C, Torres C, et al:
Evidence that molecular changes in cells occur before morphological
alterations during the progression of breast ductal carcinoma.
Breast Cancer Res. 10:R872008. View Article : Google Scholar
|
|
4.
|
Tamimi R, Baer H, Marotti J, et al:
Comparison of molecular phenotypes of ductal carcinoma in situ and
invasive breast cancer. Breast Cancer Res. 10:R672008. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Sjöblom T, Jones S, Wood L, et al: The
consensus coding sequences of human breast and colorectal cancers.
Science. 314:268–274. 2006.
|
|
6.
|
Deng L-W, Chiu I and Strominger J: MLL 5
protein forms intra-nuclear foci and overexpression inhibits cell
cycle progression. Proc Natl Acad Sci USA. 101:757–762. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Ayton P and Cleary M: Molecular mechanisms
of leukemogenesis mediated by MLL fusion proteins. Oncogene.
20:5695–5707. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Yu B, Hess J, Horning S, Brown G and
Korsmeyer S: Altered Hox expression and segmental identity in
Mll-mutant mice. Nature. 378:505–508. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
McKinnell I, Ishibashi J, Le Grand F, et
al: Pax7 activates myogenic genes by recruitment of a histone
methyltransferase complex. Nat Cell Biol. 10:77–84. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Aziz A, Liu Q-C and Dilworth F: Regulating
a master regulator: establishing tissue-specific gene expression in
skeletal muscle. Epigenetics. 5:691–695. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Ruault M, Brun M, Ventura M, Roizès G and
De Sario A: MLL3, a new human member of the TRX/MLL gene family,
maps to 7q36, a chromosome region frequently deleted in myeloid
leukaemia. Gene. 284:73–81. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Lee J, Kim D-H, Lee S, et al: A tumor
suppressive coactivator complex of p53 containing ASC-2 and histone
H3-lysine-4 methyltransferase MLL3 or its paralogue MLL4. Proc Natl
Acad Sci USA. 106:8513–8518. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Glaser S, Schaft J, Lubitz S, et al:
Multiple epigenetic maintenance factors implicated by the loss of
Mll2 in mouse development. Development. 133:1423–1432. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Emerling B, Bonifas J, Kratz C, et al:
MLL5, a homolog of Drosophila trithorax located within a
segment of chromosome band 7q22 implicated in myeloid leukemia.
Oncogene. 21:4849–4854. 2002.PubMed/NCBI
|
|
15.
|
Sebastian S, Sreenivas P, Sambasivan R, et
al: MLL5, a trithorax homolog, indirectly regulates H3K4
methylation, represses cyclin A2 expression and promotes myogenic
differentiation. Proc Natl Acad Sci USA. 106:4719–4724. 2009.
View Article : Google Scholar
|
|
16.
|
Madan V, Madan B, Brykczynska U, et al:
Impaired function of primitive hematopoietic cells in mice lacking
the Mixed-Lineage-Leukemia homolog MLL5. Blood. 113:1444–1454.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Ansari K, Kasiri S, Hussain I and Mandal
S: Mixed lineage leukemia histone methylases play critical roles in
estrogen-mediated regulation of HOXC13. FEBS J. 276:7400–7411.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Mo R, Rao S and Zhu Y-J: Identification of
the MLL2 complex as a coactivator for estrogen receptor alpha. J
Biol Chem. 281:15714–15720. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Ansari K, Hussain I, Kasiri S and Mandal
S: HOXC10 is overexpressed in breast cancer and transcriptionally
regulated by estrogen via involvement of histone methylases MLL3
and MLL4. J Mol Endocrinol. 48:61–75. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Neve R, Chin K, Fridlyand J, et al: A
collection of breast cancer cell lines for the study of
functionally distinct cancer subtypes. Cancer Cell. 10:515–527.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Finak G, Bertos N, Pepin F, et al: Stromal
gene expression predicts clinical outcome in breast cancer. Nat
Med. 14:518–527. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Karnoub AE, Dash AB, Vo AP, et al:
Mesenchymal stem cells within tumour stroma promote breast cancer
metastasis. Nature. 449:557–563. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Sims R, Nishioka K and Reinberg D: Histone
lysine methylation: a signature for chromatin function. Trends
Genet. 19:629–639. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Miremadi A, Oestergaard M, Pharoah P and
Caldas C: Cancer genetics of epigenetic genes. Hum Mol Genet.
16:R28–R49. 2007. View Article : Google Scholar
|
|
25.
|
Hamamoto R, Silva F, Tsuge M, et al:
Enhanced SMYD3 expression is essential for the growth of breast
cancer cells. Cancer Sci. 97:113–118. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Chang CJ and Hung MC: The role of EZH2 in
tumour progression. Br J Cancer. 106:243–247. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Al Sarakbi W, Sasi W, Jiang W, Roberts T,
Newbold R and Mokbel K: The mRNA expression of SETD2 in human
breast cancer: correlation with clinicopathological parameters. BMC
Cancer. 9:2902009.PubMed/NCBI
|
|
28.
|
Nishikawa N, Toyota M, Suzuki H, et al:
Gene amplification and overexpression of PRDM14 in breast cancers.
Cancer Res. 67:9649–9657. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Canaani E, Nakamura T, Rozovskaia T, et
al: ALL-1/MLL1, a homologue of Drosophila trithorax,
modifies chromatin and is directly involved in infant acute
leukaemia. Br J Cancer. 90:756–760. 2004.
|
|
30.
|
Patani N, Jiang W, Newbold R and Mokbel K:
Histone-modifier gene expression profiles are associated with
pathological and clinical outcomes in human breast cancer.
Anticancer Res. 31:4115–4125. 2011.PubMed/NCBI
|
|
31.
|
Liu H, Takeda S, Kumar R, et al:
Phosphorylation of MLL by ATR is required for execution of
mammalian S-phase checkpoint. Nature. 467:343–346. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Huntsman D, Chin S, Muleris M, et al:
MLL2, the second human homolog of the Drosophila trithorax gene,
maps to 19q13.1 and is amplified in solid tumor cell lines.
Oncogene. 18:7975–7984. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Morin RD, Mendez-Lago M, Mungall AJ, et
al: Frequent mutation of histone-modifying genes in non-Hodgkin
lymphoma. Nature. 476:298–303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Pasqualucci L, Trifonov V, Fabbri G, et
al: Analysis of the coding genome of diffuse large B-cell lymphoma.
Nat Genet. 43:830–837. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Dalgliesh G, Furge K, Greenman C, et al:
Systematic sequencing of renal carcinoma reveals inactivation of
histone modifying genes. Nature. 463:360–363. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Guo C, Chang CC, Wortham M, et al: Global
identification of MLL2-targeted loci reveals MLL2’s role in diverse
signaling pathways. Proc Natl Acad Sci USA. 109:17603–17608.
2012.PubMed/NCBI
|
|
37.
|
Wang X-X, Fu L, Li X, et al: Somatic
mutations of the mixed-lineage leukemia 3 (MLL3) gene in primary
breast cancers. Pathol Oncol Res. 17:429–433. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Parsons D, Li M, Zhang X, et al: The
genetic landscape of the childhood cancer medulloblastoma. Science.
331:435–439. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Watanabe Y, Castoro R, Kim H, et al:
Frequent alteration of MLL3 frameshift mutations in microsatellite
deficient colorectal cancer. PLoS One. 6:e233202011. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Damm F, Oberacker T, Thol F, et al:
Prognostic importance of histone methyltransferase MLL5 expression
in acute myeloid leukemia. J Clin Oncol. 29:682–689. 2011.
View Article : Google Scholar : PubMed/NCBI
|