Introduction
The MET receptor tyrosine kinase is expressed in a
variety of normal and malignant cells (1–3).
Many studies have reported that MET is overexpressed in a variety
of cancers and that MET overexpression could be the result of the
MET proto-oncogene amplification, which is notably observed
in transcriptional activation (4),
a small subset of cancers (5–8), and
has recently seen in lung cancers (9,10).
The signaling pathway of MET receptor tyrosine kinase and its
ligand, hepatocyte growth factor (HGF) (11), has been shown to play an important
role in increased cell proliferation, reduced apoptosis, altered
cytoskeletal function, increased metastasis and other biologic
responses (12,13). Like other receptor tyrosine kinases
(RTKs), upon HGF binding, MET dimerizes and autophosphorylates,
creating active docking sites for proteins that mediate downstream
signaling leading to the activation of pathways such as MAPK,
PI3K-AKT and the STAT signaling pathway (14–16).
Thus, MET activation produces a variety of biological responses
that lead to increased cell growth, cell motility, cell invasion
and angiogenesis, as well as anti-apoptotic effects. After MET
activation, HGF-MET complexes are internalized via clathrin-coated
vesicles and delivered to the early endosomal tubulovesicular
compartment commonly referred to as the sorting endosome. In the
sorting endosome, ubiquitinated receptors can be recognized by the
endosomal sorting complex required for transport (ESCRT) machinery
that generates multivesicular bodies (MVBs) by packaging cargo into
small vesicles that bud off from the limiting membrane into the
lumen of the endosomes (17). This
sorting pathway terminates HGF/MET signaling by delivering
receptors to the lysosomes for degradation, a process known as
downregulation (18).
Alternatively, MET is recycled to the plasma membrane by endosomal
recycling pathways.
The EGFR tyrosine kinase inhibitors (EGFR-TKIs),
gefitinib and erlotinib, have been shown to block the signal
transduction pathways implicated in the proliferation and survival
of cancer cells (19–22). These EGFR-TKIs are effective in
treating non-small cell lung cancers (NSCLCs) that have activating
mutations in the EGFR gene (23,24).
Most EGFR mutant NSCLCs initially respond to EGFR inhibitors;
however, most of these tumors ultimately become resistant to the
drug treatment. Engelman et al have previously demonstrated
that amplification of MET induces gefitinib resistance by
driving ERBB3 (HER3)-dependent activation of PI3K, a pathway
thought to be specific to EGFR/ERBB family receptors in lung cancer
(9). However, the mechanisms that
contribute to gefitinib resistance in the remaining tumors are
unknown.
We reported previously that an aberration in certain
steps of EGF-EGFR/phosphorylated EGFR(pEGFR) endocytic trafficking
from the early endosomes to the late endosomes/lysosomes occurs in
the gefitinib-resistant EGFR wild-type NSCLC cells, whereas
endocytosis of EGFR/pEGFR is normal in gefitinib-sensitive
EGFR mutant NSCLC cells (25,26).
We also made the novel observation that large amounts of sorting
nexin 1 (SNX1) are localized in the aggregated vesicular structures
of early endosomes where the internalized pEGFR is also accumulated
(27). Furthermore, we have
recently reported that silencing of endogenous SNX1 by siRNA
stimulates endocytosis and the ligand-induced downregulation of
EGFR/pEGFR, while increasing of pEGFR protein expression in
gefitinib-resistant cells (28).
Thereby, we postulate that SNX1 plays a negative role in the
regulation of EGF-dependent downregulation of EGFR and its phospho
rylation via the early/late endocytic pathway in human lung cancer
cells.
SNX1 is a family of sorting nexin proteins, of which
approximately 25 human sorting nexins have been identified so far
(29). They have previously been
demonstrated to interact with EGFR (30), and are localized to early endosomes
through their phospholipid-binding motifs termed as the phox
homology (PX) domain (29).
Previous studies have also revealed that SNX1 overexpression causes
enhanced EGFR degradation, and that deletion mutants of SNX1
blocked EGFR degradation but fail to inhibit receptor endocytosis
(30,31). Therefore, it was suggested that
SNX1 interacts with EGFR and enhances the degradation of the
receptor upon EGF stimulation, thereby implying that SNX1 plays a
role in endosome-lysosome trafficking. However, recent evidence has
failed to support the intracellular co-localization of SNX1 with
EGFR and its direct role in EGFR degradation, raising the
possibility that alternative mechanisms are involved in the
function of SNX1 (32,33). The detailed function of SNX1
regulating endocytic trafficking of transmembrane tyrosine kinase
receptors remains unclear.
In the present study, we analyzed the intracellular
regulatory function of SNX1 with regard to HGF-induced endocytosis
and the downregulation of MET/phosphorylated MET (pMET) by using
confocal immunofluorescence microscopy, western blot analysis, and
RNAi-mediated knockdown approaches in gefitinib-resistant NSCLC
cell lines. We demonstrated that silencing of endogenous SNX1 by
siRNA stimulates efficient endocytosis of HGF-induced MET and pMET
in gefitinib-resistant NSCLC cells. We further found that depletion
of SNX1 stimulates the ligand-induced downregulation of MET/pMET,
while increasing of pMET protein expression in gefitinib-resistant
cells. Therefore, we postulate that SNX1 plays a suppressive role
in the regulation of HGF-dependent downregulation of MET and its
phosphorylation via the early/late endocytic pathway in human lung
cancer cells.
Materials and methods
Materials
Texas red-labeled EGF, Texas red-labeled human
transferrin and SlowFade anti-fade reagent were purchased from
Molecular Probes (Eugene, OR, USA). Recombinant human HGF and EGF
were purchased from PeproTech (London, UK). Bafilomycin A1,
cycloheximide (CHX), and DAPI were obtained from Sigma (St. Louis,
MO, USA). Other chemicals were of reagent grade and were obtained
from commercial sources.
Cell culture
Cell lines PC9 and A549 (National Kyushu Cancer
Center, Fukuoka, Japan) were cultured in RPMI-1640 supplemented
with 10% fetal bovine serum (FBS). Cells were maintained under
standard cell culture conditions at 37°C and 5% CO2 in a
humid environment.
Small interfering RNA
siRNA targeting SNX1 was purchased from Dharmacon
(Boulder, CO, USA). The target sequence of the siRNA was as
follows: 5′-AAGAACAAGACCAAGA GCCAC-3′. Scramble sequence was used
as a control. The two NSCLC cell lines were transfected with
Lipofectamine 2000 (Life Technologies, Gaithersburg, MD, USA) in
the presence of 40 nM siRNA targeting SNX1 according to the
manufacturer’s protocol. Knockdown efficiency was determined by
qRT-PCR and confocal immunofluorescence microscopy analysis.
qRT-PCR analysis
The NSCLC PC9 or A549 cells transfected with
siRNA-control or siRNA-SNX1 were stimulated with EGF (100 ng/ml) at
37°C for 5, 15 or 30 min, and total RNA was extracted from each
cell line using an RNeasy RNA isolation kit (Qiagen, Hilden,
Germany) according to the manufacturer’s instructions.
Transcription into cDNA was done in a 20-μl volume using
ThermoScript RT-PCR System with random hexamer (Invitrogen,
Carlsbad, CA, USA) according to the manufacturer’s instructions.
All PCR reactions were carried out in a final volume of 25
μl and were performed in the ABI PRISM 7000 Sequence
Detection System (Applied Biosystems, Foster City, CA, USA)
according to the manufacturer’s protocol. Sequence-specific primers
were quoted from an official website ‘PrimerBank’ (http://pga.mgh.harvard.edu/primerbank/)
for the indicated genes (Table I).
The reaction mix consisted of SYBR Premix Ex Taq (2X) (Takara Bio,
Shiga, Japan) 12.5 μl, ROX Reference Dye (50X) (Takara Bio)
0.5 μl, 0.2 μM of each specific forward and reverse
primer, and 9 μl of diluted cDNA (equivalent to 0.03–2.85 ng
of total RNA). Amplifications were done under standard conditions
(10 sec at 95°C followed by 40 cycles of 5 sec at 95°C and 31 sec
at 60°C). The number of PCR cycles needed to reach the fluorescence
threshold was determined in triplicate for each cDNA, averaged and
then normalized to a reference gene (β-actin). A standard curve was
generated with serial 3-fold dilutions of a representative cDNA.
For all assays tested, the PCR reaction was linear over the range
studied (20–40 cycles of amplification). All RT-PCR reactions gave
a single band when analyzed by gel electrophoresis.
 | Table I.Primers for qRT-PCR for human MET and
β-actin. |
Table I.
Primers for qRT-PCR for human MET and
β-actin.
| PCR primer | Nucleotide
sequence |
|---|
| MET | |
| Forward |
5′-TGGTGCAGAGGAGCAATGG-3′ |
| Reverse |
5′-CATTCTGGATGGGTGTTTCCG-3′ |
| β-actin | |
| Forward |
5′-CATGTACGTTGCTATCCAGGC-3′ |
| Reverse |
5′-CTCCTTAATGTCACGCACGAT-3′ |
Antibodies
Alexa 488-labeled goat anti-mouse and goat
anti-rabbit secondary antibodies were obtained from Molecular
Probes. Normal rabbit IgG and normal mouse monoclonal IgG1 were
purchased from Imgenex (San Diego, CA, USA) and Angio-proteomie
(Boston, MA, USA), respectively. Normal goat serum was purchased
from Sigma. Mouse monoclonal antibody to SNX1 was purchased from BD
Biosciences (San Jose, CA, USA). Anti-MET, pMET, EGFR, pEGFR and
LAMP1 antibodies were obtained from Cell Signaling Technology
(Beverly, MA, USA), and anti-β-actin antibody was obtained from
Sigma. Mouse monoclonal anti-HGF α-chain antibody was purchased
from Institute of Immunology (Tokyo, Japan). Anti-cathepsin D was
affinity-purified by protein A Sepharose CL-4B (Sigma), followed by
immunoaffinity chromatography using antigen-conjugated Sepharose 4B
as described previously (34,35).
Immunofluorescence microscopy
General procedures
Immunofluorescence microscopy was described
previously (25–28). The A549 cells were grown for 2 days
on glass coverslips in 6-well plates in RPMI-1640 with 10% fetal
bovine serum. Cells were fixed with 3.7% formaldehyde in
phosphate-buffered saline (PBS), pH 7.4, permeabilized in PBS
containing 0.1% saponin. After washing with PBS, cells were blocked
with PBS-10% normal goat serum. All subsequent antibody and wash
solutions contained 0.1% saponin. The A549 cells were incubated
with specific primary antibodies (mouse monoclonal anti-HGF α-chain
antibody, mouse monoclonal anti-LAMPI antibody, mouse monoclonal
anti-EGFR antibody or mouse monoclonal anti-SNX1 antibody), for 1
h, followed by washes with PBS containing 0.1% saponin and
incubation for 1 h with the secondary antibodies at 20
μg/ml. Each cell line was stained with DAPI to reveal
nuclei. Controls for antibody specificity were non-immune normal
mouse IgG1 or non-immune normal rabbit IgG. To label early
endosomes, cells were incubated with RPMI-1640 without FBS for 3 h
at 37°C followed by 20 min incubation in culture medium containing
Texas red-conjugated transferrin, and then cells were fixed and
double-stained for MET with anti-MET monoclonal antibody or EGFR
with anti-EGFR antibody. Late endosomes/lysosomes were stained with
anti-LAMP1 antibody, since the LAMP1 protein is distributed within
endocytic organelles and is at its highest concentration in the
late endosomes/lysosomes, as observed for other lysosomal
glycoproteins, namely, lysosomal-associated membrane protein-1
(LAMP-1) and LAMP-2 (36,37). The distribution of the labeled
proteins was then analyzed by confocal immunofluorescence
microscopy of the fixed cells. Slides were mounted with SlowFade
anti-fade reagent and observed on a Zeiss LSM 510 META confocal
laser scanning microscope (Carl Zeiss, Oberkochen, Germany),
equipped with krypton/argon laser sources. Co-localization of MET
or EGFR and Texas red-labeled transferrin, HGF α-chain and
cathepsin D, or pMET and LAMP1 was quantified using Image J
software and the MacSCOPE X software (Mitani Corporation, Osaka,
Japan).
Treatment of the cells with HGF
In order to clarify MET internalization, we followed
the uptake of HGF with time in each cell line. To minimize the
contribution of recycling and/or lysosomal degradation of the
internalized HGF, we quantified the HGF uptake in each cell type
for time periods of up to 30 min. At 48 h transfection, A549 cells
treated with siRNA-control or siRNA-SNX1 were starved for 12 h with
RPMI-1640 without FBS at 37°C and the cells were incubated in the
presence of Texas red-transferrin in prewarmed medium, and the
serum-starved cells were then incubated with HGF (100 ng/ml) at
37°C for 15 or 30 min, and the distribution of internalized HGF
stained with anti-HGF α-chain antibody and lysosomes stained with
anti-cathepsin D antibody was then assessed by confocal
immunofluorescence microscopy. In some cases, cells were starved
for 3 h with RPMI-1640 without FBS at 37°C and then the
phosphorylation of MET was induced with HGF (100 ng/ml) for 15 min
on ice in binding medium (1 mg/ml BSA in RPMI-1640 medium). The
cells were then rinsed with ice-cold PBS, incubated in the presence
of Texas red-transferrin in prewarmed medium, and chased at 37°C
for 15 or 30 min. The fixed cells were double-stained for pMET with
anti-pMET antibody and LAMP1 with anti-LIMP1 antibody.
Western blot analysis
Protein samples were separated by sodium dodecyl
sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and then
transferred to polyvinylidene difluoride membranes (Millipore,
Billerica, MA, USA). Following blocking, the membrane was blotted
with the appropriate antibody, and subsequently, horseradish
peroxidase-conjugated anti-mouse or anti-rabbit IgG (GE Healthcare
Bioscience, Tokyo, Japan) was applied. The final signal was
revealed by ECL chemiluminescence (Pierce, Rockford, IL, USA).
Digital images were analyzed with NIH Image software to measure the
density of each band without a saturated signal.
HGF-stimulates phosphorylated MET
degradation
The A549 cells were starved for 12 h with RPMI-1640
without FBS at 37°C. The serum-starved cells were then preincubated
for 30 min in the presence of CHX (20 μg/ml) before
incubation with HGF (100 ng/ml) at 37°C for the indicated times.
The cells were then washed with ice-cold-PBS and lysed, followed by
SDS-PAGE and western blot analysis. Bafilomycin A1 (0.17 μM)
was added when the cells were incubated with RPMI-1640.
Statistical analysis
Data are expressed as mean ± SD unless otherwise
noted. Significance (p<0.05) was determined by using Student’s
t-test, since the data met all the assumptions for parametric
statistical analysis.
Results
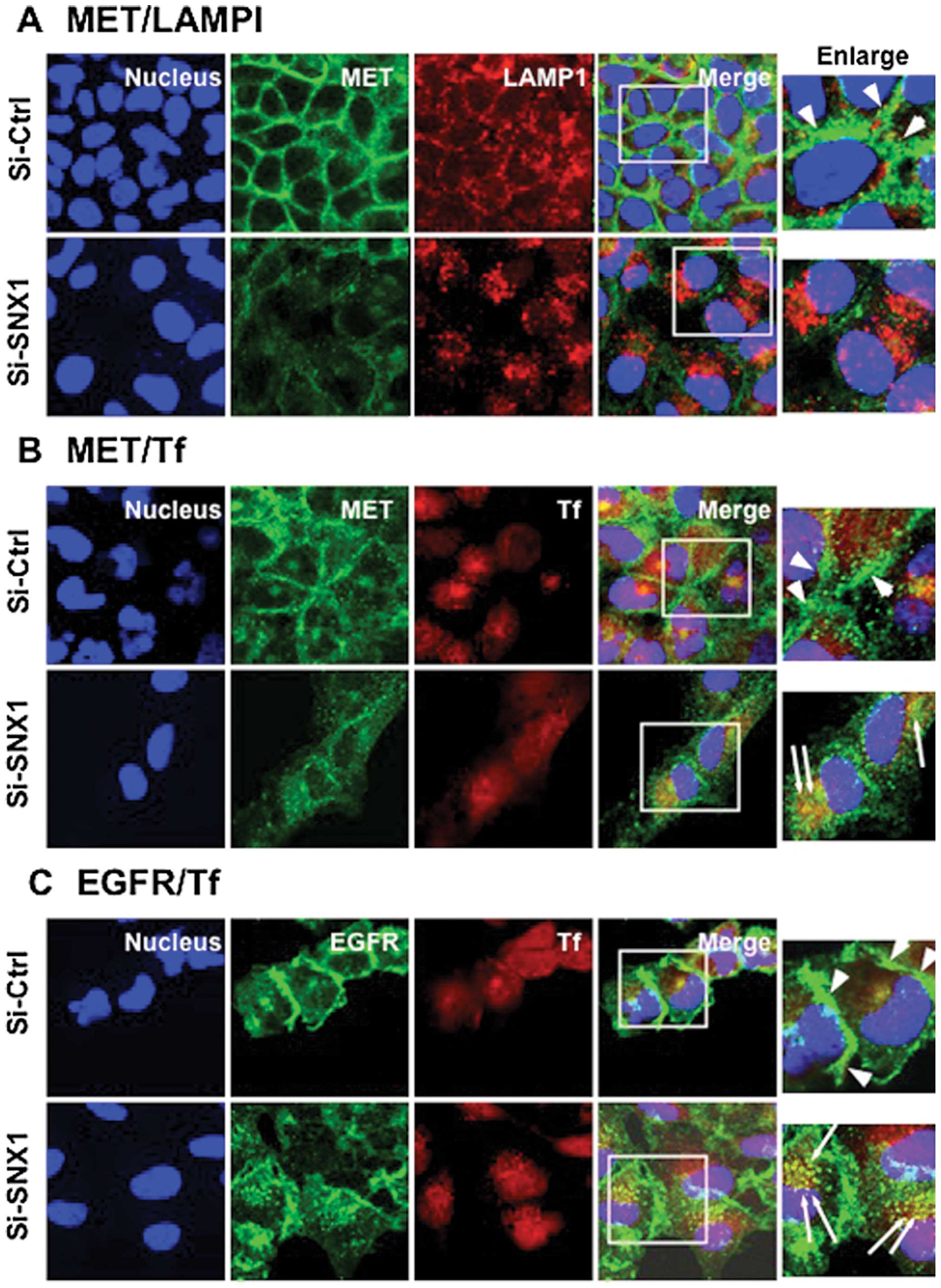
Intracellular distribution of MET in a
gefitinib-resistant NSCLC cell line
To examine the intracellular distribution of MET in
a gefitinib-resistant cell line, A549 cells were fixed and
double-labeled with antibodies specific to MET and lysosomal
integral membrane protein (LAMP1) or SNX1. We determined the
intracellular distribution of late endosomes/lysosomes by using
antibody specific to LAMP1 that is distributed within endocytic
organelles and is at the highest concentration in the late
endosomes/lysosomes, as observed for other lysosomal glycoproteins
(36,37). In addition, to examine the
intracellular distribution of MET with endocytosed transferrin, a
marker of early endosomes, Texas red-labeled transferrin was
applied to the cells for 20 min. After transferrin binds to its
receptor on the cell surface, it is internalized via
clathrin-coated vesicles and is subsequently delivered to the early
endosomes. Confocal immunofluorescence microscopy studies showed
that MET was mostly seen to be associated with plasma membranes,
and that MET-positive staining was not co-localized with SNX1- or
transferrin-positive early endosomes or with LAMP1-positive late
endosomes/lysosomes (Fig. 1).
Furthermore, to analyze the intracellular distribution of EGFR, the
cells were fixed and double-labeled with antibody to EGFR and the
endocytosed Texas red-labeled transferrin, and then the
intracellular distribution of EGFR was examined. The results showed
that most EGFR staining was mainly distributed in the plasma
membranes and that EGFR-positive staining was not co-localized with
transferrin-positive early endosomes in A549 cells.
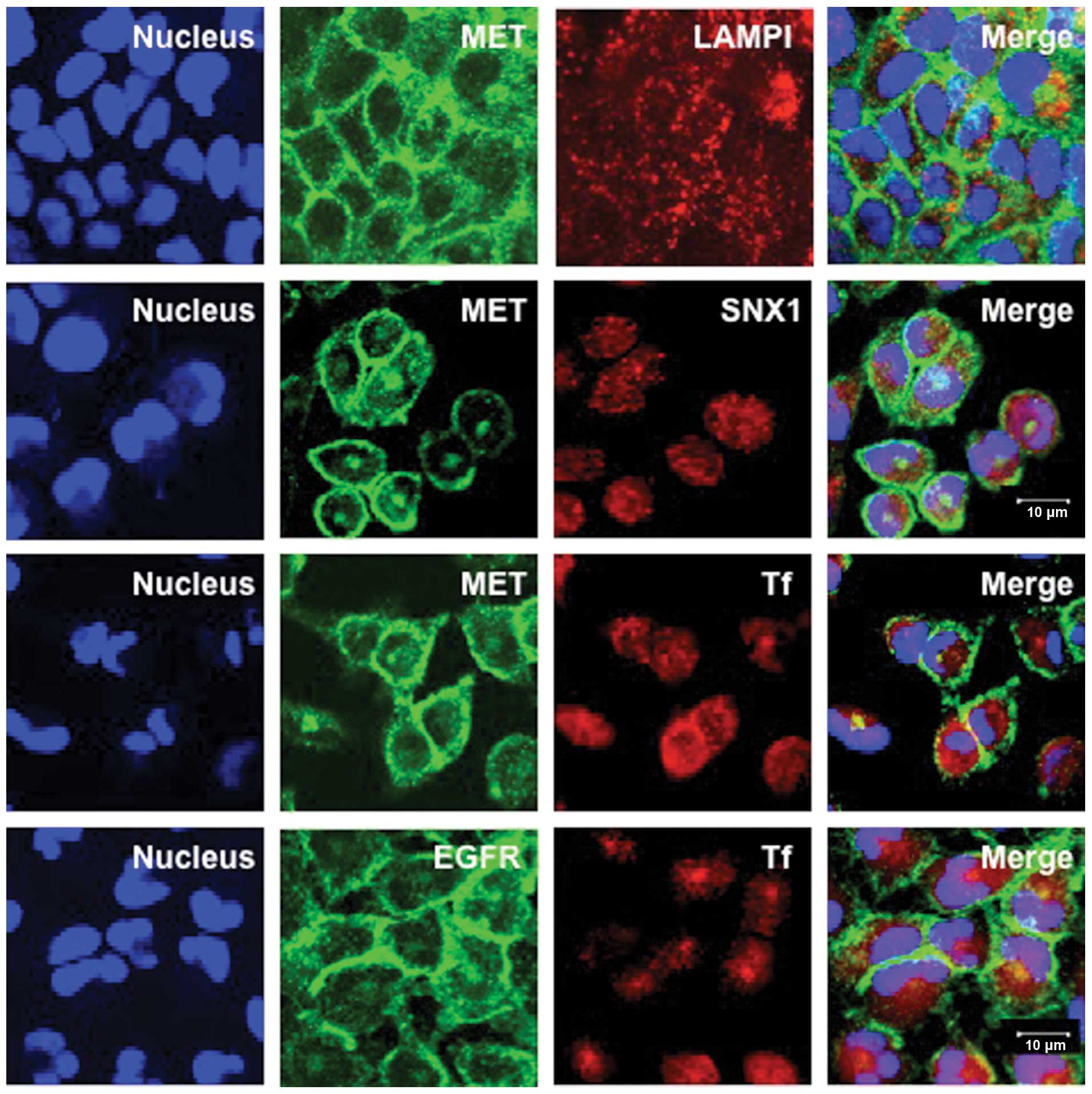 | Figure 1.Intracellular distribution of MET and
EGFR in the gefitinib-resistant A549 cells. The A549 cells were
fixed and double-stained for MET (green) and LAMP1 (red), MET
(green) and SNX1 (red), or MET (green) and the endocytosed Texas
red-labeled transferrin (red) as described in Materials and
methods. Cells were also double-stained for EGFR (green) and the
endocytosed Texas red-labeled transferrin (red). Superimposed
images of MET or EGFR and each organelle marker protein are shown.
Each cell line was stained with DAPI (blue) to reveal the nuclei.
In A549 cells, MET is exclusively localized in the plasma membrane
at a cell-cell contact sites, however, no colocalization with
organelle marker proteins such as LAMP1, SNX1 or transferrin is
observed, suggesting that MET is mainly associated with plasma
membrane, but not with the early endosomes or late endosomes in
A549 cells. EGFR is also localized in the plasma membrane, and is
not associated with the early endosomes in A549 cells. Bar, 10
μm. |
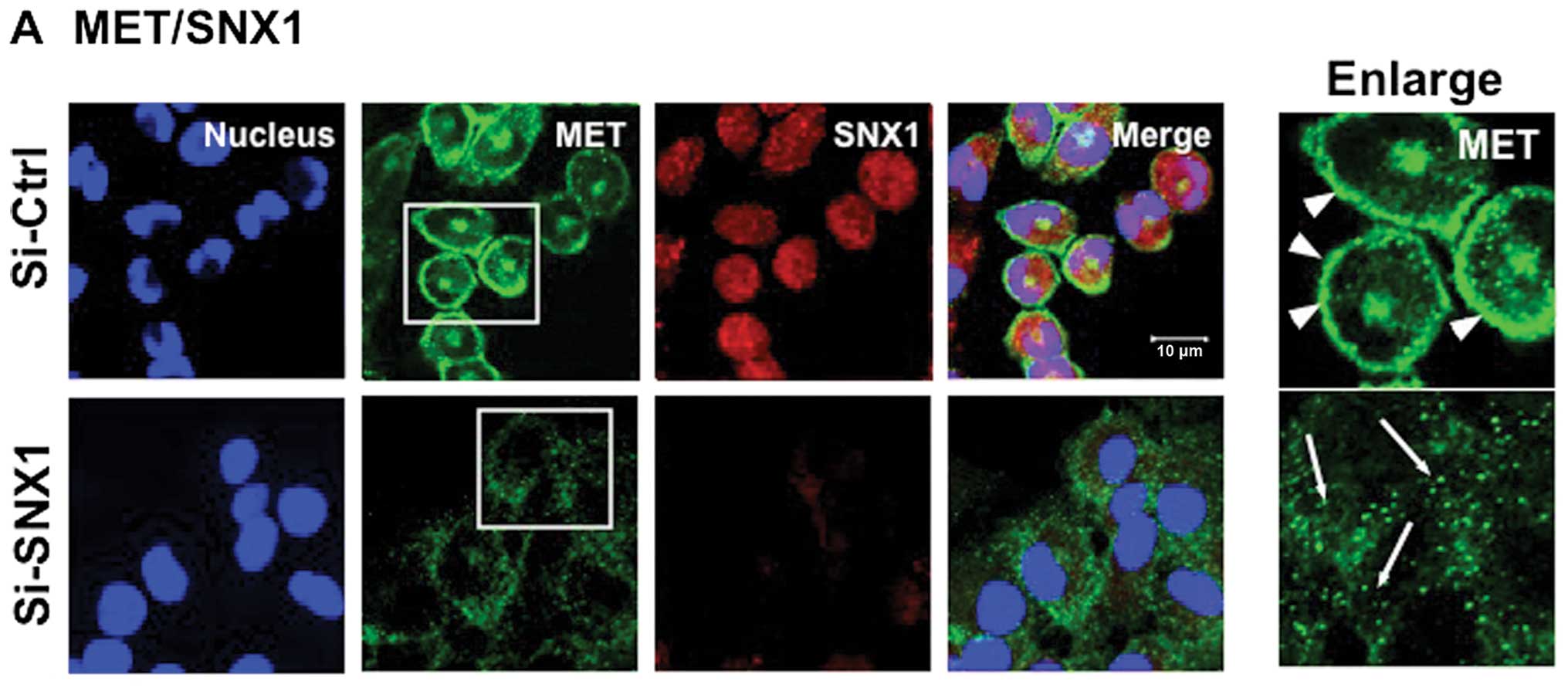
Knockdown of SNX1 by specific siRNA
effectively downregulates the expression of endogenous SNX1 protein
levels and causes a dramatic alteration in the intracellular
distribution of plasma membrane-associated MET in the
gefitinib-resistant NSCLC cell line
To examine the biological function of SNX1 in
regulating HGF-stimulated MET signaling and its endocytosis, we
used siRNA to knockdown the endogenous of SNX1 in the NSCLC cell
line A549. First, we examined the depletion of endogenous SNX1
protein levels by confocal immunofluorescence microscopy. The
results showed that the endogenous SNX1 was successfully depleted
using siRNA in A549 cells (Fig.
2A). Second, western blot analysis (Fig. 2B) revealed that endogenous MET
expression was almost completely silenced by specific siRNA in the
gefitinib-resistant cell line A549. Third, to examine the
endogenous MET mRNA transcript levels, the gefitinib-sensitive
NSCLC cell line PC9 and gefitinib-resistant NSCLC cell line A549
were transfected with siRNA-control or siRNA-SNX1 and then used in
qRT-PCR analysis. The results shown in Fig. 2C, indicate that siRNA-SNX1
suppressed SNX1 mRNA in both NSCLC cell lines. Moreover, it should
be noted that SNX1 knockdown did not change the endogenous
expression of MET transcript in the NSCLC cell lines (Fig. 2C). It was interesting to find that
the endogenous expression of MET mRNA transcripts was apparently
higher in the gefitinib-resistant cell line A549 compared to that
of the gefitinib-sensitive cell line PC9; the expression level of
MET transcripts in the gefitinib-resistant cell line A549 was
approximately 6.2-fold greater than that in the gefitinib-sensitive
cell line PC9 (Fig. 2C).
To investigate the alteration in the intracellular
distribution of MET in the gefitinib-resistant cell line A549, the
cells transfected with the siRNA-control or the siRNA-SNX1 were
double-labeled with either antibodies specific to MET or antibodies
specific to SNX1. The confocal immunofluorescence microscopy
studies showed that in A549 cells transfected with siRNA-contol,
MET was mostly seen to be associated with plasma membranes, and
MET-positive staining was not co-localized with SNX1-positive early
endosomes (Fig. 2A). This
immunostaining pattern of MET in A549 cells was similar to that in
PC9 cells (data not shown). In contrast, in A549 cells transfected
with siRNA-SNX1, large amounts of plasma-membrane-associated MET
staining were not seen, and there was increased small punctate
stainings of MET distributed throughout the cytoplasm. These
results suggest that depletion of SNX1 dramatically alters the
distribution of plasma membrane-associated MET in A549 cells.
We further analyzed a change in the intracellular
distribution of MET in the siRNA-SNX1-transfected cells. Cells in
the siRNA-control or the siRNA-SNX1-transfected groups were fixed
and double-labeled with antibodies to MET and LAMP1 or stained with
MET antibody and the endocytosed Texas red-labeled transferrin, and
the change in the intracellular distribution of MET was examined by
confocal immunofluorescence microscopy. The results showed that
considerable amounts of plasma membrane-associated MET, as observed
in the siRNA-control transfected cells, were absent from the cells
and large amounts of MET-positive small punctate vesicles were seen
to be dispersed throughout the cytoplasm, in which some
MET-positive staining obviously overlapped with the endocytosed
transferrin-positive vesicles (Fig.
3B). Co-localization of MET- and LAMP1-positive staining was
not discernible since most MET-positive staining was absent in the
cells (Fig. 3A). We also examined
the effect of siRNA-SNX1 transfection into A549 cells on the
cellular distribution of EGFR. Cells in the siRNA-control or the
siRNA-SNX1-transfected groups were fixed and double-labeled with
EGFR antibody and the endocytosed Texas red-labeled transferrin.
The results indicated that most of the plasma-membrane associated
EGFR seen in the siRNA-control-transfected cells was not present,
and that increased amounts of EGFR-positive punctate small vesicles
were co-localized with the endocytosed Texas red-labeled
transferrin that were seen to be spreading throughout the cytoplasm
(Fig. 3C). These observations were
consistent with those seen in the case of MET, indicating that the
transfection of siRNA-SNX1 in A549 cells can cause a dramatic
change in the plasma membrane-distribution of MET, leading to
increasing amounts of MET- or EGFR-positive small vesicles in the
cytoplasm, and the disappearance of these labels from the plasma
membranes. Quantitative analysis of the co-localization rate was
calculated as the percentage of the integrated density of the
endocytosed Texas red-labeled transferrin-early endosome
marker-co-localizing MET or EGFR compared with that of total MET or
EGFR (% total MET or EGFR) in A549 cells transfected with
siRNA-control or siRNA-SNX1 (Fig.
3D). Our data verified that transferrin-colocalizing MET (20.2
vs. 2.4% total MET) or EGFR (24.1 vs. 3.3% total EGFR) is markedly
higher in the siRNA-SNX-transfected cells than in the
siRNA-control-transfected cells (Fig.
3D). These results indicate that membrane trafficking of MET or
EGFR between plasma membranes and early endosomes may be
considerably suppressed in A549 cells.
Depletion of SNX1 stimulates efficient
endocytosis of HGF-induced MET and phosphorylation of MET via the
early/late endocytic pathway in a gefitinib-resistant NSCLC cell
line
We recently reported the novel evidence that
gefitinib-resistant cells show internalized EGFR accumulation in
aggregated early endosomes, and that this is associated with SNX1
(26–28). In contrast, gefitinib-sensitive
cells show efficient endocytosis of EGFR. We therefore imply that
impairment of protein function, such as SNX1 in the regulation of
EGFR trafficking in the early endocytic pathway, might cause these
perturbations in EGFR endocytosis, leading to gefitinib resistance
in NSCLC cell lines.
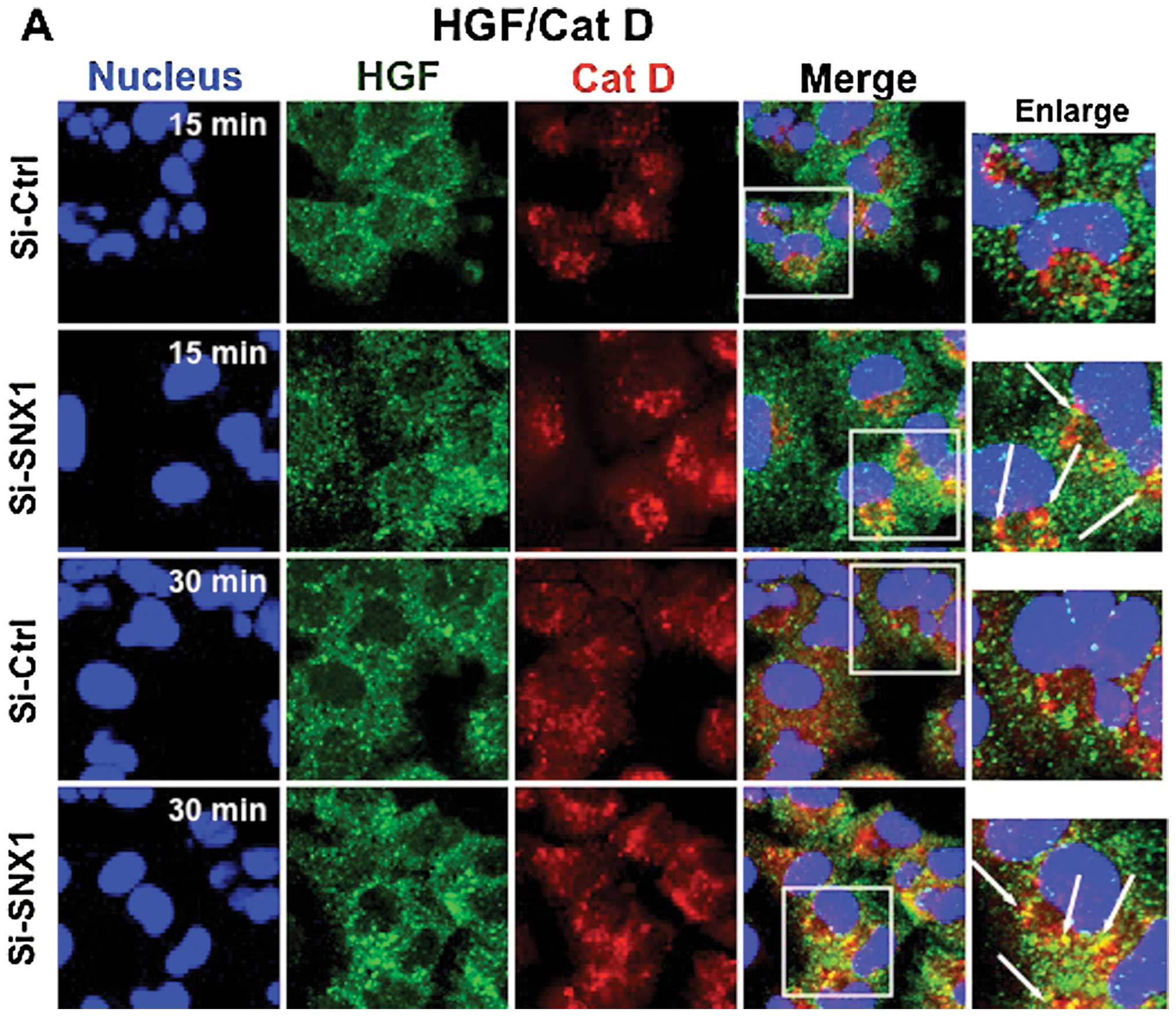
Accordingly, we next examined the effect of SNX1
silencing on HGF-induced MET endocytosis via the early/late
endocytic pathway in A549 cells. The cells transfected with
siRNA-control or siRNA-SNX1 were stimulated at 37°C with HGF for 5,
15 or 30 min, and then double-stained for the internalized HGF
(stained by anti-HGF α-chain antibody) and cathepsin D (stained by
anti-cathepsin D antibody), a marker of lysosomes, or the
internalized HGF and the endocytosed Texas red-labeled transferrin.
Confocal immunofluorescence studies demonstrated a rapid
endocytosis of HGF, and that the distribution of small punctate
vesicles that stained positive for internalized HGF overlapped with
the Texas red-labeled transferrin was seen in the cytoplasm of the
cells transfected with siRNA-SNX1 after 5-min incubation (Fig. 4B). In addition, co-localization of
the internalized HGF and cathepsin D-positive lysosomes was
observed in the perinuclear region after the 15-min incubation
(Fig. 4A). Furthermore, an
increased amount of internalized HGF co-localized with the
cathepsin D-positive lysosomes was seen after 30 min. In contrast,
in the cells transfected with siRNA-control, the internalization of
HGF was considerably suppressed and the endocytosed HGF-positive
staining was not co-localized with cathepsin D-positive vesicular
structures even after 30 min internalization (Fig. 4A). Double staining of HGF and
cathepsin D showed that HGF was rapidly internalized and
transported to lysosomes after 15 min of HGF stimulation with HGF
in the cells transfected with siRNA-SNX1, but endocytosis of HGF
was considerably suppressed and the internalized HGF remained
co-localized with early endosomes after 30 min of HGF stimulation
in the cells transfected with siRNA-control. The co-localization
rate was calculated as the percentage of the integrated density of
early endosome or lysosome marker co-localizing HGF compared with
that of total HGF (% total HGF) (Fig.
4C and D). The co-localization rate calculations confirmed that
cathepsin D-co-localizing HGF was greater in the
siRNA-SNX1-transfected cells than in the siRNA-control-transfected
cells after both 15 min (28.9±4.6 vs. 5.9±0.1% total HGF) and 30
min (39.3±5.6 vs. 11.6±2.1% total HGF) of HGF stimulation (Fig. 4C). However, the amounts of
transferrin-co-localizing HGF appeared to be similar in the cells
transfected with siRNA-SNX1 and in the cells transfected with
siRNA-control after 15 min (43.3±6.7 vs. 36.5±6.1% total HGF) of
HGF stimulation (Fig. 4D). These
results demonstrated that the endocytosed HGF was retained in early
endosomes before being sorted to late endosomes/lysosomes in the
cells transfected with siRNA-control, whereas it was quickly sorted
to late endosomes/lysosomes in the cells transfected with
siRNA-SNX1.
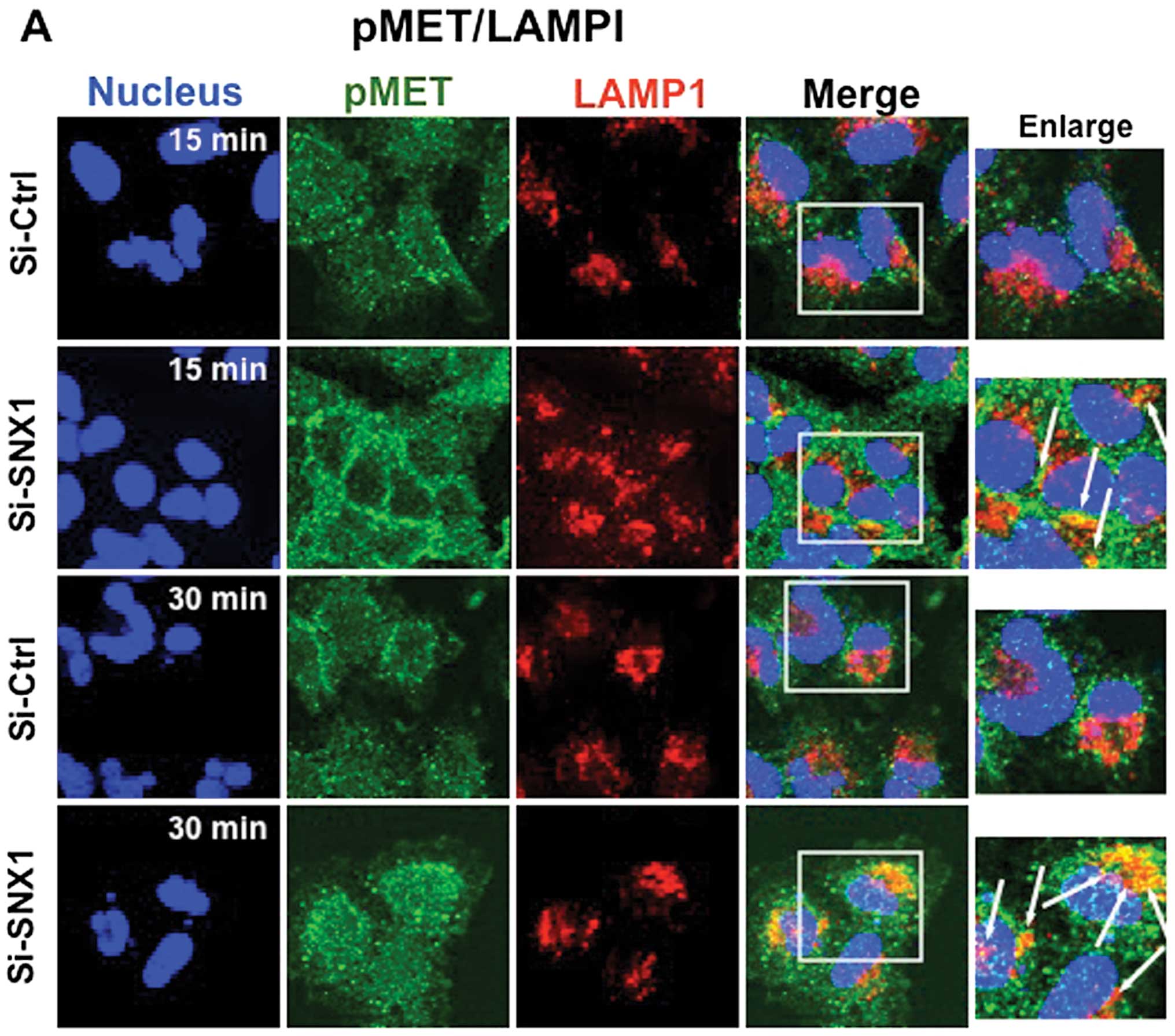
To further substantiate the effect of SNX1 depletion
on the HGF-induced phosphorylation of MET and the endocytosis of
pMET, A549 cells transfected with siRNA-control or siRNA-SNX1 were
stimulated with HGF for 15 or 30 min, and then double-stained for
pMET and LAMP1, or for pMET and the endocytosed Texas red-labeled
transferrin. Confocal immunofluorescence microscopy studies
revealed that in the cells transfected with siRNA-SNX1, a
significant increase in co-localized pMET and LAMP1 was seen after
15 or 30 min of HGF stimulation. In contrast, pMET was
predominantly colocalized with the transferrin-positive early
endosomes, and no co-localization of pMET was seen with
LAMP1-positive vesicles in the siRNA-control-transfected cells
(Fig. 5A). An increase of
co-localized pMET and transferrin was also observed both in the
siRNA-control- and siRNA-SNX1-transfected cells after 30 min of HGF
stimulation (Fig. 5B). The
co-localization rate calculations confirmed that
LAMP1-co-localizing pMET was greater in the cells transfected with
siRNA-SNX1 than in the cells transfected with siRNA-control after
both 15 min (17.2±6.1 vs. 7.7±0.9% total pMET) and 30 min (26.1±3.4
vs. 6.7±0.2% total pMET) of HGF stimulation (Fig. 5C), whereas the amounts of
transferrin-co-localizing pMET appeared to be similar in the cells
transfected with siRNA-SNX1 to that in the cells transfected with
siRNA-control after both 15 min (37.0±3.4 vs. 33.7±5.9% total pMET)
and 30 min (36.5±3.8 vs. 37.7±4.1% total pMET) of HGF stimulation
(Fig. 5D). These data demonstrated
that depletion of SNX1 by siRNA rescues the rapid endocytosis and
endocytic trafficking of MET/pMET via the early/late endocytic
pathway in gefitinib-resistant cells. In contrast, an aberration of
MET endocytosis was noted from the early to late endocytic pathway
in the gefitinib-resistant cells transfected with
siRNA-control.
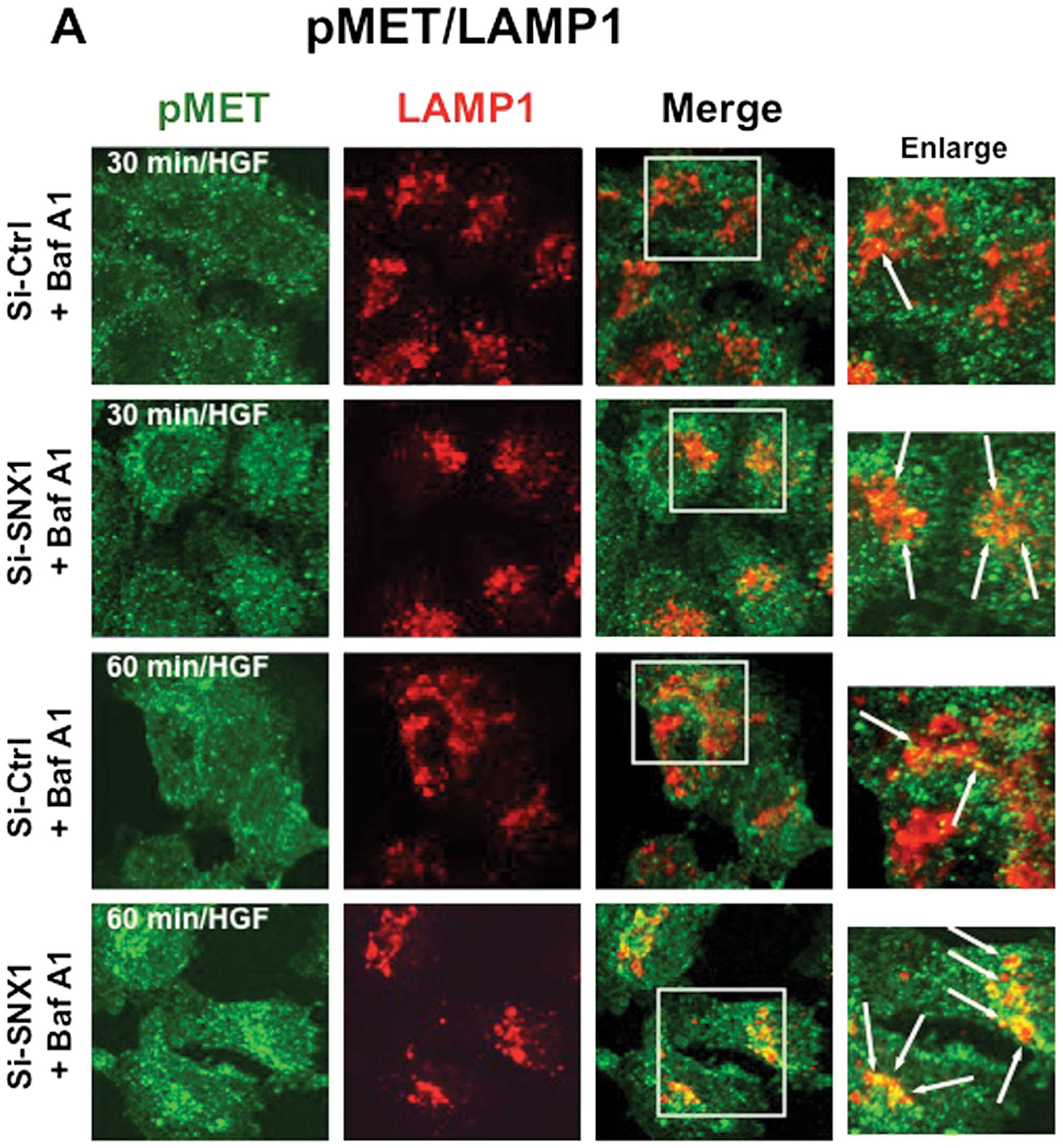
We investigated the effects of bafilomycin A1, a
lysosome inhibitor, on the HGF-induced endocytosis of pMET in the
A549 cells transfected with siRNA-control or siRNA-SNX1. Cells were
stimulated with HGF for 30 or 60 min in the presence of bafilomycin
A1 and then double-stained for pMET and LAMP1, or for pMET and the
endocytosed Texas red-labeled transferrin. The results showed that
the amount of pMET staining co-localized with LAMP1 increased after
30 or 60 min of HGF stimulation (Fig.
6A), and an increase of co-localized pMET and transferrin
staining was also observed both in the siRNA-control and siRNA-SNX1
transfected cells after 30 min of HGF stimulation (Fig. 6B). The co-localization rate
calculations demonstrated that in the presence of bafilomycin A1,
LAMP1-co-localizing pMET was greater in the cells transfected with
siRNA-SNX1 than in the cells transfected with siRNA-control after
both 30 min (29.7±3.6 vs. 11.7±4.0% total pMET) and 60 min
(38.2±8.9 vs. 17.7±4.6% total pMET) of HGF stimulation. An
increased amount of transferrinco-localizing pMET was seen in the
cells transfected with siRNA-SNX1 and in the cells transfected with
siRNA-control after 30 min (25.0±3.6 vs. 16.1±0.7% total pMET)
(Fig. 6C) and 60 min (27.1±5.3 vs.
17.7±7.4% total pMET) of HGF stimulation (Fig. 6D). These results indicate that
bafilomycin A1 treatment significantly blocked HGF-induced pMET
endocytosis, and increased amounts of pMET staining colocalized
with LAMP1-positive late endosomes/lysosomes. Therefore,
bafilomycin A1 has a stronger effect on the HGF-induced pMET
endocytosis in the cells transfected with siRNA-SNX1 than that in
cells transfected with the siRNA-control.
Depletion of SNX1 stimulates the
HGF-induced phosphorylation of MET and phosphorylated MET
degradation in the gefitinib-resistant NSCLC cell line
To analyze the effect of the silencing of SNX1
protein levels on the HGF-induced increase of pMET in the
gefitinib-resistant A549 cells, cells transfected with
siRNA-control or siRNA-SNX1 were stimulated with HGF at 37°C for
the indicated times, and then the cell lysates were analyzed by
western blotting. As shown in Fig. 7A
and D, the treatment of the cells with siRNA-SNX1 completely
suppressed the endogenous expression of SNX1 protein in the A549
cells.
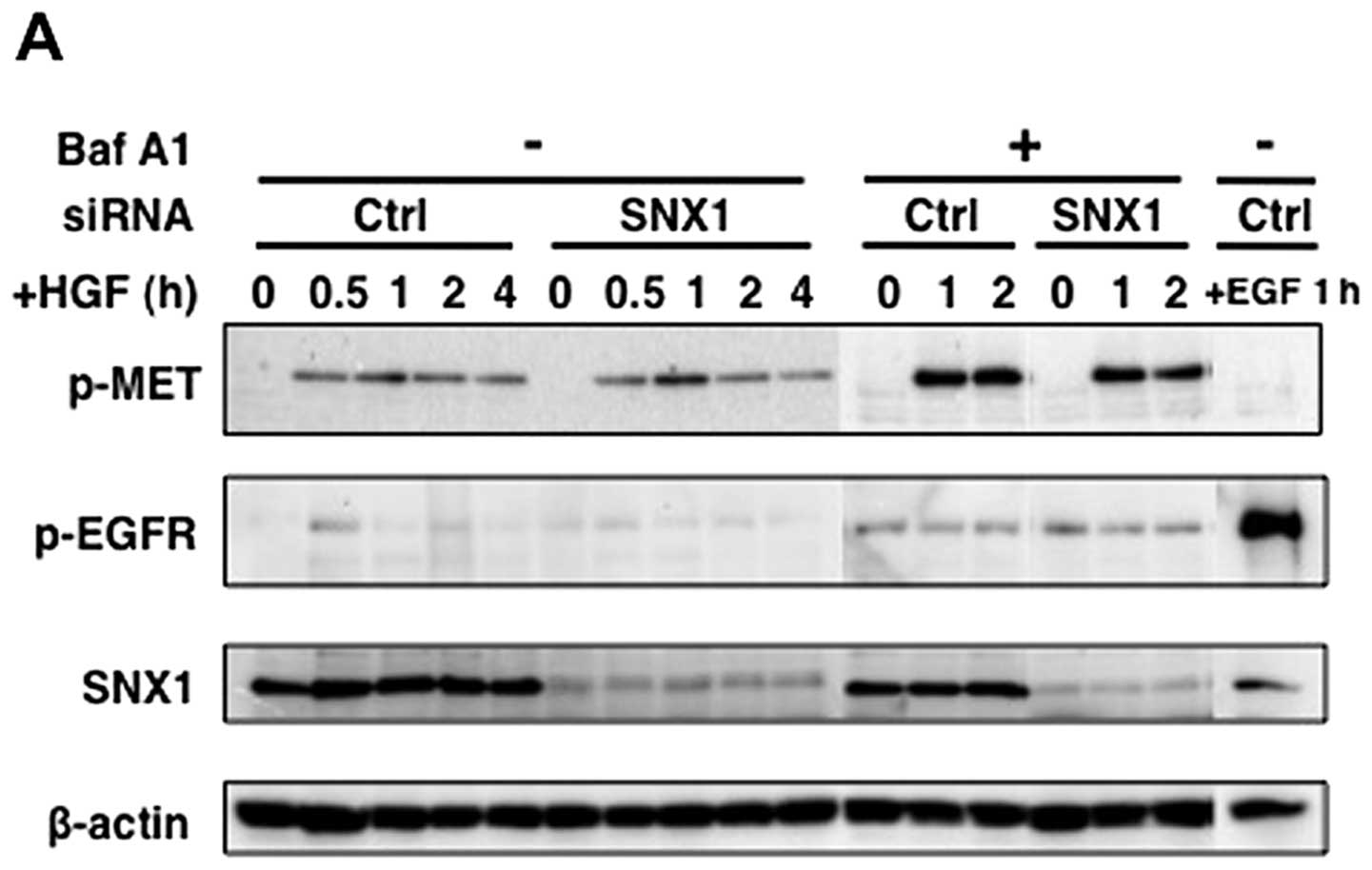 | Figure 7.Silencing of SNX1 by siRNA stimulates
HGF-induced MET phosphorylation and pMET degradation in the
gefitinib-resistant NSCLC cell line. The A549 cells transfected
with siRNA-control (si-Ctrl) or siRNA-SNX1 (si-SNX1) were starved
for 12 h, followed by incubation with CHX (20 μg/ml) and HGF
(100 ng/ml) at 37°C in the absence or presence of bafilomycin A1
for the times indicated. In some cases, the A549 cells transfected
with si-Ctrl were treated with EGF (100 ng/ml) at 37°C for 1 h.
Then cells were lysed, followed by SDS-PAGE and western blot
analysis (A) as described in Materials and methods. The amounts of
pMET, pEGFR, SNX1 and β-actin remaining in the cell lysates
following HGF or EGF stimulation were quantified using NIH Image
software, and were plotted after normalization with β-actin.
Relative expression of (B) pMET, (C) pEGFR, (D) SNX1 and (E)
β-actin in the si-Ctrltransfected cells or si-SNX1-transfected
cells is shown. The error bar denotes SD from three separate
experiments, and significance was determined using Student’s
t-test. Note that an increased expression of pMET was seen at 60
min following HGF stimulation in the si-SNX1-transfected cells, as
compared to that in the si-Ctrl-transfected cells, and the
expression levels of pMET in the siRNA-SNX1-transfected cells was
1.7-fold (at 60 min) higher than that in the
siRNA-control-transfected cells. Also the increased pMET was
rapidly degraded within 4 h in the si-SNX1-transfected cells. These
results indicate that intracellular degradation of HGF-induced pMET
proceeds efficiently via the endosomal/lysosomal pathway in the
A549 cells, since large amounts pMET were accumulated in the cells
treated with HGF in the presence of bafilomycin A1. |
Furthermore, we found novel evidence that the
expression level of pMET was considerably increased at 60 min after
HGF-stimulation in the siRNA-SNX1-transfected A549 cells compared
to the increase seen in the siRNA-control-transfected cells
(Fig. 7A). Quantitative analysis
showed that in the siRNA-SNX1-transfected A549 cells, the induced
expression level of pMET protein following HGF stimulation was
about 118-fold and 39.9-fold at 60 and 120 min, respectively
(Fig. 7B). In contrast, in the
siRNA-control-transfected A549 cells, the increase of the pMET
expression was 63.5-fold and 40.9-fold at 60 and 120 min,
respectively. It should be also noted that the expression levels of
pMET in the siRNA-SNX1-transfected cells was 1.7-fold (at 60 min)
higher than that in the siRNA-control-transfected cells after HGF
stimulation (Fig. 7B) and that an
increase in phosphorylated EGFR (pEGFR) was not observed in the
cells stimulated with HGF (Fig.
7C), thereby demonstrating that HGF treatment selectively
stimulates phosphorylation of MET within 60 min in A549 cells.
The increased pMET appeared to be rapidly degraded
within 4 h in the siRNA-SNX1-transfected cells (Fig. 7A and B), indicating that
intracellular degradation of HGF-induced pMET proceeds efficiently
via an endosomal/lysosomal pathway in A549 cells, since large
amounts of accumulated pMET were observed in the cells when the
cells were treated with HGF in the presence of bafilomycin A1.
These results indicate that depletion of SNX1 by siRNA efficiently
increases endogenous phosphorylation of MET and its degradation via
the endocytic pathway, thereby implying that SNX1 plays a
suppressive role in the phosphorylation and downregulation of MET
via the endocytic pathway in the gefitinib-resistant NSCLC cell
line A549.
Discussion
In the present study, we investigated the
intracellular regulatory function of SNX1 with regard to
HGF-induced endocytosis and downregulation of MET/pMET by using
RNAi-mediated knockdown approaches via the early/late endocytic
pathway in gefitinib-resistant NSCLC cell line A549. Using confocal
immunofluorescence microscopy and qRT-PCR techniques, we verified a
significant reduction of endogenous SNX1 protein expression and
mRNA expression in the gefitinib-sensitive cell line PC9 and the
gefitinib-resistant cell line A549 transfected with siRNA-SNX1. We
also found that endogenous expression of MET transcripts in the
gefitinib-resistant cell line A549 was significantly higher than in
the gefitinib-sensitive cell line PC9; the expression level of MET
transcript in A549 cells was approximately 6.2-fold that of the
values in PC9 cells, and knockdown of SNX1 in these NSCLC cell
lines did not change the expression of MET transcript. Therefore,
we suggest that the expression of SNX1 protein might not have any
effect on the regulation of MET mRNA expression in these NSCLC cell
lines.
We provided novel evidence by using confocal
immunofluorescence microscopy that depletion of endogenous SNX1 by
siRNA-SNX1 induced efficient endocytosis of ligand-induced MET or
pMET via the early/late endocytic pathway in the
gefitinib-resistant NSCLC cell line A549. We showed that increased
co-localization of the internalized HGF complexed with MET and
cathepsin D or of pMET and LIMPII after 15 min internalization in
the gefitinib-resistant cells transfected with siRNA-SNX1. In
contrast, in the siRNA-control-transfected cells, the
internalization of HGF was suppressed and the endocytosed
HGF-positive staining and pMET staining did not overlap with
cathepsin D-positive lysosomal vesicular structures and
LAMP1-positive late endosome vesicles, respectively, even after 30
min of HGF stimulation. Our data suggest that depletion of SNX1
stimulates HGF-induced MET/pMET endocytosis in the
gefitinib-resistant cells. We further found that substantial
inhibition of pMET degradation in A549 cells was verified by using
bafilomycin A1, a lysosomal inhibitor. Bafilomycin A1 treatment in
A549 cells substantially blocked the efficient HGF-induced
degradation of pMET, thereby confirming that the HGF-induced pMET
is trafficked to late endosomes/lysosomes where extensive
degradation for MET protein takes place.
We also showed using western blot analysis that
knockdown of SNX1 induced a dramatic increase in the expression of
pMET in the siRNA-SNX1-transfected A549 cells and that the observed
maximum increase of pMET at 60 min in the siRNA-SNX1-transfected
cells was approximately 1.7-fold higher than that in the
siRNA-control-transfected cells. Moreover, the increased amounts of
pMET at 60 min were rapidly degraded within 2 h in the
siRNA-SNX1-transfected cells as compared to that in the
siRNA-control-transfected cells. This suggests that intracellular
endocytic trafficking followed by downregulation of HGF-induced
pMET takes place rapidly via the endosomal/lysosomal pathway in the
siRNA-SNX1-transfected cells, whereas considerable amounts of pMET
are retained in the early endosomes instead of being delivered to
late endosomes/lysosomes for degradation in the
siRNA-control-transfected cells. Thus, SNX1 might mediate a
negative regulator of HGF-induced MET/pMET endocytosis, followed by
downregulation via the early/late endosomal pathway. Consequently,
we assume that an overexpression of SNX1 in A549 cells suppresses
the HGF-stimulated MET/pMET endocytic traffic out of early
endosomes for targeting the late endosomes/lysosomes where
degradation of HGF-MET/pMET proceeds.
We demonstrated recently that in the
gefitinib-resistant NSCLC cell line, early endosomes labeled with
the endocytosed Texas red-labeled transferrin formed aggregated
vesicular structures distributed in the perinuclear region, and
that considerable amounts of cytosolic SNX1 were distributed in
these aggregated early endosomal vesicles (26,27).
Conversely, no such aggregation of SNX1-positive early endosomes
was observed in the gefitinib-sensitive NSCLC cell line. Moreover,
we showed recently that the transfection of siRNA-SNX1 into the
gefitinib-resistant NSCLC cells resulted in an efficient EGFR
phosphorylation and a rapid endocytic delivery of pEGFR from early
endosomes to late endosomes as revealed by confocal
immunofluorescence microscopy. Western blot analysis demonstrated
that silencing of SNX1 expression by siRNA in the
gefitinib-resistant cells leads to an accelerated degradation of
EGFR along with a dramatic increase in the amounts of pEGFR after
EGF stimulation (28). On the
basis of these data, we postulated that membrane trafficking of
pEGFR from early endosomes to late endosomes might be significantly
impaired in the gefitinib-resistant NSCLC cells. Therefore, we
assume that abrogation of certain components of the SNX1
trafficking machinery could cause this perturbation of EGFR
endocytosis, which might lead to the acquisition of gefitinib
resistance in NSCLC cell lines.
It was reported that following EGF stimulation, the
phosphorylated EGFR is internalized by rapid clathrin-mediated
endocytosis, and the internalized EGFR is then sorted via the
endosomal-sorting complex required for transport (ESCRT)-dependent
pathway for targeting degradation or recycling pathway (18). Consequently, our present data
demonstrating the suppressive role of SNX1 in HGF-induced
phosphorylation of MET and MET endocytosis is consistent with that
in the EGF-stimulated phosphorylation and endocytosis of EGFR via
the endocytic pathway in human lung cancer cells. Therefore, it
should be noted that SNX1 indeed plays a suppressive role on the
HGF-stimulated MET endocytosis and its phosphorylation in the
gefitinib-resistant human lung cancer cells. Accordingly, we
suggest that SNX1 has a critical function in the maintenance of
tightly regulated EGFR- and MET-mediated signaling. It was reported
previously that SNX1 interacts with EGFR, and that the
overexpression of SNX1 enhanced EGF-dependent EGFR degradation in
lysosomes, thereby demonstrating that SNX1 is a likely mediator in
the intracellular sorting of EGFR for targeting the lysosomes for
degradation (30). In this
context, our present data regarding the suppressive action of SNX1
on HGF-stimulated MET endocytosis appear to be in contrast to
findings reported previously (30,31).
Engelman et al demonstrated that MET
amplification leads to acquired resistance to gefitinib or
erlotinib in lung cancer patients by activating ERBB3 signaling
(9), and another group reported
that MET amplification occurs with or without a representative
secondary T790M mutation in EGFR mutant lung tumors with acquired
resistance to gefitinib or erlotinib (38). Accordingly, it is reasonable to
speculate that depletion of SNX1 leading to the enhanced
downregulation of HGF-stimulated MET might affect the sensitivity
to EGFR-TKI gefitinib in human NSCLC cells. Further studies to
investigate the role of SNX1 on HGF/MET activation and signaling
that could affect the EGFR pathway in NSCLC cell lines will be
required.
Abbreviations:
|
SNX1
|
sorting nexin 1;
|
|
HGF
|
hepatocyte growth factor;
|
|
pMET
|
phosphorylated MET;
|
|
EGFR
|
epidermal growth factor receptor;
|
|
pEGFR
|
phosphorylated epidermal growth factor
receptor;
|
|
NSCLC
|
non-small cell lung cancer
|
References
|
1.
|
Di Renzo MF, Narsimhan RP, Olivero M,
Bretti S, Giordano S, Medico E, Gaglia P, Zara P and Comoglio PM:
Expression of the Met/HGF receptor in normal and neoplastic human
tissues. Oncogene. 6:1997–2003. 1991.
|
|
2.
|
Comoglio PM and Boccaccio C: The HGF
receptor family: Unconventional signal transducers for invasive
cell growth. Genes Cells. 1:347–354. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Jeffers M, Rong S and Vande Woude GF:
Hepatocyte growth factor/scatter factor-Met signaling in
tumorigenicity and invasion/metastasis. J Mol Med. 74:505–513.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Pennacchietti S, Michieli P, Galluzzo M,
Mazzone M, Giordano S and Comoglio PM: Hypoxia promotes invasive
growth by transcriptional activation of the met protooncogene.
Cancer Cell. 3:347–361. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Kuniyasu H, Yasui W, Kitadai Y, Yokozaki
H, Ito H and Tahara E: Frequent amplification of the c-met gene in
scirrhous type stomach cancer. Biochem Biophys Res Commun.
189:227–232. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Kijima Y, Hokita S, Yoshinaka H, Itoh T,
Koriyama C, Eizuru Y, Akiba S and Aikou T: Amplification and
overexpression of c-met gene in Epstein-Barr virus-associated
gastric carcinomas. Oncology. 62:60–65. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Nakazawa K, Dobashi Y, Suzuki S, Fujii H,
Takeda Y and Ooi A: Amplification and overexpression of c-erbB-2,
epidermal growth factor receptor, and c-met in biliary tract
cancers. J Pathol. 206:356–365. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Miller CT, Lin L, Casper AM, Lim J, Thomas
DG, Orringer MB, Chang AC, Chambers AF, Giordano TJ, Glover TW and
Beer DG: Genomic amplification of MET with boundaries within
fragile site FRA7G and upregulation of MET pathways in esophageal
adenocarcinoma. Oncogene. 25:409–418. 2006.PubMed/NCBI
|
|
9.
|
Engelman JA, Zejnullahu K, Mitsudomi T,
Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen
J, Kosaka T, Holmes AJ, Rogers AM, Cappuzzo F, Mok T, Lee C,
Johnson BE, Cantley LC and Jänne PA: MET amplification leads to
gefitinib resistance in lung cancer by activating ERBB3 signaling.
Science. 316:1039–1043. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Lutterbach B, Zeng Q, Davis LJ, Hatch H,
Hang G, Kohl NE, Gibbs JB and Pan BS: Lung cancer cell lines
harboring MET gene amplification are dependent on Met for growth
and survival. Cancer Res. 67:2081–2088. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Nakamura T, Teramoto H and Ichihara A:
Purification and characterization of a growth factor from rat
platelets for mature parenchymal hepatocytes in primary cultures.
Proc Natl Acad Sci USA. 83:6489–6493. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Birchmeier C and Gherardi E: Developmental
roles of HGF/SF and its receptor, the c-Met tyrosine kinase. Trends
Cell Biol. 8:404–410. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Birchmeier C, Birchmeier W, Gherardi E and
Vande Woude GF: Met, metastasis, motility and more. Nat Rev Mol
Cell Biol. 4:915–925. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Ullrich A and Schlessinger J: Signal
transduction by receptors with tyrosine kinase activity. Cell.
61:203–212. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Rodrigues GA and Park M:
Autophosphorylation modulates the kinase activity and oncogenic
potential of the Met receptor tyrosine kinase. Oncogene.
9:2019–2027. 1994.PubMed/NCBI
|
|
16.
|
Ma PC, Tretiakova MS, Nallasura V,
Jagadeeswaran R, Husain AN and Salgia R: Downstream signalling and
specific inhibition of c-MET/HGF pathway in small cell lung cancer:
Implications for tumour invasion. Br J Cancer. 97:368–377. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Henne WM, Buchkovich NJ and Emr SD: The
ESCRT pathway. Dev Cell. 21:77–91. 2011. View Article : Google Scholar
|
|
18.
|
Yarden Y: The EGFR family and its ligands
in human cancer signaling mechanisms and therapeutic opportunities.
Eur J Cancer. 37:3–8. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Woodburn JR: The epidermal growth factor
receptor and its inhibition in cancer therapy. Pharmacol Ther.
82:241–250. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Baselga J and Averbuch SD: ZD1839
(‘Iressa’) as an anticancer agent. Drugs. 60(Suppl 1): S33–S42.
2000.
|
|
21.
|
Arteaga CL and Johnson DH: Tyrosine kinase
inhibitors-ZD1839 (Iressa). Curr Opin Oncol. 13:491–498. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Barker AJ, Gibson KH, Grundy W, Godfrey
AA, Barlow JJ, Healy MP, Woodburn JR, Ashton SE, Curry BJ, Scarlett
L, Henthorn L and Richards L: Studies leading to the identification
of ZD1839 (IRESSA): an orally active, selective epidermal growth
factor receptor tyrosine kinase inhibitor targeted to the treatment
of cancer. Bioorg Med Chem Lett. 11:1911–1914. 2001. View Article : Google Scholar
|
|
23.
|
Lynch TJ, Bell DW, Sordella R,
Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat
SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J and
Haber DA: Activating mutations in the epidermal growth factor
receptor underlying responsiveness of non-small-cell lung cancer to
gefitinib. N Engl J Med. 350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Paez JG, Jänne PA, Lee JC, Tracy S,
Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ,
Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE and
Meyerson M: EGFR mutations in lung cancer: correlation with
clinical response to gefitinib therapy. Science. 304:1497–1500.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Nishimura Y, Bereczky B and Ono M: The
EGFR inhibitor gefitinib suppresses ligand-stimulated endocytosis
of EGFR via the early/late endocytic pathway in non-small cell lung
cancer cell lines. Histochem Cell Biol. 127:541–553. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Nishimura Y, Yoshioka K, Bereczky B and
Itoh K: Evidence for efficient phosphorylation of EGFR and rapid
endocytosis of phosphorylated EGFR via the early/late endocytic
pathway in a gefitinib-sensitive non-small cell lung cancer cell
line. Mol Cancer. 7:1–13. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Nishimura Y, Yoshioka K, Takiguchi S,
Bereczky B, Nakabeppu Y and Itoh K: A role for SNX1 in the
regulation of EGF-dependent phosphorylated EGFR endocytosis via the
early/late endocytic pathway in a gefitinib-sensitive human lung
cancer cells. Curr Signal Transduct Ther. 6:383–395. 2011.
View Article : Google Scholar
|
|
28.
|
Nishimura Y, Takiguchi S, Yoshioka K,
Nakabeppu Y and Itoh K: Silencing of SNX1 by siRNA stimulates the
ligand-induced endocytosis of EGFR and increases EGFR
phosphorylation in gefitinib-resistant human lung cancer cell
lines. Int J Oncol. 41:1520–1530. 2012.PubMed/NCBI
|
|
29.
|
Worby CA and Dixon JE: Sorting out the
cellular function of sorting nexins. Nat Rev Mol Cell Biol.
3:919–931. 2002. View
Article : Google Scholar
|
|
30.
|
Kurten RC, Cadena DL and Gill GN: Enhanced
degradation of EGF receptors by a sorting nexin, SNX1. Science.
272:1008–1010. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Zhong Q, Lasar CS, Tronchere H, Sato T,
Meerloo T, Yeo M, Songyang Z, Emr SD and Gill GN: Endosomal
localization and function of sorting nexin 1. Proc Natl Acad Sci
USA. 99:6767–6772. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Carlton J, Bujny M, Peter BJ, Oorschot VM,
Rutherford A, Mellor H, Klumperman J, McMahon HT and Cullen PJ:
Sorting nexin-1 mediates tubular endosome-to-TGN transport through
coincidence sensing of high-curvature membranes and
3-phosphoinositides. Curr Biol. 14:1791–1800. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Gullapalli A, Garrett TA, Paing MM,
Griffin CT, Yang Y and Trejo J: A role for sorting nexin 2 in
epidermal growth factor receptor down-regulation: evidence for
distinct functions of sorting nexin 1 and 2 in protein trafficking.
Mol Biol Cell. 15:2143–2155. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Nishimura Y, Higaki M and Kato K:
Identification of a precursor form of cathepsin D in microsomal
lumen: characterization of enzymatic activation and proteolytic
processing in vitro. Biochem Biophys Res Commun. 148:335–343. 1987.
View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Nishimura Y, Kawabata T and Kato K:
Identification of latent procathepsins B and L in microsomal lumen:
characterization of enzymatic activation and proteolytic processing
in vitro. Arch Biochem Biophys. 261:64–71. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Kornfeld S and Mellman I: The biogenesis
of lysosomes. Ann Rev Cell Biol. 5:483–525. 1989. View Article : Google Scholar
|
|
37.
|
Sandoval IV, Arredondo JJ, Alcalde J,
Gonzalez-Noriega A, Vandekerckhove J, Jimenez MA and Rico M: The
residues Leu (Ile) 475-Ile (Leu) 476, contained in the extended
carboxyl cytoplasmic tail, are critical for targeting of the
resident lysosomal membrane protein LIMPII to lysosomes. J Biol
Chem. 269:6622–6631. 1994.PubMed/NCBI
|
|
38.
|
Bean J, Brennan C, Shih JY, Riely G, Viale
A, Wang L, Chitale D, Motoi N, Szoke J, Broderick S, Balak M, Chang
WC, Yu CJ, Gazdar A, Pass H, Rusch V, Gerald W, Huang SF, Yang PC,
Miller V, Ladanyi M, Yang CH and Pao W: MET amplification occurs
with or without T790M mutations in EGFR mutant lung tumors with
acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci
USA. 104:20932–20937. 2007. View Article : Google Scholar : PubMed/NCBI
|