Introduction
Breast cancer is a highly heterogeneous disease that
is currently classified by the expression profiling of estrogen
receptor (ER), progesterone receptor (PgR) and the human epidermal
growth factor receptor 2 (HER2) (1,2).
Endocrine therapies such as tamoxifen and aromatase inhibitor have
produced a significant improvement in outcomes for patients with
ER-positive breast cancer. The HER2-targeting therapies, such as
trastuzumab and lapatinib, have significantly improved the outlook
for patients with HER2-positive breast cancer. However, increased
risk of endometrial cancer with long-term tamoxifen administration
and of bone fracture due to osteoporosis in postmenopausal women
undergoing aromatase inhibitor treatment are recognized side
effects (3–5). In addition, triple negative breast
cancer (TNBC), defined as tumors that are characterized by lack of
ER, PgR and HER2, accounts for ∼15% of all breast cancers and shows
significantly poorer prognosis compared with other types of breast
cancers because of a lack of clinically established targeted
therapies (6,7). Due to the emergence of these side
effects, endocrine-resistant and chemo-resistant breast cancers and
TNBC, it is necessary to search for novel molecular targets for
drugs based on well-characterized mechanisms of action.
Current ‘omics’ technologies, including
transcriptomics, are a very useful approach for identifying novel
therapeutic targets for various cancers, including breast cancer
(8–10). We previously used DNA microarray to
analyze the genome-wide gene expression profiles of TNBCs and
normal human vital organs including heart, lung, liver and kidney
(10). After comparing the
expression profiles of TNBCs and normal human tissues, we focused
on β1,3-N-acetylgalactosaminyltransferase II
(B3GALNT2), which was significantly upregulated in TNBCs
compared with normal breast ducts (10). B3GALNT2 was first identified as a
novel glycosyltransferase having β1,3-glycosyltranferase motifs,
which are highly conserved in β1,3-galactosyltranferase and
β1,3-N-acetylglucosaminyltranferase families, using a BLAST
search (11). The purified
putative catalytic domain of this protein reportedly has
N-acetylgalactosaminyltransferase in vitro activity
and β1,3-linkage as determined by NMR spectroscopic analysis.
However, to date, no reports have characterized the biologic
function of B3GALNT2 or the significance of its transactivation in
clinical breast cancer cell growth.
Glycosylation plays crucial roles in a variety of
biological functions, such as cell-cell and cell-substrate
interactions, differentiation and signal transduction in tumor
cells (12). The glycoproteins of
tumor cells are often aberrant, both in structure and in quantity,
leading to abnormal biological functions, including cell
proliferation, migration, invasion and transformation (13–15).
Although these alterations are caused by the dysregulated
expression or structure of specific glycosyltransferase (16), their overview remains poorly
understood.
In this study, we show that B3GALNT2 is
overexpressed in breast cancers including TNBC and that
downregulation of B3GALNT2 results in a significant
reduction of breast cancer cell growth due to apoptosis induction.
Moreover, we demonstrate that overexpression of B3GALNT2 results in
its secretion into culture medium. Our findings suggest that
B3GALNT2 represents a promising candidate for the development of
molecular targeting therapy and might be a suitable diagnostic
marker for breast cancer.
Materials and methods
Cell lines and specimens
Human breast cancer cell lines, BT-20, HCC1143,
HCC1395, HCC1599, MCF-7, MDA-MB-453, OCOB-F, T47D and ZR-75-1 and
human embryonic kidney fibroblast HEK293T cells were obtained from
the American Type Culture Collection (ATCC, Rockville, MD, USA).
BSY-1 cell line was a kind gift from Dr Takao Yamori of the
Division of Molecular Pharmacology, Cancer Chemotherapy Center,
Japanese Foundation for Cancer Research. All cells were cultured
under conditions recommended by the ATCC as previously described
(17). We monitored the cell
morphology of these cell lines by microscopy and confirmed that
they had maintained their morphologic states in comparison with the
original morphologic images. No mycoplasma contamination was
detected in the cultures of any of these cell lines using a
Mycoplasma Detection kit (Takara, Kyoto, Japan) in 2011. A total of
30 TNBCs and 13 normal mammary tissues were obtained with informed
consent from patients who were treated at the Tokushima Breast Care
Clinic, Tokushima, Japan, as previously described (10). This study and the use of all
clinical materials described above, was approved by the Ethics
Committee of The University of Tokushima.
Reverse transcription and real-time
PCR
Total RNA from clinical breast cancer samples and
breast cancer cell lines was isolated using a NucleoSpin RNA II
(Takara) according to the manufacturer’s instructions. The poly
A-RNA of normal human heart, liver, kidney, lung and mammary gland
(MG) (Takata Clontech) was reverse transcribed as described
previously (17,18). Real-time PCR analysis was performed
using Power SYBR Green PCR Master Mix (Life Technologies, Carlsbad,
CA, USA) using an ABI PRISM 7500 Real-Time PCR system (Life
Technologies) according to the manufacturer’s instructions.
Gene-specific primers used for real-time PCR were as follows:
5′-AAGACCTGTGAGACAGGAATGC-3′ and 5′-GTTCTGGGTGAAAGTGCCAG-3′ for
B3GALNT2 and 5′-ATTGCCGACAGGATGCAG-3′ and 5′-CTCAGGAGGA
GCAATGATCTT-3′ for ACTB as a quantitative control.
Northern blot analysis
Isolation of mRNA from breast cancer cell lines was
performed using mRNA Purification Kit 4 (GE Healthcare,
Buckinghamshire, UK) according to the manufacturer’s instructions.
The northern blot for breast cancer cell lines was prepared as
described previously (17). The
breast cancer blot and human multiple tissue blots (MTN and MTN II)
were hybridized with [α32P]-dCTP labeled PCR products of
B3GALNT2 by RT-PCR using a primer set as follows:
5′-TGATGTGGTAGTTGGCGTGT-3′ and 5′-AGAACTCCCCCTCCATCATT-3′.
Prehybridization, hybridization and washing were performed as
described previously (17). The
blots were autoradiographed with intensifying screens at −80°C for
14 days.
Gene silencing effect by siRNA
treatment
BT-20, MDA- MB-453 and ZR-75-1 cells were plated
onto 12-well plates (5×104, 5×104 and
2.5×104 cells/well, respectively). Transfection of 10 nM
siRNAs was performed using Lipofectamine RNAiMax (Life
Technologies) according to the manufacturer’s instructions. The
target sequences for three B3GALNT2 and a control
EGFP siRNAs were 5′-GUCAACGUGUGCUUGUG AA-3′ for
siB3GALNT2-1, 5′-CGAGCUUCAAUUUGUUGC U-3′ for siB3GALNT2-2,
5′-CCGGAAAGUGGCAGGAG UU-3′ for siB3GALNT2-3 and 5′-GCAGCACGACUUCUUC
AAG-3′ for siEGFP. To evaluate the knockdown effect of the siRNAs
by quantitative RT-PCR, total RNA was extracted from the
siRNA-transfected cells at 6 days after siRNA transfection. The
gene-specific primers are described above. These experiments were
performed in duplicate. To quantify cell viability, MTT assays were
performed using a Cell-Counting Kit-8 according to the
manufacturer’s recommendations (Dojindo, Kumamoto, Japan).
Absorbance at 450 nm was measured with the microplate reader
Infinite 200 (Tecan, Männedorf, Switzerland). These experiments
were performed in duplicate (MDA-MB-453 and ZR-75-1 cells) or
triplicate (BT-20 cells).
Plasmids
To construct the B3GALNT2 expression vectors, the
entire coding sequence of B3GALNT2 cDNA was amplified by
RT-PCR using KOD plus DNA polymerase (Toyobo, Osaka, Japan) and
cloned into the pCAGGSn3FC expression vector in frame with Flag-tag
at the COOH terminus. The primer sets of B3GALNT2-wild-type (WT)
was as follows: 5′-ATAAG AATGCGGCCGCATGCGAAACTGGCTGGTGC-3′
and 5′-CCGCTCGAGTCTTGCTTGACATCGACAAGG-3′
(the underlined letters indicate the NotI or XhoI
sites, respectively). We also performed conventional two-step
mutagenesis PCR to generate mutants in which N116A and N174A were
substituted to alanines, as described previously (14). The primer sets were
5′-GTAAACTACTCGCCATCACAAATC-3′ and 5′-GATTT GTGATGGCGAGTAGTTTAC-3′ for N116A,
5′-GTTTCCAGAGGGCCATCACTGTC-3′ and
5′-GACAGTGATGGCCC
TCTGGAAAC-3′ for N174A (the underlined letters indicate the
mutation sites, respectively). The DNA sequences of all constructs
were confirmed by DNA sequencing (ABI3500XL, Life
Technologies).
Fluorescence activated cell-sorting
(FACS) analysis
FACS analysis was performed as previously described
(10). Briefly, the cells were
collected at 2, 4 and 6 days after treatment of siRNAs against
B3GALNT2 or EGFP as a control. The cells were fixed
by 70% ethanol at room temperature for 30 min and then incubated at
37°C for 30 min with 1 mg/ml RNase A, followed by staining with 20
μg/ml propidium iodide at room temperature for 30 min in the
dark. The DNA content of 10,000 cells was analyzed with a
FACSCalibur flow cytometer and CellQuest software (BD Biosciences,
Franklin Lakes, NJ, USA).
Western blot analysis
Western blot analysis was performed as previously
described (19). Briefly, the
cells were lysed in lysis buffer [50 mM Tris-HCl (pH 8.0), 150 mM
NaCl, 0.5% Nonidet P-40, 0.5% CHAPS] including 0.1% protease
inhibitor cocktail III (Calbiochem, San Diego, CA, USA). The cell
lysates were incubated on ice for 30 min and centrifuged at 15,000
rpm for 15 min to remove cell debris. Then, the proteins were mixed
with SDS sample buffer [25 mM Tris-HCl (pH 6.8), 0.8% SDS, 5%
glycerol] and boiled for 5 min. After SDS-PAGE, membranes blotted
with proteins were incubated with anti-Flag M2 (Sigma-Aldrich, St.
Louis, MO, USA, F3165) or anti-β-actin (AC-15, Sigma-Aldrich,
A-5441) monoclonal antibodies diluted at 1:5,000 and PARP rabbit
polyclonal antibody (Cell Signaling Technology, 9542) diluted at
1:500, respectively. Finally, the membrane was incubated with
horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h
and protein bands were visualized by enhanced chemiluminescence
detection reagents (ECL, GE Healthcare).
Immunocytochemical staining
Immunocytochemical staining was performed as
previously described (10). To
examine the subcellular localization of the B3GALNT2 protein,
HEK293T cells were plated onto an 8-well glass slide (Thermo Fisher
Scientific, Rochester, NY, USA) at a density of 1.0×104
cells/well and transfected with 0.2 μg each of expression
plasmids using FuGENE 6 reagent (Promega, Madison, WI, USA)
according to the manufacturer’s recommendations. To detect
exogenous B3GALNT2-Flag, anti-Flag M2 mouse antibody was used at
1:1,000 and Alexa 488-conjugated anti-mouse antibody. The Golgi
apparatus were visualized by staining with anti-Golgi-58k mouse
monoclonal antibody (Sigma-Aldrich) (20). Cell morphology was analyzed by
Alexa Fluor 488 phalloidin (Molecular Probes, Eugene, OR, USA)
diluted at 1:1,000.
Inhibition of N-glycosylation
HEK293T cells were plated onto 6-well dishes at a
density of 2.0×105 cells/well and transfected with 1
μg each of expression plasmids using FuGENE 6 reagent
according to the manufacturer’s instructions. To validate the
N-glycosylation of B3GALNT2, the cells were cultured with 10
μg/ml tunicamycin (Sigma-Aldrich), an inhibitor against
N-glycosylation, for 24 h, at 4 h after transfection. Cells were
then lysed with SDS sample buffer.
Preparation of secreted B3GALNT2
To detect secreted B3GALNT2 protein, we transfected
B3GALNT2-expressing plasmids to HEK293T cells as described above.
The medium was changed to DMEM with 0.1% FBS at 48 h after
transfection. Cells were cultured for a further 48 h and then
supernatants were collected. After removing debris by
centrifugation at 15,000 rpm for 15 min, we performed acetone
precipitation at −30°C overnight. Then, the pellets were
resuspended with SDS sample buffer.
Statistical analysis
Statistical analysis was conducted using Student’s
t-test. A P-value of <0.05 was considered to be statistically
significant. Box blot analysis was performed with StatView J 5.0
software using the microarray data of 13 normal ducts and 30 TNBCs
[the Gene Expression Omnibus database accession no. GSE38959;
(10)].
Results
B3GALNT2 is upregulated in breast cancer
specimens and cell lines
To identify the novel therapeutic targets for TNBC
therapy, we previously performed genome-wide gene expression
profile analysis of TNBC and normal human tissues by DNA microarray
analysis (10). In parallel with
this approach, we attempted to search for the genes that encode
proteins containing glycosyltransferase motifs, either based on
reported information or according to prediction by the
protein-motif program SMART because we previously succeeded in
identifying and characterizing the cancer-specific
glycosyltransferase GALNT6, that plays a critical role for mammary
carcinogenesis, as a drug target (14,15).
Among the glycosyltransferase genes that are upregulated in
expression profiles of breast cancers, we focused on the
B3GALNT2 gene, which encodes a glycosyltransferase, as a
drug target for breast cancer. Gene expression profiling analysis
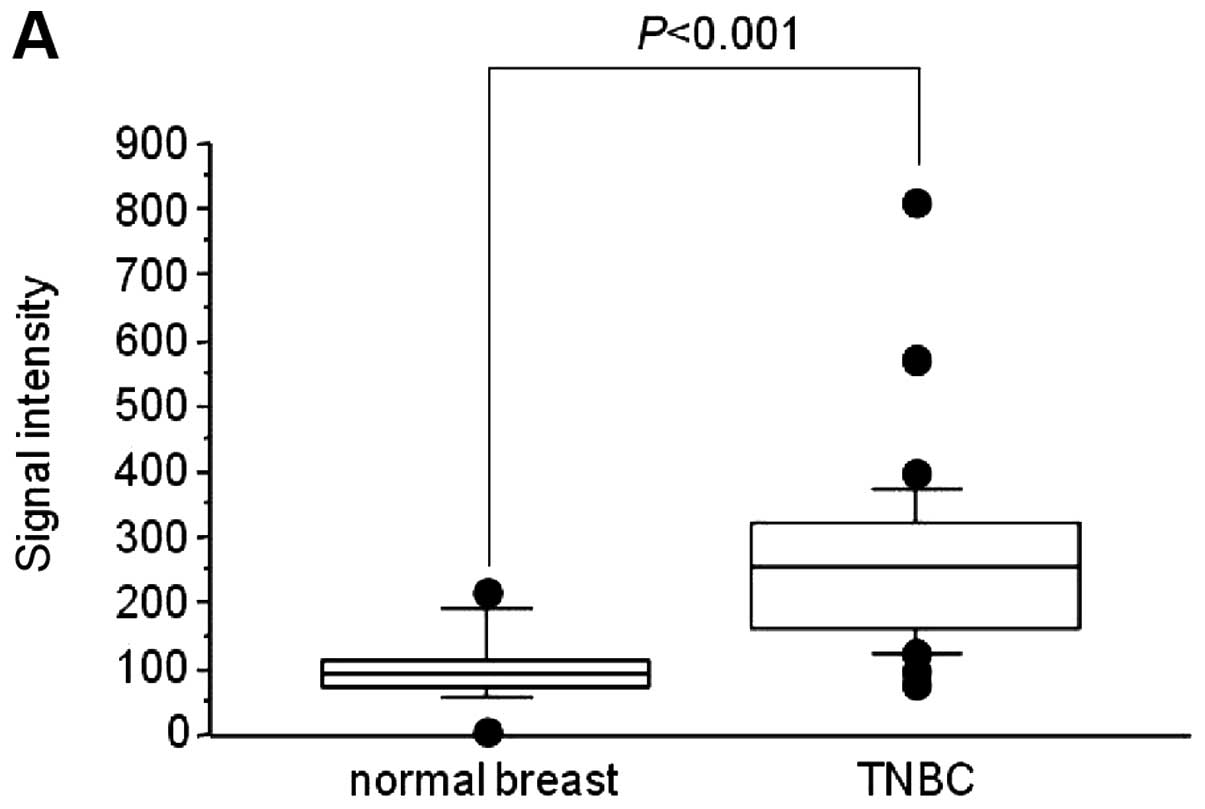
showed significant overexpression of B3GALNT2 in TNBC cases,
compared with normal ductal cells (Fig. 1A). Using quantitative RT-PCR, we
verified that B3GALNT2 was upregulated >2-fold in 5 of 10
TNBC samples compared with clinically normal breast tissues,
whereas B3GALNT2 was hardly expressed in the heart, lung,
liver, kidney and mammary gland (Fig.
1B). Subsequent northern blot analysis of 10 breast cancer cell
lines detected an approximately 4.7-kb B3GALNT2 transcript
at a high level in 8 of the 10 breast cancer cell lines that were
examined (Fig. 1C).
Furthermore, multiple tissue northern blot analysis
revealed that a 4.7-kb transcript of B3GALNT2 was slightly
expressed in heart, skeletal muscle, spleen, kidney, liver, small
intestine, placenta, lung, prostate and ovary, while a 2.4-kb
transcript was exclusively expressed in testis (Fig. 1D). According to the National Center
for Biotechnology Information (NCBI) database, two representative
transcripts of 4,755 nucleotides (B3GALNT2-V1, GenBank
accession no. NM_152490) and 2,022 nucleotides (B3GALNT2-V2,
GenBank accession no. BC029564) that share the same open reading
frame encoding a 500-amino acid protein seemed to correspond to the
two bands observed in multiple tissue northern blot analysis
(Fig. 1E).
Effect of B3GALNT2 on cell growth
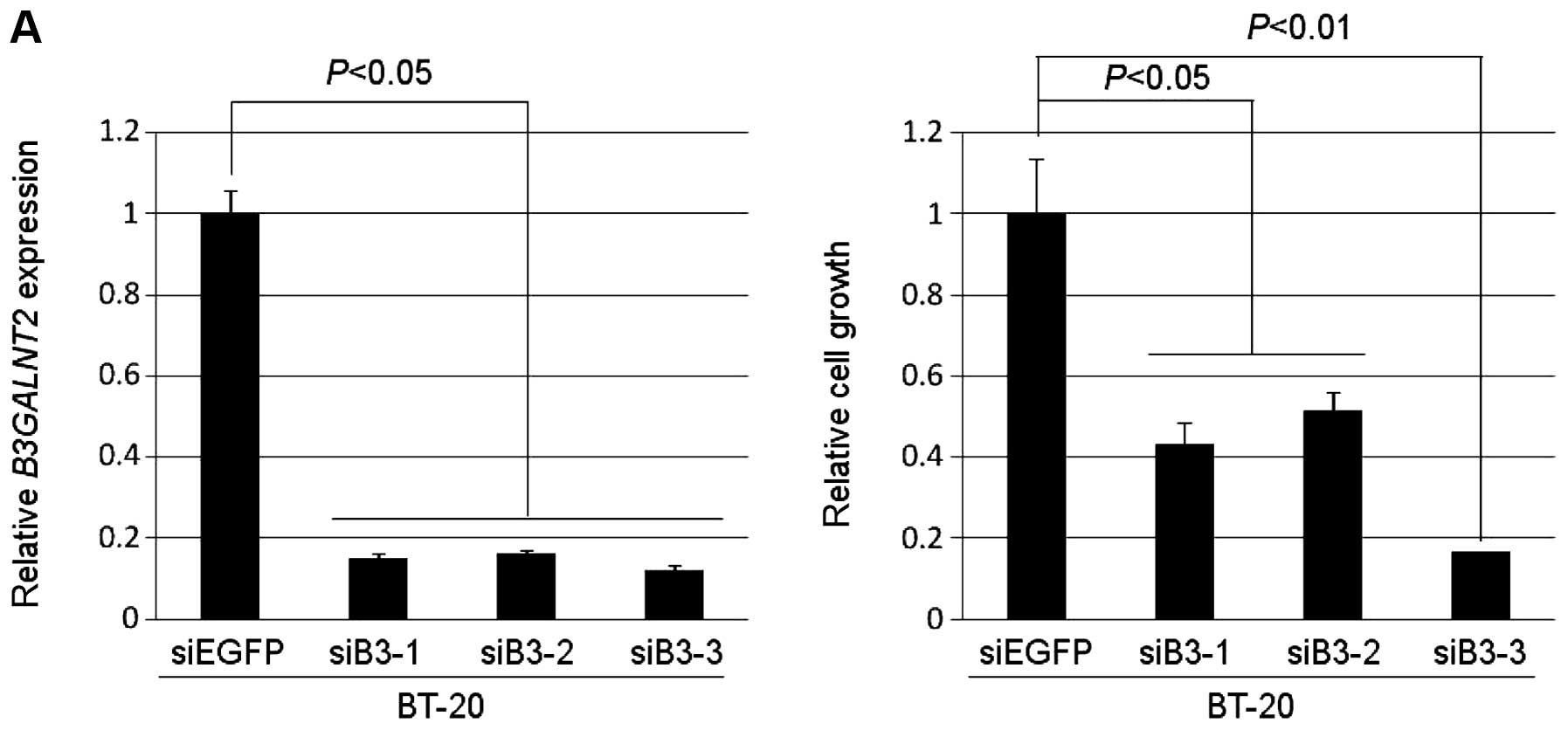
To assess whether B3GALNT2 is essential for
the growth or survival of breast cancer cells, we transfected
synthetic oligonucleotide siRNAs against B3GALNT2 into the
TNBC cell line, which consisted of BT-20 cells in which
B3GALNT2 was highly expressed. The mRNA levels of
B3GALNT2 in the cells transfected with siB3-1, -2 or -3 were
significantly downregulated in comparison with cells transfected
with siEGFP as a control (Fig.
2A). We also observed a significant decrease in the number of
viable cells measured by MTT assay (Fig. 2A). Similarly, silencing of
B3GALNT2 expression by siB3-3 markedly decreased cell
proliferation of the non-TNBC cell lines MDA-MB-453 and ZR-75-1, in
which B3GALNT2 was highly expressed (Fig. 2B and C).
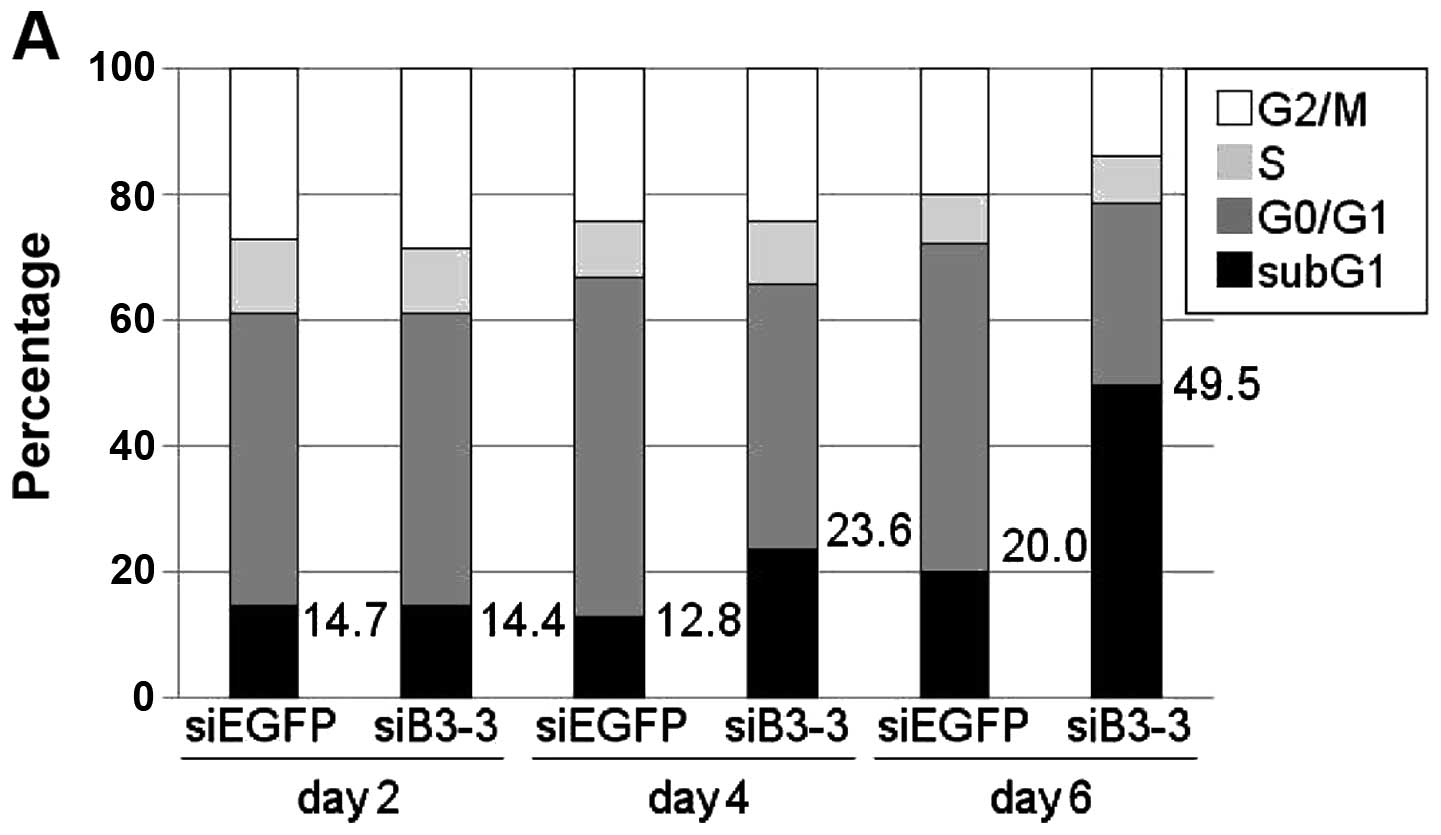
To further assess the knockdown effect of
B3GALNT2, we performed FACS analysis and found that the
percentage of sub-G1 population was clearly increased in a
time-dependent manner in B3GALNT2-depleted BT-20 cells
(Fig. 3A). In addition, we
observed an obvious cleaved PARP at day 4 after treatment of siRNA
against B3GALNT2 (siB3-3) (Fig.
3B). Interestingly, 4 days after transfection of siB3-3, severe
disrupted cytoskeletal organization was observed by
immunocytochemistry with fluorescence-labeled phalloidin (Fig. 3C), suggesting that depletion of
B3GALNT2 resulted in suppression of breast cancer cell
growth due to apoptosis. These findings suggest that
B3GALNT2 is crucial for both TNBC and non-TNBC cell
growth.
Secretory nature and N-glycosylation of
B3GALNT2
The B3GALNT2 gene is reported to encode a
type II transmembrane enzyme, which possesses a single
transmembrane domain, a stem region and a C-terminal catalytic
domain for enzyme activity (21).
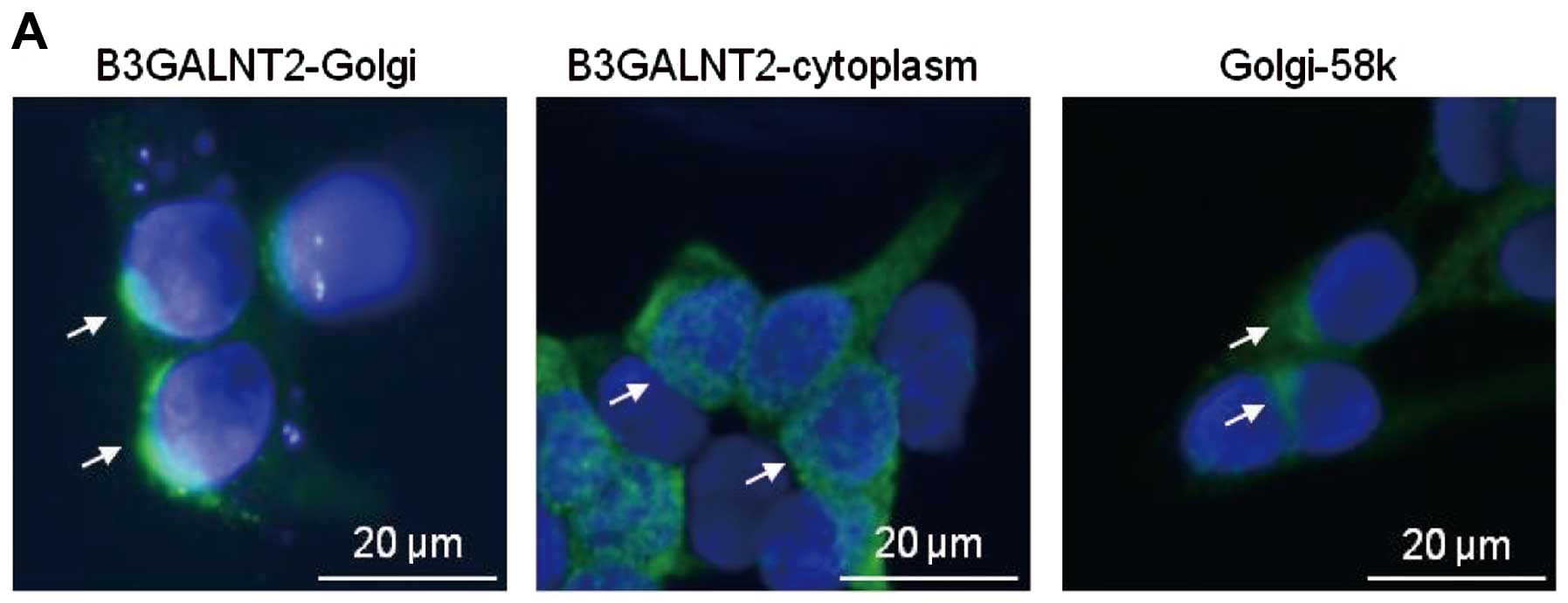
To first investigate the subcellular localization of B3GALNT2 in
mammalian cells, we transiently transfected the Flag-tagged
B3GALNT2 construct (B3GALNT2-Flag) into HEK293T cells and then
performed immunocytochemical staining analysis. B3GALNT2-Flag was
observed to have highly intense staining in the Golgi apparatus,
but it was also diffusely observed in cytoplasm (Fig. 4A). Furthermore, because many type
II glycosyltransferases are found as secreted soluble enzymes
through proteolytic cleavage of the stem region (22), we hypothesized that B3GALNT2 has a
secretory nature. To investigate this possibility, HEK293T cells
were transiently transfected with B3GALNT2-Flag and then western
blot analysis with anti-Flag antibody was performed using cell
lysates and culture media. We detected a band of B3GALNT2-Flag in
both cell lysates and culture media, but its molecular weight in
culture media was smaller than that in cell lysate, suggesting the
possibility that the B3GALNT2 protein was cleaved during its
secretion into culture media (Fig.
4B).
N-glycosylation on many glycosyltransferases is
known to be associated with its biological functions, especially
its secretion (22,23). In addition, the NetNGlyc 1.0 server
(http://cbs.dtu.dk/services/NetNGlyc)
and NCBI database predict that B3GALNT2 possesses two potential
N-linked glycosylation at positions Asn-116 and Asn-174 that are
completely conserved among other species, such as mouse, rat and
Xenopus laevis (Fig. 4C).
To confirm whether B3GALNT2 is N-glycosylated, we treated HEK293T
cells transfected with B3GALNT2-wild-type (B3GALNT2-WT) with
tunicamycin, which is an N-glycosylation inhibitor and then
analyzed the molecular weight of B3GALNT2-WT protein by western
blot analysis. As expected, tunicamycin treatment resulted in a
size reduction of B3GALNT2-WT (Fig.
4D, WT). Next, to assess the N-glycosylated amino acids of
B3GALNT2, we constructed the expression plasmids of N116A or N174A
single mutation and N116AN174A double mutation, in which the
conserved asparagine residues at position 116 or 174 were replaced
by alanine residues. When HEK293T cells were transfected with these
plasmids without tunicamycin treatment, the molecular weight of
N116A and N174A was smaller than that of B3GALNT2-WT, respectively
and the molecular weight of N116AN174A double mutant was
significantly smaller. In addition, tunicamycin treatment induced a
decreased molecular mass of N116A and N174A, although the
N116AN174A double mutant mass was not decreased (Fig. 4D).
To evaluate the effect of N-glycosylation on the
secretion of the B3GALNT2 protein, we examined its secretion in
HEK293T cells transfected with B3GALNT2-WT, -N116A, -N174A and
N116AN174A, respectively. The secretion of B3GALNT2-N116A or -N174A
was reduced compared with WT, while the secretion of N116AN174A
mutant in culture media was drastically reduced (Fig. 4E). However, all of the mutants were
mainly localized to the Golgi apparatus as well as B3GALNT2-WT
(Fig. 4F), thus suggesting no
effects of these mutants on its Golgi retention. Taken together,
both Asn-116 and Asn-174 amino acids of B3GALNT2 are
N-glycosylation sites and their N-glycosylation is necessary for
B3GALNT2 secretion.
Discussion
Glycosylation is a posttranslational modification
and is associated with various physiologic events. The aberrant
expression of glycosyltransferase and the immature glycan structure
of proteins and lipids are observed in many cancers. These
phenomena are also involved in the development and progression of
cancers (13–16,24).
Abnormalities of the glycan structure of glycoproteins are
frequently observed in breast cancer cells (13–15).
In particular, we previously identified and characterized the
oncogenic roles of a cancer-specific glycosyltransferase,
UDP-N-acetyl-α-D-galactosamine (GalNAc): polypeptide
N-acetylgalactosaminyltransferase 6 (GALNT6) that regulated
cell proliferation and cytoskeleton structure through aberrant
O-glycosylation and stabilization of an oncoprotein mucin 1
(MUC1) (14) and fibronectin
(15), which indicated that the
development of GALNT6 inhibitors would be valuable for breast
cancer therapy. To further elucidate the oncogenic role of aberrant
glycosyltransferase expression, we attempted to identify
cancer-specific glycosyltransferases that are exclusively
upregulated in breast cancers through the analysis of comprehensive
gene expression profiles of TNBC and normal human tissues. In this
study, we focused on a breast cancer-specific glycosyltransferase,
B3GALNT2 and showed its potential as a druggable target by
showing its critical roles in breast cancer cell growth.
B3GALNT2 was indicated to be the member of the
β1,3-glycosyltransferase (β3GT) family by having three β3GT motifs
and its function was shown by in vitro analyses to be a
synthesis of GalNAcβ1–3GlcNAcβ1-R structure on both N-glycans and
O-glycans of proteins (11).
However, the biological and biochemical functions of B3GALNT2 have
not been clarified in mammalian cells, including human cancer
cells, primarily because the GalNAc β1–3GlcNAcβ1-R structure has
been reported only in α-dystroglycan in mammalian cells (25). Recently, mutations in the
B3GALNT2 gene were identified in individuals with
dystroglycanopathy by whole-exome and Sanger sequencing
technologies, suggesting that α-dystroglycan is the potential
substrate of B3GALNT2 (26). In
contrast, the expression of α-dystroglycan has been reported to be
frequently downregulated in breast cancers (27). Moreover, Stevens et al
(26) showed that exogenous
V5-tagged B3GALNT2 was mainly localized in the endoplasmic
reticulum (ER) of C2C12 myoblasts, which is not concordant with our
results indicating that exogenous B3GALNT2-Flag was mainly
localized in the Golgi apparatus (Fig.
4A). This discrepancy may be due to differences in the
experimental procedures employed, including the type of cell lines
or the different expression vector constructs. Indeed, both
previous findings and our results showed only exogenous expression
of B3GALNT2 in mammalian cells. Hence, further study is necessary
to clarify the biological roles or exact subcellular-localization
of glycosylated α-dystroglycan in endogenous
B3GALNT2-overexpressing breast cancer cells.
We first identified B3GALNT2 to be
upregulated in TNBCs, but demonstrated that the silencing of
B3GALNT2 expression by siRNA resulted in significant
suppression of the growth of non-TNBC and TNBC cell lines (Fig. 2). Nevertheless, the overexpression
of B3GALNT2 into HEK293 or NIH3T3 cells could not enhance
cell proliferation (data not shown), indicating that
B3GALNT2 is indispensable for the survival of breast cancer
cells, but B3GALNT2 alone may not be sufficient for
transformation activity. Furthermore, B3GALNT2 was shown to be a
secreted protein (Fig. 4), but
addition of the conditioned media of the B3GALNT2-transfected
HEK293T cells into the culture media of HEK293A cells could not
enhance cell growth (data not shown). These results suggest that
the intrinsic glycosyltransferase activity of B3GALNT2 might be
critical for breast cancer cell growth. However, it has been
reported that GnT-V secreted from WiDr colon cancer cells is
directly involved in tumor angiogenesis in a
glycosylation-independent manner, thus providing biological
importance for the secretion of this glycosyltransferase (28). Therefore, further studies are
needed to clarify the precise biological roles of the secreted form
of B3GALNT2.
Furthermore, we demonstrated that N-glycosylation at
Asn-116 or Asn-174 of B3GALNT2 is critical for its efficient
secretion, but the effects of these posttranslational modifications
on its biological functions, including enzyme activity, are
unknown. Some studies have shown that N-glycosylation on
glycosyltransferases is required for their proper-folding and/or
enzymatic activities (29–31). Therefore, further study is
necessary to clarify the precise pathophysiological roles of
B3GALNT2 in mammary carcinogenesis through identification and
characterization of its specific substrates and to screen
inhibitors targeting glycosyltransferase activity of B3GALNT2 for
potential breast cancer therapeutic applications.
In conclusion, we demonstrated that overexpression
of cancer-specific glycosyltransferase B3GALNT2 is critical for the
growth or survival in breast cancers, including TNBC and
ER-positive breast cancer. Our findings showed the usefulness of
B3GALNT2 as a promising diagnostic and/or therapeutic target for
breast cancer.
Acknowledgements
We thank Drs Kaoru Takegawa
(Department of Bioscience and Biotechnology, Faculty of
Agriculture, Kyushu University) and Tomoya Fukawa (The University
of Tokushima) for helpful discussions and Ms. Hinako Koseki for
technical assistance. This study was supported by a Grant-in-Aid
for Young Scientists (B) (grant nos. 23790369 and 25860240) from
MEXT.
References
|
1.
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J,
Murray T and Thun MJ: Cancer Statistics, 2008. CA Cancer J Clin.
58:71–96. 2008. View Article : Google Scholar
|
|
2.
|
Di Cosmo S and Baselga J: Management
breast cancer with targeted agents: Importance of heterogeneity.
Nat Rev Clin Oncol. 7:139–147. 2010.PubMed/NCBI
|
|
3.
|
Berry DA, Cronin KA, Plevritis SK, Fryback
DG, Clarke L, Zelen M, Mandelblatt JS, Yakovlev AY, Habbema JD and
Feuer EJ: Cancer Intervention and Surveillance Modeling Network
(CISNET) Collaborators: Effect of screening and adjuvant therapy on
mortality from breast cancer. N Engl J Med. 353:1784–1792. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Early Breast Cancer Trialists’
Collaborative Group (EBCTCG): Effects of chemotherapy and hormonal
therapy for early breast cancer on recurrence and 15-year survival:
an overview of the randomised trials. Lancet. 365:1687–1717.
2005.PubMed/NCBI
|
|
5.
|
Adamo V, Iorfida M, Montalto E, Festa V,
Garipoli C, Scimone A, Zanghì M and Caristi N: Overview and new
strategies in metastatic breast cancer (MBC) for treatment of
tamoxifen-resistant patients. Ann Oncol. 18:53–57. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Foulkes WD, Smith IE and Reis-Filho JS:
Triple-negative breast cancer. N Engl J Med. 363:1938–1948. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Liedtke C, Mazouni C, Hess KR, André F,
Tordai A, Mejia JA, Symmans WF, Gonzalez-Angulo AM, Hennessy B,
Green M, Cristofanilli M, Hortobagyi GN and Pusztai L: Response to
neoadjuvant therapy and longterm survival in patients with
triple-negative breast cancer. J Clin Oncol. 26:1275–1281. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Petricoin EF III, Hackett JL, Lesko LJ,
Puri RK, Gutman SI, Chumakov K, Woodcock J, Feigal DW Jr, Zoon KC
and Sistare FD: Medical applications of microarray technologies: a
regulatory science perspective. Nat Genet. 32:474–479. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Nishidate T, Katagiri T, Lin ML, Mano Y,
Miki Y, Kasumi F, Yoshimoto M, Tsunoda T, Hirata K and Nakamura Y:
Genome-wide gene-expression profiles of breast-cancer cells
purified with laser microbeam microdissection: identification of
genes associated with progression and metastasis. Int J Oncol.
25:797–819. 2004.
|
|
10.
|
Komatsu M, Yoshimaru T, Matsuo T, Kiyotani
K, Miyoshi Y, Tanahashi T, Rokutan K, Yamaguchi R, Saito A, Imoto
S, Miyano S, Nakamura Y, Sasa M, Shimada M and Katagiri T:
Molecular features of triple negative breast cancer cells by
genome-wide gene expression profiling analysis. Int J Oncol.
42:478–506. 2013.PubMed/NCBI
|
|
11.
|
Hiruma T, Togayachi A, Okamura K, Sato T,
Kikuchi N, Kwon YD, Nakamura A, Fujimura K, Gotoh M, Tachibana K,
Ishizuka Y, Noce T, Nakanishi H and Narimatsu H: A novel human
beta1,3-N-acetylgalactosaminyltransferase that synthesizes a unique
carbohydrate structure, GalNAcbeta1-3GlcNAc. J Biol Chem.
279:14087–14095. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Hart GW and Copeland RJ: Glycomics hits
the big time. Cell. 143:672–676. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Fuster MM and Esko JD: The sweet and sour
of cancer: glycans as novel therapeutic targets. Nat Rev Cancer.
5:526–542. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Park JH, Nishidate T, Kijima K, Ohashi T,
Takegawa K, Fujikane T, Hirata K, Nakamura Y and Katagiri T:
Critical roles of mucin 1 glycosylation by transactivated
polypeptide N-acetylgalactosaminyltransferase 6 in mammary
carcinogenesis. Cancer Res. 70:2759–2769. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Park JH, Katagiri T, Chung S, Kijima K and
Nakamura Y: Polypeptide N-acetylgalactosaminyltransferase 6
disrupts mammary acinar morphogenesis through O-glycosylation of
fibronectin. Neoplasia. 13:320–326. 2011.PubMed/NCBI
|
|
16.
|
Potapenko IO, Haakensen VD, Lüders T,
Helland A, Bukholm I, Sørlie T, Kristensen VN, Lingjaerde OC and
Børresen-Dale AL: Glycan gene expression signatures in normal and
malignant breast tissue; possible role in diagnosis and
progression. Mol Oncol. 4:98–118. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Park JH, Lin ML, Nishidate T, Nakamura Y
and Katagiri T: PDZ-binding kinase/T-LAK cell-originated protein
kinase, a putative cancer/testis antigen with an oncogenic activity
in breast cancer. Cancer Res. 66:9186–9195. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Kim JW, Akiyama M, Park JH, Lin ML, Shimo
A, Ueki T, Daigo Y, Tsunoda T, Nishidate T, Nakamura Y and Katagiri
T: Activation of an estrogen/estrogen receptor signaling by BIG3
through its inhibitory effect on nuclear transport of PHB2/REA in
breast cancer. Cancer Sci. 100:1468–1478. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Shimo A, Nishidate T, Ohta T, Fukuda M,
Nakamura Y and Katagiri T: Elevated expression of protein regulator
of cytokinesis 1, involved in the growth of breast cancer cells.
Cancer Sci. 98:174–181. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Nogueira E, Fidalgo M, Molnar A, Kyriakis
J, Force T, Zalvide J and Pombo CM: SOK1 translocates from the
Golgi to the nucleus upon chemical anoxia and induces apoptotic
cell death. J Biol Chem. 283:16248–16258. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Paulson JC and Colley KJ:
Glycosyltransferases. Structure, localization, and control of cell
type-specific glycosylation. J Biol Chem. 264:17615–17618.
1989.PubMed/NCBI
|
|
22.
|
El-Battari A, Prorok M, Angata K, Mathieu
S, Zerfaoui M, Ong E, Suzuki M, Lombardo D and Fukuda M: Different
glycosyltransferases are differentially processed for secretion,
dimerization, and autoglycosylation. Glycobiology. 13:941–953.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Kato T, Suzuki M, Murata T and Park EY:
The effects of N-glycosylation sites and the N-terminal region on
the biological function of beta1,3-N-acetylglucosaminyltransferase
2 and its secretion. Biochem Biophys Res Commun. 329:699–705. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Brockhausen I: Pathways of O-glycan
biosynthesis in cancer cells. Biochim Biophys Acta. 1473:67–95.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Yoshida-Moriguchi T, Yu L, Stalnaker SH,
Davis S, Kunz S, Madson M, Oldstone MB, Schachter H, Wells L and
Campbell KP: O-mannosyl phosphorylation of alpha-dystroglycan is
required for laminin binding. Science. 327:88–92. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Stevens E, Carss KJ, Cirak S, Foley AR,
Torelli S, Willer T, Tambunan DE, Yau S, Brodd L, Sewry CA, Feng L,
Haliloglu G, Orhan D, Dobyns WB, Enns GM, Manning M, Krause A,
Salih MA, Walsh CA, Hurles M, Campbell KP, Manzini MC; UK10K
Consortium; Stemple D, Lin YY and Muntoni F: Mutations in B3GALNT2
cause congenital muscular dystrophy and hypoglycosylation of
α-dystroglycan. Am J Hum Genet. 92:354–365. 2013.PubMed/NCBI
|
|
27.
|
Sgambato A, Migaldi M, Montanari M,
Camerini A, Brancaccio A, Rossi G, Cangiano R, Losasso C, Capelli
G, Trentini GP and Cittadini A: Dystroglycan expression is
frequently reduced in human breast and colon cancers and is
associated with tumor progression. Am J Pathol. 162:849–860. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Saito T, Miyoshi E, Sasai K, Nakano N,
Eguchi H, Honke K and Taniguchi N: A secreted type of
β1,6-N-acetylglucosaminyltransferase-V (GnT-V) induces tumor
angiogenesis without mediation of glycosylation. A novel function
of GnT-V distinct from the original glycosyltransferase activity. J
Biol Chem. 277:17002–17008. 2002.
|
|
29.
|
Toki D, Sarkar M, Yip B, Reck F, Joziasse
D, Fukuda M, Schachter H and Brockhausen I: Expression of stable
human O-glycan core 2 beta-1,6-N-acetylglucosaminyltransferase in
Sf9 insect cells. Biochem J. 325:63–69. 1997.PubMed/NCBI
|
|
30.
|
Barbier O, Girard C, Breton R, Bélanger A
and Hum DW: N-glycosylation and residue 96 are involved in the
functional properties of UDP-glucuronosyltransferase enzymes.
Biochemistry. 39:11540–11552. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Christensen LL, Bross P and Ørntoft TF:
Glycosylation of the N-terminal potential N-glycosylation sites in
the human alpha1,3-fucosyltransferase V and -VI (hFucTV and -VI).
Glycoconj J. 17:859–865. 2000. View Article : Google Scholar : PubMed/NCBI
|