Introduction
Pancreatic cancer (PC) is one of the most lethal
malignant diseases with a poor prognosis. PC is the fourth leading
cause of cancer-related deaths in Western countries and has the
lowest patient survival rate among all solid cancers. It has been
estimated that 43,920 people were newly diagnosed with PC in 2012
(1). Although great efforts have
been made in PC treatment using surgery, radiation therapy and
chemotherapy, the 5-year survival rate of PC patients is still
disappointing. Therefore, it is important to explore the molecular
mechanisms that regulate PC development in order to develop
effective therapies for PC.
Transmembrane-4-L-six-family-1 (TM4SF1), a small 22
kDa four-transmembrane-domain protein also known as L6-Ag, is a
surface protein highly expressed in human lung, breast, colon,
ovarian, renal and prostate carcinomas, and it is weakly expressed
in normal vascular endothelium (2). Due to its unique expression pattern,
TM4SF1 has attracted much attention as a therapeutic target for
monoclonal antibody-based cancer therapy (2). TM4SF1 belongs to a distinct family
that includes five other structurally similar proteins:
TM4SF4/IL-TMP, TM4SF5/L6H, TM4SF18/L6D, TM4SF19/OCTM4 and
TM4SF20/TCCE518 (3). TM4SF1 has
been shown to be associated with the growth, motility, invasion and
metastasis of tumor cells (4–7). In
particular, recent studies have shown that TM4SF1 is highly
expressed in PC tissues and cell lines and that downregulation of
TM4SF1 can decrease migration, invasion and chemoresistance of PC
cells in vitro; increase the effectiveness of gemcitabine
treatment; and inhibit tumor angiogenesis and metastasis in
orthotopic tumor models in vivo (8). However, the molecular mechanisms that
regulate TM4SF1 expression and function in PC remain unclear.
MicroRNAs (miRNAs) are a new class of endogenous,
non-coding and short (19–24 nucleotides) single-stranded RNAs.
miRNAs regulate gene expression by binding to the 3′-untranslated
region (UTR) of the target gene leading to either downregulation of
the mRNA transcript or inhibition of the protein translation
process (9). miRNAs can regulate
many cellular processes, such as apoptosis, cell cycle progression,
proliferation, differentiation, invasion and migration, and either
promote or inhibit tumorigenesis, depending on the genes they
target and their differential expression in normal and cancer
tissues (10–17).
hsa-miR-141, a member of the miR-200 family, is
overexpressed in ovarian cancer, colorectal cancer, thyroid
papillary carcinoma, pancreatic ductal adenocarcinoma,
nasopharyngeal carcinoma, prostate tumor, cholangiocarcinoma and
endo metrial carcinoma, but it is downregulated in gastric cancer,
renal cell carcinoma and breast cancer (18–28).
These results lead to a controversial issue regarding the function
of hsa-miR-141 in cancer progression.
In this study, we first investigated the correlation
between the expression level of TM4SF1 and miR-141 in PC cells.
Next, we demonstrated whether TM4SF1 is a direct target of miR-141.
In addition, we determined whether the miR-141 mimic could affect
invasion, migration, cell proliferation, cell cycle progression or
apoptosis of PC cells.
Materials and methods
Cell culture
The human PC cell lines SW1990, PANC-1, BxPC-3 and
CFPAC-1 cells were obtained from the Shanghai Cell Bank (Shanghai,
China) and cultured in Dulbecco’s modified Eagle’s medium (DMEM,
Wisent, St. Bruno, QC, Canada) supplemented with 10% fetal bovine
serum (FBS, Wisent), 2 mM glutamine, 100 μg/ml penicillin,
and 100 μg/ml streptomycin in a humidified chamber at 37°C
with 5% CO2.
miRNAs transfection
Inhibitor miR-141 (141I), the inhibitor negative
control (141I-NC), the miR-141 mimic (141M) and the mimic negative
control (141M-NC) were designed and synthesized by GenePharma
(Shanghai, China). SW1990 and BxPc-3 cells were seeded in 6-well
plates at a density of 50%, 24 h later the cells were transfected
with miRNAs using Lipofectamine 2000 (Invitrogen, Carlsbad, CA,
USA) following the manufacturer’s instructions.
Quantitative real-time polymerase chain
reaction (qRT-PCR)
Total RNA was extracted from SW1990 and BxPc-3 cells
by using TRIzol (Invitrogen). Primescript RT Reagent (Takara,
Dalian, China) was used to synthesize cDNA. qRT-PCR was performed
using SYBR-Green (Takara) on a 7500 Real-Time PCR System (Applied
Biosystems, Foster City, CA, USA). The primers were as follows:
TM4SF1 forward, 5′-ACCACTATG TCTTGATTCCCTC-3′; and reverse,
5′-ATTGTGGCTCTG TCCTGGGT-3′; GAPDH forward, 5′-TCACCCACACTGTG
CCCATCTACGA-3′; and reverse, 5′-CAGCGGAACCGC TCATTGCCAATGG-3′;
hsa-miR-141 forward, 5′-CGCTAA CACTGTCTGGTAAAG-3′; and reverse,
5′-GTGCAGGGT CCGAGGT-3′; U6 forward, 5′-ATTGGAACGATACAGAGA
AGATT-3′; and reverse, 5′-GGAACGCTTCACGAATTTG-3′. The conditions
were as follows: 95°C for 3 min; 35 cycles of 94°C for 30 sec, 60°C
for 30 sec and 72°C for 30 sec; and 72°C for 5 min. GAPDH mRNA and
U6 were used as internal controls for determining the relative
expression level of TM4SF1 mRNA and hsa-miR-141, respectively. The
comparative ΔΔCt method was used to calculate relative expression
levels of mRNAs and miRNAs, and the fold changes were analyzed by
2−ΔΔCt.
Western blot analysis
Total protein was extracted from SW1990 and BxPc-3
cells using RIPA buffer supplemented with 1% phenylmethylsulfonyl
fluoride (PMSF), and the protein concentration was estimated using
a BCA kit (Keygen, Nanjing, China). Protein was separated by 12%
sodium dodecyl sulfate polyacrylamide gel electrophoresis
(SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF)
membranes. The membranes were blocked in Tris-buffered saline (TBS)
with 5% non-fat dry milk at 4°C for 15 h then incubated with
primary antibodies against TM4SF1 (Abcam, Cambridge, MA, USA) or
GAPDH (Beyotime, Jiangsu, China) at 4°C overnight. Membranes were
incubated with anti-rabbit (or mouse) IgG-horseradish
peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology,
Santa Cruz, CA, USA) for 2 h at room temperature, washed three
times, developed with an electrochemiluminescence kit (Pierce,
Rockford, IL, USA), and exposed to X-ray film to visualize the
images. GAPDH served as a loading control.
Luciferase reporter assay
Four oligonucleotides corresponding to the 3′UTR of
TM4SF1 were synthesized as follows: wild-type forward,
5′-CTAGATAAAGACTGGCATCTTCACAGGAT GTCAGTGTTTAAATTTAGTAGGCCGG-3′; and
reverse, 5′-CCTACTAAATTTAAACACTGACATCCTGTGAAGAT GCCAGTCTTTAT-3′;
and mutant-type forward, 5′-CTAGAT
AAAGACTGGCATCTTCACAGGATGTTGATGCTTAAA TTTAGTAGGCCGG-3′; and reverse,
5′-CCTACTAAATTT AAGCATCAACATCCTGTGAAGATGCCAGTCTTTAT-3′. The
oligonucleotides were cloned into the Xbal site of the pGL3
luciferase reporter gene (Promega, Madison, WI, USA) to generate
pGL3-TM4SF1-3′UTR and pGL3-TM4SF1-3′UTR-mut vector. SW1990 and
BxPc-3 cells were cultured in 24-well plates and co-transfected
with 200 ng of pGL3-TM4SF1 or pGL3-TM4SF1-mut and 20 ng of pRL-SV40
(Promega) containing Renilla luciferase and 20 pmol of 141M
or 141NC. At 48 h after transfection, cells were collected and
luciferase activity was measured by using a Dual-Luciferase
Reporter assay kit (Promega) following the manufacturer’s
instructions. All experiments were repeated three times.
Cell invasion and migration assay
Invasion and migration of cells were measured by a
Matrigel invasion chamber assay, using a chamber of 6.5 mm in
diameter with 8-μm pore size Transwell chambers (Corning,
Corning, NY, USA). SW1990 and BxPc-3 cells were seeded into the
upper chamber (1.0×105 cells per Transwell) pre-coated
with 1 mg/ml Matrigel for the invasion assay or without Matrigel
for the migration assay, and the bottom wells were filled with 500
μl of 10% FBS-DMEM. After incubation for 24 h at 37°C,
non-invading or non-migrating cells were removed with cotton swabs,
and cells that had invaded or migrated to the underside of the
membrane were stained with 0.1% crystal violet for 15 min at 37°C.
Then, the membranes were washed with phosphate-buffered saline
(PBS), and the invaded or migrated cells were counted under an
inverted microscope. All experiments were carried out in
triplicate.
Cell proliferation assay
SW1990 and BxPc-3 cells were seeded in 96-well
culture plates (Costar, Cambridge, UK) at a density of
2×103 cells/well, 24 h later the cells were transfected
with miRNAs, and 48 h later cell proliferation was detected by
using an MTT kit (Keygen) following the manufacturer’s instructions
daily for 5 days. Briefly, 20 μl of MTT solution (5 mg/ml)
was added to each well, cells were incubated for 4 h at 37°C, then
the medium in each well was replaced with 150 μl of dimethyl
sulfoxide (DMSO), and the plate was oscillated for 10 min. The
optical density (OD) was detected by a microplate reader (Tecan,
Shanghai, China) at 490 nm with 650 nm as a reference wavelength.
Each assay was performed in triplicate and repeated independently
three times.
Flow cytometry analysis of cell cycle
progression and apoptosis
Cell cycle progression and apoptosis were assessed
by flow cytometry (Becton-Dickinson, San Jose, CA, USA). SW1990 and
BxPc-3 cells were grown and transfected with miRNAs. For cell cycle
analysis, cells were collected and washed twice with PBS and fixed
with 70% ethanol at −20°C overnight. Cells were washed twice with
PBS and resuspended in 500 μl of PBS with 0.2% Triton X-100,
10 mM EDTA, 100 μg/ml RNase A, and 50 μg/ml propidium
iodide (PI). The samples were incubated at room temperature for 30
min. For apoptosis, cells were collected and washed twice with PBS,
suspended in 100 μl of 1X binding buffer, and stained with 5
μl of Annexin V fluorescein isothiocyanate (FITC) and 5
μl of PI at room temperature for 15 min in the dark. Then,
the samples were analyzed by flow cytometry (Becton-Dickinson). All
experiments were carried out in triplicate.
Statistical analysis
Each experiment was conducted at least three times.
All data were expressed as mean ± standard deviation (SD).
Differences between each group were analyzed by a Student’s t-test.
Statistical analysis was performed with SPSS software (version
16.0, SPSS Inc., Chicago, IL, USA). P<0.05 was considered
statistically significant.
Results
The TM4SF1 protein level negatively
correlates with the hsa-miR-141 level in PC cell lines
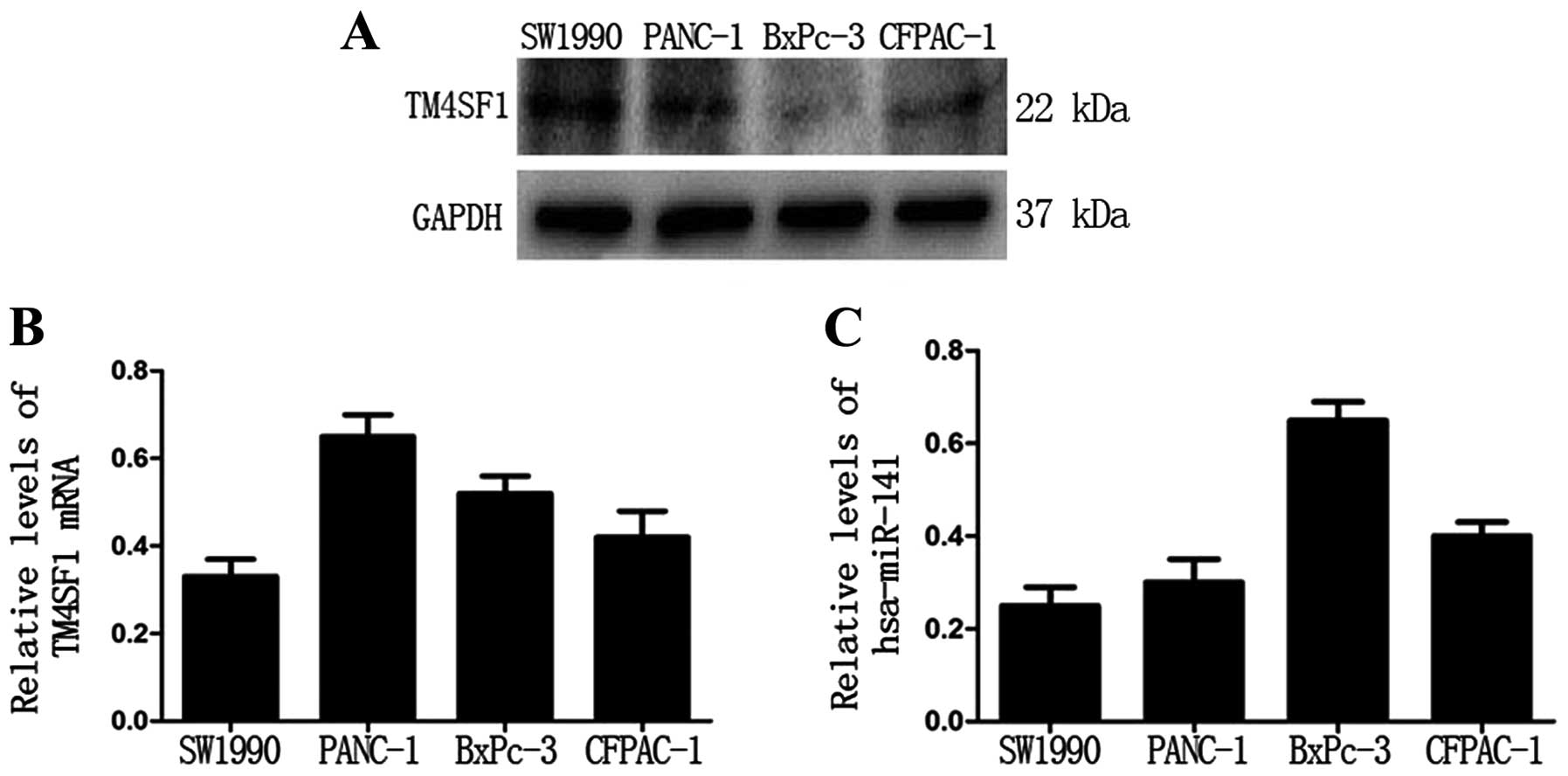
First, we examined the protein level of TM4SF1 in
four PC cell lines (SW1990, PANC-1, BxPC-3 and CFPAC-1). Western
blot analysis showed that the protein level of TM4SF1 was the
highest in SW1990 cells and the lowest in BxPC-3 cells (Fig. 1A). However, qRT-PCR analysis showed
that there was no significant relationship between TM4SF1 protein
and mRNA levels (Fig. 1B).
Notably, qRT-PCR analysis showed that the expression level of
miR-141 was the highest in BxPc-3 cells and the lowest in SW1990
cells (Fig. 1C). These data
suggest that the TM4SF1 protein level is negatively correlated with
the hsa-miR-141 level in PC cells.
TM4SF1 is a target of hsa-miR-141 in PC
cells
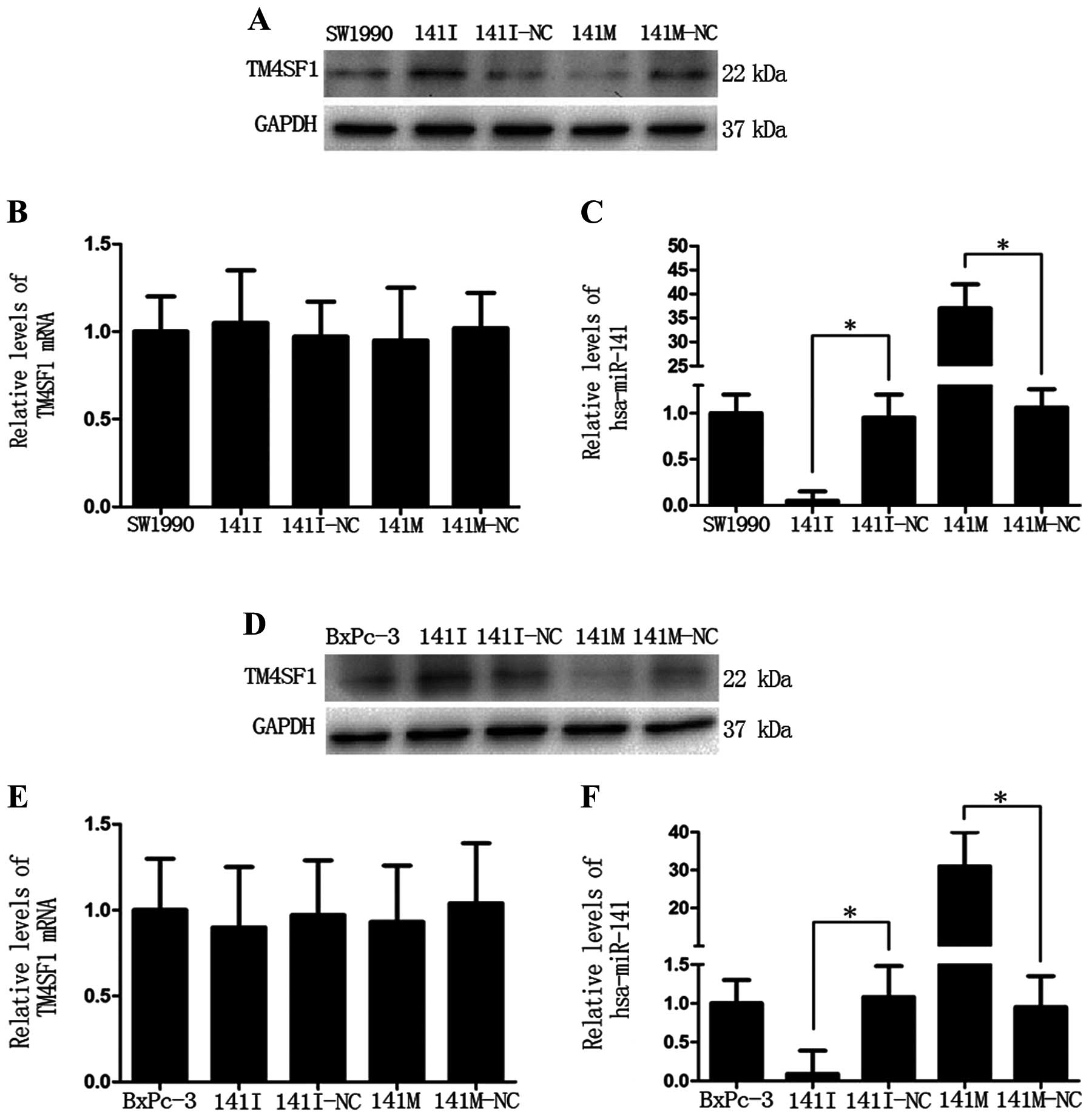
Next, we detected protein and mRNA expression levels
of TM4SF1 in SW1990 and BxPc-3 cells transfected with miR-141 mimic
(141M), miR-141 inhibitor (141I), or the corresponding negative
control (141M-NC or 141I-NC). Western blot analysis showed that the
TM4SF1 protein level was lower in the 141M group and higher in the
141I group (Fig. 2A and D),
compared to the negative controls, respectively. In addition, the
TM4SF1 protein level negatively correlated with the hsa-miR-141
level (*P<0.05, Fig. 2C
and F). qRT-PCR analysis showed that there was no obvious
change in the TM4SF1 mRNA level (Fig.
2B and E). These data indicate that miR-141
post-transcriptionally inhibits TM4SF1 expression.
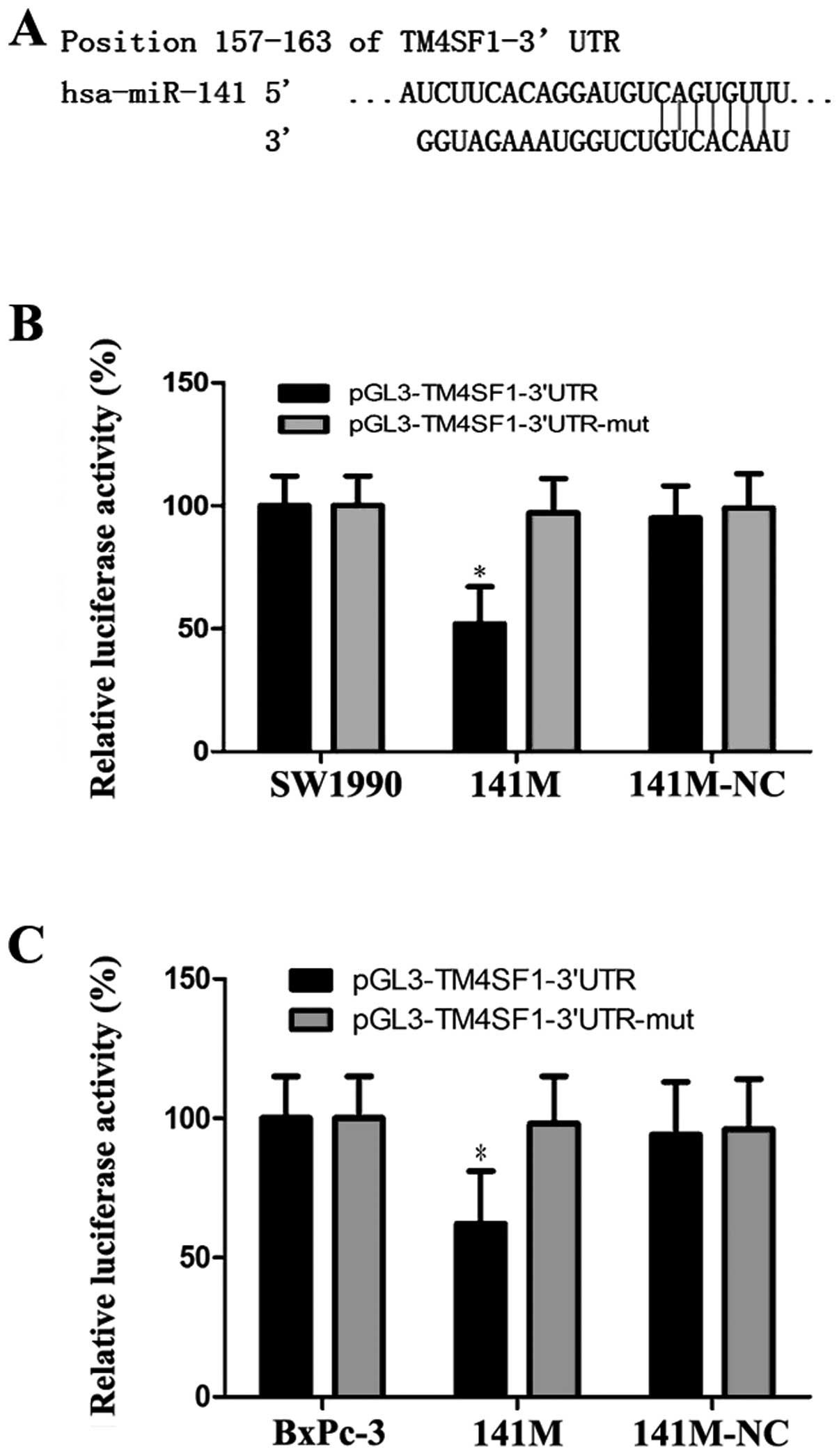
To confirm that hsa-miR-141 directly targets the
3′UTR of the TM4SF1 gene, we used TargetScan to predict the 3′UTR
of TM4SF1 and the binding site of miR-141 (Fig. 3A). Based on this program, we
generated pGL3-TM4SF1 and pGL3-TM4SF1-mut vectors as the luciferase
reporter and control, respectively, and transfected them into
SW1990 and BxPc-3 cells, together with 141M or 141M-NC. The
luciferase assay showed that luciferase activity was approximately
48% and 43% less in the 141M group compared with the control
(*P<0.05, Fig. 3B and
C). These results suggest that miR-141 directly targets TM4SF1
via the binding site in its 3′UTR region.
hsa-miR-141 inhibits invasion and
migration of PC cells in vitro
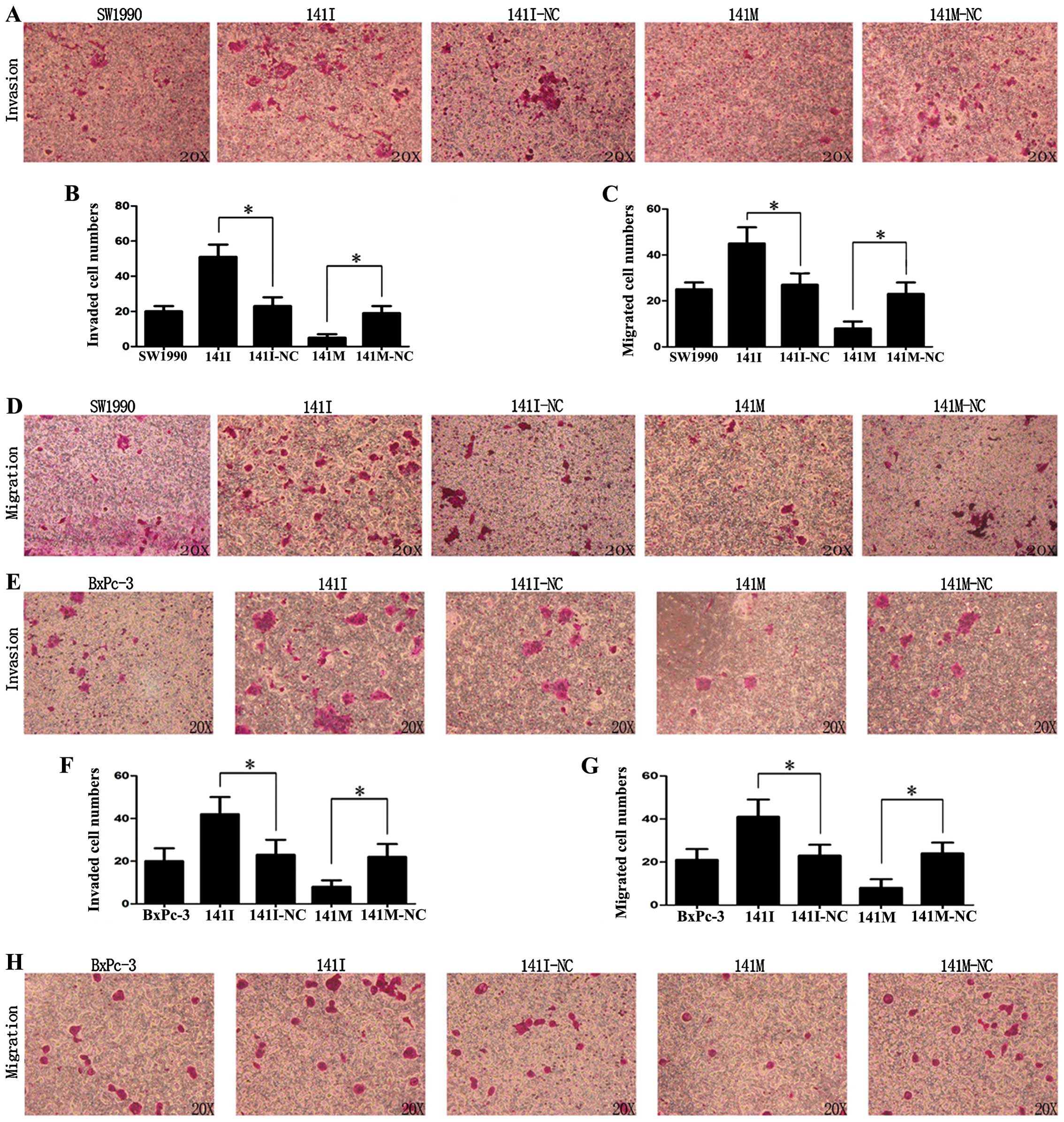
Matrigel invasion and Transwell assays were used to
detect the effects of hsa-miR-141 on the invasion and migration of
SW1990 and BxPc-3 cells in vitro. As shown in Fig. 4A, B, E and F, transfection with
141I could promote invasion, while 141M inhibited invasion of
SW1990 and BxPc-3 cells (P<0.05). Similarly, as shown in
Fig. 4C, D, G and H, transfection
with 141I could promote migration, while 141M could inhibit
migration of SW1990 and BxPc-3 cells (P<0.05). These data
suggest that hsa-miR-141 was able to inhibit invasion and migration
of PC cells in vitro.
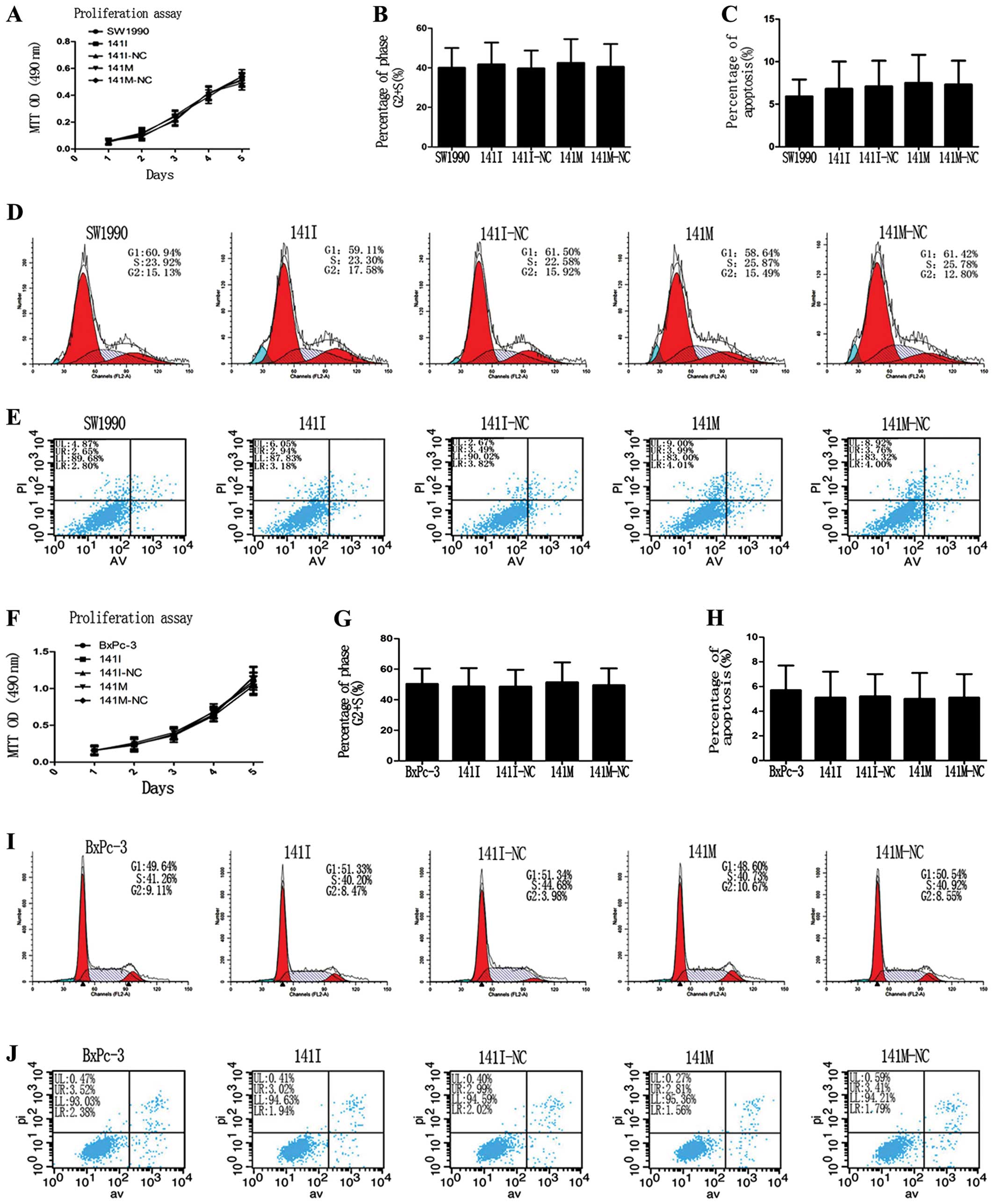
miR-141 has no effects on proliferation,
cell cycle progression, and apoptosis of PC cells in vitro
To clarify whether hsa-miR-141 could affect cell
proliferation, cell cycle progression, or apoptosis in SW1990 and
BxPc-3 cells, we performed an MTT assay and found that transfection
of 141M or 141I caused no significant difference in cell
proliferation in each group (Fig. 5A
and F). Flow cytometry analysis of cell cycle progression and
apoptosis showed that the percentage of cells in the S+G2 phase and
the total apoptosis rate were not significantly different in each
group (Fig. 5D, I, E and J).
Discussion
TM4SF1 is a member of the tetraspanin superfamily
and was first described as a tumor-specific antigen in many human
epithelial malignancies such as lung, breast, colon, ovarian, renal
and prostate carcinomas (2,4).
Janes and Watt found that TM4SF1 could interact with integrin
family members to form transmembrane complexes that affect cell
adhesion, migration and tumor metastasis (29). TM4SF1 is overexpressed in the
endothelium of human cancers, and it has been proposed that TM4SF1
acts as a ‘molecular organizer’ to facilitate the gathering of
specific cell surface proteins and the formation and stability of
functional signaling complexes in tumor angiogenesis (30,31).
The specific mechanism by which TM4SF1 is
overexpressed in epithelial tumors remains unclear. By using
TargetScan, we predicted that hsa-miR-141 could target TM4SF1 and
regulate its expression. First, we detected the levels of TM4SF1
and hsa-miR-141 in four PC cell lines and found that TM4SF1 protein
levels negatively correlated with hsa-miR-141 levels in different
PC cell lines. Furthermore, western blot analysis showed that the
TM4SF1 protein level was lower in the miR-141 mimic group and
higher in the miR-141 inhibitor group, compared to the negative
controls, respectively. Importantly, in these cells the TM4SF1
protein level but not the TM4SF1 mRNA level negatively correlated
with the hsa-miR-141 level. These data suggest that hsa-miR-141
down-regulates TM4SF1 expression at the post-transcriptional level.
Next, we performed a luciferase assay and provided evidence that
TM4SF1 is a direct target gene of hsa-miR-141.
hsa-miR-141 belongs to the miR-200 family, which
consists of the following members: miR-141, miR-200a, miR-200b,
miR-200c and miR-429 (32).
Overexpression of hsa-miR-141 has been shown to inhibit invasion
and migration of breast cancer, colorectal cancer and pancreatic
cancer (33–35). Consistent with these previous
studies, in this study we employed Matrigel invasion and Transwell
migration assays to demonstrate that the miR-141 mimic resulted in
a significant decrease of cell invasion and migration, while the
miR-141 inhibitor led to a significant increase of cell invasion
and migration. Given the crucial role of the cell surface protein
TM4SF1 in tumor invasion and metastasis (29), it is reasonable to expect that
miR-141 could directly target and downregulate the expression of
TM4SF1, leading to loss of oncogenic function of TM4SF1. Our
findings satisfactorily explain the downregulation of miR-141 and
overexpression of TM4SF1 in PC and support current opinions that
TM4SF1 is an oncoprotein and that miR-141 is a tumor-suppressive
miRNA.
Notably, the effects of hsa-miR-141 on cancer cell
proliferation have been studied, but the role of hsa-miR-141 in
cell proliferation of different cancers is controversial. The
miR-200 family has been reported to be overexpressed in pancreatic
ductal adenocarcinoma (PDAC) cells and enhance cell proliferation
(21). Similar results have been
reported in cholangiocarcinoma, ovarian carcinoma and
choriocarcinoma (24,36,37).
However, overexpression of hsa-miR-141 can significantly inhibit
the proliferation of gastric cancer cells (26). In this study, we performed an MTT
assay and flow cytometry analysis and found that miR-141 had no
obvious effects on cell proliferation, cell cycle progression, or
apoptosis in our experimental settings. SW1990 and BxPC-3 cells are
derived from pancreatic adenocarcinoma, while CFPAC and PANC-1
cells originate from PDAC. The different sources of PC cells may
lead to different results with regard to the role of hsa-miR-141 in
the regulation of cell proliferation and apoptosis. Further studies
that employ a variety of different PC cell lines and in vivo
xenograft mouse models will help clarify the controversial
results.
In conclusion, in this study we showed that the
miR-141 level negatively correlated with TM4SF1 protein in PC
cells. By using gain and loss of function approaches, we
demonstrated that miR-141 downregulated TM4SF1 expression to
inhibit the metastatic potential of PC cells but had no effects on
cell proliferation, cell cycle progression or apoptosis.
Furthermore, for the first time, we identified TM4SF1 as a direct
target of miR-141. Our findings that TM4SF1 expression is inhibited
by miR-141 provide new insight into the oncogenic function
mechanism of TM4SF1 and suggest that miR-141 represents a novel
molecular target for PC therapy.
Acknowledgements
This study was supported by the
National Nature Science Foundation of China (Nos. 81170336 and
81272239) and the Research Special Fund for Public Welfare Industry
of Health of China (201202007). We thank Medjaden Bioscience
Limited for assisting in the preparation of this manuscript.
References
|
1.
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar
|
|
2.
|
Wright MD, Ni J and Rudy GB: The L6
membrane proteins - a new four-transmembrane superfamily. Protein
Sci. 9:1594–1600. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Allioli N, Vincent S, Vlaeminck-Guillem V,
Decaussin-Petrucci M, Ragage F, Ruffion A and Samarut J: TM4SF1, a
novel primary androgen receptor target gene over-expressed in human
prostate cancer and involved in cell migration. Prostate.
71:1239–1250. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Hellstrom I, Horn D, Linsley P, Brown JP,
Brankovan V and Hellstrom KE: Monoclonal mouse antibodies raised
against human lung carcinoma. Cancer Res. 46:3917–3923.
1986.PubMed/NCBI
|
|
5.
|
Chang YW, Chen SC, Cheng EC, Ko YP, Lin
YC, Kao YR, Tsay YG, Yang PC, Wu CW and Roffler SR: CD13
(amino-peptidase N) can associate with tumor-associated antigen L6
and enhance the motility of human lung cancer cells. Int J Cancer.
116:243–252. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Lekishvili T, Fromm E, Mujoomdar M and
Berditchevski F: The tumour-associated antigen L6 (L6-Ag) is
recruited to the tetraspanin-enriched microdomains: implication for
tumour cell motility. J Cell Sci. 121:685–694. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Kao YR, Shih JY, Wen WC, Ko YP, Chen BM,
Chan YL, Chu YW, Yang PC, Wu CW and Roffler SR: Tumor-associated
antigen L6 and the invasion of human lung cancer cells. Clin Cancer
Res. 9:2807–2816. 2003.PubMed/NCBI
|
|
8.
|
Cao J, Ramachandran V, Arumugam T, Nast F
and Logsdon C: TM4SF1 stimulates pancreatic cancer cell migration
and invasion. Pancreas. 38:9862009.
|
|
9.
|
Ying SY, Chang DC and Lin SL: The microRNA
(miRNA): overview of the RNA genes that modulate gene function. Mol
Biotechnol. 38:257–268. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Wang F, Xue X, Wei J, An Y, Yao J, Cai H,
Wu J, Dai C, Qian Z, Xu Z and Miao Y: hsa-miR-520h downregulates
ABCG2 in pancreatic cancer cells to inhibit migration, invasion,
and side populations. Br J Cancer. 103:567–574. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Aguda BD, Kim Y, Piper-Hunter MG, Friedman
A and Marsh CB: MicroRNA regulation of a cancer network:
consequences of the feedback loops involving miR-17-92, E2F, and
Myc. Proc Natl Acad Sci USA. 105:19678–19683. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Grady WM, Parkin RK, Mitchell PS, Lee JH,
Kim YH, Tsuchiya KD, Washington MK, Paraskeva C, Willson JK, Kaz
AM, Kroh EM, Allen A, Fritz BR, Markowitz SD and Tewari M:
Epigenetic silencing of the intronic microRNA hsa-miR-342 and its
host gene EVL in colorectal cancer. Oncogene. 27:3880–3888. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Furuta M, Kozaki KI, Tanaka S, Arii S,
Imoto I and Inazawa J: miR-124 and miR-203 are epigenetically
silenced tumor-suppressive microRNAs in hepatocellular carcinoma.
Carcinogenesis. 31:766–776. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Gandellini P, Folini M, Longoni N, Pennati
M, Binda M, Colecchia M, Salvioni R, Supino R, Moretti R, Limonta
P, Valdagni R, Daidone MG and Zaffaroni N: miR-205 exerts
tumor-suppressive functions in human prostate through
down-regulation of protein kinase Cepsilon. Cancer Res.
69:2287–2295. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Hoffman AE, Zheng T, Yi C, Leaderer D,
Weidhaas J, Slack F, Zhang Y, Paranjape T and Zhu Y: microRNA
miR-196a-2 and breast cancer: a genetic and epigenetic association
study and functional analysis. Cancer Res. 69:5970–5977. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Lee KH, Chen YL, Yeh SD, Hsiao M, Lin JT,
Goan YG and Lu PJ: MicroRNA-330 acts as tumor suppressor and
induces apoptosis of prostate cancer cells through E2F1-mediated
suppression of Akt phosphorylation. Oncogene. 28:3360–3370. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Aqeilan RI, Calin GA and Croce CM: miR-15a
and miR-16-1 in cancer: discovery, function and future
perspectives. Cell Death Differ. 17:215–220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Iorio MV, Visone R, Di Leva G, Donati V,
Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H,
Calin GA, Menard S and Croce CM: MicroRNA signatures in human
ovarian cancer. Cancer Res. 67:8699–8707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Bandres E, Cubedo E, Agirre X, Malumbres
R, Zarate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzo M and
Garcia-Foncillas J: Identification by Real-time PCR of 13 mature
microRNAs differentially expressed in colorectal cancer and
non-tumoral tissues. Mol Cancer. 5:292006. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Pallante P, Visone R, Ferracin M, Ferraro
A, Berlingieri MT, Troncone G, Chiappetta G, Liu CG, Santoro M,
Negrini M, Croce CM and Fusco A: MicroRNA deregulation in human
thyroid papillary carcinomas. Endocr Relat Cancer. 13:497–508.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Kent OA, Mullendore M, Wentzel EA,
Lopez-Romero P, Tan AC, Alvarez H, West K, Ochs MF, Hidalgo M,
Arking DE, Maitra A and Mendell JT: A resource for analysis of
microRNA expression and function in pancreatic ductal
adenocarcinoma cells. Cancer Biol Ther. 8:2013–2024.
2009.PubMed/NCBI
|
|
22.
|
Zhang L, Deng T, Li X, Liu H, Zhou H, Ma
J, Wu M, Zhou M, Shen S, Niu Z, Zhang W, Shi L, Xiang B, Lu J, Wang
L, Li D, Tang H and Li G: microRNA-141 is involved in a
nasopharyngeal carcinoma-related genes network. Carcinogenesis.
31:559–566. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Amaral FC, Torres N, Saggioro F, Neder L,
Machado HR, Silva WA Jr, Moreira AC and Castro M: MicroRNAs
differentially expressed in ACTH-secreting pituitary tumors. J Clin
Endocrinol Metab. 94:320–323. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Meng F, Henson R, Lang M, Wehbe H,
Maheshwari S, Mendell JT, Jiang J, Schmittgen TD and Patel T:
Involvement of human micro-RNA in growth and response to
chemotherapy in human cholangiocarcinoma cell lines.
Gastroenterology. 130:2113–2129. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Snowdon J, Zhang X, Childs T, Tron VA and
Feilotter H: The microRNA-200 family is upregulated in endometrial
carcinoma. PLoS One. 6:e228282011. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Du Y, Xu Y, Ding L, Yao H, Yu H, Zhou T
and Si J: Down-regulation of miR-141 in gastric cancer and its
involvement in cell growth. J Gastroenterol. 44:556–561. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Nakada C, Matsuura K, Tsukamoto Y,
Tanigawa M, Yoshimoto T, Narimatsu T, Nguyen LT, Hijiya N, Uchida
T, Sato F, Mimata H, Seto M and Moriyama M: Genome-wide microRNA
expression profiling in renal cell carcinoma: significant
down-regulation of miR-141 and miR-200c. J Pathol. 216:418–427.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Gregory PA, Bert AG, Paterson EL, Barry
SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y and Goodall GJ:
The miR-200 family and miR-205 regulate epithelial to mesenchymal
transition by targeting ZEB1 and SIP1. Nat Cell Biol. 10:593–601.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Janes SM and Watt FM: New roles for
integrins in squamous-cell carcinoma. Nat Rev Cancer. 6:175–183.
2006. View
Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Shih SC, Zukauskas A, Li D, Liu G, Ang LH,
Nagy JA, Brown LF and Dvorak HF: The L6 protein TM4SF1 is critical
for endothelial cell function and tumor angiogenesis. Cancer Res.
69:3272–3277. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Maecker HT, Todd SC and Levy S: The
tetraspanin superfamily: molecular facilitators. FASEB J.
11:428–442. 1997.PubMed/NCBI
|
|
32.
|
Baffa R, Fassan M, Volinia S, O’Hara B,
Liu CG, Palazzo JP, Gardiman M, Rugge M, Gomella LG, Croce CM and
Rosenberg A: MicroRNA expression profiling of human metastatic
cancers identifies cancer gene targets. J Pathol. 219:214–221.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Neves R, Scheel C, Weinhold S, Honisch E,
Iwaniuk KM, Trompeter HI, Niederacher D, Wernet P, Santourlidis S
and Uhrberg M: Role of DNA methylation in miR-200c/141 cluster
silencing in invasive breast cancer cells. BMC Res Notes.
3:2192010. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Hu M, Xia M, Chen X, Lin Z, Xu Y, Ma Y and
Su L: MicroRNA-141 regulates Smad interacting protein 1 (SIP1) and
inhibits migration and invasion of colorectal cancer cells. Dig Dis
Sci. 55:2365–2372. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Burk U, Schubert J, Wellner U, Schmalhofer
O, Vincan E, Spaderna S and Brabletz T: A reciprocal repression
between ZEB1 and members of the miR-200 family promotes EMT and
invasion in cancer cells. EMBO Rep. 9:582–589. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Mateescu B, Batista L, Cardon M, Gruosso
T, de Feraudy Y, Mariani O, Nicolas A, Meyniel JP, Cottu P,
Sastre-Garau X and Mechta-Grigoriou F: miR-141 and miR-200a act on
ovarian tumorigenesis by controlling oxidative stress response. Nat
Med. 17:1627–1635. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Morales-Prieto DM, Schleussner E and
Markert UR: Reduction in miR-141 is induced by leukemia inhibitory
factor and inhibits proliferation in choriocarcinoma cell line
JEG-3. Am J Reprod Immunol. 66(Suppl 1): 57–62. 2011. View Article : Google Scholar : PubMed/NCBI
|