Introduction
Medicinal plants have proven to be a rich source of
bioactive compounds for therapeutic agents, and currently 75% of
prescribed drugs worldwide are derived from plant sources (1). Feroniellin A (FERO), a novel
furanocoumarin is isolated from the roots of Feroniella
lucida. The chemical structure of furanocoumarins consists of a
furan ring fused with coumarin, which is present in many plants.
Coumarins possess anticoagulant, antimicrobial, antioxidant,
anti-inflammatory, anti-allergic and anticancer properties
(2). Some furanocoumarin
derivatives isolated from plants show anticancer activity by
functioning as topoisomerase I inhibitors (3), efflux transport inhibitors or drug
metabolism inhibitors (4,5). However, the specific molecular
mechanisms by which FERO shows anticancer activity have not been
revealed.
Multidrug resistance (MDR) is a major problem in
cancer therapy and is often the result of overexpression of the
drug efflux protein P-glycoprotein (P-gp). P-gp is a 170-kDa
protein that belongs to the ATP-binding cassette superfamily of
membrane transporter proteins (6,7).
P-gp is an energy-dependent drug efflux pump that maintains
intracellular drug concentrations below cytotoxic levels, thereby
decreasing the cytotoxic effects of a variety of chemotherapeutic
agents (6–9). P-gp also plays a role in inhibition
of drug accumulation and caspase activation in MDR tumors (10,11).
Of special note, recent lines of evidence have shown that NF-κB- or
SIRT1-mediated regulation of P-gp plays a critical role in
anticancer drug resistance (12,13).
Autophagy, an ancient system necessary to maintain
homeostasis in eukaryotic cells, degrades long-lived cytoplasmic
proteins and organelles and provides nutrients during starvation or
stress conditions (14) through
programmed processing involving the sequential activity of
autophagy-related gene (ATG) products. As autophagy is necessary
for cellular homeostasis, it is involved in biological processes
including development, aging and degeneration (15). However, aberrant regulation of
autophagy is related to many diseases, including cancer and
neurodegenerative disorders (16).
As a specific example, the first report connecting autophagy to
cancer showed that allelic loss of the essential autophagy gene
Beclin-1 (BECN1) is prevalent in human breast, ovarian, and
prostate cancers (17), and that
Becn1+/− mice develop mammary gland hyperplasia
and lymphomas as well as lung and liver tumors (18). Subsequent studies demonstrated that
ATG5−/− and ATG7−/− livers give
rise to adenomas (19). These
lines of evidence suggest that autophagy acts as a tumor suppressor
in cancer development. Contrary to this, many other reports have
shown that autophagy exerts a pro-survival function in tumor cells
(20–22). Additional studies have demonstrated
that the inhibition of autophagy by pharmaceutical drugs sensitized
cells to apoptotic cell death, and that combination therapies using
autophagy inhibitors plus chemotherapy led to faster tumor cell
death than did chemotherapy alone (23). These findings indicate that
pro-survival autophagy may represent a major hindrance to
successful cancer therapy.
The present study was initiated to screen small
molecules derived from plants grown in Thailand in order to reverse
MDR in A549RT-eto cells. We have identified that FERO induces
autophagy, which is necessary for FERO-induced apoptosis in
A549RT-eto cells. Therefore, we propose that FERO represents a
powerful candidate for the treatment of multidrug-resistant lung
cancer.
Materials and methods
Cell cultures
A549RT-eto cells were developed and kindly provided
by the Laboratory of Biochemistry, Chulabhorn Research Institute,
Thailand and have been described elsewhere (24). A549RT-eto cells were cultured in
RPMI-1640 medium (Gibco, Grand Island, NY, USA) supplemented with
10% FBS and 1% penicillin, and streptomycin (Gibco) at 37°C in a
humidified atmosphere of 5% CO2 in air.
Antibodies and reagents
For immunoblotting, antibodies against ATG5, LC3B
(D11), mTOR, phospho-mTOR (Ser2448) and cleaved PARP (Asp214) were
acquired from Cell Signaling Biotechnology (Beverly, MA, USA).
Anti-P-gp (Calbiochem; San Diego, CA, USA) and β-actin (C4), BECN1
(H300), BID (FL-195), caspase-9 p35 (H-170), NF-κB p65 (F-6), P53
(DO-1), SIRT1 (H-300) and Sp1 (1C6) antibodies were purchased from
Santa Cruz Biotechnology (Santa Cruz, CA, USA). Rapamycin, Z-VAD
and BAY11-7082 were purchased from Sigma-Aldrich (St. Louis, MO,
USA). Feroniellin A was isolated from the roots of Feroniella
lucida and was kindly provided by the Natural Products Research
Unit, Department of Chemistry, Faculty of Science, Chulalongkorn
University, Thailand and have been described elsewhere (25).
Immunoblotting
Cells were harvested and lysed with lysis buffer
[150 mM NaCl, 1% NP-40, 50 mM Tris-HCl (pH 7.5)] containing 0.1 mM
Na2VO3, 1 mM NaF and protease inhibitors
(Sigma). For immunoblotting, proteins from whole cell lysates were
resolved by 10 or 12% SDS-polyacrylamide gel electrophoresis (PAGE)
and then transferred to nitrocellulose membranes. Primary
antibodies were used at 1:1,000 or 1:2,000 dilutions, and secondary
antibodies conjugated with horseradish peroxidase were used at
1:2,000 dilutions in 5% non-fat dry milk. After the final washing,
nitrocellulose membranes were exposed using enhanced
chemiluminescence assay and visualized on a LAS 4000 mini (Fuji,
Tokyo, Japan).
Nuclear NF-κB pull-down assay
A549RT-eto cells (1×106 cells/ml) were
incubated with FERO or DMSO as a control for 12 h and nuclear
extracts were prepared. Cells were pelleted and resuspended in 0.4
ml hypotonic lysis buffer [20 mM HEPES (pH 7.9), 10 mM KCl, 1 mM
EDTA, 0.2% Triton X-100, and 1 mM Na2VO3 plus
protease inhibitors] and kept on ice for 20 min. After
centrifugation at 14 000 g for 5 min at 4°C, the nuclear pellet was
extracted with 0.1 ml hypertonic lysis buffer on ice for a further
20 min. After centrifugation at 14,000 g for 5 min at 4°C, the
supernatants were diluted to 100 mM NaCl and incubated with 25
μl of agarose beads conjugated to a consensus NF-κB binding
oligonucleotide (Santa Cruz Biotech) for 1 h at 4°C. After 3
washes, sample buffer was added and boiled for 5 min. The binding
NF-κB (p65) protein to the oligonucleotide conjugated with agarose
was detected by immunoblotting using an anti-p65 NF-κB Ab (Santa
Cruz Biotech).
Small interfering RNA (siRNA)
transfection
Cells were trypsinized and incubated overnight to
achieve 60–70% confluency before siRNA transfection.
Beclin-1 human siRNA (pre-made at Bioneer, Daejeon, Korea;
100 nM; sense: 5′-UGG AAU GGA AUG AGA UUA A(dTdT)-3′; antisense:
5′-UUA AUC UCA UUC CAU UCC A(dTdT)-3′ or negative control siRNA
(Bioneer); 100 nM; sense: 5′-CCU ACG CCA CCA UUU CGU (dTdT)-3′;
antisense: 5′-ACG AAA UUG GUG GCG UAG G (dTdT)-3′ were mixed with
Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). The cells were
incubated with the transfection mixture for 24 h and then rinsed
with RPMI-1640 medium containing 10% fetal bovine serum. The cells
were incubated for 48 h before harvest.
Reverse-transcription polymerase chain
reaction (RT-PCR)
Total RNA was extracted from cells using the RNeasy
mini kit (Qiagen, Valencia, CA, USA) in accordance with the
manufacturer’s instructions. Three micrograms of total RNA was
converted to cDNA using Superscript II reverse transcriptase
(Invitrogen), and PCR was performed using the specific primers:
human MDR1, sense: 5′-CCC ATC ATT GCA ATA GCA GG-3′ and
antisense: 5′-GTT CAA ACT TCT GCT CCT GA-3′; MRP2, sense:
5′-ACA GAG GCT GGT GGC AAC C-3′ and antisense: 5′-ACC ATT ACC TTG
TCA CTG TCC-3′; BCRP, sense: 5′-GAT CAC AGT CTT CAA GGA GAT
C-3′ and antisense: 5′-CAG TCC CAG TAC GAC TGT GAC A-3′ were used.
The cDNAs from each sample were diluted, and PCR was run at an
optimized cycle number. β-actin mRNA was measured as an
internal standard. After amplification, the products were subjected
to electrophoresis on 2.0% agarose gels and detected by ethidium
bromide staining.
Luciferase reporter assay
A549RT-eto cells were transfected with
hMDR1-luciferase or control pGL3 empty luciferase vector. To
normalize transfection efficiency, a pGK-β-gal vector that
expresses galactosidase from a phosphoglucokinase promoter was
included in the transfection mixture. FERO was added to the
transfected cells at 12 h before harvest. At 48 h
post-transfection, cells were washed with cold PBS and lysed in
lysis solution [25 mM Tris (pH 7.8), 2 mM EDTA, 2 mM DTT, 10%
glycerol and 1% Triton X-100]. Luciferase activity was measured
with a luminometer using a luciferase kit (Promega, Madison, WI,
USA).
Cell viability assay
The
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay was performed to measure cell survival as described
previously (26). Dye solution
containing tetrazolium was added to cells in a 96-well plate format
and incubated for 2 h. The absorbance of the formazan produced by
living cells was measured at 570 nm. The relative percentage of
cell survival was calculated by the mean absorbance of the treated
cells (ODT) and the mean absorbance of control cells
(ODC) following the formula: % Cell survival =
(ODT / ODC).
Statistical analysis
Data are presented as a mean ± standard deviation
(SD). Student’s t-test was used for statistical analysis, with
significance defined as a p-value <0.05.
Results
Feroniellin A induces apoptosis in human
A549 lung cells resistant to etoposide
Previous observations have revealed that human
etoposide resistant A549 lung cancer cells (A549RT-eto) exhibit
upregulation of Stat1 and HDAC4, leading to elevated levels of
P-glycoprotein (P-gp) encoded by multidrug resistance (MDR)
1 (unpublished data). Thus, we performed a screen for small
molecules derived from medicinal plants in Thailand that reverse
MDR. We found that a small compound known as Feroniellin A (FERO;
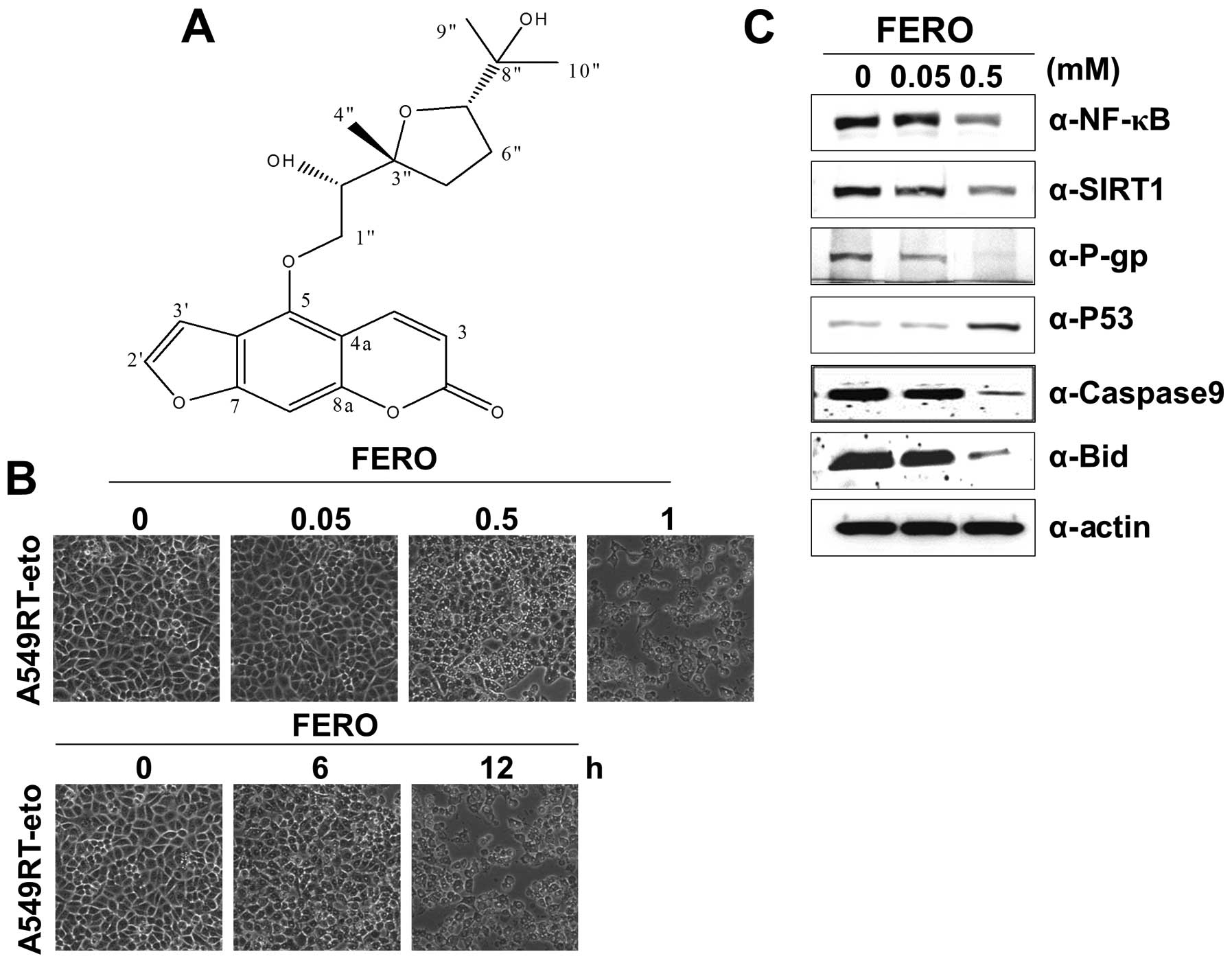
molecular weight = 388) (molecular structure shown in Fig. 1A) that efficiently induced death in
A549RT-eto cells in a dose- and time-dependent manner (Fig. 1B). We found that parental A549
cells showed more cytotoxicity toward FERO as well (data not
shown). Since it has been reported that NF-κB and SIRT1 are
involved in the regulation of MDR through P-gp protein levels
(27), we sought to examine
protein levels of NF-κB, SIRT1 and P-gp in A549RT-eto cells treated
with 0.05 and 0.5 mM FERO 12 h post-treatment. Our findings show
that FERO treatment reduces expression levels of NF-κB, SIRT1 and
P-gp and enhances expression levels of P53, a representative tumor
suppressor protein (Fig. 1C). In
addition, we detected reduced pre-caspase-9 and pre-Bid levels in
the same lysates of A549RT-eto cells, indicating induction of
intrinsic apoptosis (Fig. 1C).
Feroniellin A reduces MDR1 transcript
levels and promoter activity in A549RT-eto cells
In addition to the MDR1 gene, other genes
such as multidrug resistance-associated protein (MRP)
2 and breast cancer resistance protein (BCRP) are
known to be involved in drug resistance (28,29).
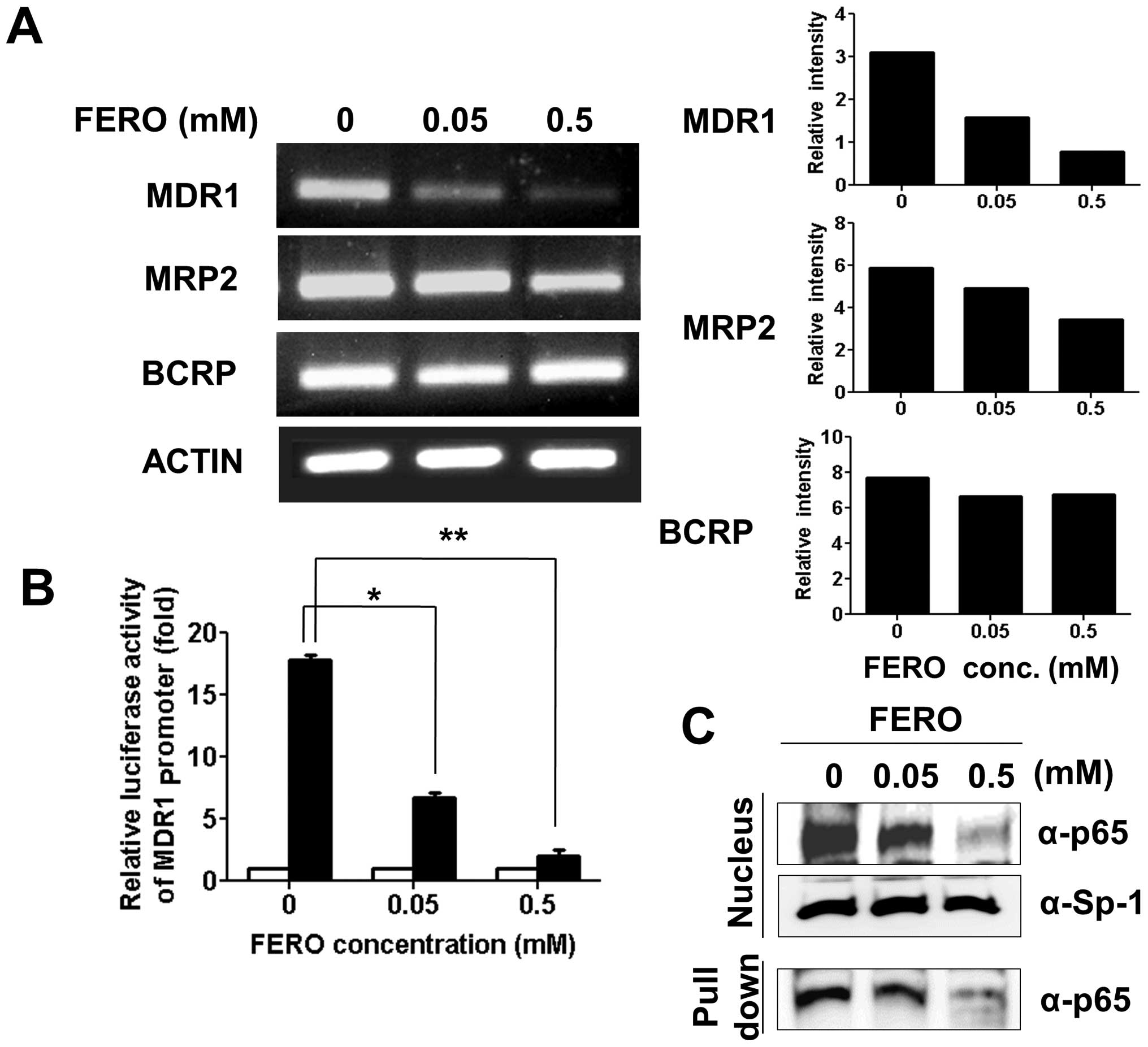
Therefore, we examined transcript levels of MDR1, MRP2 and
BCRP in A549RT-eto cells during FERO treatment. RNA was
isolated and cDNA generated from A549RT-eto cells treated with 0.05
and 0.5 mM FERO for 12 h and genes were amplified using an
optimized number of cycles. We found a significant reduction in
MDR1 transcript levels with slight reductions in MRP2
levels following FERO treatment in a dose-dependent manner
(Fig. 2A). However, transcript
levels of BCRP were not reduced by FERO treatment (Fig. 2B). In addition to transcript
levels, we also examined MDR1 promoter activity in
A549RT-eto cells during FERO treatment using an
MDR1-promoter lucif-erase reporter vector (12). FERO treatment induced a dramatic
decrease of MDR1-mediated luciferase activity in A549RT-eto
cells (Fig. 2B), indicating that
FERO reduces MDR1 promoter activity. Since it has been
reported that the transcription factor NF-κB is involved in the
regulation of P-gp (30,31), we next examined the nuclear
localization of NF-κB and its DNA binding activity during FERO
treatment. We found that FERO treatment inhibits translocation of
NF-κB from the cytoplasm to the nucleus, which results in lowered
levels of NF-κB at its binding site (Fig. 2C). On the basis of these results,
we suggest that FERO inhibits MDR1 transcription, resulting
in a decrease in P-gp protein levels, which leads to sensitization
to apoptosis in A549RT-eto cells.
Inhibition of NF-κB accelerates
FERO-induced apoptosis of A549RT-eto cells through downregulation
of P-gp
Because we observed enhanced NF-κB protein levels
and activity in A549RT-eto cells, we next explored whether NF-κB is
involved in resistance to etoposide in A549 cells through
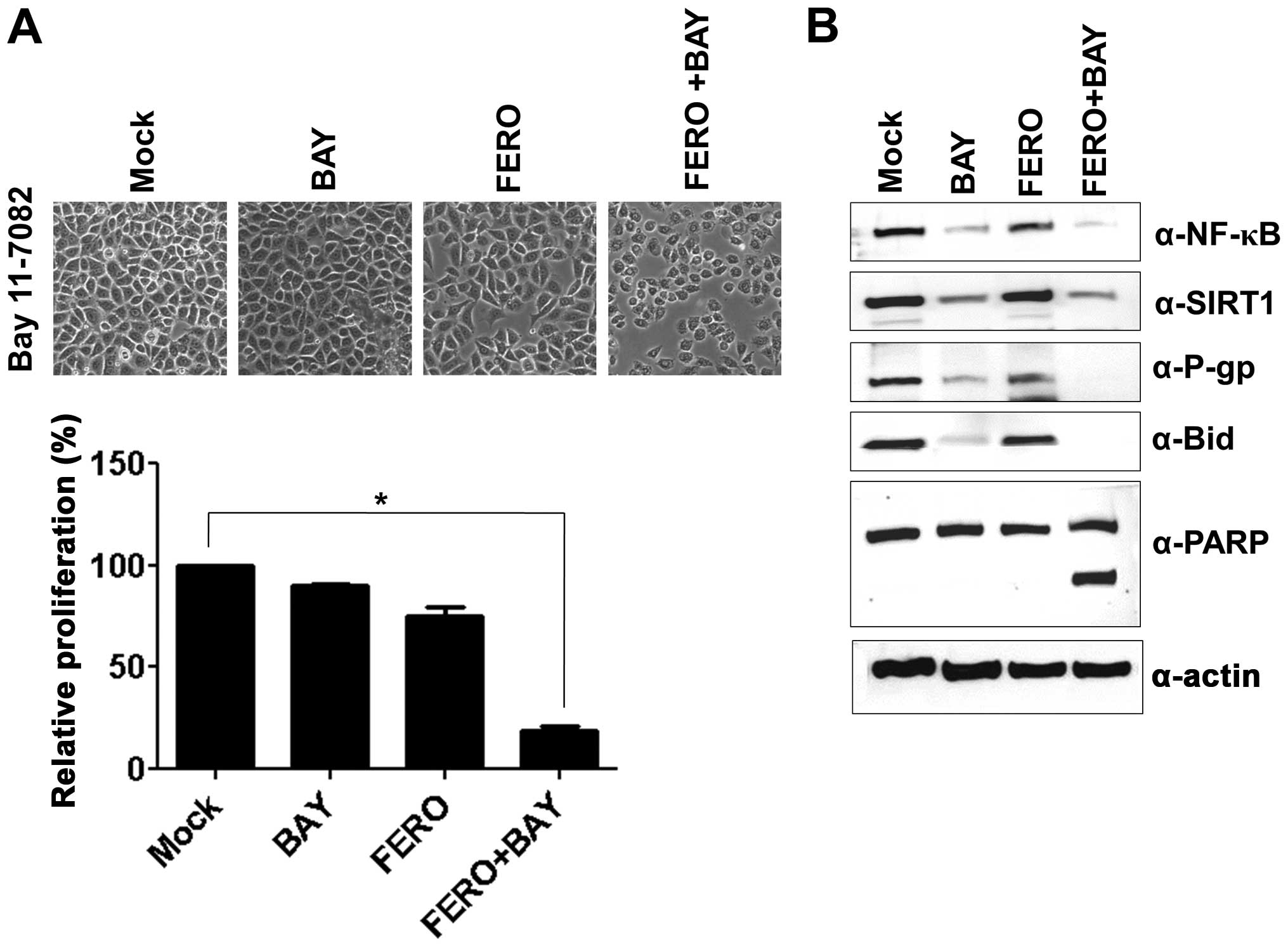
upregulation of P-gp. A549RT-eto cells were treated with 10
μM BAY11-7082 (BAY), an inhibitor of NF-κB, and examined for
cellular viability using the MTT assay. BAY treatment alone did not
influence inhibition of cell growth in A549RT-eto cells, while 0.05
mM FERO treatment inhibited cell growth by ∼30% at 24 h
post-treatment compared to DMSO controls (Fig. 3A). Furthermore, we found that a
combined treatment of FERO and BAY accelerated FERO-mediated
apoptosis in A549RT-eto cells (Fig.
3A), which was confirmed by observations of cleaved PARP and
pre-Bid (Fig. 3B). Moreover, when
we examined protein levels of NF-κB, SIRT1 and P-gp after treatment
with BAY alone, FERO alone, or BAY plus FERO, we found that BAY
treatment alone decreased not only expression levels of NF-κB but
also P-gp, but did not influence protein levels of SIRT1 in the
cells (Fig. 3B). FERO treatment
alone was shown to drastically diminish protein levels of NF-κB,
SIRT1 and P-gp in A549RT-eto cells (Fig. 1C). We also observed that the
combined treatment resulted in more significantly reduced protein
levels of NF-κB, SIRT1 and P-gp (Fig.
3B). These results suggest that NF-κB is involved in MDR in
A549 cells by upregulation of P-gp, resulting in resistance to
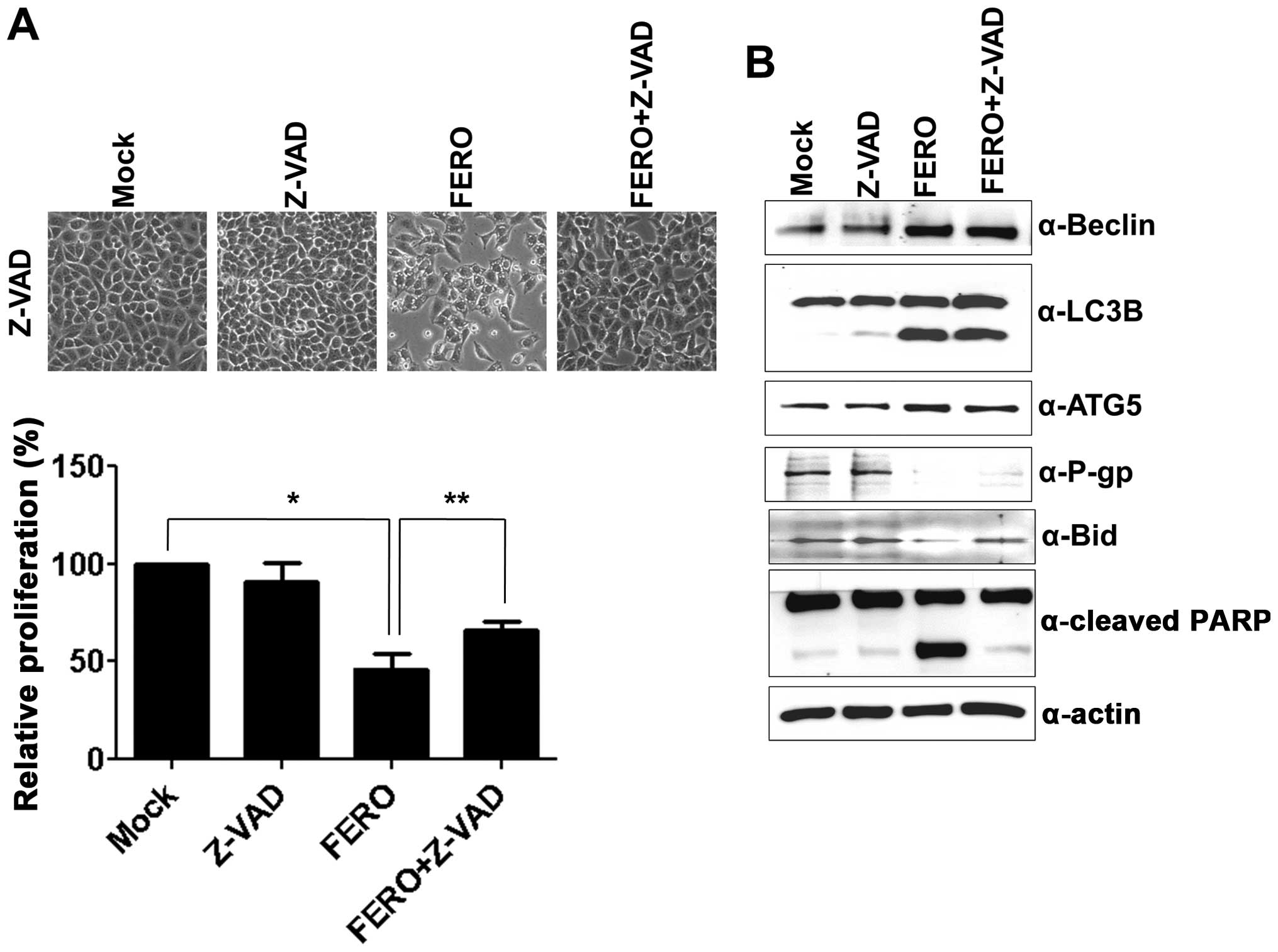
etoposide. To examine whether activation of caspase caused by FERO
treatment is involved in apoptotic cell death in A549RT-eto cells,
a pan-caspase inhibitor, Z-VAD, was used. FERO treatment alone at
0.5 mM resulted in a significant induction of apoptotic cell death
at 24 h post-treatment, while the combined treatment with FERO and
Z-VAD blocked FERO-induced apoptotic cell death (Fig. 4A), which was confirmed by a lack of
cleavage of PARP and pre-Bid proteins (Fig. 4B). These results indicate that
activation of caspases is involved in FERO-induced apoptotic cell
death in A549RT-eto cells. However, Z-VAD treatment did not block
downregulation of P-gp expression induced by FERO, indicating that
a decrease of P-gp expression is necessary but not sufficient for
apoptotic cell death.
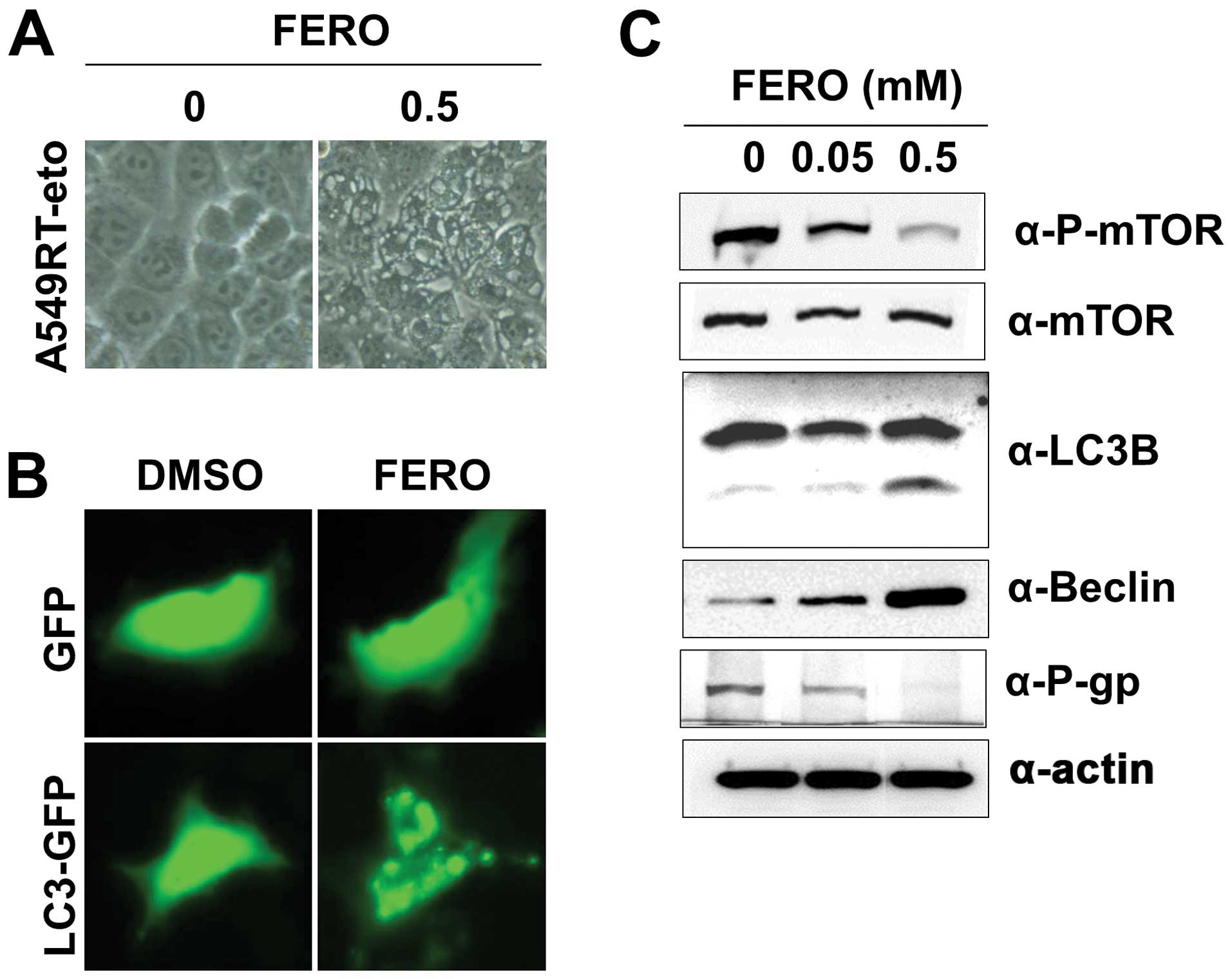
FERO also induces autophagy in A549RT-eto
cells
Auto-phagy has a dual role in cancer, as a tumor
suppressor and a pro-tumor cell survival mechanism (32,33).
Therefore, we investigated whether FERO could induce autophagy,
which is characterized by the formation of vacuoles, GFP-LC3
puncta, and LC3 I to LC3 II conversion. The presence of vacuoles
was apparent in A549RT-eto cells treated with 0.5 mM FERO (Fig. 5A). There was also a clearly
observable accumulation of GFP-LC3 II puncta in A549RT-eto cells
treated with FERO, while no GFP-LC3 puncta were present in
A549RT-eto cells treated with DMSO (Fig. 5B). Moreover, we observed LC3 I to
LC3 II conversion in FERO-treated A549RT-eto cells in a
dose-dependent manner (Fig. 5C).
Lastly, we found induction of Beclin-1, required for the formation
of autophagic vesicles (34), and
reduction of phospho-mTOR levels, an inhibitor of autophagy,
indicating that FERO treatment clearly induced autophagy in
A549RT-eto cells (Fig. 5C).
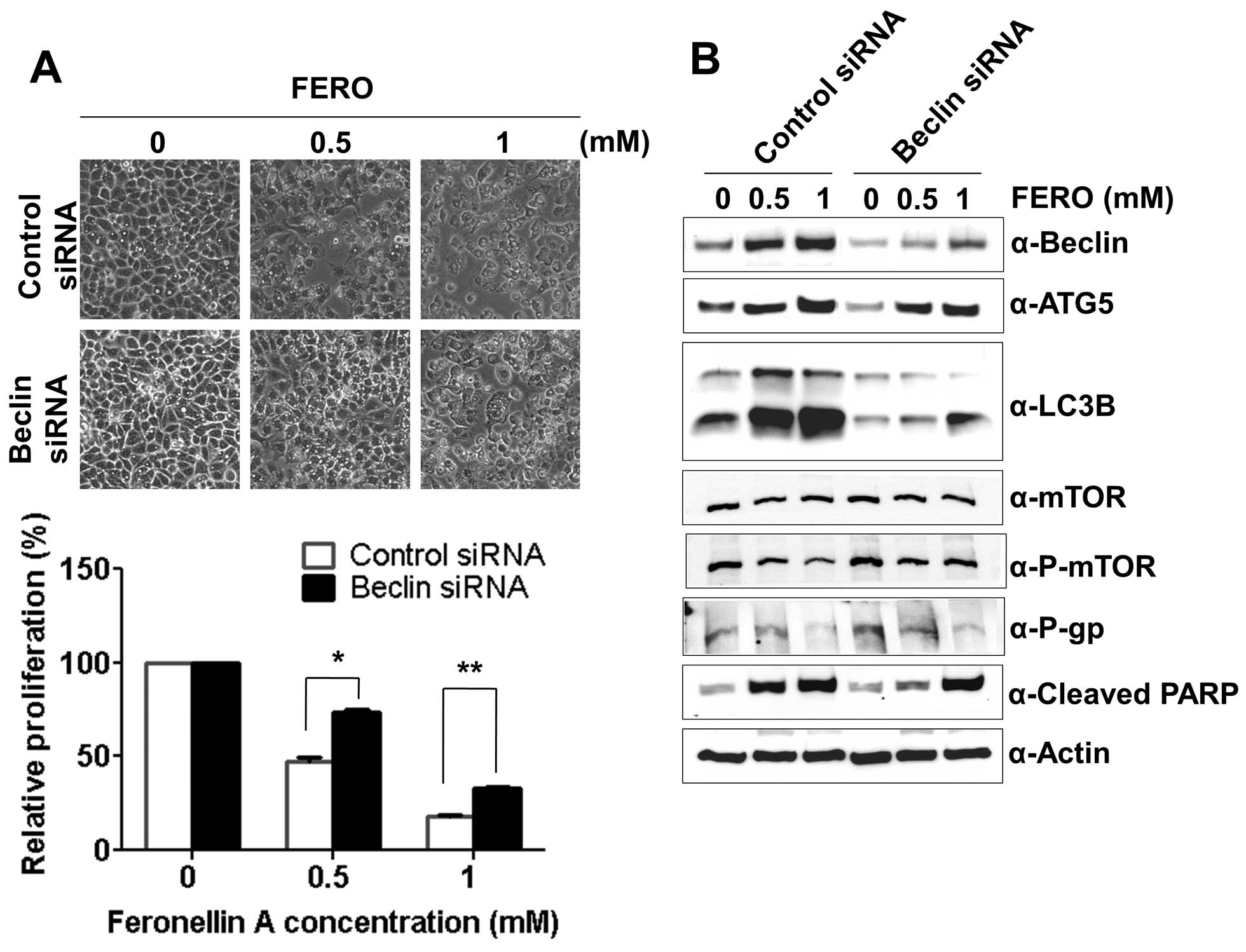
Modulation of autophagy regulates
FERO-induced apoptosis in A549RT-eto cells
Expression of Beclin-1 is known to be essential for
double-membrane autophagosome formation required during the initial
steps of autophagy (34).
Therefore, we tested whether autophagy impairment via the
suppression of Beclin-1 hinders or accelerates FERO-induced
apoptosis. We first determined the optimal siRNA concentration for
suppression of Beclin-1 expression, which was found to be 100 nM
(data not shown). FERO treatment with control siRNA induced
apoptosis by activation of caspase-3 or -7 due to cleavage of PARP
in a dose-dependent manner (Fig.
6A). Concomitantly, FERO induced autophagy coincided with an
upregulation of Beclin-1 and ATG5, increased LC3 I to LC3 II
conversion, and a reduction of phospho-mTOR levels in A549RT-eto
cells. However, suppression of Beclin-1 with siRNA inhibited
FERO-induced apoptosis, indicating that autophagy is necessary for
FERO-induced apoptosis. Interestingly, we found that suppression of
Beclin-1 by siRNA inhibited the conversion of LC3 I to LC3
II but did not inhibit upregulation of ATG5 expression induced by
FERO. These results indicate that conversion of LC3 I to LC3 II
occurs later and ATG5 mediated processes take place earlier than
Beclin-1 mediated autophagy following FERO treatment (Fig. 6B). In addition, similar to the
observation that Z-VAD suppresses FERO-induced apoptosis but does
not block downregulation of P-gp, we also found that suppression of
Beclin-1 does not block FERO-mediated decrease of P-gp expression
despite the inhibition of FERO-induced apoptosis. This result
indicates that downregulation of P-gp expression is necessary but
not sufficient for the progression of apoptosis.
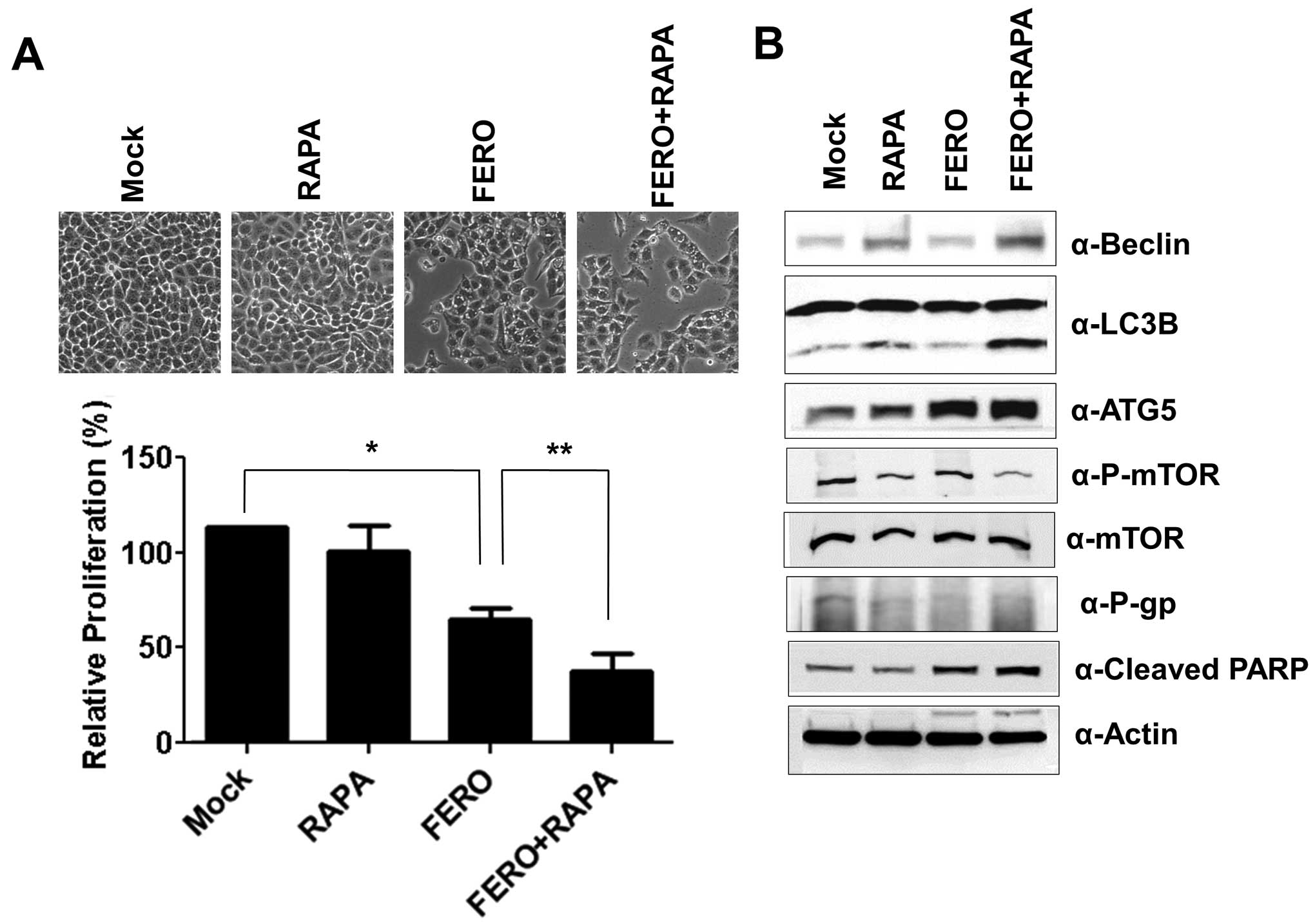
To verify that autophagy is necessary for
FERO-induced apoptosis, we administered rapamycin (RAPA), an
inhibitor of mammalian target of rapamycin (mTOR), in order to
accelerate A549RT-eto cell autophagy in the presence of FERO
(35,36). FERO induced apoptosis in A549RT-eto
cells as well as dual administration of FERO and RAPA was observed
to enhance apoptosis (Fig. 7A).
Co-treatment of FERO and RAPA induced autophagy by upregulating
Beclin-1 and ATG5 expression, and converting LC3 I to LC3 II
(Fig. 7B). These results suggest
that FERO-induced autophagy promotes apoptotic cell death in
A549RT-eto cells.
Discussion
Presently, identification of effective
chemotherapeutic agents in phytochemicals of great interest to
cancer biologists, because of MDR in cancer cells, which are
developed following prolonged treatment with anticancer drug.
Feroniellin A (FERO) was reported to possess in vitro
cytotoxicity in human KB carcinoma and HeLa carcinoma cells
(25). However, the molecular
mechanism by which this compound exerts cytotoxicity in these
cancer cell lines is unknown. For the first time, we have provided
a line of evidence showing that FERO reverses MDR1 activity by
downregulation of NF-κB, which leads to enhanced apoptotic
susceptibility in A549RT-eto cells. Consistent with our results,
other studies have shown that metformin, mollugin, and puerarin,
which are derived from plants also reverse MDR1 activity
effectively through downregulation of NF-κB in human breast cancer
cells resistant to adriamycin (30,37,38).
Moreover, our study shows that inhibition of apoptosis with Z-VAD
does not decrease P-gp expression, indicating that downregulation
of P-gp is necessary but not sufficient for apoptosis. As SIRT1,
β-catenin and HIF-1α have also been associated with MDR in many
cancer models (39–41), we are currently investigating
whether FERO affects the expression levels and activity of these
proteins, rendering A549RT-eto cells susceptible to apoptosis.
Detailed reviews of autophagic progress can be found
as described elsewhere (33,42).
Briefly, autophagy is initiated by activation of the ATG1 complex,
which includes ATG1/ATG13/ATG17, among other components. Next,
autophagosome nucleation occurs, which requires class III
phsophatidylinositol-3-kinase plus Beclin-1/ATG6 in addition to
several other factors to recruit proteins and lipids involved in
autophagosome formation. Vesicle elongation and completion are
mediated by two-ubiquitin-like systems: ATG7 (E1-like) and ATG3
(E2-like), which are necessary for the lipid modification of LC3
(phosphatidylethanolamine; PE), which requires initial cleavage of
LC3. Subsequently, the ATG12/ATG5/ATG16 complex mediates LC3-PE
binding to the autophagosome membrane. Finally, the completed
autophagosome fuses with lysosomes, where the autophago-some
contents are degraded. We observed that suppression of Beclin-1 in
the presence of FERO results in the inhibition of LC3 I to LC3 II
conversion but does not inhibit ATG5 expression. Considering the
sequential progress of autophagy, we assume that LC3 I to LC3 II
conversion is very closely linked to Beclin-1 activity but that
suppression of Beclin-1 itself does not affect FERO-induced ATG5
expression. Although we have observed that autophagy impairment
such as Beclin-1 suppression with siRNA generated reactive
oxygen species (ROS) (data not shown) as reported elsewhere
(43,44), we also found that autophagy
impairment with Beclin-1 suppression in fact protects cells from
FERO-induced apoptosis. Based on this finding, we speculate that
the amount of ROS generated by autophagy impairment does not
influence aggravation of A49RT-eto cell survival. In addition,
apoptosis inhibition with Z-VAD did not block FERO-induced
autophagic progress such as upregulation of Beclin1 and ATG5, and
converting LC3 I to LC3 II (Fig.
4B), indicating that autophagic progress is the upstream event
of apoptosis.
Recently, autophagy inhibitors such as chloroquine
(CQ) or hydroxychloroquine have been used in clinical trials
because autophagy is believed to affect tumor survival (45–47).
Therefore, it is possible that conventional chemotherapy with
autophagy inhibitors may increase tumor cytotoxicity. In a mouse
prostate cancer model, co-treatment with an Src family kinase
inhibitor and CQ increased tumor cell numbers in vitro but
reduced tumor cell growth in vivo (48). Conversely, since it has been
suggested that autophagy also plays a role as a tumor suppressor,
introduction of autophagy stimulants such as mTOR inhibitors with
conventional chemotherapeutic drugs leads to accelerated tumor cell
death compared to treatment with anti-cancer drugs alone (49,50).
Since there are conflicting reports as to the positive and negative
effects of autophagy on tumor cell death (32,33),
the field needs to be cautious when attempting to manipulate
autophagy to improve clinical outcomes. In conclusion, our study
has demonstrated that FERO-induced autophagy is required for
apoptotic progression and suggests that autophagy plays a role as a
tumor suppressor in our model.
Acknowledgements
This study was supported by a grant
from the World Class University Program (R31-2008-000-20004-0)
through the National Research Foundation funded by the Korean
government, and a grant from the Biomedical Fusion Technology
Development Project of the Korea Research Institute of Bioscience
and Biotechnology (KGM1231312). This study was also supported by
the Office of the Higher Education Commission, Thailand, under the
Strategic Scholarships Fellowships Frontier Research Networks
(Specific for Southern region) for the Join PhD Program Thai
Doctoral degree program; a CHE-SSR-PhD SW Scholarship to C.K.
References
|
1.
|
Tan G, Gyllenhaal C and Soejarto DD:
Biodiversity as a source of anticancer drugs. Curr Drug Targets.
7:265–277. 2006.PubMed/NCBI
|
|
2.
|
Bronikowska J, Szliszka E, Jaworska D,
Czuba ZP and Krol W: The coumarin psoralidin enhances anticancer
effect of tumor necrosis factor-related apoptosis-inducing ligand
(TRAIL). Molecules. 17:6449–6464. 2012. View Article : Google Scholar
|
|
3.
|
Diwan R and Malpathak N: Furanocoumarins:
novel topoisom-erase I inhibitors from Ruta graveolens L.
Bioorg Med Chem. 17:7052–7055. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Ohnishi A, Matsuo H, Yamada S, et al:
Effect of furanocoumarin derivatives in grapefruit juice on the
uptake of vinblastine by Caco-2 cells and on the activity of
cytochrome P450 3A4. Br J Pharmacol. 130:1369–1377. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Paine MF, Widmer WW, Hart HL, et al: A
furanocoumarin-free grapefruit juice establishes furanocoumarins as
the mediators of the grapefruit juice-felodipine interaction. Am J
Clin Nutr. 83:1097–1105. 2006.PubMed/NCBI
|
|
6.
|
Biedler JL: Drug resistance: genotype
versus phenotype - Thirty-second G.H.A. Clowes Memorial Award
Lecture. Cancer Res. 54:666–678. 1994.PubMed/NCBI
|
|
7.
|
Bosch I and Croop J: P-glycoprotein
multidrug resistance and cancer. Biochim Biophys Acta. 9:37–54.
1996.
|
|
8.
|
Goldstein LJ, Galski H, Fojo A, et al:
Expression of a multidrug resistance gene in human cancers. J Natl
Cancer Inst. 81:116–124. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Gottesman MM, Fojo T and Bates SE:
Multidrug resistance in cancer: role of ATP-dependent transporters.
Nat Rev Cancer. 2:48–58. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Friedrich K, Wieder T, Von Haefen C, et
al: Overexpression of caspase-3 restores sensitivity for
drug-induced apoptosis in breast cancer cell lines with acquired
drug resistance. Oncogene. 20:2749–2760. 2001. View Article : Google Scholar
|
|
11.
|
Ruefli AA, Tainton KM, Darcy PK, Smyth MJ
and Johnstone RW: P-glycoprotein inhibits caspase-8 activation but
not formation of the death inducing signal complex (disc) following
Fas ligation. Cell Death Differ. 9:1266–1272. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Bentires-Alj M, Barbu V, Fillet M, et al:
NF-kappaB transcription factor induces drug resistance through
MDR1 expression in cancer cells. Oncogene. 22:90–97. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Chu F, Chou PM, Zheng X, Mirkin BL and
Rebbaa A: Control of multidrug resistance gene mdr1 and cancer
resistance to chemotherapy by the longevity gene sirt1. Cancer Res.
65:10183–10187. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Klionsky DJ and Emr SD: Autophagy as a
regulated pathway of cellular degradation. Science. 290:1717–1721.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Levine B and Klionsky DJ: Development by
self-digestion: molecular mechanisms and biological functions of
autophagy. Dev Cell. 6:463–477. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Levine B and Kroemer G: Autophagy in the
pathogenesis of disease. Cell. 132:27–42. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Qu X, Yu J, Bhagat G, et al: Promotion of
tumorigenesis by heterozygous disruption of the beclin 1 autophagy
gene. J Clin Invest. 112:1809–1820. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Levine B: Cell biology: autophagy and
cancer. Nature. 446:745–747. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Takamura A, Komatsu M, Hara T, et al:
Autophagy-deficient mice develop multiple liver tumors. Genes Dev.
25:795–800. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Han J, Hou W, Goldstein LA, et al:
Involvement of protective autophagy in TRAIL resistance of
apoptosis-defective tumor cells. J Biol Chem. 283:19665–19677.
2008. View Article : Google Scholar
|
|
21.
|
Katayama M, Kawaguchi T, Berger MS and
Pieper RO: DNA damaging agent-induced autophagy produces a
cytoprotective adenosine triphosphate surge in malignant glioma
cells. Cell Death Differ. 14:548–558. 2007. View Article : Google Scholar
|
|
22.
|
Li X and Fan Z: The epidermal growth
factor receptor antibody cetuximab induces autophagy in cancer
cells by downregulating HIF-1alpha and Bcl-2 and activating the
beclin 1/hVps34 complex. Cancer Res. 70:5942–5952. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Ogata M, Hino S, Saito A, et al: Autophagy
is activated for cell survival after endoplasmic reticulum stress.
Mol Cell Biol. 26:9220–9231. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Kanintronkul Y, Worayuthakarn R, Thasana
N, et al: Overcoming multidrug resistance in human lung cancer with
novel benzo[α] quinolizin-4-ones. Anticancer Res. 31:921–927.
2011.PubMed/NCBI
|
|
25.
|
Phuwapraisirisan P, Surapinit S, Sombund
S, Siripong P and Tip-pyang S: Feroniellins A–C, novel cytotoxic
furanocoumarins with highly oxygenated C10 moieties from
Feroniella lucida. Tetrahedron Lett. 47:3685–3688. 2006.
|
|
26.
|
Kaewpiboon C, Lirdprapamongkol K,
Srisomsap C, et al: Studies of the in vitro cytotoxic, antioxidant,
lipase inhibitory and antimicrobial activities of selected Thai
medicinal plants. BMC Complement Altern Med. 12:2172012. View Article : Google Scholar
|
|
27.
|
Jung YJ, Lee JE, Lee AS, et al: SIRT1
overexpression decreases cisplatin-induced acetylation of NF-kappaB
p65 subunit and cytotoxicity in renal proximal tubule cells.
Biochem Biophys Res Commun. 419:206–210. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Sugimoto Y, Tsukahara S, Ishikawa E and
Mitsuhashi J: Breast cancer resistance protein: molecular target
for anticancer drug resistance and
pharmacokinetics/pharmacodynamics. Cancer Sci. 96:457–465. 2005.
View Article : Google Scholar
|
|
29.
|
Young LC, Campling BG, Cole SPC, Deeley RG
and Gerlach JH: Multidrug resistance proteins MRP3, MRP1, and MRP2
in lung cancer: correlation of protein levels with drug response
and messenger RNA levels. Clin Cancer Res. 7:1798–1804.
2001.PubMed/NCBI
|
|
30.
|
Kim HG, Hien TT, Han EH, et al: Metformin
inhibits P-glycoprotein expression via the NF-kappaB pathway and
CRE transcriptional activity through AMPK activation. Br J
Pharmacol. 162:1096–1108. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Sun J, Yeung CA, Co NN, et al: Clitocine
reversal of P-glycoprotein associated multi-drug resistance through
down-regulation of transcription factor NF-kappaB in R-HepG2 cell
line. PLoS One. 7:e407202012. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Levy JM and Thorburn A: Targeting
autophagy during cancer therapy to improve clinical outcomes.
Pharmacol Ther. 131:130–141. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Yang ZJ, Chee CE, Huang S and Sinicrope
FA: The role of autophagy in cancer: therapeutic implications. Mol
Cancer Ther. 10:1533–1541. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Jia YL, Li J, Qin ZH and Liang ZQ:
Autophagic and apoptotic mechanisms of curcumin-induced death in
K562 cells. J Asian Nat Prod Res. 11:918–928. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Dai ZJ, Gao J, Ma XB, et al: Antitumor
effects of rapamycin in pancreatic cancer cells by inducing
apoptosis and autophagy. Int J Mol Sci. 14:273–285. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Rubinsztein DC and Nixon RA: Rapamycin
induces autophagic flux in neurons. Proc Natl Acad Sci USA.
107:E1812010. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Hien TT, Kim HG, Han EH, Kang KW and Jeong
HG: Molecular mechanism of suppression of MDR1 by puerarin from
Pueraria lobata via NF-kappaB pathway and cAMP-responsive
element transcriptional activity-dependent up-regulation of
AMP-activated protein kinase in breast cancer MCF-7/adr cells. Mol
Nutr Food Res. 54:918–928. 2010.PubMed/NCBI
|
|
38.
|
Tran TP, Kim HG, Choi JH, Na MK and Jeong
HG: Reversal of P-glycoprotein-mediated multidrug resistance is
induced by mollugin in MCF-7/adriamycin cells. Phytomedicine.
20:622–631. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Katerere DR and Eloff JN: Antibacterial
and antioxidant activity of Sutherlandia frutescens
(Fabaceae), a reputed anti-HIV/AIDS phytomedicine. Phytother Res.
19:779–781. 2005.PubMed/NCBI
|
|
40.
|
Zhang H, Zhang X, Wu X, et al:
Interference of Frizzled 1 (FZD1) reverses multidrug resistance in
breast cancer cells through the Wnt/beta-catenin pathway. Cancer
Lett. 323:106–113. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Zhu H, Xia L, Zhang Y, et al: Activating
transcription factor 4 confers a multidrug resistance phenotype to
gastric cancer cells through transactivation of SIRT1 expression.
PLoS One. 7:e314312012. View Article : Google Scholar
|
|
42.
|
Denton D, Nicolson S and Kumar S: Cell
death by autophagy: facts and apparent artefacts. Cell Death
Differ. 19:87–95. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Kang HT, Lee KB, Kim SY, Choi HR and Park
SC: Autophagy impairment induces premature senescence in primary
human fibroblasts. PLoS One. 6:e233672011. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Tal MC, Sasai M, Lee HK, Yordy B, Shadel
GS and Iwasaki A: Absence of autophagy results in reactive oxygen
species-dependent amplification of RLR signaling. Proc Natl Acad
Sci USA. 106:2770–2775. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45.
|
Selvakumaran M, Amaravadi RK, Vasilevskaya
IA and O’Dwyer PJ: Autophagy inhibition sensitizes colon cancer
cells to antiangiogenic and cytotoxic therapy. Clin Cancer Res.
19:2995–3007. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46.
|
Shen J, Zheng H, Ruan J, et al: Autophagy
inhibition induces enhanced proapoptotic effects of ZD6474 in
glioblastoma. Br J Cancer. 109:164–171. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47.
|
Yang PM, Liu YL, Lin YC, Shun CT, Wu MS
and Chen CC: Inhibition of autophagy enhances anticancer effects of
atorvastatin in digestive malignancies. Cancer Res. 70:7699–7709.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
48.
|
Wu Z, Chang PC, Yang JC, et al: Autophagy
blockade sensitizes prostate cancer cells towards Src family kinase
inhibitors. Genes Cancer. 1:40–49. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49.
|
Josset E, Burckel H, Noel G and Bischoff
P: The mTOR inhibitor RAD001 potentiates autophagic cell death
induced by temozolomide in a glioblastoma cell line. Anticancer
Res. 33:1845–1851. 2013.PubMed/NCBI
|
|
50.
|
Takeuchi H, Kondo Y, Fujiwara K, et al:
Synergistic augmentation of rapamycin-induced autophagy in
malignant glioma cells by phosphatidylinositol 3-kinase/protein
kinase B inhibitors. Cancer Res. 65:3336–3346. 2005.PubMed/NCBI
|