Introduction
MicroRNAs (miRNAs) are endogenous expressed small
non-coding RNAs with important functions in almost all biological
processes (1). Usually these
miRNAs negatively influence gene expression by destabilizing mRNAs
or inhibiting their translation (2). MiRNAs recognize their substrates by
binding sequences which are perfectly complementary to the so
called seed regions compromising nucleotides 2–8. The targeted
regions usually lie in the 3’UTR of the mRNAs and are present in
multiple copies (2). A single
miRNA can modulate the expression of more than hundred genes.
MiRNAs are processed from long capped and
polyadenylated primary transcripts (pri-miRNA) produced by RNA
polymerase II. Before exported to the cytoplasma a nuclear
processing step synthesizes the ∼70 nucleotides (nt) long precursor
miRNA (pre-miR). Further maturation includes cleavage to a duplex
of ∼20 nt and loading of one strand to a ribonucleotide protein
complex able to function as miRNA-induced silencing complexes
(miRISCs) (1,2). Expression patterns of specific miRNAs
vary within different cell and tissue types (3). Their expression is often dynamically
changing during developmental processes and in diseases (1). Characterization of miRNA expressions
indicates that several miRNAs may have a crucial role in cancer
progression of various tumors. They can function as
tumor-suppressors or as oncogenes (termed oncomirs) (4).
MiRNA signature in different cancers identified
microRNA-21 (miR-21) as one of the most abundant oncomirs (5). Elevated miR-21 expression in cancer
cells compared to the normal control cells were observed in almost
all solid tumors (6) including
head and neck (7), colorectal
(8,9), lung (6,10),
breast (11,12), esophageal (13) and liver cancer (14).
Consistent with a postulated function as an oncomir,
miR-21 targets mRNAs coding for proteins with functions as
inhibitor of cell signaling such as phosphatase and tensin homolog
(PTEN) tumor suppressor (14),
Sprouty1 (Spry1) (15) or Sprouty2
(Spry2) (16) or of cell cycle
progression like cell division cycle 25 homolog A (CDC25A)
(17) as well as genes involved in
regulation of cell death like programmed cell death 4 (PDCD4)
(18,19).
In this investigation we analyzed the expression of
miR-21 in 36 NSCLC-derived tumor tissue and 14 cell lines and
compared it to the expression in normal lung tissue. We further
analyzed if elevated miR-21 expression in the malignant tissue is
influenced by environmental growth conditions.
Materials and methods
Patients
Tumor and normal lung tissue samples were derived
from patients with histological confirmed NSCLC who underwent
surgical resection at the Otto Wagner Hospital. At the time of
surgery the patients had not undergone chemotherapy. Only tissues
with a macroscopic visible tumor were chosen. All tumors were
histologically confirmed. The normal tissue samples were taken from
the surrounding normal tissue. The distance between the normal
tissue to the tumor was ≥2 cm. If the interspace between the tumor
and the normal lung tissue specimens was >5 cm, the normal
sample was classified as distal, if the distance to the tumor was
<5 cm it was characterized as proximal. All tissue samples were
immediately flash frozen in liquid nitrogen and stored at −80°C
until use. Information on the pathologic stage of each tumor
according to the WHO classifications was available for 33 of 36
patients. Samples were derived from 6 patients staged as IA and 12
as IB. Only two of the tumor samples were obtained from patients
classified as stage IIA. Furthermore, samples from 4 IIB-staged and
8 IIIA staged patients were investigated. One of the patients had
lung cancer classified as stage IV. Concerning histology, 23 of the
tumors were adenocarcinoma and 13 were squamous cell carcinoma. The
use of the patient sections for the study was approved by the
ethics committee of the City of Vienna (EK 07-176-VK) according to
legal Austrian regulations.
Cell culture
Six of the cancer-derived cell lines and the normal
embryonic lung fibroblasts WI-38 as well as immortalised bronchial
epithelial cells (Beas2B, CRL9606) were purchased from the American
Type Culture Collection (ATCC) and cultured in the recommended
medium containing 10% fetal bovine serum (FBS) (GE Healthcare,
Chalfont St. Giles, Buckinghamshire, UK) and supplemented with
penicillin (100 U/ml) and streptomycin (100 μg/ml). Three of
these cell lines (A549, A427, SK-LU-1) harbor a K-RasG12
mutation and one (Calu-6) a K-RasQ61 mutation. Two of
these NSCLC-derived cell lines (CRL2868 and CRL5883) carry an
acquired exon 19 in frame deletion (E746-A750) of EGFR.
Additionally, eight NSCLC cell lines were
established at our institute as described (20). These cell lines were cultured using
DMEM growth medium (GE Healthcare) containing 10% FBS supplemented
with penicillin, streptomycin and pyruvate. Cell lines were
authenticated by array-comparative genomic hybridization or DNA
fingerprinting and regularly checked for Mycoplasma
contamination.
Synchronization of cells in certain phases of the
cell cycle was achieved by serum starvation as described (21). Propidium iodide-based DNA content
analysis (PI-staining) was performed as described (22). Hypoxic conditions were achieved by
incubating the cells in a Heracell 150i incubator (Thermo
Scientific, Waltham, MA, USA) at 1% oxygen, 5% CO2 and
37°C. To standardize pH, 25 mM HEPES, pH 8.0 was added to the
medium. Anchorage-independent growth was achieved by cultivating
the cells in plates coated with 1% noble agar (Sigma-Aldrich, St.
Louis, MO, USA).
Isolation of human primary lung
fibroblasts and tumor-associated fibroblasts
During lobectomy operations a surgical specimen was
obtained directly from the tumor site as well as from a distant
tumor-free part of the removed lung lobe. Samples were immediately
transferred into the laboratory and washed with DMEM containing
antibiotic and anti-mycotic solution (penicillin, streptomycin and
amphotericin B, Sigma-Aldrich). The tissue piece was minced by a
scalpel and incubated with trypsin solution (0.25 wt/vol% trypsin
in PBS, Sigma). After 30 min at 37°C, the samples were triturated
with a pipette while in DMEM with 10% FCS. Following centrifugation
the suspension and clumps were plated in a tissue culture flask in
DMEM with 10% FCS. Following 48 h of culture the non-adhering
connective fiber dense pieces were transferred into a new culture
flask. After a few days these clumps adhered and fibroblasts
radially migrated out from the tissue pieces. At confluence, the
cultures were trypsinized (0.25 wt/vol% trypsin in PBS) and the
cells were transferred into tissue-culture flasks containing DMEM
with 10% FCS. Following four passages, the lung fibroblasts were
frozen in 10% dimethyl sulphoxid-10%FCS-DMEM solution and stored in
liquid N2 for later use. Procedures to isolate lung tissue
associated fibroblasts were approved by the ethics committee of the
Medical University of Vienna (MUW#904-2009).
RNA isolation and northern blotting
RNA isolation was performed as described (23). For the tissue, prior to the
isolation procedure samples were cut into small pieces and
transferred to homogenization tubes prefilled with lysis buffer
plus 5–6 ceramic beads. Homogenization was performed twice at 5500
for 2×20 sec, with 10-sec breaks in a Precellys 24 homogenisator
(PEQLAB, Erlangen, Germany).
RNA (10 or 15 μg) was separated by an 18%
denaturing polyacrylamide gel electrophoresis containing 7.6 M urea
using TBE as running buffer. Prior to loading RNA was mixed with
equal volumes of loading buffer (formamide plus 10 mM EDTA pH 8.0
and bromophenol blue) and incubated for 5 min at 70°C. Separated
RNA was blotted onto nylon membrane (GeneScreen plus from
Perkin-Elmer, Waltham, MA, USA) using 0.5X TBE as buffer in a
Bio-Rad transfer apparatus (Bio-Rad, Hercules, CA, USA) at constant
150 mA overnight at 4°C. The transferred RNA was crosslinked to the
membrane with UV (Stratalinker; auto-crosslinking: 1200 μJ
energy). After methylene blue staining the membrane was stored at
−20°C. Hybridisation was performed as described (24). As probes oligo-nucleotides from
Microsynth AG (Balgach, Switzerland) were used: miR-21 probe:
TCAACATCAGTCTGATAAGCTA; U6 probe [probe for Homo sapiens U6
small nuclear 2 RNA (RNU6-2)]: CACGAATTTGCGTGTCATCCTT; 5SrRNA probe
ATT CCCAGGCGGTCTCCCATCC. Probes were labeled by polynucleotide
kinase (New England Biolabs, Ipswich, MA, USA) according to the
manufacturer’s instructions using 5 μl γ[32P]-ATP
(3,000 Ci/mmole) (Hartmann Analytic, Braunschweig, Germany).
Washing of the membranes was performed at 50°C in a non-stringent
washing buffer (3X SSC, 25 mM sodium phosphate pH 7.5, 5% SDS) and
a stringent washing buffer (1X SSC, 1% SDS). Intensity of the
signals was detected using a Typhoon phosphorimager (GE
Healthcare).
Immunoblotting
Western blotting was performed as described
(21). Primary antibodies against
β-actin (Novus Biologicals, CO, USA) and Au5 epitope (Bethyl
Laboratories, TX, USA) were used.
Results
MiR-21 levels in NSCLC-derived tissue are
elevated
As an initial experiment we compared miR-21 levels
of normal and tumor tissue from patients with histological
confirmed NSCLC. Endogenous miR-21 levels were determined by
northern blotting of total RNA isolated from malignant and the
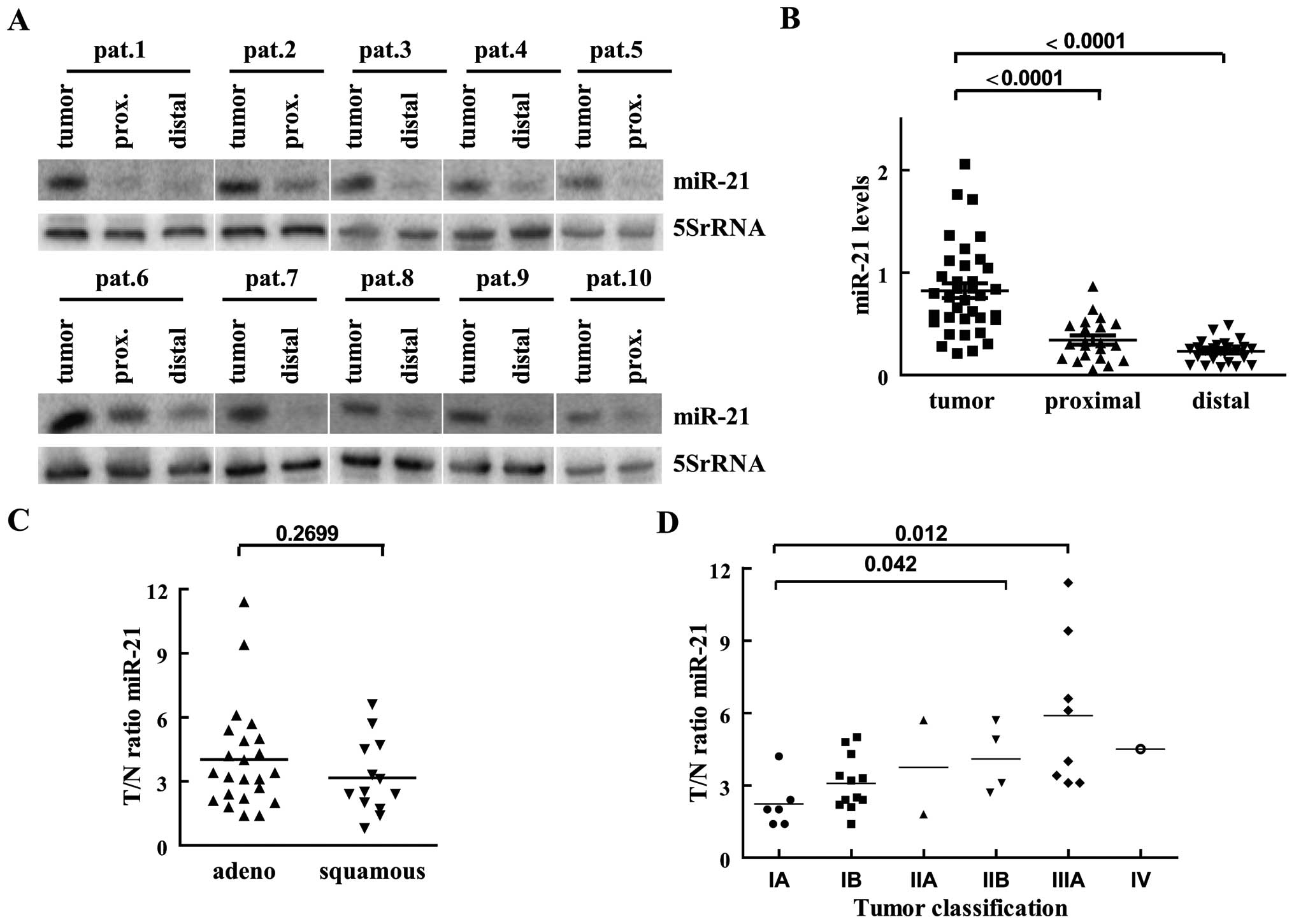
corresponding normal tissue sections of the same patient (Fig. 1A). Mature miR-21 was visible in all
samples analyzed, while we failed to detect pre-miR-21 RNA in most
cases and therefore it was excluded from the analysis. As loading
control probes recognizing U6 and 5SrRNA were used. Since in the
samples from a few patients the U6 levels were strongly reduced
(data not shown), we normalized the detected miR-21 signal to the
one obtained for 5SrRNA. For the first analysis, miR-21 levels were
calculated in reference to an internal standard value loaded on
every northern blot and grouped into malignant tissue and normal
samples derived from tissue proximal or distal to the tumor
section. As depicted in Fig. 1B,
miR-21 levels in normal lung tissue isolated distal to the tumor
(n=26) were constantly low and showed little fluctuation. In
sections proximal to the tumor (n=21), miR-21 expression was on
average increased and showed more variability. In the tumor-derived
samples low and high miR-21 expressions were observed, leading to
strong variations of miR-21 levels. In the tumor compartments
miR-21 levels were on average 4- and 3-fold increased in relation
to distal and proximal analyzed tissue, respectively (Fig. 1B). For the consecutive data
analysis, each tumor sample was normalized to the value obtained in
the unaffected lung tissue of the same patient. In 30 of 36 sample
pairs the miR-21 level was increased >2-fold in the
tumor-derived RNA in comparison to the one obtained from the
corresponding healthy tissue. In only six patients the miR-21
amount in the tumor section was comparable (less than 2-fold
changed) to the normal tissue sample. None of the investigated
tumor tissues showed lower miR-21 levels in comparison to the
normal lung. Compared to the corresponding normal lung tissue,
miR-21 elevation varied from 1.4- to 11-fold in the tested patient.
In the majority of tumors miR-21 levels were raised >2-fold and
in one sixth of the tumors we observed even a 5–10-fold increase of
miR-21 levels when compared to the expression levels of the normal
lung tissue of the same patient (Fig.
1A, C and D). Concerning histology, in adenocarcinoma miR-21
upregulation was not significantly different from the one
calculated for squamous cell carcinoma, although on average
adenocarcinoma expressed more miR-21 than squamous cell carcinoma
(Fig. 1C). With respect to
differentiation, we observed that upregulation of miR-21 in the
malignant section was stronger in less differentiated tumors (mean
G1=3.1, G2=3.5, G3=4.7), but the differences were not significant
(data not shown). Regarding tumor classification according to the
TNM system, in higher malignant stages a more pronounced
upregulation of miR-21 in the tumor compared to normal tissue was
observed (Fig. 1D). Due to the
small sample size a correlation between tumor staging and miR-21
increase cannot be calculated, but using a Mann-Whitney U test
miR-21 increased significant between tumor stage IA and IIB (U=2,
p=0.042) and between IA and IIIA (U=4, p=0.012).
MiR-21 levels in NSCLC-derived cell lines
are comparable to those measured in normal lung cells
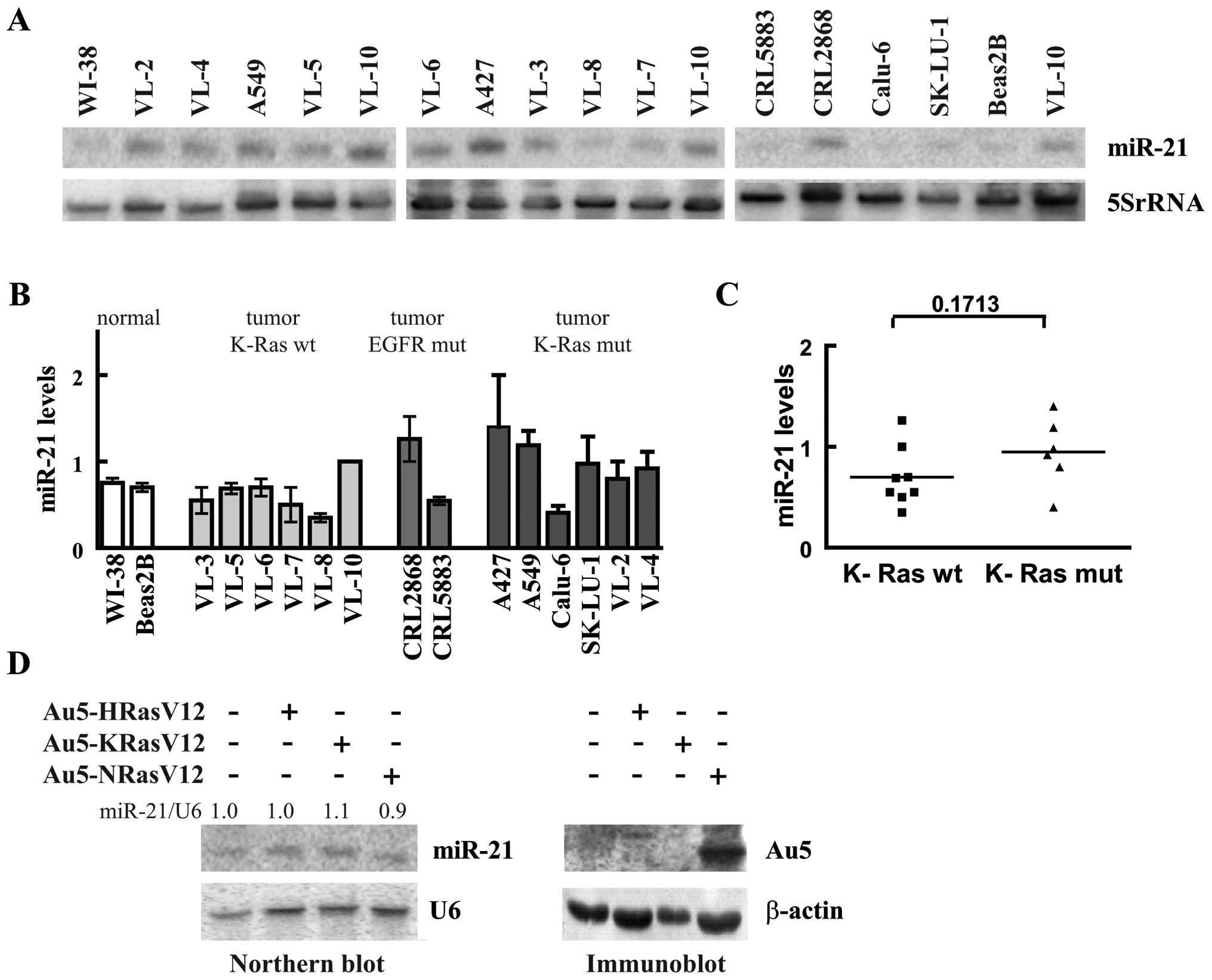
Next we analyzed miR-21 in 14 NSCLC-derived cell
lines by northern blots. As shown in Fig. 2A all cell lines expressed miR-21,
but the endogenous levels showed only modest variation. For
quantitative analysis one of the RNAs (the one of VL-10) was loaded
on all northern blots and used as a reference. When compared to
normal cells (WI-38 and Beas2B) miR-21 levels in lung tumor cell
lines varied between 0.6- and 1.8-fold indicating that in none of
the tested cell lines miR-21 was clearly overexpressed. Oncogenic
mutations like an activating mutation of the epidermal growth
factor receptor (EGFR) and K-Ras mutation had no obvious influence
on miR-21 expression. The two cell lines with the activated EGFR,
CRL2868 and CRL5883, expressed ∼1.6- and 0.7-fold of the level
measured in normal lung cells, respectively (Fig. 2B). Additionally, the cell lines
were grouped into cell lines harboring a K-Ras mutation and cell
lines with unaffected K-Ras alleles (20). The cell lines with the mutated
K-Ras had on average higher miR-21 levels compared to the cell
lines with the wild-type gene (Fig.
2C). However, possibly due to the small sample size the
increase failed to be significant. To further evaluate the
influence of oncogenic Ras on miR-21 levels, we expressed dominant
active versions of H-Ras, K-Ras and N-Ras in WI-38 cells using the
adenoviral system (21). As
illustrated in Fig. 2D oncogenic
Ras had no influence on miR-21 expression, indicating that Ras is
not an important determinant of miR-21 expression.
MiR-21 levels are constant throughout the
cell cycle
Although the differences of miR-21 within the
measured collection of cell lines varied ∼3-fold, in context of the
data obtained from the analysis of the tissue sections, it is
surprising that none of the cell lines exhibited a >2-fold
increase of miR-21 compared to a normal cell line. This difference
could be due to the fact that the miRNA levels of logarithmically
growing cells were analyzed, whereas the proportion of
proliferating cells may differ fundamentally in the tissue samples.
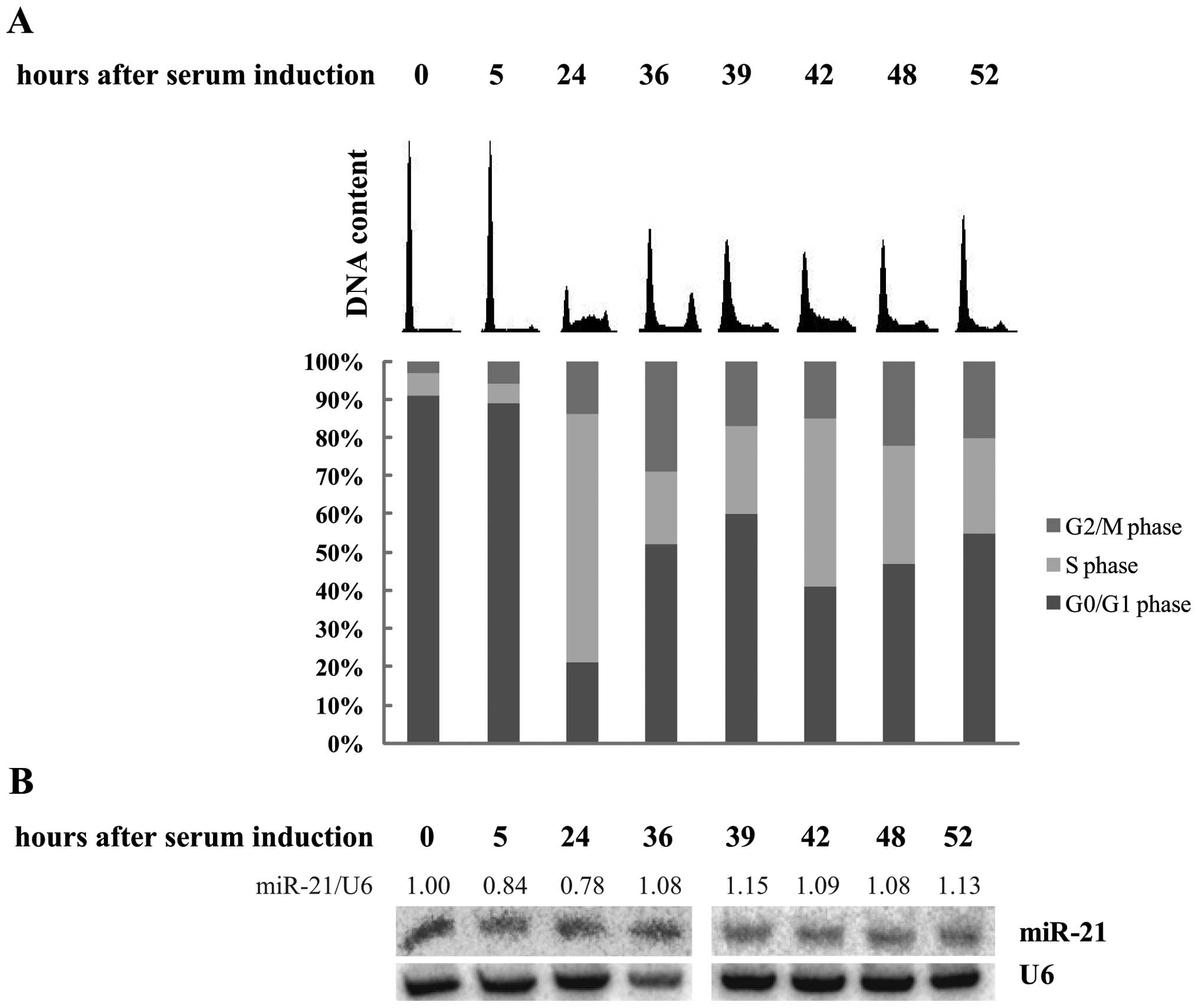
In order to investigate if miR-21 levels are dependent on
proliferation status, we arrested WI-38 cells by serum starvation
for 3 days and induced cell cycle by addition of medium containing
20% serum. At different time-points cell synchrony was monitored
and miR-21 levels were determined. Cells were efficiently blocked
in G0 phase in serum-free medium, and ∼80% of the cells re-entered
the cell cycle after serum addition. Five hours after cell cycle
induction, when cells are expected in G1 phase, DNA content did not
change, while 24 h after serum release, most of the cells were
clearly in S phase (65% of the cells), and 12 h later the majority
of cells had passed through DNA replication and accumulated with 4N
DNA content, and then again showed a G1 peak, a DNA distribution
typically for cells around mitosis (Fig. 3A). At later time-points cells were
growing rather asynchronously. Northern blot analysis revealed that
miR-21 is constantly expressed throughout the cell cycle (Fig. 3B). These data indicate that the
observed differences in the results obtained from tissue and cell
line analyses were not due to variations in proliferation
status.
MiR-21 levels are increased in NSCLC
tissue, but not in tumor-derived cells
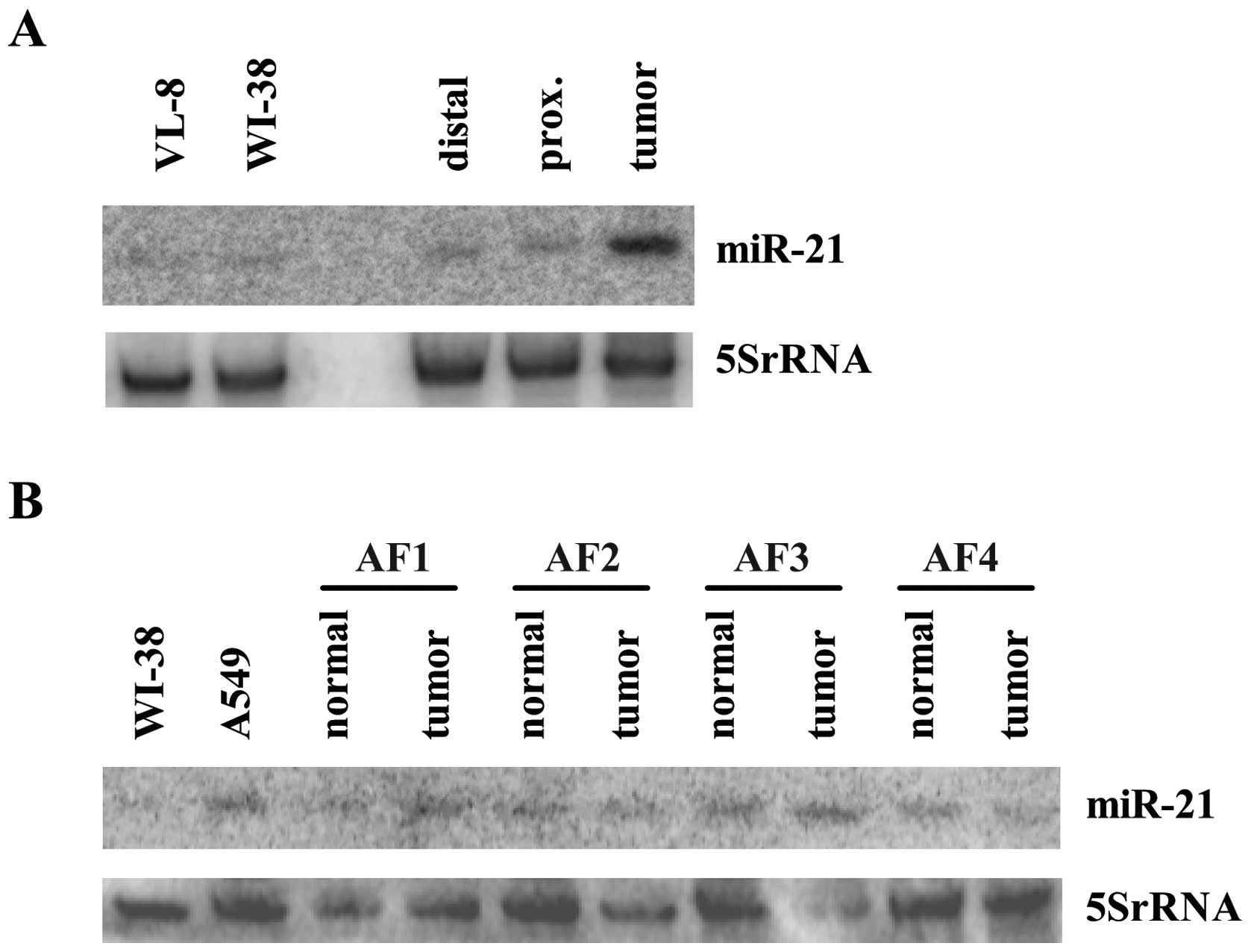
To directly compare the miR-21 levels in cell lines
with those in tissue, we performed a northern blot loaded with RNA
from the tissue samples of a patient together with RNA from two
logarithmically growing cell lines. As demonstrated in Fig. 4A, miR-21 expression measured in
cell lines is comparable to the one measured in normal tissue. In
contrast, miR-21 is strongly induced in the sample derived from the
tumor tissue. Therefore, we conclude that the observed miR-21
increase in the tissue is not primarily caused by tumor
cell-specific alterations. To investigate if the stromal cells of
the tumor rather than the cancer cells are responsible for the
increased miR-21 expression, fibroblasts from the tumor and the
normal lung section of 4 patients diagnosed with NSCLC were
isolated. In none of the 4 patients the miR-21 levels in the
tumor-associated fibroblasts (TAFs) were clearly increased compared
to the levels measured in the corresponding sample of the
fibroblasts isolated from the normal lung tissue of the patient
(Fig. 4B). In relation to cultured
primary fibroblasts (WI-38) and to one of the high expressing tumor
cell line (A549) the fibroblasts show comparable endogenous levels
of miR-21 (Fig. 4B) indicating
that the high miR-21 expression detected in tumor tissue is not
caused by the stromal compartment but instead is due to conditions
specific for tumor tissue. These special circumstances are
obviously not mimicked by standardized cell culture.
MiR-21 levels are not influenced by
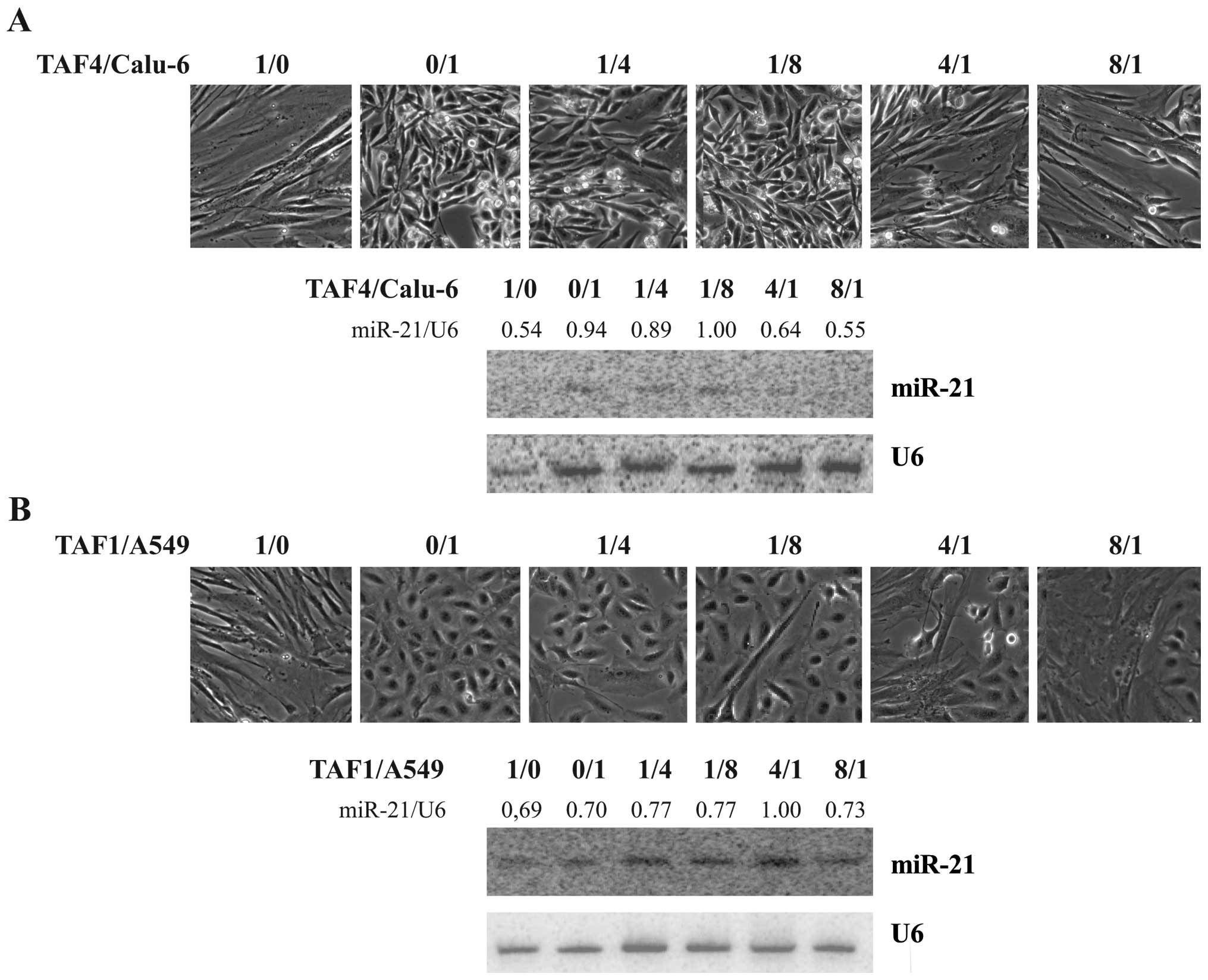
co-cultivation of tumor-derived cells with fibroblasts
Recently, cancer is not only seen as a disease of
transformed cells but as the consequence of an interaction of the
cancer cells with stromal components. Fibroblasts are important
components of the tumor micro-environment and paracrine stimulation
pathways between cancer cells and the associated fibroblasts are
thought to influence gene expression in both cell types (25). To mimic tumor stromal interactions,
we co-cultivated TAFs with NSCLC-derived cell lines for 3 days. The
two cell lines (A549 and Calu-6) were chosen and seeded as a
mixture in varying ratios as demonstrated in Fig. 6. The growth performance of the
chosen cells was not strongly influenced by co-cultivation (upper
panel of Fig. 5A and B).
Endogenous miR-21 level in TAF4 is on average about half of the one
detected in Calu-6 cells, and miR-21 expression of the co-culture
reflects the level measured in the predominant cell line (lower
panel Fig. 5A). Endogenous miR-21
expression in A549 is comparable to the levels detected in TAF1
cells, and co-cultivation had no influence on miR-21 expression
(Fig. 5B). These data indicate
that the signals secreted by the stromal fibroblasts are not
responsible for augmentation of miR-21 expression in cancer
cells.
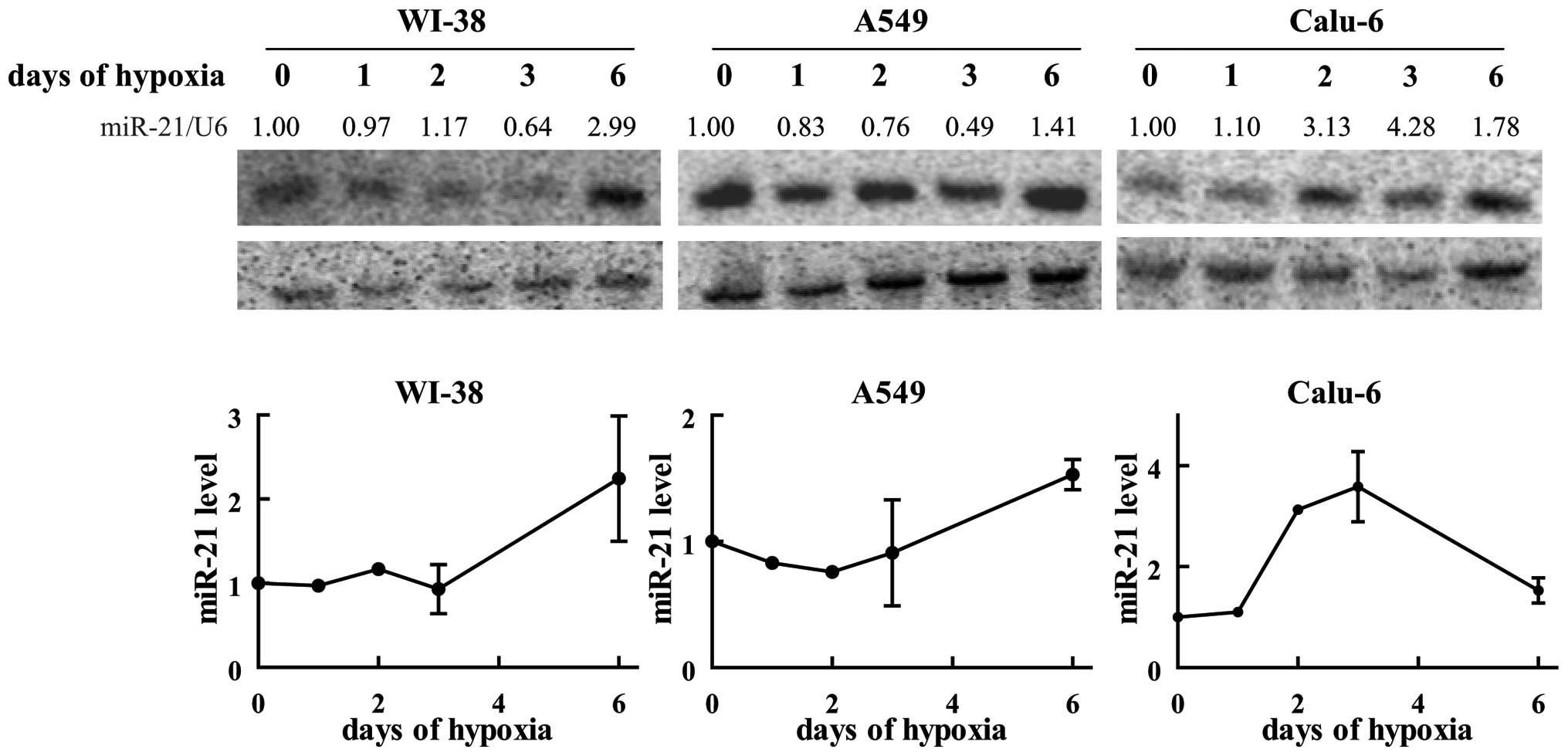
Hypoxic conditions increase miR-21
levels
Low oxygen availability is a characteristic growth
condition of many fast growing tumor masses (25). In order to investigate the role of
hypoxia on miR-21 expression, WI-38 as well as two NSCLC-derived
cell lines (A549 and Calu-6) were incubated for different time
periods at low oxygen levels (1%) and the miR-21 levels were
compared to the one of cells growing under normoxic conditions.
While at earlier time-points hypoxia failed to influence miR-21
expressions, in WI-38 as well as in the cancer cell line A459
miR-21 levels increased ∼1.5-3-fold when cells were incubated for 6
days in low oxygen (Fig. 6). In
Calu-6, miR-21 levels increased already after 2 days in hypoxia and
reached their peak (a 3–4-fold increase compared to normoxic
levels) at day 3, while at day 6 the levels were diminished again
(Fig. 6). In contrast to the other
two tested cell lines, an obvious fraction of Calu-6 cells was
already dying at day 6 (data not shown), indicating that these
cells are more sensitive to oxygen deficiency. Therefore, we cannot
exclude that the decrease of miR-21 levels after 6 days is
connected to cell death induced by the lowered oxygen availability.
Nonetheless, these data demonstrate that hypoxic environment is one
determinant responsible for the detected increase of miR-21 levels
in the tumor tissue.
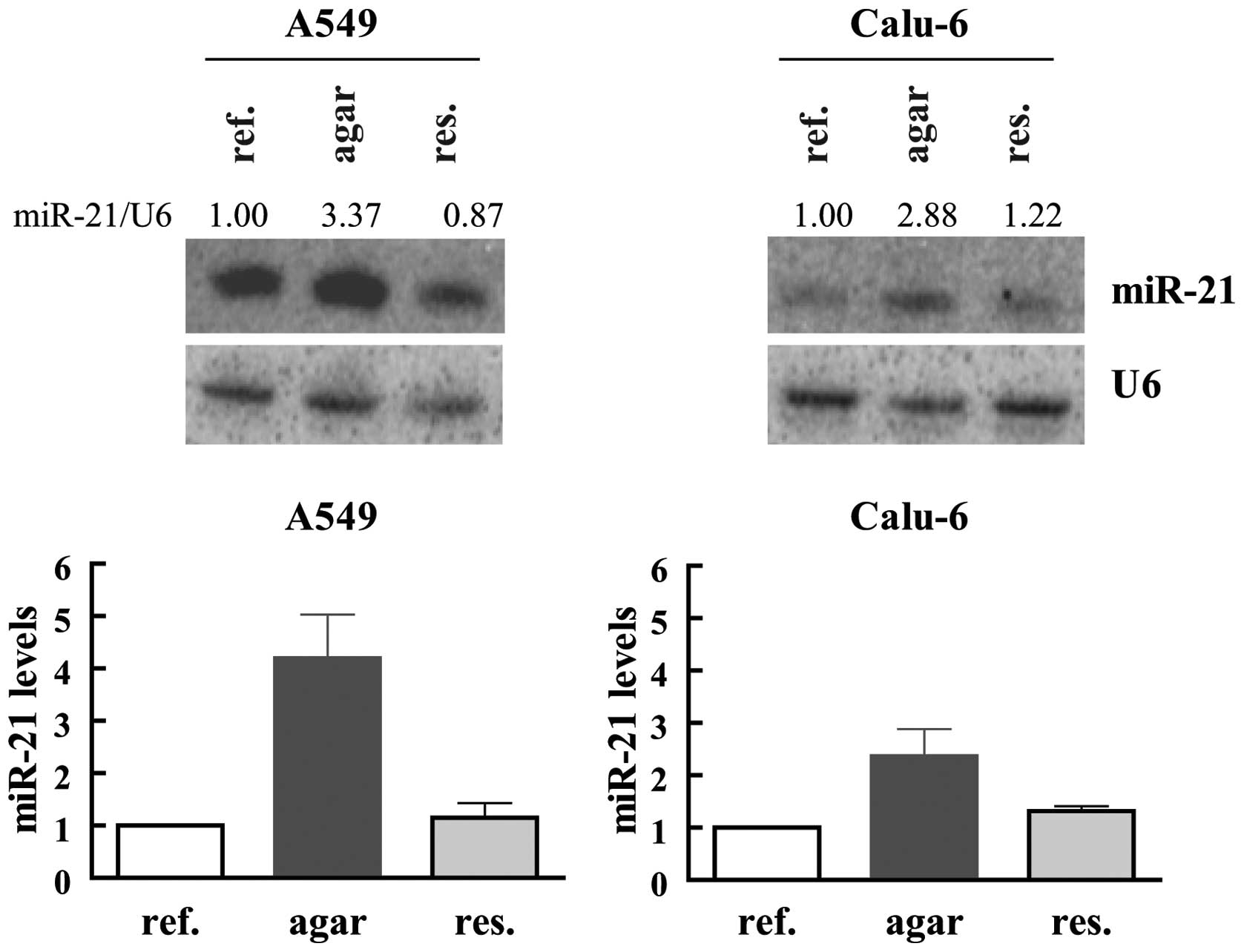
Anchorage-independent growth causes
augmented miR-21 levels
In order to invade adjacent tissues and to
disseminate through the body, tumor cells need to acquire the
ability to avoid cell detachment-induced apoptosis and to become
anchorage-independent (26). To
analyze the influence of anchorage-independent growth conditions on
miR-21 expression, the tumor cells were transferred into agar
coated plates preventing cellular attachment to the plate. While
most of the Calu-6 cells formed clearly visible multicellular
spheroids after 5 days of cultivation, in A549 cell line, at this
time only few growing spheroid–like structures were visible.
Anchorage-independent growing spheroids of Calu-6 and A549 were
further cultivated for 1 and 3 weeks, respectively, before RNA was
isolated. In both cases miR-21 was clearly increased when cells
were forced to detach (Fig. 7).
The increase was much more pronounced in the A549 cells (∼3–5-fold)
as compared to Calu-6 cells (∼2–3-fold). Although at the time when
cells were harvested both cell lines formed nicely growing spheres
without visible single cells, it is possible that the difference in
the amplitude of miR-21 increase is caused by a stronger selection
of especially high miR-21 expressing A549 cells during the longer
adaption process to anchorage-independent growth. To test this
hypothesis, spheroids were re-seeded and cultivated to monolayers
before cells were harvested. In both cell lines miR-21 was
downregulated to the levels detected in logarithmically growing
cells (Fig. 7). These data show
that anchorage-independent growth conditions result in a distinct
and reversible increase of miR-21 expression.
Discussion
MicroRNAs are important regulators of gene
expression. Since one miRNA can target hundreds of mRNAs, changing
its abundance can have an important influence in reprogramming
expression systems and adapting to new circumstances. Therefore
miRNAs have important functions in many cellular processes as well
as in development and diseases including cancer. One of the miRNAs
frequently found to be deregulated in malignant tissue is
miR-21.
In line with previous reports (27,28),
we demonstrated that in lung tissues derived from patients with
NSCLC, miR-21 is significantly increased in the tumor tissue when
compared with non-malignant lung samples. While in these earlier
studies in a huge percentage of samples miR-21 was undetectable, in
our study miR-21 was clearly detectable in all patients and
variations in the normal lung tissue were minimal. In most of the
tumor sections miR-21 levels were clearly increased and the extent
of miR-21 elevation increased with malignancy. Tumors staged as IIB
and IIIA showed significantly higher miR-21 expression than tumors
staged as IA. Possibly due to the differences in detection, the
former studies found no connection between staging and miR-21
levels (27,28). In accordance with the earlier
investigations (28), we observed
a tendency of higher miR-21 amounts in adenocarcinoma compared to
squamous cell carcinoma, although the difference was not
significant.
In contrast to the observations in the tissue
analysis, none of the analyzed NSCLC-derived cell lines (n=14)
expressed clearly elevated miR-21 level compared to normal lung
derived cells. This discrepancy cannot be explained by a possible
difference in the proliferating status of the cells within the
tissue sample and cell culture, since our data demonstrate that
miR-21 levels do not fluctuate throughout the cell cycle. Earlier
reports described that EGFR (29)
and K-Ras mutations (30,31) can influence miR-21 levels in lung
cancer-derived tissue. Our cell line panel included also cell lines
harboring activating mutations in K-Ras or EGFR, but both mutations
failed to influence miR-21 levels. This can be due the fact that
our sample size is not representative, but it is possible that
tumor tissue specific factors are necessary for miR-21
elevation.
In line with this assumption, we observed that all
investigated tumor-derived cells cultured under standardized
conditions express levels of miR-21 comparable to the one measured
in normal lung tissue-derived samples, while the majority of tissue
samples clearly increased miR-21 expression levels. Additional to
the cancer cells, isolated tumor-associated fibroblasts from lung
cancer patients were investigated and also these cells express low
levels of miR-21 comparable to the one isolated from the normal
lung tissue. This is in agreement with reported in situ
hybridization studies demonstrating that in lung cancer tissue
miR-21 is predominantly expressed in the cancer cells (32).
Our data demonstrating that tumor-derived cell lines
cultured under standardized conditions fail to overexpress miR-21
could explain why in colon cancer two studies in cell lines found
that the tumor/normal ratio of miR-21 is negative (33) or low (34), while in contrast reports analyzing
miR-21 in colorectal cancer tissue showed that miR-21 levels are
significantly increased in the tumor (8,9).
Therefore, we conclude that the tissue-specific tumor environment
might be necessary for miR-21 elevation.
Although the reprogrammed tumor-associated
fibroblasts are expressing low miR-21 amounts, we cannot exclude
that miR-21 levels in the cancer cells may be influenced by the
tumor-adapted fibroblasts via released factors different from the
one secreted by normal lung fibroblasts (35). In our experimental set-up
co-cultivation with fibroblasts fails to influence miR-21
expression, and thereby fail to support the hypothesis that in
cancer cells paracrine signals from the stromal compartment are
responsible for the miR-21 increase.
Apart from the interplay between the cancer cells
and the associated fibroblasts, low oxygen levels is a
characteristic microenviromental feature especially of larger
tumors. Hypoxic conditions induced miR-21 levels in all tested lung
cell lines. Corroborating, analysis of colon (36) and breast cell lines revealed that
miR-21 is also induced in these tissues although with other
kinetics (37). Elevated miR-21 in
response to lowered oxygen level was shown to target CDC25A and
thereby interferes with cell proliferation (36). Additionally it was shown that
miR-21 elevation positively influences VEGF expression. A function
in facilitating angiogenesis could be another role of miR-21
(38).
Furthermore we demonstrated that miR-21 levels are
elevated when cells lose their usual extracellular matrix and are
forced to grow anchorage-independent in spheres. In line with our
data it is reported that ectopic miR-21 expression facilitates
sphere and tumor formation in SCID mice (39). Corroborating, blocking of miR-21
activity interferes with the metastatic behavior of melanoma cells
(40) and breast cells (41). Identified targets, which would
explain an important role of miR-21 in invasion and metastasis, are
Tropomyosin (42), Reck and TIMP3
(43). Since in our studies miR-21
elevation in spheres was a reversible process we can not
substantiate earlier studies showing that cancer stem cells
selected as consequence of chemo-resistance exhibit strongly
upregulated miR-21 levels (39).
In conclusion, our data show that in lung cancer
miR-21 is enriched especially in higher graded tumors and suggest
that this upregulation is an adaption of the cancer cells to the
tumor-specific environment.
Acknowledgements
We thank Andreas Baierl for helpful
support in statistical questions. Furthermore we are thankful to
Daniel Drev for skilful technical assistance, and Rosana Kral,
Christoph-Erik Mayer and Florian Jantscher for supporting tissue
sample collection. This study was supported by Jubiläumsfonds der
Österreichischen Nationalbank No. 13720 and by Herzfelder’sche
Familienstiftung 09.
References
|
1.
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Landgraf P, Rusu M, Sheridan R, et al: A
mammalian microRNA expression atlas based on small RNA library
sequencing. Cell. 129:1401–1414. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar
|
|
5.
|
Jazbutyte V and Thum T: MicroRNA-21: from
cancer to cardiovascular disease. Curr Drug Targets. 11:926–935.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Volinia S, Calin GA, Liu CG, et al: A
microRNA expression signature of human solid tumors defines cancer
gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Avissar M, McClean MD, Kelsey KT and
Marsit CJ: MicroRNA expression in head and neck cancer associates
with alcohol consumption and survival. Carcinogenesis.
30:2059–2063. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Slaby O, Svoboda M, Fabian P, et al:
Altered expression of miR-21, miR-31, miR-143 and miR-145 is
related to clinicopathologic features of colorectal cancer.
Oncology. 72:397–402. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Yamamichi N, Shimomura R, Inada K, et al:
Locked nucleic acid in situ hybridization analysis of miR-21
expression during colorectal cancer development. Clin Cancer Res.
15:4009–4016. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Yanaihara N, Caplen N, Bowman E, et al:
Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Iorio MV, Ferracin M, Liu CG, et al:
MicroRNA gene expression deregulation in human breast cancer.
Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Yan LX, Huang XF, Shao Q, et al: MicroRNA
miR-21 overexpression in human breast cancer is associated with
advanced clinical stage, lymph node metastasis and patient poor
prognosis. RNA. 14:2348–2360. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Feber A, Xi L, Luketich JD, et al:
MicroRNA expression profiles of esophageal cancer. J Thorac
Cardiovasc Surg. 135:255–260. 2008. View Article : Google Scholar
|
|
14.
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Thum T, Gross C, Fiedler J, et al:
MicroRNA-21 contributes to myocardial disease by stimulating MAP
kinase signalling in fibroblasts. Nature. 456:980–984. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Sayed D, Rane S, Lypowy J, et al:
MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Mol
Biol Cell. 19:3272–3282. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Wang P, Zou F, Zhang X, et al: microRNA-21
negatively regulates Cdc25A and cell cycle progression in colon
cancer cells. Cancer Res. 69:8157–8165. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Frankel LB, Christoffersen NR, Jacobsen A,
Lindow M, Krogh A and Lund AH: Programmed cell death 4 (PDCD4) is
an important functional target of the microRNA miR-21 in breast
cancer cells. J Biol Chem. 283:1026–1033. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Asangani IA, Rasheed SA, Nikolova DA, et
al: MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor
suppressor Pdcd4 and stimulates invasion, intravasation and
metastasis in colorectal cancer. Oncogene. 27:2128–2136. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Sutterluty H, Mayer CE, Setinek U, et al:
Down-regulation of Sprouty2 in non-small cell lung cancer
contributes to tumor malignancy via extracellular signal-regulated
kinase pathway-dependent and -independent mechanisms. Mol Cancer
Res. 5:509–520. 2007. View Article : Google Scholar
|
|
21.
|
Mayer CE, Haigl B, Jantscher F, et al:
Bimodal expression of Sprouty2 during the cell cycle is mediated by
phase-specific Ras/MAPK and c-Cbl activities. Cell Mol Life Sci.
67:3299–3311. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Jantscher F, Pirker C, Mayer CE, Berger W
and Sutterluety H: Overexpression of Aurora-A in primary cells
interferes with S-phase entry by diminishing Cyclin D1 dependent
activities. Mol Cancer. 10:282011. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Haigl B, Mayer CE, Siegwart G and
Sutterluty H: Sprouty4 levels are increased under hypoxic
conditions by enhanced mRNA stability and transcription. Biol Chem.
391:813–821. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Sutterluety H, Bartl S, Doetzlhofer A,
Khier H, Wintersberger E and Seiser C: Growth-regulated antisense
transcription of the mouse thymidine kinase gene. Nucleic Acids
Res. 26:4989–4995. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: the next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Taddei ML, Giannoni E, Fiaschi T and
Chiarugi P: Anoikis: an emerging hallmark in health and diseases. J
Pathol. 226:380–393. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Markou A, Tsaroucha EG, Kaklamanis L,
Fotinou M, Georgoulias V and Lianidou ES: Prognostic value of
mature microRNA-21 and microRNA-205 overexpression in non-small
cell lung cancer by quantitative real-time RT-PCR. Clin Chem.
54:1696–1704. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Voortman J, Goto A, Mendiboure J, et al:
MicroRNA expression and clinical outcomes in patients treated with
adjuvant chemotherapy after complete resection of non-small cell
lung carcinoma. Cancer Res. 70:8288–8298. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Seike M, Goto A, Okano T, et al: MiR-21 is
an EGFR-regulated anti-apoptotic factor in lung cancer in
never-smokers. Proc Natl Acad Sci USA. 106:12085–12090. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Frezzetti D, De Menna M, Zoppoli P, et al:
Upregulation of miR-21 by Ras in vivo and its role in tumor growth.
Oncogene. 30:275–286. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Hatley ME, Patrick DM, Garcia MR, et al:
Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21.
Cancer Cell. 18:282–293. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Sempere LF, Preis M, Yezefski T, et al:
Fluorescence-based codetection with protein markers reveals
distinct cellular compartments for altered MicroRNA expression in
solid tumors. Clin Cancer Res. 16:4246–4255. 2010. View Article : Google Scholar
|
|
33.
|
Gaur A, Jewell DA, Liang Y, et al:
Characterization of microRNA expression levels and their biological
correlates in human cancer cell lines. Cancer Res. 67:2456–2468.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Bandres E, Cubedo E, Agirre X, et al:
Identification by real-time PCR of 13 mature microRNAs
differentially expressed in colorectal cancer and non-tumoral
tissues. Mol Cancer. 5:292006. View Article : Google Scholar
|
|
35.
|
Bremnes RM, Donnem T, Al-Saad S, et al:
The role of tumor stroma in cancer progression and prognosis:
emphasis on carcinoma-associated fibroblasts and non-small cell
lung cancer. J Thorac Oncol. 6:209–217. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
de Oliveira PE, Zhang L, Wang Z and Lazo
JS: Hypoxia-mediated regulation of Cdc25A phosphatase by p21 and
miR-21. Cell Cycle. 8:3157–3164. 2009.PubMed/NCBI
|
|
37.
|
Kulshreshtha R, Ferracin M, Wojcik SE, et
al: A microRNA signature of hypoxia. Mol Cell Biol. 27:1859–1867.
2007. View Article : Google Scholar
|
|
38.
|
Liu LZ, Li C, Chen Q, et al: MiR-21
induced angiogenesis through AKT and ERK activation and HIF-1alpha
expression. PLoS One. 6:e191392011. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Yu Y, Kanwar SS, Patel BB, et al:
MicroRNA-21 induces stemness by downregulating transforming growth
factor beta receptor 2 (TGFbetaR2) in colon cancer cells.
Carcinogenesis. 33:68–76. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Yang CH, Yue J, Pfeffer SR, Handorf CR and
Pfeffer LM: MicroRNA miR-21 regulates the metastatic behavior of
B16 melanoma cells. J Biol Chem. 286:39172–39178. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Zhu S, Wu H, Wu F, Nie D, Sheng S and Mo
YY: MicroRNA-21 targets tumor suppressor genes in invasion and
metastasis. Cell Res. 18:350–359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42.
|
Zhu S, Si ML, Wu H and Mo YY: MicroRNA-21
targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol
Chem. 282:14328–14336. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Gabriely G, Wurdinger T, Kesari S, et al:
MicroRNA 21 promotes glioma invasion by targeting matrix
metalloproteinase regulators. Mol Cell Biol. 28:5369–5380. 2008.
View Article : Google Scholar : PubMed/NCBI
|