Introduction
Each year, 600,000 new cases of head and neck
squamous cell carcinoma (HNSCC) are diagnosed worldwide (1). Common risk factors for most forms of
HNSCC include heavy consumption of tobacco and/or alcohol (2), although the oropharyngeal squamous
cell carcinoma (OPSCC) is less likely to be associated with tobacco
and alcohol exposure and more often correlated with human
papillomavirus (HPV) infection (3). The incidence of OPSCC associated with
HPV infection is increasing; for example, among cases of tonsillar
cancer in Stockholm, HPV-positive cases rose from 23% in the 1970s
to 57% in the 1990s and 79% from 2000 to 2007 (4). Moreover, alongside tobacco and
alcohol, high-risk HPV variants (HR-HPVs) have emerged as risk
factor for HNSCC, including OPSCC (5).
HNSCC patients who are HPV positive have
substantially better prognosis than those who are HPV negative
(6–10). Although the detection of
E6/E7 mRNA transcripts is regarded as the gold standard for
the presence of clinically relevant (active) HPV (11), the requirement of unfixed (fresh
frozen) tissue and the cost of polymerase chain reaction (PCR) make
direct detection of E6/E7 impractical for cancer diagnostics
at present. Accordingly, many studies have attempted to identify an
easily measured surrogate maker for the diagnosis of HPV-associated
HNSCC.
Expression of the tumor suppressor
p16INK4a has been proposed as a surrogate marker for HPV
infection: its over-expression is thought to reflect the presence
of biologically active HPV infection given that functional
inactivation of pRb by viral E7 induces p16INK4a
upregulation. Detection of p16INK4a expression can also
be performed using formalin-fixed, paraffin-embedded (FFPE) samples
(11–13). However, there is controversy as to
whether p16INK4a expression reliably indicates HPV
infection (11,12).
Klaes et al classified p16INK4a
staining as negative (<1% of cells positive), sporadic (<5%
of cells positive), focal (<25% of cells positive) or diffuse
(>25% of cells positive) (14).
Other studies have defined p16INK4a expression in tumors
as strong and diffuse when ≥70% of cells (cytoplasm and nuclei) are
stained (15–17), while Fischer et al assessed
tumors as p16INK4a positive when ≥5% of cells were
immunopositive (18). These
diverse scoring systems may lead to significant discrepancies
across studies in the relationship between HPV infection and
p16INK4a expression. Furthermore, p16INK4a
expression has been observed in tumor-free tonsillar tissue without
HPV infection, implicating other mechanisms in p16INK4a
upregulation (19). Bussu et
al concluded that it is unnecessary to measure a surrogate,
such as p16INK4a expression, for objective, reliable,
and direct diagnosis because HPV nucleic acids can be detected by
PCR without requiring subjective assessments by histopathologists
(17).
In this study, we evaluated the relationship between
HPV infection and p16INK4a expression and the value of
both HPV-positive status and p16INK4a expression levels
for HNSCC prognosis using tissue samples from a well-characterized
cohort of Japanese patients with HNSCC receiving curative
treatment. We measured the presence of HPV DNA, HPV E6/E7
mRNA expression using fresh-frozen samples and measured
p16INK4a expression using FFPE samples.
Materials and methods
Subjects and study design
The eligibility criteria for this study were as
follows: the presence of previously untreated, pathologically
confirmed primary HNSCC without distant metastasis (M0); receiving
curative treatment of surgery alone, surgery combined with
radiation therapy (RT) or chemoradiotherapy (CRT), concurrent
chemoradiotherapy (CCRT) or RT alone with >66 Gy of total
dosage; and complete remission after primary treatment. The
treatment modalities were determined according to tumor location,
tumor stage, response to induction chemotherapy, and general
physical condition, not by the results of HPV status and
p16INK4a in tumor tissue. Based on these criteria, 150
patients treated by the Department of Otorhinolaryngology, Head and
Neck Surgery, University of the Ryukyus, Japan were recruited
between October 2006 and June 2013. In contrast to our previous
study on HPV status and squamous cell carcinoma antigen (SCCA),
this study involved a greater total number of cases who met the
inclusion criteria and we also updated the prognostic information
of some of the patients reported previously (8). Each patient gave written informed
consent before enrolment. The research protocol was approved by the
Ethics Committee of the University of the Ryukyus. Tissue samples
were snap-frozen in liquid nitrogen during biopsy or surgical
excision and stored in liquid nitrogen until further analysis.
Demographic and clinicopathologic parameters for each patient were
collected at scheduled intervals during the follow-up period.
Cell lines and culture
The cervical cancer cell lines CaSki (harboring ∼600
copies of integrated HPV-16 DNA/genome) and SiHa (1–2 copies of
integrated HPV-16 DNA/genome) were purchased from the European
Collection of Animal Cell Cultures (Salisbury, UK) and the American
Type Culture Collection (Tokyo, Japan), respectively, and cultured
according to the suppliers’ instructions.
DNA and RNA extraction
Genomic DNA and total RNA were extracted from frozen
tumor samples, SiHa cells and CaSki cells using the Gentra
Purification Tissue kit (Qiagen, Germantown, MD) and the ToTALLY
RNA™ kit (Ambion, Austin, TX), respectively, according
to the manufacturers’ protocols. The extracted RNA was suspended in
50 μl ultra-high quality diethyl pyrocarbonate-treated
water.
PCR for detection of HPV DNA
The presence and integrity of the DNA in all samples
was verified by PCR β-globin gene amplification using the primers
PC04 and GH20 (20). Water
(negative control) and DNA from HPV-16-positive CaSki cells
(positive control) were included in each amplification series. The
presence of HPV DNA was analyzed by PCR using the general consensus
primer sets GP5+/GP6+ and MY09/11 (21,22).
DNA samples that were negative for HPV using
GP5+/GP6+ or MY09/11 PCR were re-amplified by
(auto-) nested PCR using the GP5+/GP6+ primer
pair as previously described (23). PCR products were purified and
directly sequenced with an ABI PRISM 3130xl Genetic Analyzer
(Applied Biosystems, Foster City, CA). Obtained sequences were
aligned and compared with those of known HPV types in the GenBank
database using the BLAST program.
Detection of HPV E6/E7 mRNA by reverse
transcription PCR
Before cDNA synthesis, any residual DNA was removed
by incubation with 1 U DNase I (Ambion) at room temperature for 25
min. cDNA was then synthesized from DNA-free total RNA using the
RETROscript® kit (Ambion) according to the
manufacturer’s instructions. To examine the presence of
contaminating DNA in RNA samples, all the assays were performed
both with and without reverse transcriptase.
To detect high-risk E6/E7 mRNA transcripts,
PCR was performed with the cDNA from HPV DNA-positive samples using
the Takara PCR Human Papillomavirus Typing Set (Takara, Bio Inc.,
Otsu, Shiga, Japan), which can identify high-risk HPV types 16, 18,
31, 33, 35, 52 and 58. To verify the HPV-16 E6/E7 mRNA
transcripts, the HPV-16 DNA-positive samples were also examined
using a half-nested PCR approach with cDNA as previously described
by Wiest et al (24).
Positive PCR products were purified and directly sequenced using an
ABI PRISM 3130xl Genetic Analyzer (Applied Biosystems).
Immunohistochemistry for
p16INK4a and scoring of results
Serial 4-μm-thick sections from FFPE tumor
samples were deparaffinized in a graded alcohol series. Epitope
retrieval was performed by heating at 95–99° C for 10 min in
Tris/EDTA buffer (pH 9.0). Endogenous peroxidase activity was
quenched by incubating the sections in 3% hydrogen peroxide plus 15
mM sodium azide for 5 min. The sections were subsequently incubated
overnight at 4°C with primary monoclonal mouse
anti-p16INK4a antibody (MTM Laboratories AG, Heidelberg,
Germany). After extensive washing in phosphate-buffered saline, the
slides were incubated for 30 min at room temperature with a
horseradish peroxidase-conjugated goat anti-mouse secondary
antibody (MTM Laboratories). Immunolabeling was visualized by
incubation in 3-3′-diaminobenzidine for 10 min. Stained slides were
counterstained with hematoxylin.
Cases were considered p16INK4a-positive
when intense nuclear and/or cytoplasmic reactivity was present. The
scoring criteria for p16INK4a immunoreactivity
(p16INK4a expression) were defined for this study based
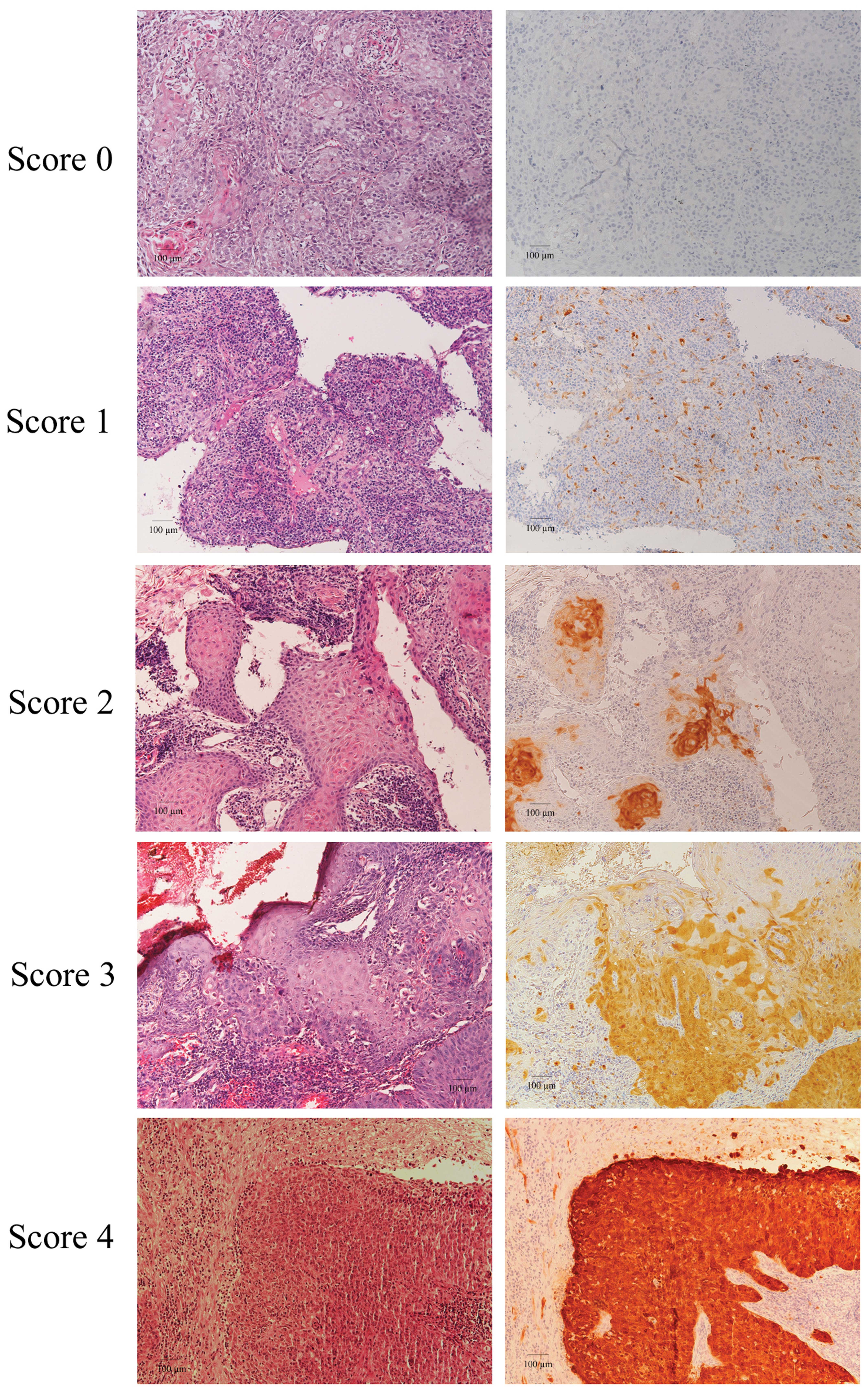
on previous scoring methods (14,15):
0 (no staining), 1 (1–10% of tumor cells positive), 2 (11–40%
positive), 3 (40–70% positive) and 4 (>70% positive). The term
‘p16INK4a Overexpression’ is defined as a score of 3 or
4.
Survival analysis
Descriptive statistics were used to characterize
patient baseline characteristics. The Mann-Whitney U-test or
Kruskal-Wallis test was used for continuous variables, and
Pearson’s χ2 test or Fisher’s exact test was used for
categorical variables. The inter-rater agreements between HPV-DNA
presence and p16INK4a expression and between HPV mRNA
expression and p16INK4a expression were measured by
calculating Cohen’s κ coefficient. A κ-value <0.20 was
considered slight agreement, 0.21–0.40 as fair, 0.41–0.60 as
moderate, 0.61–0.80 as good and 0.81–0.99 as excellent agreement
(17,25–27).
Locoregional control was defined as complete and
persistent disappearance of disease at the primary tumor (T site)
and regional lymph nodes (N site) after treatment. Recurrence-free
survival was defined as the time from the end of treatment to
cancer recurrence or last follow-up. Disease-specific survival was
defined as the time from the end of treatment to subsidence of
disease or last follow-up. Survival curves were evaluated by the
Kaplan-Meier method, and survival distributions were compared using
the log-rank test. Multivariate Cox proportional hazard analysis
was used to identify prognostic parameters and treatments
associated with risk of recurrence and disease-specific death.
P-values <0.05 were considered significant. All statistical
analyses were performed using the SPSS statistical package (SPSS
for Windows version 12.0; SPSS, Inc., Chicago, IL).
Results
Characteristics of eligible patients and
follow-up
Primary tumor location was the nasopharynx in 10
patients (6.7%), oropharynx in 53 (35.3%), hypopharynx in 39
(26.0%), larynx in 24 (16.0%) and oral cavity in 24 (16.0%). The
follow-up period ranged from 6 to 77 months, with a median of 38
months for patients whose data were censored. Sixty-four patients
were treated with CCRT, 45 with surgery and postoperative RT or CRT
(radiation dosage, 50–54 Gy), 22 with surgery alone and 19 with RT
alone. Demographic and clinical characteristics are summarized in
Table I.
 | Table I.Demographic and clinical
characteristics. |
Table I.
Demographic and clinical
characteristics.
| Characteristic | Total no. |
HPV+ |
HPV− | P-value | p16INK4a
overexpression | P-value |
|---|
|
|
|---|
| n=47 | n=103 | (+) n=30 | (-) n=120 |
|---|
| Gender, n
(%)* | | | | | | | |
| Male | 127 | 38 (29.9) | 89 (70.1) | 0.381 | 24 (18.9) | 103 (81.1) | 0.408 |
| Female | 23 | 9 (39.1) | 14 (60.9) | | 6 (26.1) | 17 (73.9) | |
| Age (years) | | | | | | | |
| Mean | 64.1 | 62.6 | 64.8 | 0.286 | 61.8 | 64.7 | 0.216 |
| Range | 28–89 | 39–89 | 28–83 | | 39–89 | 28–86 | |
| ≤50, n (%) | 20 | 9 (45.9) | 11 (55.0) | 0.157 | 7 (35.0) | 13 (65.0) | 0.128 |
| >50, n
(%) | 130 | 38 (29.2) | 92 (70.8) | | 23 (17.7) | 107 (82.3) | |
| Smoking, n
(%)a | | | | 0.066 | | | 0.097 |
| Never | 30 | 14 (46.7) | 16 (53.3) | | 10 (33.3) | 20 (66.7) | |
| ≤400 | 21 | 8 (38.1) | 13 (61.9) | | 5 (23.8) | 16 (76.2) | |
| >400 | 99 | 25 (25.3) | 74 (74.7) | | 15 (15.2) | 84 (84.8) | |
| Alcohol use, n
(%)b | | | | 0.592 | | | 0.138 |
| Never | 25 | 10 (40.0) | 15 (60.0) | | 6 (24.0) | 19 (76.0) | |
| ≤50 | 51 | 15 (29.4) | 36 (70.6) | | 14 (27.5) | 37 (72.5) | |
| >50 | 74 | 22 (29.7) | 52 (70.3) | | 10 (13.5) | 64 (86.5) | |
| T classification, n
(%) | | | | 0.089 | | | 0.052 |
| T1 | 18 | 2 (11.1) | 16 (88.9) | | 1 (5.6) | 17 (94.4) | |
| T2 | 58 | 22 (37.9) | 36 (62.1) | | 17 (29.3) | 41 (70.7) | |
| T3 | 41 | 10 (24.4) | 31 (75.6) | | 5 (12.2) | 36 (87.8) | |
| T4 | 33 | 13 (39.4) | 20 (60.6) | | 7 (21.2) | 26 (78.8) | |
| Node status, n
(%) | | | | | | | |
| N0 or N1 | 83 | 24 (28.9) | 59 (71.1) | 0.477 | 17 (20.5) | 66 (79.5) | 0.870 |
| N2 or N3 | 67 | 23 (34.3) | 44 (65.7) | | 13 (19.4) | 54 (80.5) | |
| TNM stage, n
(%) | | | | | | | |
| Early (I and
II) | 42 | 9 (21.4) | 33 (78.6) | 0.103 | 8 (19.0) | 34 (81.0) | 0.856 |
| Advanced (III and
IV) | 108 | 38 (35.2) | 70 (64.8) | | 22 (20.4) | 86 (79.6) | |
| Differentiation, n
(%) | | | | < 0.001 | | | 0.030 |
| Well | 68 | 13 (19.1) | 55 (80.9) | | 8 (42.1) | 11 (57.9) | |
| Moderate | 63 | 21 (33.3) | 42 (66.7) | | 12 (19.0) | 51 (81.0) | |
| Poor | 19 | 13 (68.4) | 6 (31.6) | | 10 (14.7) | 58 (85.3) | |
| Tumor location, n
(%) | | | | 0.017 | | | 0.002 |
| Hypopharynx | 39 | 7 17.9) | 32 (82.1) | | 3 (7.7) | 36 (92.3) | |
| Oropharynx | 53 | 25 (47.2) | 28 (52.8) | | 20 (37.7) | 33 (62.3) | |
| Oral cavity | 24 | 8 (33.3) | 16 (66.7) | | 2 (8.3) | 22 (91.7) | |
| Larynx | 24 | 4 (16.7) | 20 (83.3) | | 3 (12.5) | 21 (87.5) | |
| Nasopharynx | 10 | 3 (30.0) | 7 (70.0) | | 2 (20.0) | 8 (80.0) | |
HPV DNA status and HPV E6/E7 mRNA
expression
HPV DNA was detected by PCR in 47 of 150 (31.3%)
primary untreated HNSCC specimens, including 30.0% (3/10) of
nasopharyngeal, 47.2% (25/53) of oropharyngeal (18/30 of palatine
tonsil), 17.9% (7/39) of hypopharyngeal, 16.7% (4/24) of laryngeal
and 33.3% (8/24) of oral cavity cases. Among HPV-positive HNSCC
samples, 39 (83.0 %) were infected with HPV-16 and the others were
infected with other high-risk types (4 with HPV-33, 1 with HPV-35,
2 with HPV-58 and 1 with HPV-56).
As two of these HPV-positive samples were
insufficient for RNA assay, E6/E7 mRNA expression by HPV-16,
HPV-33, HPV-35, HPV-58 and HPV-56 was examined by reverse
transcription PCR in 45 samples. The E6 and E7 mRNA
transcripts were detected in 21 of 45 (46.7%) specimens, the
majority from OPSCC cases (18/25 HPV-positive cases), as shown in
Table II.
 | Table II.mRNA expression in HPV-DNA-positive
HNSCC samples from various sites. |
Table II.
mRNA expression in HPV-DNA-positive
HNSCC samples from various sites.
| HPV | Site
| Total (%)a |
|---|
| NP (%) | OP (%) | HP (%)a | LC (%)a | OC (%) |
|---|
|
DNA+/mRNA+ (%) | 1 (33.3) | 18 (72.0) | 0 (0) | 1 (33.3) | 1 (12.5) | 21 (46.7) |
|
DNA+/mRNA− (%) | 2 (66.7) | 7 (28.0) | 6 (100) | 2 (66.7) | 7 (87.5) | 24 (53.3) |
Between the HPV DNA-positive and -negative groups,
there were significant differences in the distribution of
histological differentiation and tumor location; for example, the
HPV DNA-positive group showed poor differentiation in histology and
has a higher occurrence of oropharyngeal carcinoma compared with
HPV DNA-negative group. The p16INK4a overexpression
group showed similar clinical characteristics (Table I).
p16INK4a expression and
correlation with HPV status
In this study, the p16INK4a expression
scoring system (0–4) was based on the percentage of
p16INK4a-positive cells (Fig. 1). Expression of p16INK4a
was observed in 40 of 150 (26.7%) HNSCC samples (Table III). The highest frequency of
p16INK4a expression was found in OPSCC samples, with 22
of 53 (41.5%) samples demonstrating a p16INK4a staining
score ≥1. The sensitivity of p16INK4a staining for
detection of HPV DNA in HNSCC was just 61.7%, as only 29 of 47
HPV-positive cases also had detectable p16INK4a
staining. The specificity was 89.3% (92/103 HPV-negative samples
had a p16INK4a staining score of 0) for all HNSCC cases
(Table III). Although the
correlation between p16INK4a expression and HPV DNA and
the correlation between p16INK4a expression and
E6/E7 mRNA expression in all HNSCC cases proved to be
significant (both P<0.001), the inter-rater agreements were
relatively low (κ=0.53 and 0.54, respectively).
 | Table III.Scoring of p16INK4a
overexpression and its association with HPV DNA in HNSCC and
OPSCC. |
Table III.
Scoring of p16INK4a
overexpression and its association with HPV DNA in HNSCC and
OPSCC.
| Scoring | p16INK4a
expression
| Total |
|---|
p16INK4a
no or lower expression (<3)
| p16INK4a
overexpression (≥3)
|
|---|
| 0 | 1 | 2 | 3 | 4 |
|---|
| HNSCC | HPV
DNA+ | 18 | 3 | 1 | 1 | 24 | 47 |
| HPV
DNA− | 92 | 3 | 3 | 1 | 4 | 103 |
| OPSCC | HPV
DNA+ | 4 | 1 | 0 | 1 | 19 | 25 |
| HPV
DNA− | 27 | 0 | 1 | 0 | 0 | 28 |
In contrast to general p16INK4a
expression, p16INK4a over-expression demonstrated high
specificity for detection of HPV DNA in HNSCC (98/103 HPV-DNA
negative cases did not overexpress p16INK4a) and OPSCC
(28/28 cases). The sensitivity of p16INK4a
overexpression was only 53.2% (25/47 cases) for detection of HPV
DNA in HNSCC, but was considerably better for detection of HPV DNA
in OPSCC at 80% (20/25 cases) (Table
III). However, p16INK4a overexpression indicated the
presence of HPV E6/E7 mRNA expression with high sensitivity
at 90.5% (19/21) and high specificity at 91.3% (116/127) in HNSCC,
and was even more accurate for OPSCC, with sensitivity at 94.4%
(17/18) and specificity at 91.4% (32/35) (Table IV). Moreover, the inter-rater
agreement of p16INK4a overexpression for HPV
E6/E7 mRNA expression status was good for HNSCC (κ=0.69) and
excellent for OPSCC (κ=0.84).
 | Table IV.Correlation between
p16INK4a overexpression and HPV mRNA in HNSCC and
OPSCC. |
Table IV.
Correlation between
p16INK4a overexpression and HPV mRNA in HNSCC and
OPSCC.
| p16INK4a
overexpression | HPV mRNA
| Sensitivity
(%) | Specificity
(%) | PPV (%) | NPV (%) |
|---|
| + | - |
|---|
| HNSCC | | | | | | |
| + (≥3) | 19 | 11 | 90.5 | 91.3 | 63.3 | 98.3 |
| − (<3) | 2 | 116 | | | | |
| OPSCC | | | | | | |
| + (≥3) | 17 | 3 | 94.4 | 91.4 | 85.0 | 97.0 |
| − (<3) | 1 | 32 | | | | |
Based on a combination of HPV DNA status and
p16INK4a overexpression, subjects with OPSCC were
divided into two groups: a double positive group of patients who
were HPV DNA-positive with p16INK4a overexpression and a
single positive-negative or double negative group of patients who
were either HPV DNA-positive without p16INK4a
overexpression, HPV DNA-negative with p16INK4a
overexpression, or HPV-DNAnegative without p16INK4a
overexpression. The combination of HPV DNA status and
p16INK4a overexpression had both high sensitivity
(94.4%) and specificity (100%) for detecting HPV E6/E7 mRNA
expression in OPSCC (Table V).
 | Table V.Relationship between HPV E6/E7
mRNA expression and HPV DNA/p16INK4a expression status
in OPSCC. |
Table V.
Relationship between HPV E6/E7
mRNA expression and HPV DNA/p16INK4a expression status
in OPSCC.
| HPV mRNA expression
| P-value |
|---|
| HPV
DNA/p16INK4a expression status | + | − |
|---|
| HPV DNA+
with | 17 | 0 | <0.001 |
| p16
overexpression | | | |
| Others | 1 | 33 | |
| HPV
DNA+ without p16 overexpression | 1 | 5 | |
| HPV
DNA− with p16 overexpression | 0 | 0 | |
| HPV
DNA− without p16 overexpression | 0 | 28 | |
Prognostic value of HPV status and
p16INK4a overexpression in OPSCC
Within the observation period, 7 of the 53 patients
(13.2%) developed recurrent disease, including 2 of 10 (20.0%)
patients with stage I/II OPSCC and 5 of 43 (11.6%) patients with
advanced stage OPSCC. However, no patient died with disease during
the follow-up period. Since the disease specific rate and overall
survival rate in OPSCC were quite fair in the present study, the
prognostic analysis of OPSCC focused on recurrence-free survival.
Since there were few recurrence-free survival events in OPSCC
cases, multivariate analysis of recurrence-free survival was
carried out instead for HNSCC cases overall.
i) Prognostic value of HPV DNA status, E6/E7
mRNA expression and p16INK4a overexpression in OPSCC.
HPV DNA-positive patients with OPSCC had better recurrence-free
survival than HPV DNA-negative patients with OPSCC (P=0.008)
(Fig. 2). The recurrence-free
survival after 3 years was 72.7% for patients with HPV DNA-negative
OPSCC and 100% for patients with HPV DNA-positive OPSCC. On the
contrary, there were no significant differences in recurrence-free
survival and disease-specific survival between HPV DNA-positive and
-negative patients with non-OPSCC (P=0.139 and 0.144, respectively;
Kaplan-Meier curves not shown).
HPV mRNA-positive OPSCC patients exhibited a trend
toward improved recurrence-free survival compared with HPV
mRNA-negative patients with OPSCC (P=0.051) (Fig. 2). The recurrence-free survival
after 3 years was 78.3% for patients with HPV mRNA-negative OPSCC
and 100% for patients with HPV mRNA-positive OPSCC.
OPSCC patients with p16INK4a
overexpression showed significantly improved recurrence-free
survival compared with OPSCC patients without p16INK4a
overexpression (P=0.034) (Fig. 2).
The recurrence-free survival after 3 years was 100 and 77.1%,
respectively.
ii) Comprehensive evaluation of survival according
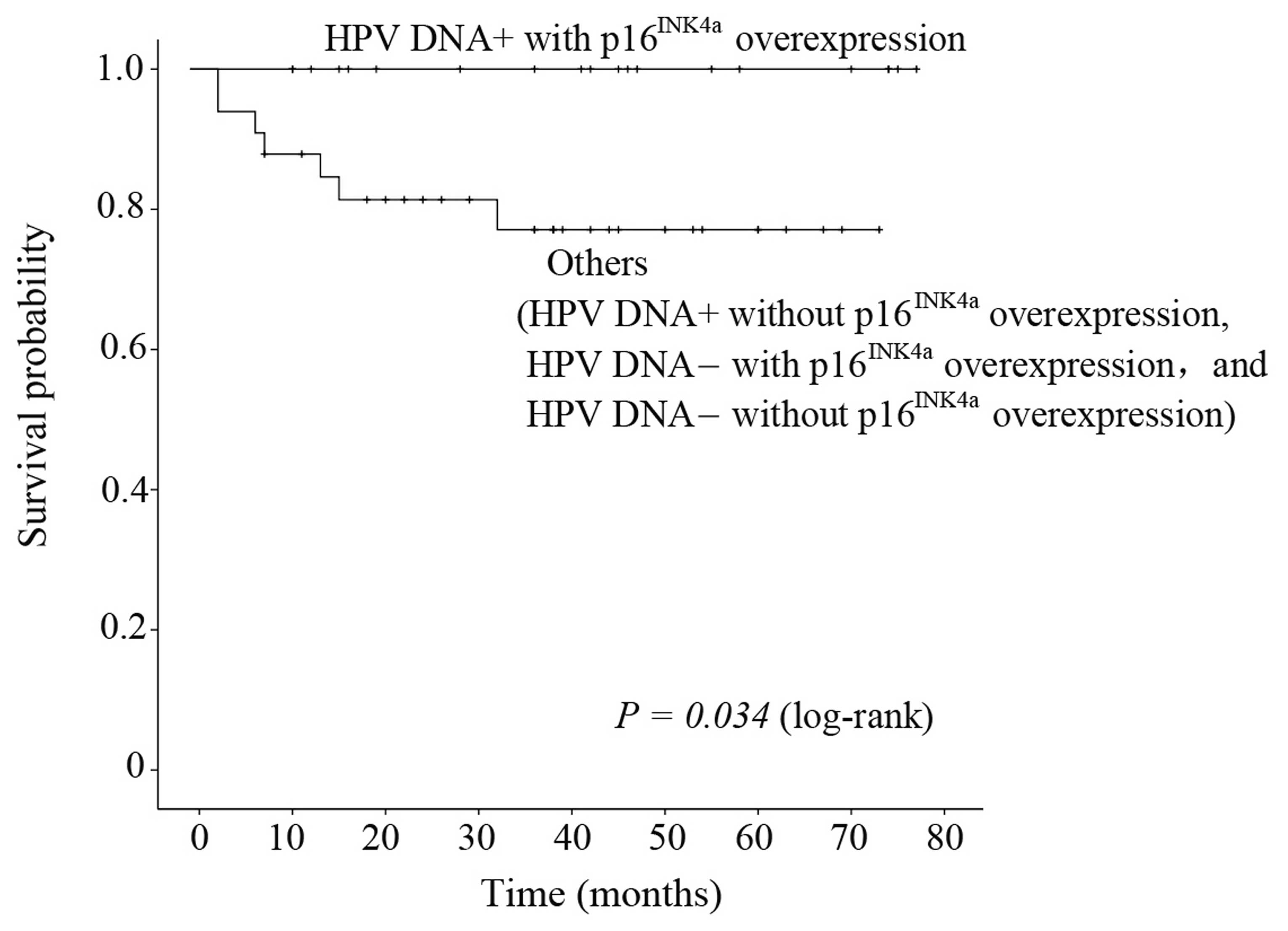
to HPV DNA status and p16INK4a overexpression in OPSCC.
Patients in the double positive group had better recurrence-free
survival than those in a single negative groups or the double
negative group (P=0.034) (Fig. 3),
with recurrence-free survival after 3 years of 100% and 77.1%,
respectively.
iii) Multivariate analysis using Cox
proportional-hazard model in HNSCC. To assess the independent
predictive value of all these factors for recurrence-free survival
in HNSCC, multivariate analysis using Cox proportional-hazards
models was performed. Both HPV DNA presence and p16INK4a
over-expression was modeled as one factor. Since no patients with
positive HPV E6/E7 mRNA expression had any recurrent lesion,
the influence of HPV mRNA expression on recurrence-free survival
could not be evaluated.
In univariate analysis, the HNSCC patients with HPV
DNA-positive status and p16INK4a overexpression
demonstrated significantly higher recurrence-free survival
(P=0.015; hazard ratio (HR) = 8.61; 95% confidence interval (CI) =
1.12–66.18) compared with other HNSCC patients (Table VI). The HNSCC patients categorized
in T stage 4 and those with oral cavity SCC had significantly lower
recurrence-free survival (T4, P=0.003, HR=2.47, 95% CI=1.6–5.76;
oral cavity SCC, P=0.031, HR=3.94, 95% CI=1.25–12.41) compared with
patients categorized in T stages 1–3 and with OPSCC,
respectively.
 | Table VI.Univariate and multivariate analysis
demonstrating the prognostic impact of HPV DNA and
p16INK4a overexpression on recurrence-free survival in
HNSCC. |
Table VI.
Univariate and multivariate analysis
demonstrating the prognostic impact of HPV DNA and
p16INK4a overexpression on recurrence-free survival in
HNSCC.
| Variable | P-value | Univariate analysis
(n=150)
HR (95% CI) | P-value | Multivariate
analysis (n=150)
HR (95% CI) |
|---|
| HPV
DNA/p16INK4a expression (i.e., HPV+ with
p16INK4a vs. others) | 0.015 | 8.61
(1.12–66.18) | 0.043 | 7.81
(1.07–57.19) |
| Age (≤50 vs. >50
years) | 0.399 | 0.64
(0.23–1.82) | | |
| Gender (male vs.
female) | 0.334 | 0.62
(0.23–1.65) | | |
| T stage (T4 vs.
T1–T3) | 0.033 | 2.47
(1.06–5.76) | 0.008 | 2.61
(1.29–5.29) |
| Nodal stage (N2 or
N3 vs. N0 or N1) | 0.942 | 0.97
(0.45–2.10) | | |
| Smoking (yes vs.
never) | 0.559 | 0.76
(0.30–1.91) | | |
| Alcohol consumption
(yes vs. never) | 0.222 | 0.56
(0.22–1.44) | | |
|
Differentiation | | | | |
| Well | | | | |
| Moderate | 0.312 | 0.65
(0.29–1.50) | | |
| Poor | 0.770 | 0.74
(0.22–2.53) | | |
| Tumor location | | | | |
| Oropharynx | | Reference | | Reference |
| Nasopharynx | 0.063 | 4.38
(0.98–19.52) | 0.317 | 1.92
(0.54–5.89) |
| Hypopharynx | 0.217 | 1.97
(0.66–5.86) | 0.865 | 1.09
(0.40–3.02) |
| Larynx | 0.500 | 1.73
(0.49–6.13) | 0.870 | 1.10
(0.35–3.50) |
| Oral cavity | 0.031 | 3.94
(1.25–12.41) | 0.107 | 2.27
(0.84–6.17) |
The final model of multivariate analysis using a Cox
proportional hazards model for identification of fair
recurrence-free survival of HNSCC showed that T1–3 stage (P=0.008;
adjusted HR=2.61; 95% CI=1.29–5.29) and the combination of HPV
DNA-positive status and p16INK4a overexpression
(P=0.043; adjusted HR=7.81; 95% CI=1.07–57.19) predicted
significantly better recurrence-free survival (Table VI).
Discussion
Over the past two decades, HR-HPV has been firmly
established as a common etiologic factor in OPSCC and is now widely
used as a prognostic marker for OPSCC. Although there are a number
of studies on the epidemiologic role and prognostic value of HPV in
OPSCC, some did not distinguish between patients receiving curative
treatment from those receiving palliative care (28–31).
Moreover, there are few studies on the prognostic value of HPV
infection in non-oropharyngeal SCC. In this study, we analyzed the
prognostic value of HPV infection in a retrospectively selected
cohort of HNSCC patients receiving curative treatment. While the
oropharynx was the site of highest prevalence (47.2%), this cohort
also included patients with nasopharyngeal, hypopharyngeal,
laryngeal and oral cavity tumors. The relatively high prevalence of
HPV in cases of nasopharynx and oral cavity SCC suggests that HPV
may play an important role in these non-oropharynx HNSCCs as well
as in OPSCC.
A significant correlation was found between the
presence of HPV DNA and improved recurrence-free survival in OPSCC,
with HPV DNA-negative patients demonstrating an apparent greater
risk of recurrence compared with HPV DNA-positive patients. OPSCC
patients with HPV mRNA expression (active infection) also displayed
improved recurrence-free survival compared with OPSCC patients
without HPV mRNA expression (P=0.051), in line with previous
studies (6,9,32–35).
Although HPV DNA-positive patients with nonoropharyngeal SCCs also
showed better recurrence-free and disease-free survival, the
prognostic value did not reach statistical significance. Since
OPSCC patients accounted for 34% of subjects in our series, the
fair prognostic significance of HPV status for non-OPSCC HNSCC
patients may be influenced by the generally excellent prognosis of
patients with OPSCC. Indeed, Isayeva et al systematically
reviewed the published data regarding the prognostic significance
of HPV in SCCs of the oral cavity, larynx, sinonasal tract and
nasopharynx and found no association between HPV status and
treatment outcome (36). Further
studies are needed to clarify the influence of HPV infection on
prognosis in non-OPSCC cases.
Several studies have suggested that
p16INK4a expression can be used as a surrogate marker
for HPV infection in OPSCC (14,37).
Klaes et al defined p16INK4a staining as
negative, sporadic, focal or diffuse and found a significant
correlation between the presence of HR-HPV and strong diffuse
p16INK4a expression (14). However, there are also several
contradictory reports on the value of p16INK4a as a
biomarker. Smith et al found no concordance between
p16INK4a expression and HPV detection in 20% of head and
neck cancers (38), possibly due
to transcriptionally inactive infection or an alternate pathway of
p16INK4a activation (39). Using the same histological criteria
as Klaes et al (14),
Hoffmann et al reported that 81.2% of HPV DNA-positive HNSCC
patients were also p16INK4a positive, compared with only
48.2% of HPV DNA-negative cases, including 3 cases with strong and
diffuse p16INK4a staining. Moreover, no
p16INK4a expression could be detected in 3 of 14 HPV
DNA+/RNA+ HNSCC lesions in their series. They
concluded that p16INK4a expression status alone is
inadequate for identifying biological active or inactive HPV
infections in HNSCC (40).
In the present study, the sensitivity of general
p16INK4a expression for detecting HPV DNA in HNSCC was
also low, and there was generally low rate of agreement between
p16INK4a-positive status and HR-HPV E6/E7 mRNA
expression in both HNSCC (κ=0.56) and OPSCC (κ=0.57). These results
indicate that p16INK4a expression alone is not suitable
for identifying HPV-related tumors. However, p16INK4a
over-expression (p16INK4a expression score ≥3) was a
sensitive and specific marker for detecting HR-HPV mRNA expression
in both HNSCC and OPSCC. This result also underscores the potential
of our scoring system for evaluating p16INK4a expression
and determining prognosis.
Recent studies have demonstrated a significant
correlation between p16INK4a expression as a surrogate
marker of HPV infection and fair prognosis in OPSCC. Lassen et
al reported that p16INK4a-positive HNSCC showed a
better response to conventional radiotherapy than
p16INK4a-negative HNSCC, and ascribed this survival
benefit to a better locoregional control rate (13). In a study by Fischer et al
(18),
p16INK4a-negative OPSCC patients demonstrated a more
than 2-fold greater risk of death compared with
p16INK4a-positive patients. Although locoregional OPSCC
relapse was independent of p16INK4aexpression,
multivariate survival analysis indicated that p16INK4a
expression was an independent prognostic indicator for OPSCC, but
not HNSCC, after adjustment for clinical T classification, clinical
N classification, and treatment modality (18). In the present study, the combined
evaluation of HPV DNA status and p16INK4a overexpression
could predict HPV E6/E7 mRNA expression in OPSCC with both
high specificity (100%) and sensitivity (94.4%). Furthermore, the
combined evaluation of HPV DNA status and p16INK4a
overexpression was an independent prognostic indicator for HNSCC in
multivariate analysis. Given the time and expense of E6/E7
mRNA analysis, this combination may be particularly useful for
larger scale clinical studies.
The combined evaluation of HPV DNA status and
p16INK4a overexpression was strongly correlated with HPV
mRNA expression in OPSCC. Since the combination of HPV DNA-positive
status and p16INK4a overexpression showed a close
relation with fair recurrence-free survival in HNSCC in
multivariate analysis, the combination can serve as an accurate
surrogate marker for biologically active HPV infection. This
combined evaluation appears to be a useful and reliable method for
detecting HPV-related HNSCC and determining its prognosis.
Abbreviations:
|
CCRT
|
concurrent chemoradiotherapy;
|
|
CI
|
confidence interval;
|
|
CRT
|
chemoradiotherapy;
|
|
FFPE
|
formalin-fixed, paraffin-embedded;
|
|
HNSCC
|
head and neck squamous cell
carcinoma;
|
|
HPV
|
human papillomavirus;
|
|
HR
|
hazard ratio;
|
|
HR-HPVs
|
high-risk HPV variants;
|
|
OPSCC
|
oropharyngeal squamous cell
carcinoma;
|
|
PCR
|
polymerase chain reaction;
|
|
RT
|
radiation therapy
|
Acknowledgements
This study was supported by grant
KAKENHI 23592535 (Japan Society for the Promotion of Science) to
M.S., grant KAKENHI 22791614 (Japan Society for the Promotion of
Science) to M.H. and a grant from the Japan-China Medical
Association to Z.D. This study was also supported and conducted in
cooperation with the Ryukyu Society for the Promotion of
Oto-Rhino-Laryngology.
References
|
1.
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2.
|
Hashibe M, Brennan P, Chuang SC, Boccia S,
Castellsague X, Chen C, Curado MP, Dal Maso L, Daudt AW, Fabianova
E, Fernandez L, Wunsch-Filho V, Franceschi S, Hayes RB, Herrero R,
Kelsey K, Koifman S, La Vecchia C, Lazarus P, Levi F, Lence JJ,
Mates D, Matos E, Menezes A, McClean MD, Muscat J, Eluf-Neto J,
Olshan AF, Purdue M, Rudnai P, Schwartz SM, Smith E, Sturgis EM,
Szeszenia-Dabrowska N, Talamini R, Wei Q, Winn DM, Shangina O,
Pilarska A, Zhang ZF, Ferro G, Berthiller J and Boffetta P:
Interaction between tobacco and alcohol use and the risk of head
and neck cancer: pooled analysis in the international head and neck
cancer epidemiology consortium. Cancer Epidemiol Biomarkers Prev.
18:541–550. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Gillison ML, Koch WM, Capone RB, Spafford
M, Westra WH, Wu L, Zahurak ML, Daniel RW, Viglione M, Symer DE,
Shah KV and Sidransky D: Evidence for a causal association between
human papillomavirus and a subset of head and neck cancers. J Natl
Cancer Inst. 92:709–720. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Näsman A, Attner P, Hammarstedt L, Du J,
Eriksson M, Giraud G, Ahrlund-Richter S, Marklund L, Romanitan M,
Lindquist D, Ramqvist T, Lindholm J, Sparén P, Ye W, Dahlstrand H,
Munck-Wikland E and Dalianis T: Incidence of human papillomavirus
(HPV) positive tonsillar carcinoma in Stockholm, Sweden: an
epidemic of viral-induced carcinoma? Int J Cancer. 125:362–366.
2009.PubMed/NCBI
|
|
5.
|
Deng Z, Hasegawa M, Matayoshi S, Kiyuna A,
Yamashita Y, Maeda H and Suzuki M: Prevalence and clinical features
of human papillomavirus in head and neck squamous cell carcinoma in
Okinawa, southern Japan. Eur Arch Otorhinolaryngol. 268:1625–1631.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Ang KK, Harris J, Wheeler R, Weber R,
Rosenthal DI, Nguyen-Tan PF, Westra WH, Chung CH, Jordan RC, Lu C,
Kim H, Axelrod R, Silverman CC, Redmond KP and Gillison ML: Human
papillomavirus and survival of patients with oropharyngeal cancer.
N Engl J Med. 363:24–35. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Ragin CC and Taioli E: Survival of
squamous cell carcinoma of the head and neck in relation to human
papillomavirus infection: review and meta-analysis. Int J Cancer.
121:1813–1820. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Deng Z, Hasegawa M, Yamashita Y, Matayoshi
S, Kiyuna A, Agena S, Uehara T, Maeda H and Suzuki M: Prognostic
value of human papillomavirus and squamous cell carcinoma antigen
in head and neck squamous cell carcinoma. Cancer Sci.
103:2127–2134. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Fakhry C, Westra WH, Li S, Cmelak A, Ridge
JA, Pinto H, Forastiere A and Gillison ML: Improved survival of
patients with human papillomavirus-positive head and neck squamous
cell carcinoma in a prospective clinical trial. J Natl Cancer Inst.
100:261–269. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Heath S, Willis V, Allan K, Purdie K,
Harwood C, Shields P, Simcock R, Williams T and Gilbert DC:
Clinically significant human papilloma virus in squamous cell
carcinoma of the head and neck in UK practice. Clin Oncol (R Coll
Radiol). 24:e18–23. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Smeets SJ, Hesselink AT, Speel EJ,
Haesevoets A, Snijders PJ, Pawlita M, Meijer CJ, Braakhuis BJ,
Leemans CR and Brakenhoff RH: A novel algorithm for reliable
detection of human papillomavirus in paraffin embedded head and
neck cancer specimen. Int J Cancer. 121:2465–2472. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Hoffmann M, Ihloff AS, Gorogh T, Weise JB,
Fazel A, Krams M, Rittgen W, Schwarz E and Kahn T: p16(INK4a)
overexpression predicts translational active human papillomavirus
infection in tonsillar cancer. Int J Cancer. 127:1595–1602. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Lassen P, Eriksen JG, Hamilton-Dutoit S,
Tramm T, Alsner J and Overgaard J: Effect of HPV-associated
p16INK4Aexpression on response to radiotherapy and
survival in squamous cell carcinoma of the head and neck. J Clin
Oncol. 27:1992–1998. 2009.
|
|
14.
|
Klaes R, Friedrich T, Spitkovsky D, Ridder
R, Rudy W, Petry U, Dallenbach-Hellweg G, Schmidt D and von Knebel
Doeberitz M: Overexpression of p16(INK4A) as a specific marker for
dysplastic and neoplastic epithelial cells of the cervix uteri. Int
J Cancer. 92:276–284. 2001. View Article : Google Scholar
|
|
15.
|
Singhi AD and Westra WH: Comparison of
human papilloma-virus in situ hybridization and p16
immunohistochemistry in the detection of human
papillomavirus-associated head and neck cancer based on a
prospective clinical experience. Cancer. 116:2166–2173. 2010.
|
|
16.
|
Schache AG, Liloglou T, Risk JM, Jones TM,
Ma XJ, Wang H, Bui S, Luo Y, Sloan P, Shaw RJ and Robinson M:
Validation of a novel diagnostic standard in HPV-positive
oropharyngeal squamous cell carcinoma. Br J Cancer. 108:1332–1339.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Bussu F, Sali M, Gallus R, Vellone VG,
Zannoni GF, Autorino R, Dinapoli N, Santangelo R, Martucci R,
Graziani C, Micciche F, Almadori G, Galli J, Delogu G, Sanguinetti
M, Rindi G, Valentini V and Paludetti G: HPV infection in squamous
cell carcinomas arising from different mucosal sites of the head
and neck region. Is p16 immunohistochemistry a reliable surrogate
marker? Br J Cancer. 108:1157–1162. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Fischer CA, Zlobec I, Green E, Probst S,
Storck C, Lugli A, Tornillo L, Wolfensberger M and Terracciano LM:
Is the improved prognosis of p16 positive oropharyngeal squamous
cell carcinoma dependent of the treatment modality? Int J Cancer.
126:1256–1262. 2010.
|
|
19.
|
Klingenberg B, Hafkamp HC, Haesevoets A,
Manni JJ, Slootweg PJ, Weissenborn SJ, Klussmann JP and Speel EJ:
p16 INK4A overexpression is frequently detected in tumour-free
tonsil tissue without association with HPV. Histopathology.
56:957–967. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Saiki RK, Scharf S, Faloona F, Mullis KB,
Horn GT, Erlich HA and Arnheim N: Enzymatic amplification of
beta-globin genomic sequences and restriction site analysis for
diagnosis of sickle cell anemia. Science. 230:1350–1354. 1985.
View Article : Google Scholar
|
|
21.
|
de Roda Husman AM, Walboomers JM, van den
Brule AJ, Meijer CJ and Snijders PJ: The use of general primers GP5
and GP6 elongated at their 3’ ends with adjacent highly conserved
sequences improves human papillomavirus detection by PCR. J Gen
Virol. 76:1057–1062. 1995.
|
|
22.
|
Manos MM, Ting Y, Wright DK, Lewis AJ,
Broker TR and Wolinsky SM: Use of polymerase chain reaction
amplification for the detection of genital human papillomaviruses.
Cancer Cells. 7:209–214. 1989.
|
|
23.
|
Remmerbach TW, Brinckmann UG, Hemprich A,
Chekol M, Kuhndel K and Liebert UG: PCR detection of human
papilloma-virus of the mucosa: comparison between MY09/11 and
GP5+/6+primer sets. J Clin Virol. 30:302–308.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Wiest T, Schwarz E, Enders C,
Flechtenmacher C and Bosch FX: Involvement of intact HPV16 E6/E7
gene expression in head and neck cancers with unaltered p53 status
and perturbed pRb cell cycle control. Oncogene. 21:1510–1517. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Landis JR and Koch GG: The measurement of
observer agreement for categorical data. Biometrics. 33:159–174.
1977. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Viera AJ and Garrett JM: Understanding
interobserver agreement: the kappa statistic. Fam Med. 37:360–363.
2005.PubMed/NCBI
|
|
27.
|
Bland JM and Altman DG: Statistical
methods for assessing agreement between two methods of clinical
measurement. Lancet. 1:307–310. 1986. View Article : Google Scholar
|
|
28.
|
Charfi L, Jouffroy T, de Cremoux P, Le
Peltier N, Thioux M, Freneaux P, Point D, Girod A, Rodriguez J and
Sastre-Garau X: Two types of squamous cell carcinoma of the
palatine tonsil characterized by distinct etiology, molecular
features and outcome. Cancer Lett. 260:72–78. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Hafkamp HC, Manni JJ, Haesevoets A, Voogd
AC, Schepers M, Bot FJ, Hopman AH, Ramaekers FC and Speel EJ:
Marked differences in survival rate between smokers and nonsmokers
with HPV 16-associated tonsillar carcinomas. Int J Cancer.
122:2656–2664. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Hannisdal K, Schjolberg A, De Angelis PM,
Boysen M and Clausen OP: Human papillomavirus (HPV)-positive
tonsillar carcinomas are frequent and have a favourable prognosis
in males in Norway. Acta Otolaryngol. 130:293–299. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Klussmann JP, Mooren JJ, Lehnen M,
Claessen SM, Stenner M, Huebbers CU, Weissenborn SJ, Wedemeyer I,
Preuss SF, Straetmans JM, Manni JJ, Hopman AH and Speel EJ: Genetic
signatures of HPV-related and unrelated oropharyngeal carcinoma and
their prognostic implications. Clin Cancer Res. 15:1779–1786. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Schwartz SR, Yueh B, McDougall JK, Daling
JR and Schwartz SM: Human papillomavirus infection and survival in
oral squamous cell cancer: a population-based study. Otolaryngol
Head Neck Surg. 125:1–9. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Evans M, Newcombe R, Fiander A, Powell J,
Rolles M, Thavaraj S, Robinson M and Powell N: Human
papillomavirus-associated oropharyngeal cancer: an observational
study of diagnosis, prevalence and prognosis in a UK population.
BMC Cancer. 13:2202013. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Jung AC, Briolat J, Millon R, de Reynies
A, Rickman D, Thomas E, Abecassis J, Clavel C and Wasylyk B:
Biological and clinical relevance of transcriptionally active human
papillomavirus (HPV) infection in oropharynx squamous cell
carcinoma. Int J Cancer. 126:1882–1894. 2010.PubMed/NCBI
|
|
35.
|
Lindquist D, Romanitan M, Hammarstedt L,
Nasman A, Dahlstrand H, Lindholm J, Onelov L, Ramqvist T, Ye W,
Munck-Wikland E and Dalianis T: Human papillomavirus is a
favourable prognostic factor in tonsillar cancer and its oncogenic
role is supported by the expression of E6 and E7. Mol Oncol.
1:350–355. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Isayeva T, Li Y, Maswahu D and
Brandwein-Gensler M: Human papillomavirus in non-oropharyngeal head
and neck cancers: a systematic literature review. Head Neck Pathol.
6(Suppl 1): S104–S120. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Wang SS, Trunk M, Schiffman M, Herrero R,
Sherman ME, Burk RD, Hildesheim A, Bratti MC, Wright T, Rodriguez
AC, Chen S, Reichert A, von Knebel Doeberitz C, Ridder R and von
Knebel Doeberitz M: Validation of p16INK4aas a marker of
oncogenic human papillomavirus infection in cervical biopsies from
a population-based cohort in Costa Rica. Cancer Epidemiol
Biomarkers Prev. 13:1355–1360. 2004.PubMed/NCBI
|
|
38.
|
Smith EM, Wang D, Kim Y, Rubenstein LM,
Lee JH, Haugen TH and Turek LP: p16INK4aexpression,
human papilloma virus, and survival in head and neck cancer. Oral
Oncol. 44:133–142. 2008.
|
|
39.
|
Thomas J and Primeaux T: Is p16
immunohistochemistry a more cost-effective method for
identification of human papilloma virus-associated head and neck
squamous cell carcinoma? Ann Diagn Pathol. 16:91–99. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Hoffmann M, Tribius S, Quabius ES, Henry
H, Pfannenschmidt S, Burkhardt C, Gorogh T, Halec G, Hoffmann AS,
Kahn T, Rocken C, Haag J, Waterboer T and Schmitt M: HPV DNA,
E6*I-mRNA expression and
p16INK4Aimmunohistochemistry in head and neck cancer -
how valid is p16INK4Aas surrogate marker? Cancer Lett.
323:88–96. 2012.
|