Introduction
MicroRNAs (miRNAs) are a class of small non-coding
RNAs whose mature products are approximately 22 nucleotides long.
MiRNAs may regulate diverse biological processes, including
development, cell proliferation, differentiation and apoptosis,
through negatively regulating target gene expression at the
post-transcriptional and/or translational level (1–3). Lim
et al showed that each vertebrate miRNA has hundreds of
different conserved or non-conserved targets (4). It has also been estimated that
approximately 30% of genes are regulated by at least one miRNA
(5). Thus, a single miRNA may have
substantial biological consequences. Recently, miRNAs have also
been reported as key factors in cancer, playing both oncogenic and
tumor-suppressing roles (2,6). In
2002, miR-126 was identified in a tissue-specific mouse screen
(7). miR-126 is encoded by intron
7 of the egfl7 gene in all vertebrates, and has been shown
to mediate angiogenesis and vascular integrity (8,9).
However, the functions of miR-126 in cancers appear to be diverse
and remain largely unknown. miR-126 was reported to be
downregulated and to act as a tumor suppressor in different cancers
of the lung (10,11), stomach (12,13),
cervix (14), bladder and prostate
(15). miR-126 was also
significantly downregulated in ectopic endometria (EC) versus
eutopic endometria (EU), suggesting that it may play an initial
role in the development and progression of endometriosis (Ems)
(16). miR-126 might disturb
vascular integrity, leading to a disorganized and abnormal tumor
vasculature (17).
To date, the target genes of miR-126 that have been
confirmed are PI3KR2 (18),
IRS-1 (19), Crk
(12), SOX2 (13), ADAM9, MMP7 (20) and SCL7A5 (21). Except for SOX2, which is a
tumor suppressor, other genes are involved in tumor formation and
progression. miR-126 plays a tumor suppressor role in human cancer
through the direct or indirect repression of several key oncogenic
molecules, such as PI3KR2, IRS-1 and Crk. At present, some data
exist showing that miR-126 plays a tumor suppressor role in gastric
cancer SGC-7901 cells through targeting Crk (12). However, Otsubo et al
demonstrated that miR-126 acts as an oncogene by targeting
SOX2 in gastric cancer cells (13). Thus, it is controversial as to
whether miR-126 is a tumor suppressing or oncogenic miRNA in
gastric cancer cells.
PLK2 is a member of Polo-like kinase (PLK) family,
which is involved in regulation of cell cycle progression or
mitotic progression (22,23). It is well known that PLK2 regulates
centriole duplication in mammalian cells (24). PLK2 is also activated by a spindle
checkpoint in a p53-dependent manner, and thereby may prevent a
mitosis catastrophe following spindle damage (25). Thus, PLK2 seems to inhibit
oncogenic transformation (26). A
previous study showed that PLK2 is a target gene of miR-126
(21,27), which is also as a tumor suppressor
(12). However, the inhibitory
effects of a specific miRNA on a target gene appear to be cell
type-specific. For example, miR-126 overexpression does not
suppress PLK2 expression in SCLC cells (21). TOM1 is a target gene of
miR-126 in cystic fibrosis (CF) airway epithelium cells, but not in
MCF7 cells (28). Similarly,
SPRED1 is a target gene of miR-126 in HUVEC cells, but not
in acute myeloid leukemia (AML) cells (8,27).
Materials and methods
Collection and analysis of clinical
samples
Forty-four gastric cancer and adjacent normal
tissues were obtained from patients undergoing surgery for gastric
cancer at The First and Second Affiliated Hospital of Medical
College of Xi’an Jiao Tong University. The samples were placed in
liquid nitrogen immediately after excision from the patients and
subsequently frozen at −80°C until RNA extraction. Informed consent
was obtained from each patient and was approved by the Institute
Research Ethics Committee at the Cancer Center, Xi’an Jiaotong
University.
RNA extraction and quantitative reverse
transcription-polymerase chain reaction (qRT-PCR) analysis
RNA from gastric cancer tissue samples was extracted
using TRIzol (Invitrogen, Life Technologies) according to the
manufacturer’s recommendations. miR-126 expression was assessed
using qRT-PCR. Human U6 small nuclear RNA (snRNA) was used as an
internal standard. Primer sequences are listed in Table I. In order to verify that
PLK2 was regulated by miR-126, we collected cells and
extracted RNA using TRIzol, according to manufacturer
recommendations, after transfecting the miR-126, miR-control and
anti-miR-126 individually at 24 h. The β-actin mRNA level
was used as an internal standard. The PLK2 and
β-actin qRT-PCR primers are shown in Table I. Reverse transcription was
performed using PrimerScript™ RT reagent kit perfect Real-Time and
qRT-PCR using SYBR® Premix Ex Taq™ II (Takara). The
qRT-PCR program used for amplification was: 95°C for 30 sec,
followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec.
 | Table IPrimer sequences. |
Table I
Primer sequences.
| Gene | Reverse
transcription primers | Real-time PCR
primers |
|---|
| U6 |
5′-CGCTTCACGAATTTGCGTGTCAT-3′ | F:
5′-GCTTCGGCAGCACATATACTAAAAT-3′
R: 5′-CGCTTCACGAATTTGCGTGTCAT-3′ |
| miR-126 |
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGGGGTCGTACCGTGAGT-3′ | GSP:
5′-GCAATTGCACTGGATACGACCGCATTA-3′
R: 5′-CAGTGCGTGTCGTGGAGT-3′ |
| PLK2 | | F:
5′-GACCCTATGGGACTCCTCTTT-3′
R: 5′-GTATGCCTTAGCCTGTTCTGG-3′ |
| β-actin | | F:
5′-TGGCACCCAGCACAATGAA-3′
R: 5′-CTAAGTCATAGTCCGCCTAGAAGCA-3′ |
pcDNA6.2-GW/EmGFP-miR-126 vector
construction
The pcDNA6.2-GW/EmGFP-miR vector was purchased from
Invitrogen. The linear structure was converted to a double
link-shaped structure by cloning multiple cloning sites (MCS)
(using EcoRI, PstI, NheI, KpnI and
HindIII) into pcDNA6.2-GW/EmGFP-miR via the vector’s joint.
The Hsa-miR-126 clone vector was constructed by Jie Rui Biology
Ltd. (Shanghai, China). Hsa-miR-126 mature sequences were cloned
into pcDNA6.2-GW/EmGFP-miR, using EcoRI and HindIII,
and named pcDNA6.2-GW/EmGFP-miR-126 (miR-126). Finally, for
miR-control vector, the miR-126 mature sequence was transformed to
clone MCS (short miR-control).
Cell culture and transfection
SGC-7901 cells were obtained from The Central
Laboratory of Biomedical Research of Xi’an Jiaotong University
Medical School. The cells were grown in Dulbecco’s modified Eagle
medium (Gibco) with 10% fetal bovine serum (FBS; Gibco), 100 U/ml
penicillin and 100 μg/ml streptomycin (Invitrogen), and incubated
at 37°C in 5% CO2. There were four experimental groups
(control, anti-miR-126, miR-control and miR-126). Anti-miR-126
sequence: 5′-GCATTATTACTCACGGTACGA-3′ (Sangon Biotech). Anti-126,
miR-control and miR-126 were transfected at a final concentration
of 100 nmol/l by Lipofectamine 2000 (Invitrogen), according to the
manufacturer’s instructions, in all experiments.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay
SGC-7901 cells were seeded at 1×104
cells/well in 96-well plates and transfected the following day with
miR-126, miR-control and anti-miR-126 using Lipofectamine 2000; the
control group was transfected with Lipofectamine 2000 only. MTT was
added to cells (Sigma, USA) at 24, 48, 72 and 96 h, respectively,
and these continued incubating for 3 h at 37°C in 5%
CO2, then the supernatant was discarded and dimethyl
sulfoxide was added to the cells. Sample absorbance was measured at
490 nm by a high-throughput universal microplate assay (BMG Lab
Technologies).
Cell cycle analysis
SGC-7901 cells were seeded at 1×105
cells/well in a 6-well plate and transfected with miR-126,
miR-control and anti-miR-126 using Lipofectamine 2000. Cells were
harvested at 24 and 48 h post-transfection, stained for DNA content
using propidium iodide (PI) and analyzed on a FACSCalibur flow
cytometer (Becton-Dickinson). Briefly, cells were harvested by
trypsinization and washed once with phosphate-buffered saline (PBS)
before fixing overnight in 70% EtOH. For DNA staining, cells were
pelleted and stained for 30 min with 300 μl PI solution (0.05 mg/ml
PI, 20 μg/ml RNAse A in 0.1% bovine serum albumin).
Cell apoptosis analysis
SGC-7901 cells were seeded at 1×105
cells/well in a 6-well plate and transfected with miR-126,
miR-control and anti-miR-126 using Lipofectamine 2000. Cells were
harvested 24 and 48 h post-transfection and washed twice with
ice-cold PBS before being stained with 7-AAD/PI, and later analyzed
on a FACSCalibur flow cytometer (Becton-Dickinson).
Colony formation assay in vitro
One thousand cells, which were post-transfected with
miR-126, miR-control or anti-miR-126 at 24 h, were seeded into
6-well plates and cultured in common media. Approximately 2 weeks
later, colonies that appeared were fixed with pre-chilled methanol
and stained with 2% Giemsa solution (Sigma, St. Louis, MO, USA).
Experiments were conducted in triplicate.
Tumor xenograft model and tumorigenicity
assay
SGC-7901 cells (1×106 cells/mouse) stably
transfected with miR-126 or miR-control lentivirus vector were
subcutaneously injected into 6-week-old male nude mice. Tumor size
was measured every 3 days. For end-point experiments, the
bioluminescence images captured in vivo were obtained using
a photobiology system (Xenogen). The mice were euthanized 22 days
after injection, and the tumors were removed and weighed. The tumor
tissues were fixed in 4% paraformaldehyde, embedded in paraffin,
sectioned at 5 μm and stained with hematoxylin and eosin
(H&E).
Western blot analysis
SGC-7901 cells were seeded at 1×106
cells/6-cm culture dish, and transfected with miR-126, miR-control
or anti-miR-126, using Lipofectamine 2000 according to the
manufacturer instructions (Invitrogen). Cells were harvested by
trypsinization, washed once with ice-cold PBS and lysed in RIPA
buffer (150 mM NaCl, 0.5% sodium deoxycholate, 0.1% sodium dodecyl
sulfate, 1% Igepal, 50 mM Tris-HCl pH 8.0, 2 mM ethylenediamine
tetraacetic acid) supplemented with 1X complete mini protease
inhibitor cocktail tablets. Then, 25 μg of protein/lane was
resolved on 10% polyacrylamide gel electrophoresis gels and
transferred to a nitrocellulose membrane (Roche Diagnostics GmbH).
Primary antibodies used were PI3KR2 (Cell Signal 9402), Crk (Santa
Cruz Sc-53923), Bcl-2 (Sigma V9131), Bax (Santa Cruz Sc-126), PLK2
(sc-25421; Santa Cruz Biotechnology) and β-actin (Bioworld,
AP0060). The membranes were further probed with horseradish
peroxidase-conjugated secondary antibodies (ZSGB-BIO). Working
solutions of the Pierce ECL Substrate were prepared and added to
polyvinyldifluoride membranes for 1 min. The membranes were removed
from the substrates and exposed to ChemiDoc-It 510 (UVP; LLC).
Luciferase assay
The wild-type 3′ untranslated regions (UTRs) or
mutant 3′UTRs of PLK2 carrying miR-126-binding sites were
cloned downstream of the luciferase reporter in a pmir-GOL vector
system (Promega; E1330) through SacI and XhoI
digestion. The wild-type PLK2 3′UTR PCR primer was as
follows: sense primer, 5′-GAGCTCGACCCTATGGGACTCCTCTTT-3′ and
antisense primer, 5′-CTCGAGGTATGCCTTAGCCTGTTCTGG-3′. The
PLK2 mutant sequence was chemically synthesized by Beijing
AuGCT DNA-SYN Biotechnology Co., Ltd., as follows: PLK2 Mut
Top: 5′-cgagtatgttgaagaagatggacatgtggtgatcctaaaacaattccccc-3′ and
PLK2 Mut Bottom:
5′-tcgagggggaattgttttaggatcaccacatgtccatcttcttcaacatactcgagct-3′.
The wild-type PLK2 reporter vector and mutation reporter
vector were named pmir-GOL-PLK2 and pmir-GOL-mutPLK2, respectively.
SGC-7901 cells were seeded in 96-well plates and transfected with
miR-126 or miR-control, and pmir-GOL-PLK2 or pmir-GOL-mutPK2, using
Lipofectamine 2000. Cells were harvested 24 h post-transfection and
luciferase activity was measured using a Dual-Glo Luciferase Assay
System (Promega; E2920) according to the manufacturer’s
recommendations.
Statistical analysis
Quantitative results are presented as the mean ±
standard deviation (SD). Statistical analysis was performed by
using a t-test, and P<0.05 was taken as the level of
significance. All results were analyzed using the statistical
software SPSS10.0.
Results
Downregulated expression of miR-126 in
gastric cancer and gastric cancer cells
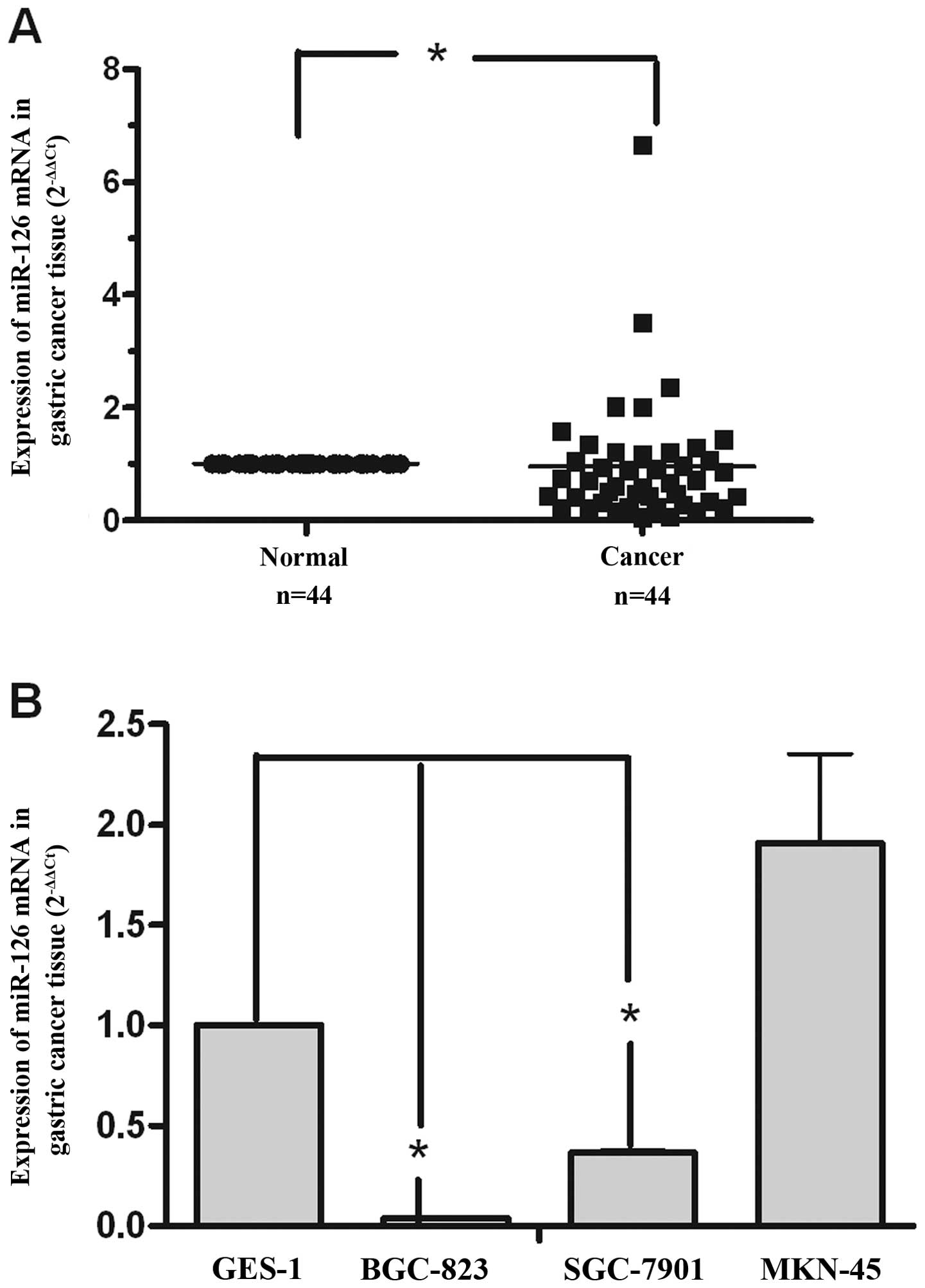
To explore miR-126 expression levels in gastric
cancer, we collected 44 paired gastric cancer and adjacent normal
tissue samples. We used qRT-PCR to determine the miR-126 expression
level in gastric cancer. In the 44 paired clinical gastric samples,
there were 29 samples with downregulation of miR-126 compared with
the matched non-tumor tissue samples (relative expression ratio
<1.0), and 18 gastric cancer tissues showed miR-126 expression
with significant downregulation (relative expression ratio <0.5)
(Fig. 1A). Furthermore, analysis
of miR-126 expression in 4 gastric cancer cell lines (GES-1,
BGC-823, SGC-7901 and MKN-45) revealed that miR-126 was
downregulated in the BGC-823 and SGC-7901 cell lines (Fig. 1B). These data suggest that miR-126
might act as a tumor suppressor in gastric cancer.
miR-126 expression suppresses gastric
cancer cell proliferation through inducing apoptosis in vitro
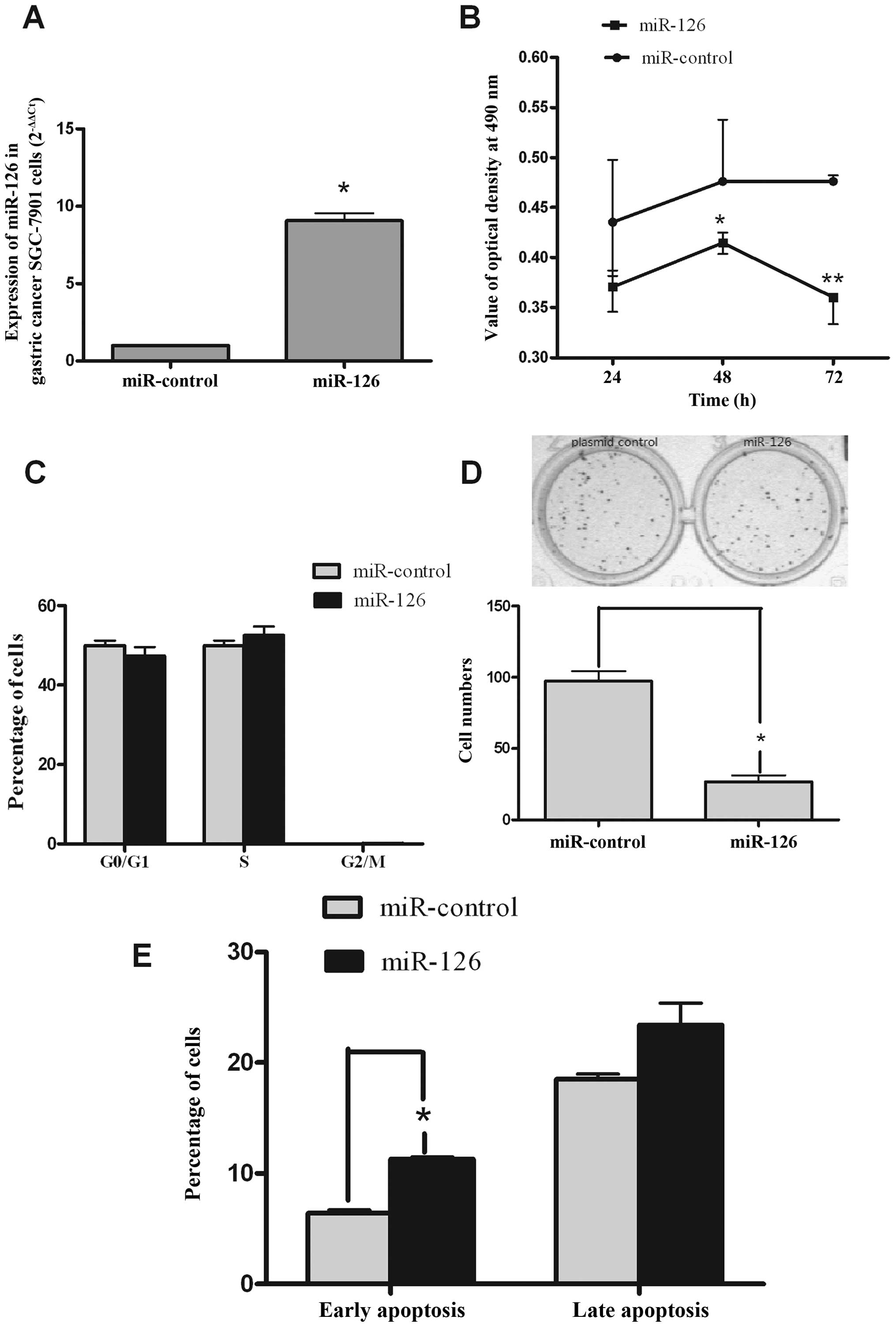
To explore the tumor suppressor role of miR-126 in
gastric cancer, SGC-7901 cells were transfected with miR-126 or
miR-control. qRT-PCR was performed to examine the expression levels
of miR-126 after transfection of the miR-126 or miR-control. As
depicted in Fig. 2A, an
approximate 10-fold increase in the expression of miR-126 was
observed in SGC-7901 cells that were transfected with miR-126
relative to the cells transfected with miR-control. To study the
role of miR-126 in SGC-7901 cell proliferation, an MTT assay was
used. The results showed that transient overexpression of miR-126
inhibited the proliferation of SGC-7901 cells at 24, 48 and 72 h
after transfection (Fig. 2B).
However, the cell cycle of SGC-7901 cells was not obviously
affected by miR-126 at 24 h (Fig.
2C). In order to further examine the inhibitory role of miR-126
in SGC-7901 cells, a colony formation assay was conducted after
similar transient transfection. miR-126-transfected cells displayed
fewer colonies compared with the control (Fig. 2D). To further investigate the
mechanisms by which miR-126 inhibits the growth of SGC-7901 cells,
we tested the apoptosis of SGC-7901 cells induced by miR-126. We
found that cells in early apoptosis and late apoptosis (at 24 h)
were increased in the miR-126 group. Thus, the inhibitory effect of
miR-126 on cell growth may occur through the induction of apoptosis
in SGC-7901 cells (Fig. 2E).
Altogether, these results indicate that miR-126 was able to inhibit
the proliferation of SGC-7901 cells in vitro.
miR-126 expression suppresses gastric
cancer cell proliferation in vivo
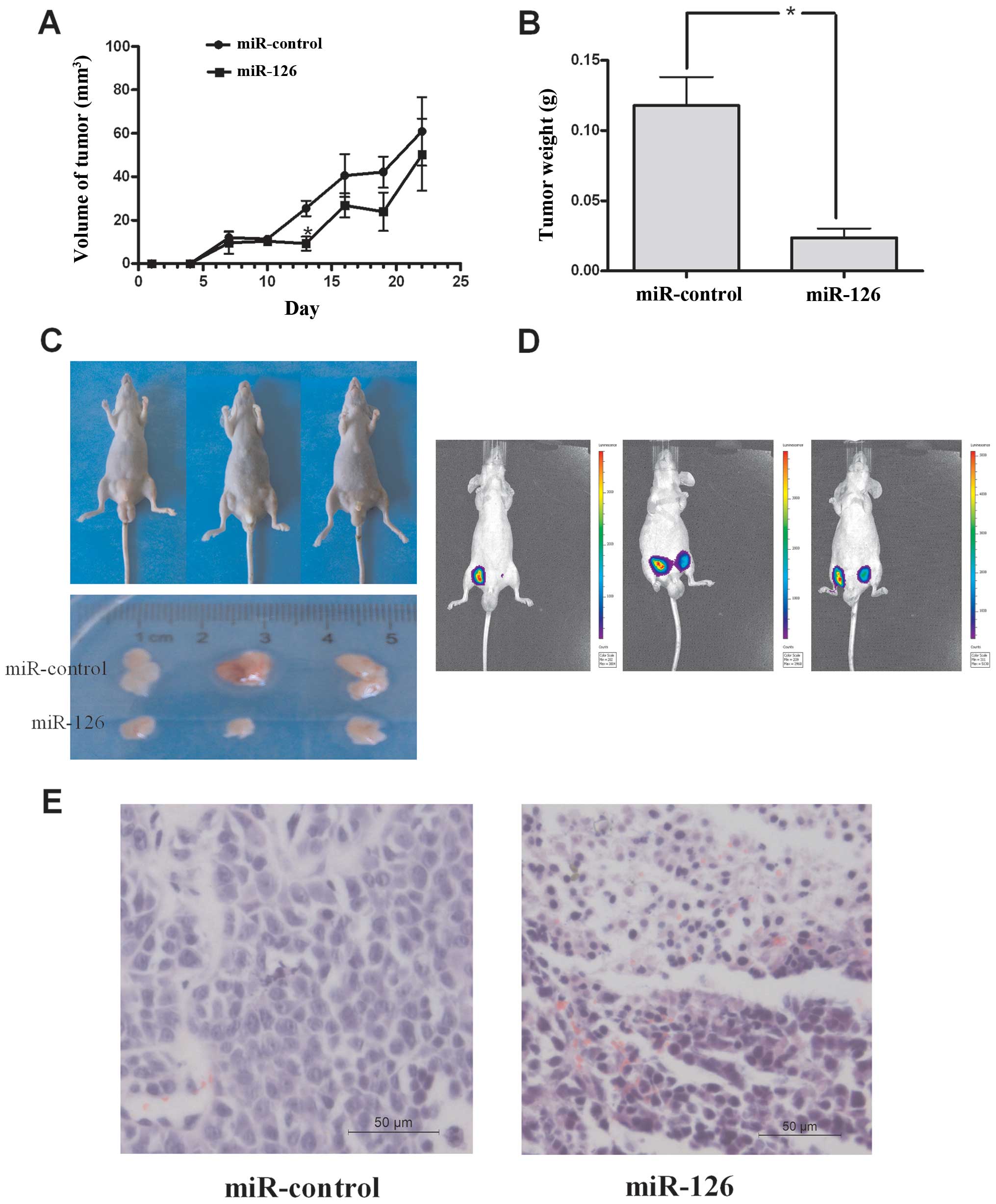
We further confirmed the growth inhibitory role of
miR-126 in vivo in a xenograft model. First, we tested the
effects of miR-126 on tumor growth in the in vivo xenograft
model. miR-126 and miR-control-transfected SGC-7901 cells were
injected subcutaneously into either posterior flank of nude mice,
and xenograft growth was observed for 22 days. As shown in Fig. 3A and C, the tumors from mice that
were injected with miR-126 were significantly smaller than those in
control mice on the 13th day after the first injection. On the 22nd
day, the tumors were tested using IVIS Spectrum before the mice
were euthanized (Fig. 3D), and the
tumors were removed and weighed (Fig.
3B). The tumor tissue was fixed in 4% paraformaldehyde,
embedded in paraffin and stained with H&E (Fig. 3E). Although, both tissues of
miR-126 and control grew tumors, the degree of malignancy in the
miR-126 groups was lower than in the miR-control group. These data
indicate that miR-126 could inhibit tumorigenicity of SGC-7901
cells in the nude mouse xenograft model.
Inhibition of miR-126 moderately promotes
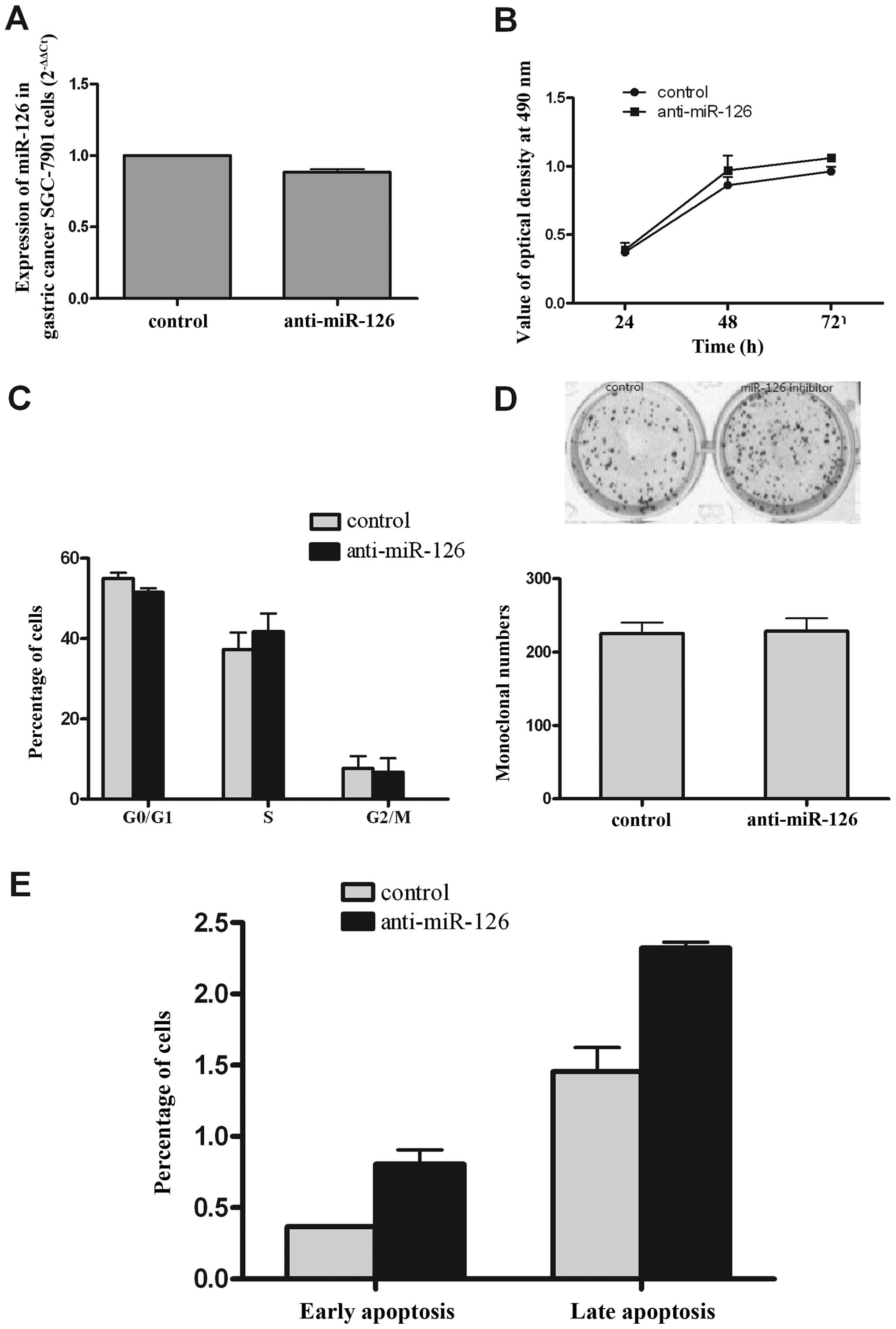
growth of SGC-7901 cells in vitro
The preceding observations demonstrated that miR-126
expression inhibited the growth and increased apoptosis of SGC-7901
cells. We next asked whether downregulated expression miR-126
promotes the growth of SGC-7901 cells. To accomplish this, we
transiently inhibited miR-126 in SGC-7901 cells with antisense
oligonucleotides. qRT-PCR was performed to examine the expression
levels of miR-126 after transfection of anti-miR-126 and control.
As shown in Fig. 4A, the
expression of miR-126 decreased by ~12% compared with the control.
The MTT assay results showed that anti-miR-126 slightly improved
the ability of growth at 48 and 72 h (Fig. 4B), but the cell cycle and colony
forming ability were unaffected by the inhibitor (Fig. 4C and D). Interestingly,
inhibitor-induced apoptosis of SGC-7901 cells was higher than in
the control (Fig. 4E), but there
was no significant difference, and the percent of apoptosis was
very low in both groups. This phenomenon is worth exploring further
in the future.
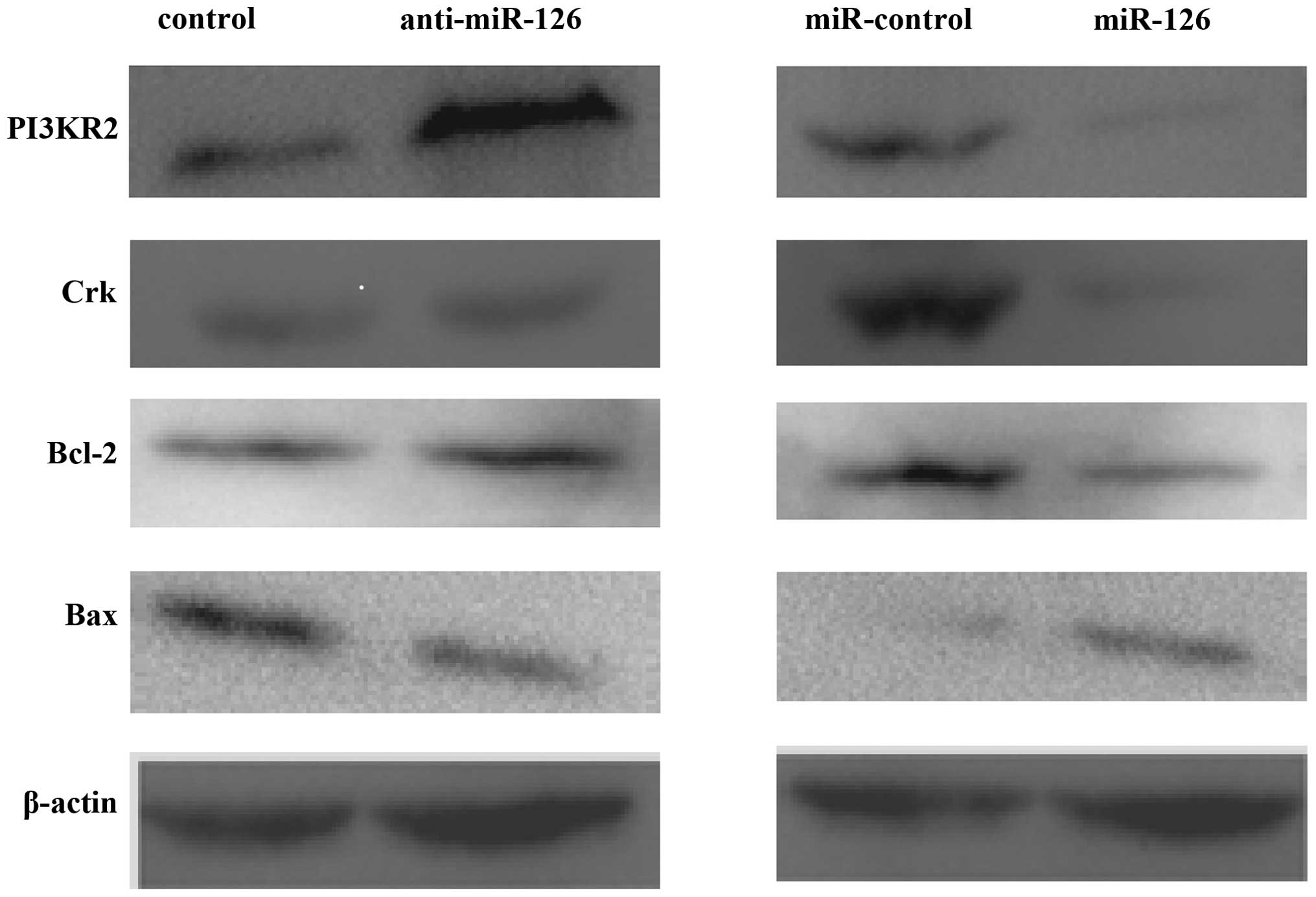
miR-126 regulates its target genes Crk
and PI3K2 in SGC-7901 cells
Feng et al reported that miR-126 functions as
a tumor suppressor in gastric cancer, with Crk as a direct
target (12). We wanted to know
whether any other target genes of miR-126 might also contribute to
the function of miR-126 in gastric cancer. Crk and PI3KR2 protein
(p85) levels, targets of miR-126, were assayed in
miR-126-expressing SGC-7901 cells. As shown in Fig. 5, miR-126 repressed levels of PI3KR2
and Crk protein expression. P13KR2 was upregulated, while the
expression of Crk was not affected by the inhibitor of miR-126.
These data indicated that miR-126 directly regulates Crk and
PI3K2 in SGC-7901 cells. In addition, in overexpressing
miR-126 SGC-7901 cells, the Bcl-2 and Bax protein levels were
changed; Bax was upregulated and Bcl-2 was downregulated in the
miR-126 group. However, protein levels of Bcl-2 and Bax were
unchanged by the inhibitor of miR-126. These data were compatible
with the results of the apoptosis analysis.
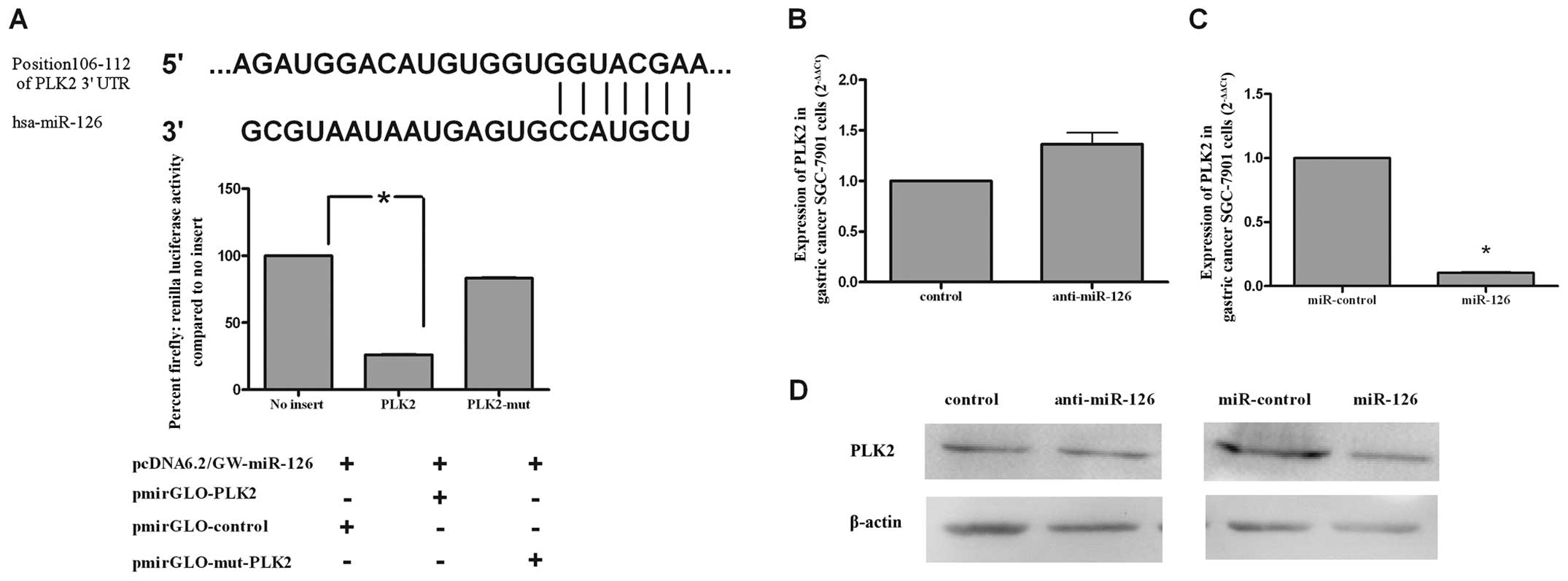
PLK2 is a target gene of miR-126 in
SGC-7901 cells
Through the miRBase (http://www.mirbase.org) and TargetScan (http://www.targetscan.org/) databases, we found
matching bases between the 3′UTR of PLK2 and miR-126 (top of
Fig. 6A). In order to determine
whether miR-126 regulates PLK2 in gastric cancer SGC-7901
cells, we performed a luciferase assay to evaluate the relationship
between miR-126 and PLK2. SGC-7901 cells were co-transfected
with miR-126 together with pmir-GOL-PLK2 or pmir-GOL-mut-PLK2.
miR-126 strongly reduced the expression of PLK2 (27.73%),
but showed almost no effect on the expression of pmir-GOL-mut-PLK2
(81.81%) (bottom of Fig. 6A). As
shown in Fig. 6B and C, qRT-PCR
revealed that PLK2 mRNA was decreased in SGC-7901 cells
overexpressing miR-126, while it was upregulated in cells treated
with anti-miR-126. These results were also confirmed in SGC-7901
cells transfected with miR-126, which showed a lower amount of PLK2
protein when compared to miR-control-treated cells (right of
Fig. 6D). The PLK2 protein level
did not change significantly between the control and anti-miR-126
groups (Fig. 6D, left). Together,
these data indicate that PLK2 is regulated by miR-126 in
SGC-7901 cells.
Discussion
Gastric cancer is the fourth most common human
malignant disease and the second most frequent cause of
cancer-related death worldwide (29). Improvement of treatment has
resulted in good long-term survival for patients with gastric
cancer (30). miRNAs, as a new
class of small non-coding RNAs, play an important role in cancer
development, and have become a new target for cancer therapy. In
our previous miRNA microarray study, we found that miR-126 was
downregulated in clinical gastric cancer samples. In the present
study, we first examined the expression of miR-126 in gastric
cancer tissues and cell lines, and confirmed the previous result
that miR-126 was downregulated in gastric cancer. Next, we
confirmed that miR-126 regulates PLK2 in SGC-7901 cells.
Moreover, we observed a decrease of PLK2 at both the mRNA and
protein levels after transfection with miR-126. These data suggest
that miR-126 is able to directly regulate PLK2 in gastric
cancer SGC-7901 cells.
It is well known that miRNA mainly performs its
regulatory functions through its targets (31). miRNAs also contribute to
maintaining the balance among genes that regulate the cell fate, as
well as their deregulation, which is a frequent hallmark in a
variety of human malignancies. Although the identification and
validation of miRNA targets have greatly progressed in the last few
years, little is known about the specific cellular and molecular
pathways and mechanism involved in these effects (3). Cancer occurrence is tightly linked to
a series of genes encoding oncogenic and tumor-suppressor proteins.
In general, oncogenic miRNAs downregulate the tumor-suppressor
proteins, while tumor-suppressor miRNAs downregulate oncogenic
proteins. It was reported that miR-126 acts as a tumor suppressor
in gastric cancer by regulating the oncogene Crk (12). However, Otsubo et al
reported that miR-126 functions as an oncogene in gastric cancer by
downregulating the expression of SOX2, which is a tumor
suppressor (13). Thus, it
appeared that the function of miR-126 was mainly determined by one
target gene in gastric cancer. Furthermore, previous reports
suggested that PLK2 is a target gene of miR-126, which is
also a tumor suppressor. It has been reported that miR-126 inhibits
apoptosis of AML cells and enhances the colony-forming ability of
mouse bone marrow progenitor cells through targeting PLK2
(27). Therefore, if miR-126
regulates PLK2 in SGC-7901 cells, then it must function as
an oncogene. However, we showed that the role of miR-126 remains
antitumorigenic in SGC-7901 cells. Thus, we inferred that miR-126
must regulate other oncogenes in SGC-7901 cells, and the role of
these oncogenes might be more important than that of
PLK2.
Individual miRNAs each have several hundred
different target genes, including oncogenes and anti-oncogenes. The
function of a miRNA is not dependent on a single target gene, but
rather on the competition or balance among its target genes in
specific types of cancer (32).
Therefore, there may be a balanced relationship among the oncogenes
or tumor suppressor target genes of miR-126 in SGC-7901 cells.
PI3KR2 is a target gene of miR-126 in endothelial cells, and
PI3KR2 protein is one of the regulatory subunits of the class IA
PI3K enzyme that is activated by tyrosine kinase receptors. PI3KR2
is involved in the PI3K-Akt-mediated survivin signaling pathway
(33–35). Crk protein has crucial functions in
the signaling pathways regulating cell adhesion, proliferation and
migration. Feng et al found that miR-126 functions as a
tumor suppressor in gastric cancer by targeting Crk
(12). Thus, we proceeded to
measure the protein level expression of PI3KR2 and Crk. Western
blot results showed reduced expression of PI3KR2 and Crk in
SGC-7901 cells through overexpression of miR-126. We found that
miR-126 function was accomplished by regulation of PI3KR2,
Crk and PLK2 simultaneously. However, the detailed
pathway involved in PI3KR2, Crk and PLK2 inhibition by miR-126 in
gastric cancer needs to be confirmed in future studies.
Collectively, our study indicates that the ability
of miR-126 to inhibit growth of SGC-7901 cells is attributable to
the balancing effect through its capacity to inhibit PLK2,
PI3KR2 and Crk expression. This provides new insight
for studies of miRNA function in the future. Of particular note,
the function of miRNA is a result of the combined action of many
target genes, and not simply a result of a particular gene, as was
previously believed.
Acknowledgements
This study was supported by Fundamental Research
Funds for the Central Universities of Xi’an Jiaotong University
(No. 08143014).
References
|
1
|
Li M, Li J, Ding X, He M and Cheng S:
microRNA and cancer. AAPS J. 12:309–317. 2010. View Article : Google Scholar
|
|
2
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar
|
|
3
|
Iorio MV and Croce CM: MicroRNAs in
cancer: small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lim LP, Lau NC, Garrett-Engele P, et al:
Microarray analysis shows that some microRNAs downregulate large
numbers of target mRNAs. Nature. 433:769–773. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim YK, Yu J, Han TS, et al: Functional
links between clustered microRNAs: suppression of cell-cycle
inhibitors by microRNA clusters in gastric cancer. Nucleic Acids
Res. 37:1672–1681. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lagos-Quintana M, Rauhut R, Yalcin A,
Meyer J, Lendeckel W and Tuschl T: Identification of
tissue-specific microRNAs from mouse. Curr Biol. 12:735–739. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fish JE, Santoro MM, Morton SU, et al:
miR-126 regulates angiogenic signaling and vascular integrity. Dev
Cell. 15:272–284. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang S, Aurora AB, Johnson BA, et al: The
endothelial-specific microRNA miR-126 governs vascular integrity
and angiogenesis. Dev Cell. 15:261–271. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yanaihara N, Caplen N, Bowman E, et al:
Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cho WC, Chow AS and Au JS: Restoration of
tumour suppressor hsa-miR-145 inhibits cancer cell growth in lung
adenocarcinoma patients with epidermal growth factor receptor
mutation. Eur J Cancer. 45:2197–2206. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Feng R, Chen X, Yu Y, et al: miR-126
functions as a tumour suppressor in human gastric cancer. Cancer
Lett. 298:50–63. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Otsubo T, Akiyama Y, Hashimoto Y, Shimada
S, Goto K and Yuasa Y: MicroRNA-126 inhibits SOX2 expression and
contributes to gastric carcinogenesis. PLoS One. 6:e166172011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang X, Tang S, Le SY, et al: Aberrant
expression of oncogenic and tumor-suppressive microRNAs in cervical
cancer is required for cancer cell growth. PLoS One. 3:e25572008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Saito Y, Friedman JM, Chihara Y, Egger G,
Chuang JC and Liang G: Epigenetic therapy upregulates the tumor
suppressor microRNA-126 and its host gene EGFL7 in human cancer
cells. Biochem Biophys Res Commun. 379:726–731. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu S, Gao S, Wang XY and Wang DB:
Expression of miR-126 and Crk in endometriosis: miR-126 may affect
the progression of endometriosis by regulating Crk expression. Arch
Gynecol Obstet. 285:1065–1072. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Meister J and Schmidt MH: miR-126 and
miR-126*: new players in cancer. Sci World J.
10:2090–2100. 2010.
|
|
18
|
Sessa R, Seano G, di Blasio L, et al: The
miR-126 regulates angiopoietin-1 signaling and vessel maturation by
targeting p85beta. Biochim Biophys Acta. 1823:1925–1935. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang J, Du YY, Lin YF, et al: The cell
growth suppressor, mir-126, targets IRS-1. Biochem Biophys Res
Commun. 377:136–140. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Felli N, Felicetti F, Lustri AM, et al:
miR-126&126* restored expressions play a tumor
suppressor role by directly regulating ADAM9 and MMP7 in melanoma.
PLoS One. 8:e568242013.
|
|
21
|
Miko E, Margitai Z, Czimmerer Z, et al:
miR-126 inhibits proliferation of small cell lung cancer cells by
targeting SLC7A5. FEBS Lett. 585:1191–1196. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Glover DM, Hagan IM and Tavares AA:
Polo-like kinases: a team that plays throughout mitosis. Genes Dev.
12:3777–3787. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nigg EA: Polo-like kinases: positive
regulators of cell division from start to finish. Curr Opin Cell
Biol. 10:776–783. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Warnke S, Kemmler S, Hames RS, et al:
Polo-like kinase-2 is required for centriole duplication in
mammalian cells. Curr Biol. 14:1200–1207. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Burns TF, Fei P, Scata KA, Dicker DT and
El-Deiry WS: Silencing of the novel p53 target gene Snk/Plk2 leads
to mitotic catastrophe in paclitaxel (taxol)-exposed cells. Mol
Cell Biol. 23:5556–5571. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Eckerdt F, Yuan J and Strebhardt K:
Polo-like kinases and oncogenesis. Oncogene. 24:267–276. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li Z, Lu J, Sun M, et al: Distinct
microRNA expression profiles in acute myeloid leukemia with common
translocations. Proc Natl Acad Sci USA. 105:15535–15540. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Oglesby IK, Bray IM, Chotirmall SH, et al:
miR-126 is downregulated in cystic fibrosis airway epithelial cells
and regulates TOM1 expression. J Immunol. 184:1702–1709. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar
|
|
30
|
Ueda T, Volinia S, Okumura H, et al:
Relation between microRNA expression and progression and prognosis
of gastric cancer: a microRNA expression analysis. Lancet Oncol.
11:136–146. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shi H, Xu J, Zhang G, et al: Walking the
interactome to identify human miRNA-disease associations through
the functional link between miRNA targets and disease genes. BMC
Syst Biol. 7:1012013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang M, Zhou S, Zhang L, et al: miR-518b
is down-regulated, and involved in cell proliferation and invasion
by targeting Rap1b in esophageal squamous cell carcinoma. FEBS
Lett. 586:3508–3521. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Abell K, Bilancio A, Clarkson RW, et al:
Stat3-induced apoptosis requires a molecular switch in PI (3) K
subunit composition. Nat Cell Biol. 7:392–398. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
De Gregorio G, Coppa A, Cosentino C, et
al: The p85 regulatory subunit of PI3K mediates TSH-cAMP-PKA growth
and survival signals. Oncogene. 26:2039–2047. 2007.PubMed/NCBI
|
|
35
|
Asano T, Yao Y, Zhu J, Li D, Abbruzzese JL
and Reddy SA: The PI 3-kinase/Akt signaling pathway is activated
due to aberrant Pten expression and targets transcription factors
NF-kappaB and c-Myc in pancreatic cancer cells. Oncogene.
23:8571–8580. 2004. View Article : Google Scholar : PubMed/NCBI
|