Introduction
Fanconi anemia (FA) is a chromosomal instability
disorder inherited as an autosomal- or X-chromosomal recessive
trait due to germline mutations in one of 15 FA genes
(FANCA/B/C/D1/D2/E/F/G/I/J/L/M/N/O/P) involved in the DNA repair
pathway (1). Clinically, FA is
characterized by various congenital malformations, increased risk
of malignancies and progressive bone marrow failure (2). Head and neck squamous cell carcinoma
(HNSCC) is the most frequently diagnosed solid cancer in FA
patients. Some FA patients may not exhibit bone marrow failure due
to hypomorphic mutations, or reversions or mosaicism in the
hematopoietic tissue and the diagnosis of HNSCC usually precedes
the diagnosis of FA in these patients (3). Oral cavity is the predominant site of
HNSCC in FA patients, frequently occurring in the tongue and
gingiva (3). The risk of HNSCC
among FA patients is 800-fold higher than in the general population
(4,5). HNSCC in FA patients occurs at a
younger age (median age: 27 years) than the general population and
FA patients as young as 10 years have developed HNSCC.
Hematopoietic stem cell transplantation (HSCT),
which greatly extends life-expectancy, is the treatment of choice
for FA patients. FA patients who survive into adulthood after HSCT
are at a higher risk for developing HNSCC, significantly reducing
their quality of life and overall life-expectancy (3–5). FA
patients (~50%) who have not undergone HSCT are expected to develop
HNSCC by 45 years of age, whereas the cumulative incidence of HNSCC
in bone marrow-transplanted FA patients is estimated to be 100% at
the same age (6). Increased risk
for HNSCC in FA patients after HSCT is attributed to the
conditioning regimen and the occurrence of chronic graft-vs.-host
disease in the oral cavity (3).
HNSCC in FA patients occurs without exposure to any of the known
risk factors namely tobacco and alcohol (3,7).
Although an association between oncogenic human papillomavirus
(HPV) infection and FA-HNSCC was suggested in previous studies
(8), recent studies suggest that
HPV infection is not the cause for HNSCC in FA patients (9). Moreover, molecular profiles of
FA-HNSCC cells are not significantly different from those of
sporadic HNSCC (10). These
findings indicate increased susceptibility to a variety of mutagens
and resultant non-lethal DNA damage that promotes oral mucosal
tumorigenesis in FA patients (11).
Gammon et al reported the presence of a
stem-like cell population in FA oral cancer cell lines based on the
difference in the colony morphologies between sporadic and FA-HNSCC
cell lines (12). Stem-like cells,
known as cancer stem cells (CSC), that initiate and sustain tumor
growth and spread have been identified in a number of solid
malignancies (13). A
subpopulation of cells within a tumor that has a higher-tumor
repopulating potential is identified as CSC (14,15).
CSC have the capacity to self-renew and to give rise to
heterogeneous lineages of cancer cells that populate the tumors
(15). CSC share gene expression
profiles and phenotypic characteristics with embryonic and somatic
stem cells including a slow proliferation rate and resistance to
standard chemotherapy and radiation therapy (16). Tumors with a higher fraction of CSC
exhibit therapeutic resistance and increased risk for local
recurrence and distant spread (16,17).
CSC can be identified and isolated using various markers and CD44
expressing tumor cells isolated from HNSCC were identified as CSC
based on increased clonogenic potential and tumor-forming ability
(18,19). Recently, HNSCC cells expressing
high levels of aldehyde dehydrogenase (ALDH) were identified as CSC
(20). The Aldefluor assay is
considered a reliable method to enrich and propagate CSC in various
solid cancers including HNSCC (20). The Aldefluor assay measures ALDH
activity by quantifying the conversion of ALDH substrate, BODIPY
aminoacetaldehyde to a fluorescent reaction product BODIPY
aminoacetate (21).
Aldefluor-treated tumor cells with high ALDH isoform 1 (ALDH1)
activity turn brightly fluorescent and two subpopulations
(ALDHpos and ALDHneg cells) can be enumerated
by standard flow cytometer or isolated by fluorescence-assisted
cell sorting (FACS) for further analysis. Similarly,
immunohistochemical staining using an ALDH1-specific antibody has
been used successfully to identify and quantify CSC in
formalin-fixed paraffin-embedded tumor sections. Aldefluor assay
and ALDH1 immunohistochemistry are widely used for detection and
enumeration of CSC in tumor cell lines and tumor samples,
respectively (22–24). In this study, we used the Aldefluor
assay, ALDH1 immunohistochemistry and tumorsphere-formation to
quantify and characterize CSC populations in FA and sporadic HNSCC
cell lines and tumor samples. We analyzed the expression patterns
of 14 stemness-related genes in ALDH1pos and
ALDH1neg cells isolated from FA-HNSCC cells using
reverse transcription-polymerase chain reaction (RT-PCR).
Materials and methods
Cell culture
The human FA-HNSCC cell lines VU-1365 and VU-1131
were kindly donated by Dr Ruud H. Brakenhoff (Vrije University
Medical Center, Amsterdam, The Netherlands) and OHSU-974 cell line
was obtained from Dr Laura Hayes (Oregon Health and Science
University, Portland, OR, USA). UMSCC-22A, a human sporadic HNSCC
cell line, was obtained from Dr Thomas E. Carey, University of
Michigan. Molecular phenotypes of these cell lines have been
defined in published reports and are shown in Table I (10,25).
These cell lines were grown in adherent conditions using the
recommended culture medium (10,25).
 | Table IClinical and molecular
characteristics of FA and sporadic HNSCC cell lines. |
Table I
Clinical and molecular
characteristics of FA and sporadic HNSCC cell lines.
| Cell line | Source |
Characteristics |
|---|
| UMSCC-22A | 58YOF | Sporadic OSCC |
| Oropharynx | TP53 mutation:
Yes |
| T2N1M0 | |
| OHSU-974 | 29YOM | FA OSCC |
| Tongue | FA Type: FA-A |
| TNM: unknown
(8) | TP53 mutation:
Yes |
| VU-1131 | 34YOF | FA OSCC |
| Floor of the
mouth | FA Type: FA-C |
| T4N2M0 (8) | TP53 mutation:
Yes |
| VU-1365 | 22YOM | FA OSCC |
| Oral cavity | FA Type: FA-A |
| TNM: unknown
(8) | TP53 mutation:
Yes |
Human and xenograft FA-HNSCC tumor
samples
Formalin-fixed paraffin-embedded tissue sections of
FA-HNSCC specimen and its corresponding orthotopic tongue
xenografts were kindly gifted by Dr Susanne Wells (Cincinnati
Children’s Hospital, Cincinnati, OH, USA). FA-HNSCC tumor sample of
a FANC-B deficient 56-year old female was obtained through the
National Disease Research Interchange (NDRI no. 0066421;
PD-RD-000237). Tumor cells derived from the fresh tumor of the same
patient (FAHNSCC-2) were implanted into the tongue of NOD/SCID mice
to generate the orthotopic tumor xenografts.
Aldefluor assay and FACS of
ALDHpos and ALDHneg cells
Tumor cell fractions with high (ALDHpos)
and low (ALDHneg) ALDH activity among FA (VU-1131,
VU-1365 and OHSU-974) and sporadic (UMSCC-22A) HNSCC cell lines
were quantified using the Aldefluor kit (StemCell Technologies,
Vancouver, BC, Canada) according to the manufacturer’s protocol.
Briefly, FA and sporadic HNSCC cells (1×106 cells/ml)
were resuspended in Aldefluor assay buffer containing ALDH1
substrate BAAA without (test sample) and with ALDH1 inhibitor DEAB
(negative control). Test sample and negative control were incubated
for 45 min at 37°C and then the cells were centrifuged and
resuspended in an Aldefluor assay buffer and kept on ice for FACS.
The amount of fluorescent ALDH1 reaction product produced in the
cells is proportional to their ALDH1 activity and only viable cells
with intact cell membrane will retain the fluorescent reaction
product. The intensely fluorescent (ALDHpos) cells were
detected in the green fluorescent channel (FITC; 520–540 nm) and
calculated as the percent of ALDHpos in each cell line.
The sorting gates were generated using the DEAB-treated
Aldefluor-stained negative control cells.
The FA-HNSCC cell line VU-1365 with the highest
ALDHpos fraction was used for isolating
ALDHpos and ALDHneg cells by FACS using a
FACSAria II (BD Biosciences, San Jose, CA, USA). Isolated Aldefluor
positive and negative fractions were used for RNA extraction and
RT-PCR analysis of a set of genes linked to ‘stemness’
phenotype.
RT-PCR analysis of stemness gene
expression in ALDHpos and ALDHneg cells
Expression patterns of 14 genes that are commonly
used as molecular markers for undifferentiated (DPPA5/ESG1, Nanog,
Oct-3/4, SOX2) and lineage-committed (Nestin, Otx2, TP63, AFB,
GATA-4, PDX-1, SOX17, HNF-3β, Brachyury, Stella) embryonic stem
cells (ESC) and induced pluripotent stem (iPS) cells were examined
using the Human Pluripotent Stem Cell Assessment Primer Pair Panel
kit (R&D Systems, Minneapolis, MN, USA) according to the
manufacturer’s protocol. Briefly, total RNA was extracted from
ALDHpos and ALDHneg cells isolated from
FA-HNSCC cell line VU-1365 using RNeasy® Mini kit
(Qiagen, Valencia, CA, USA). RNA was treated with DNase I (Ambion,
Austin, TX, USA) to remove DNA contamination and absence of genomic
DNA was further confirmed by running a control reaction of PCR of
total RNA without reverse transcription (RT). RNA concentration was
measured using a NanoDrop ND-1000 spectrophotometer (NanoDrop
Technologies, Houston, TX, USA) and 1 μg RNA of each sample was
used for cDNA synthesis using the Cells-to-cDNA™ II kit
(Ambion/Applied Biosystems, Austin, TX, USA). cDNA (1 μl) was used
for each primer pair, and PCR amplification was performed according
to the manufacturer’s recommended parameters. Amplified PCR
products were analyzed on a 1.8% agarose gel and the predicted
sizes of the PCR products range from 230–591 bp. Synthetic
double-stranded DNA provided in the kit (positive control 57) was
used as a positive control for PCR amplification for each pair of
primers. Human GAPDH primer is used as a control for successful
cDNA synthesis.
Tumorsphere formation assay
Tumorsphere-forming potential of UMSCC-22A and
VU-1365 cells were assayed using the MammoCult™ kit (Catalog no.
05620; StemCell Technologies) according to the manufacturer’s
protocol. Briefly, 80–90% confluent tumor cells were harvested
using a sterile cell scraper without trypsinization and resuspended
in complete MammoCult® medium and centrifuged the cells
at 500 × g for 3 min at room temperature. Cell pellets were
resuspended in complete MammoCult®, counted and diluted
in the same media to a predetermined concentration. Cells were
plated in triplicates (5,000 cells/well) in a 6-well ultra-low
adherent plate (Catalog no. 27145; StemCell Technologies). Cells
were grown at 37°C and 5% CO2 for 7 days in a tissue
culture incubator and monitored daily for tumorsphere formation.
After 7 days, photomicrographs of tumorspheres were taken using an
inverted phase-contrast microscope and used for counting the number
of tumorspheres per high power field (HPF).
ALDH1 immunohistochemistry
Archival specimens of formalin-fixed
paraffin-embedded sporadic HNSCC (n=5), FA-HNSCC (n=1) and
orthotopic FA-HNSCC xenografts (n=2) generated in the tongue of
NOD/SCID mice were used to evaluate the expression of ALDH1 in
tumor cells. Representative tissue sections were deparaffinized and
rehydrated and antigen retrieval was performed by boiling in
Antigen Decloaker (Biocare Medical, Concord, CA, USA) solution for
2 min. Endogenous peroxidase activity was blocked by incubating the
tissue section with 3% H2O2 in methanol for
10 min. Non-specific binding sites were blocked by incubating the
tissue sections in Background Terminator (Biocare Medical) solution
for 10 min. Tissue sections were treated with normal horse serum
for 10 min to block non-specific binding sites and then incubated
with anti-human ALDH-1 antibody (1:100; BD Transduction
Laboratories, mAb Clone no. 44) overnight at room temperature.
ALDH1-antibody binding sites were detected using the Vectastain
Elite ABC kit-Mouse (Vector Laboratories, Inc., Burlingame, CA,
USA) according to the protocol provided with this kit. Peroxidase
reactivity was visualized using 3-3′-diaminobenzidine tetrachloride
as chromogenic substrate and counterstained with hematoxylin. For
negative control, tissue sections were incubated with mouse IgG1
isotype control.
Results
ALDH1-hi subfraction is
enriched in FA-HNSCC cell lines
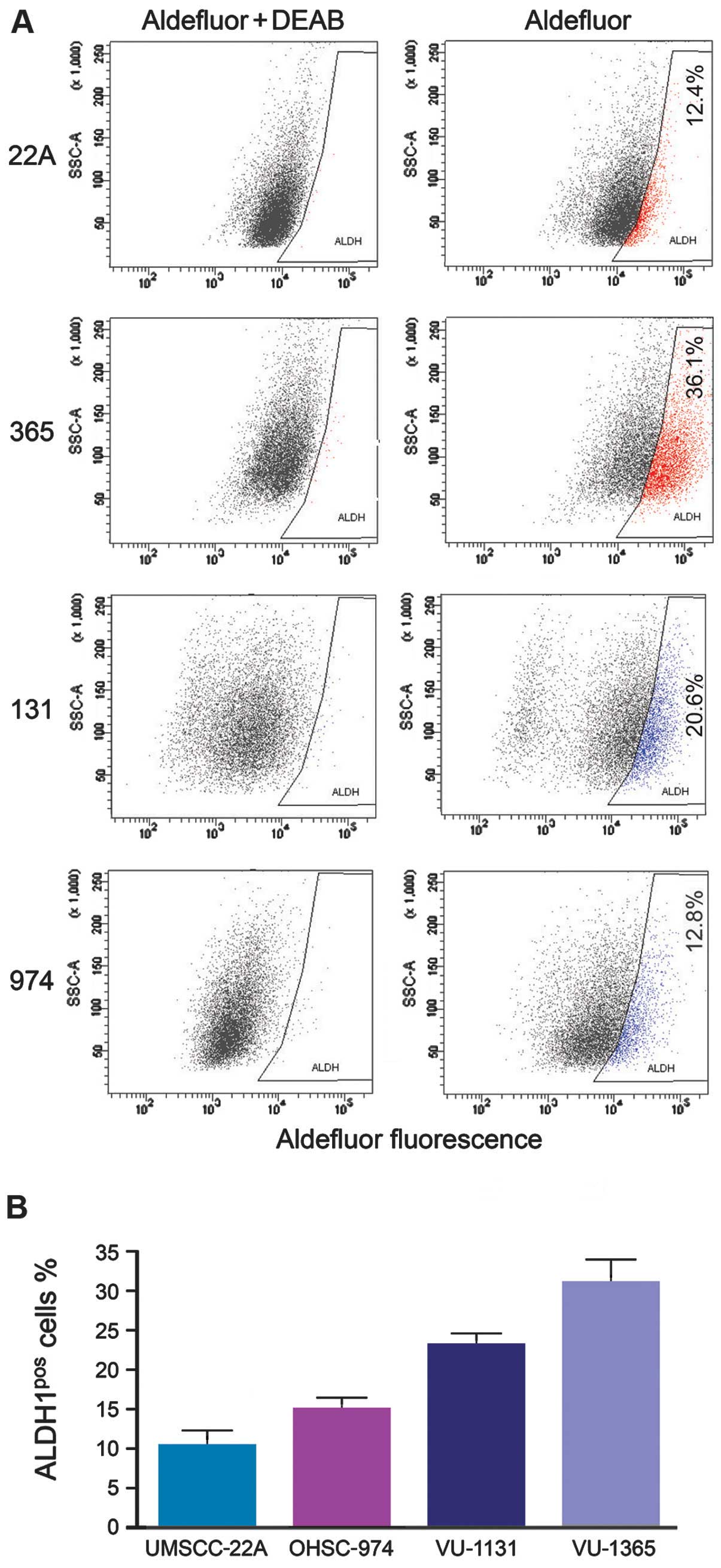
Using the Aldef luor assay, we examined the
ALDHpos and ALDHneg fractions in sporadic
(UMSCC-22A) and FA (VU-1365, VU-1131 and OHSU-974) cell lines. FACS
analysis demonstrated variable ALDH1 expression among sporadic and
FA-HNSCC cell lines. FA-HNSCC cell lines consistently demonstrated
a greater fraction of ALDHpos cells (VU-1365=31±2.9%,
VU-1131=23±2.7%, OHSU-974=15±2.0%) compared to sporadic HNSCC cell
line (UMSCC-22A=10.33±3.0%) (Fig.
1). The FA-HNSCC cell line VU-1365 demonstrated the highest
percentage of ALDHpos CSC. We selected this cell line to
isolate the ALDHpos and ALDHneg fraction to
confirm the stemness phenotype of ALDHpos cells by
RT-PCR.
ALDH1pos and
ALDH1neg cells isolated from FA-HNSCC differentially
express stemness-related genes
To determine whether ALDH1pos isolated
from FA-HNSCC cells represent tumor cells with stem cell-like
properties, we compared the expression profiles of genes linked to
undifferentiated and lineage-committed ECS and iPS cells between
ALDH1pos an ALDH1neg cells using RT-PCR. The
ALDH1pos cells consistently express mRNA transcripts of
genes specific for undifferentiated ESC and iPS cells (Nanog,
Oct-3/4) and germ cells (Stella), whereas these genes are not
expressed in ALDH1neg (Fig.
2). Both ALDH1pos and ALDH1neg express
SOX2, Nestin and TP63 genes that are linked to ectodermal
lineage-committed ECS and iPS cells. Neither of these fractions
express genes linked to either endodermal-(AFB, GATA-4, PDX-1,
SOX17 and HNF-3β) or mesodermal-(Brachyury) lineage-committed ES
and iPS cells (Fig. 2).
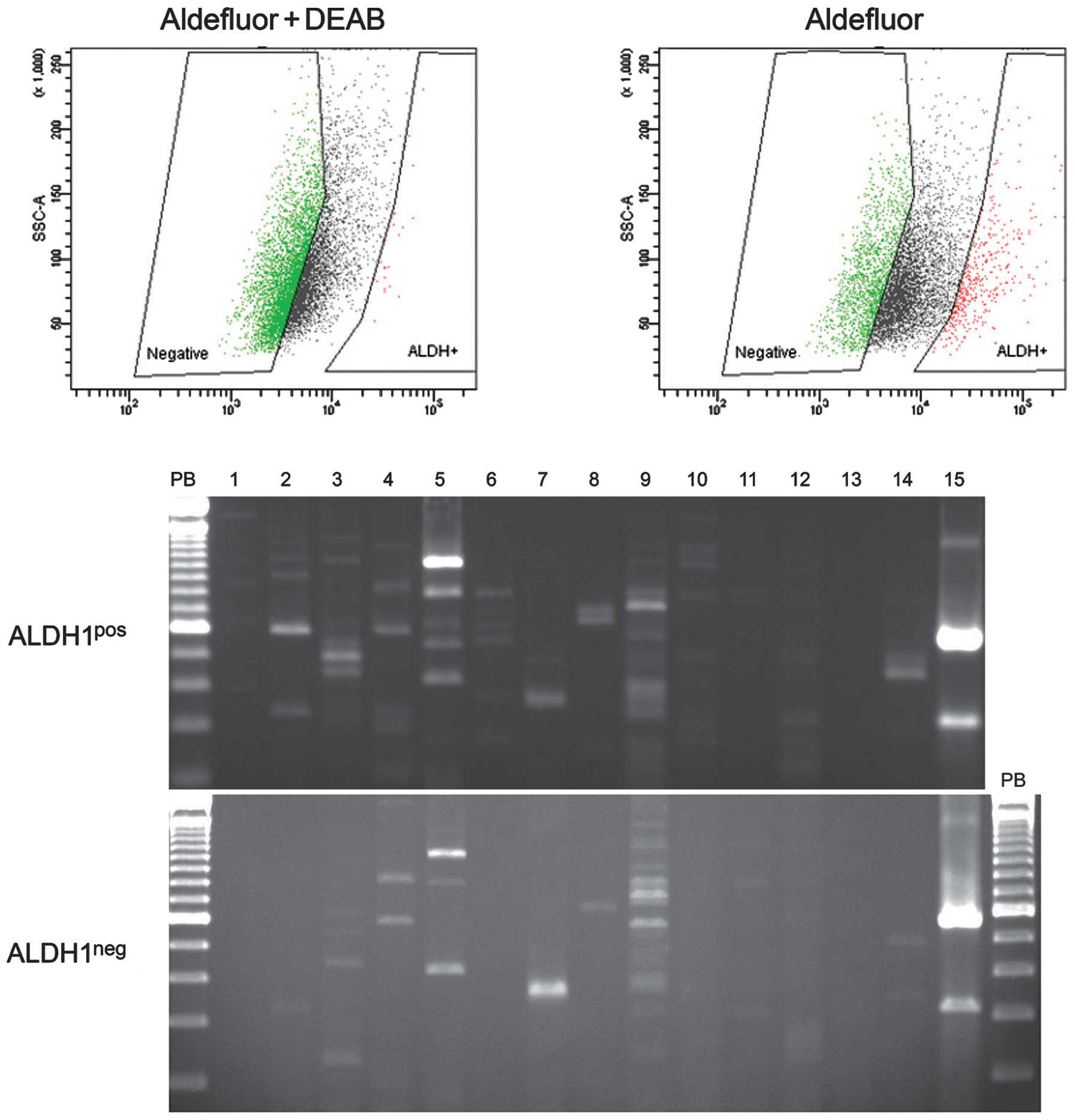 | Figure 2The expression profiles of
stemness-genes in ALDH1pos and ALDH1neg cell
populations in FA-HNSCC. The Aldefluor assay was used to isolate
ALDH1pos and ALDH1neg cells in VU1365 cell
line and used for RNA isolation and reverse
transcription-polymerase chain reaction (RT-PCR) array analysis.
The human pluripotent stem cell assessment primer pairs were used
to amplify the mRNA transcripts of 14 genes that are frequently
used as molecular markers of undifferentiated embryonic stem (ES)
cells (Lane 2, DPPA5/ESG1; Lane 3, Nanog; Lane 4, Oct-3/4; Lane 5,
SOX2), ectodermal lineage-committed stem cells (Lane 5, SOX2; Lane
6, Nestin; Lane 7, Otx2; Lane 8, TP63), endodermal
lineage-committed stem cells (Lane 9, AFB; Lane 10, GATA-4; Lane
11, SOX17; Lane 12, HNF-3β), mesodermal lineage-committed germ
cells (Lane 13, Brachury) and germs cells (Lane 14, Stella). Lane
15, GAPDH-loading control. |
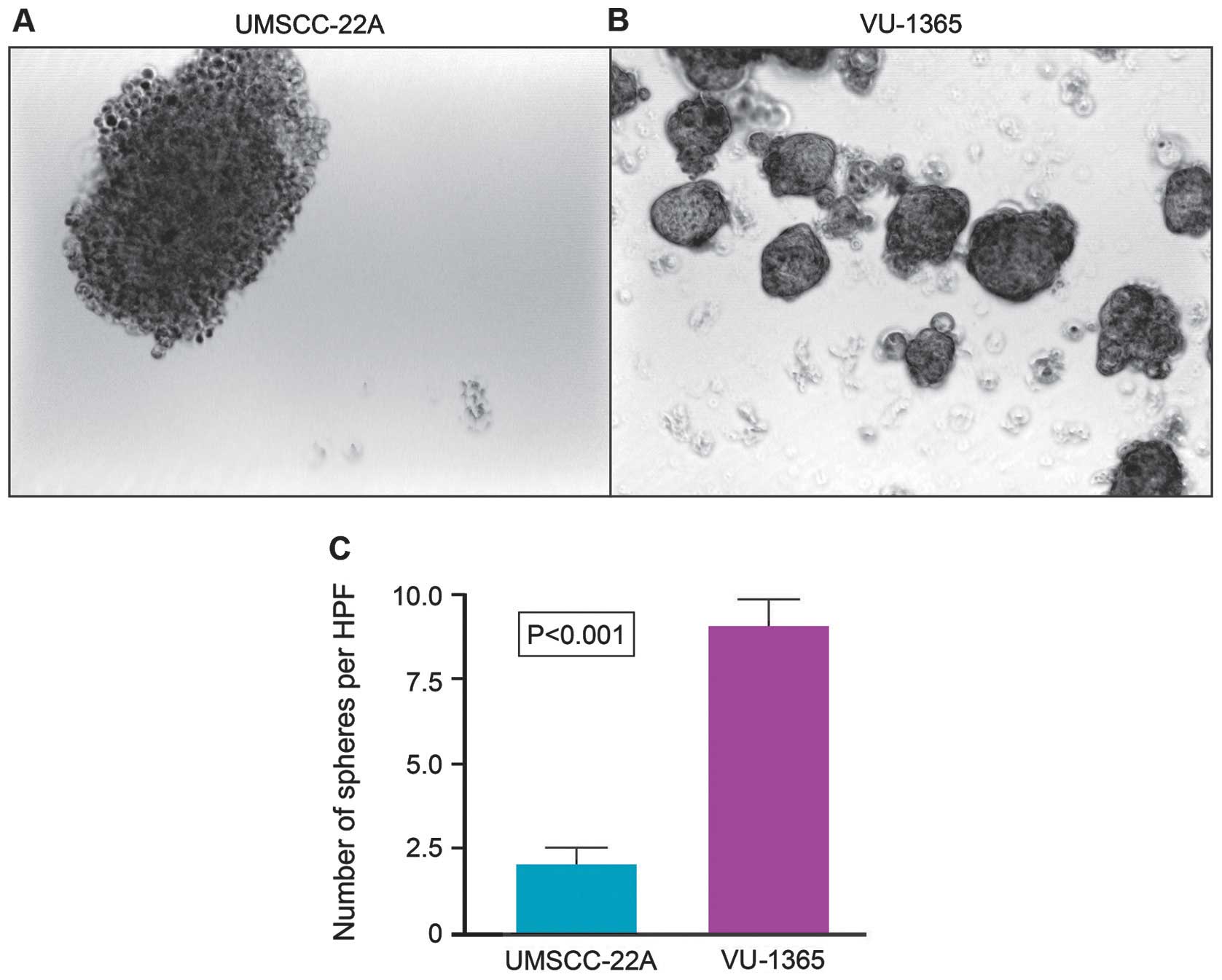
FA-HNSCC cells reveal higher tumorsphere
formation capacity than the sporadic HNSCC cells
Sphere-forming capacity of tumor cells correlates
directly with CSC phenotype and thus tumorigenic efficiency of the
cancer cells can be determined based on the number of spheres that
originate from specific number of seeded cells. We compared the
tumorsphere-forming capacities of tumor cell lines derived from
FA-HNSCC (VU-1365) and sporadic HNSCC (UMSCC-22A). Although both
cell lines formed stable tumorspheres at day 7, there were
significant quantitative and qualitative differences in their
sphere-forming potential (Fig. 3).
The number of tumorspheres formed by VU-1365 cells are
significantly (p<0.001) higher than UMSCC-22A cells (Fig. 3). Tumorspheres formed by VU-1365
cells appear to be solid and round whereas UMSCC-22A spheres are
looser, less cohesive and irregular in shape (Fig. 3).
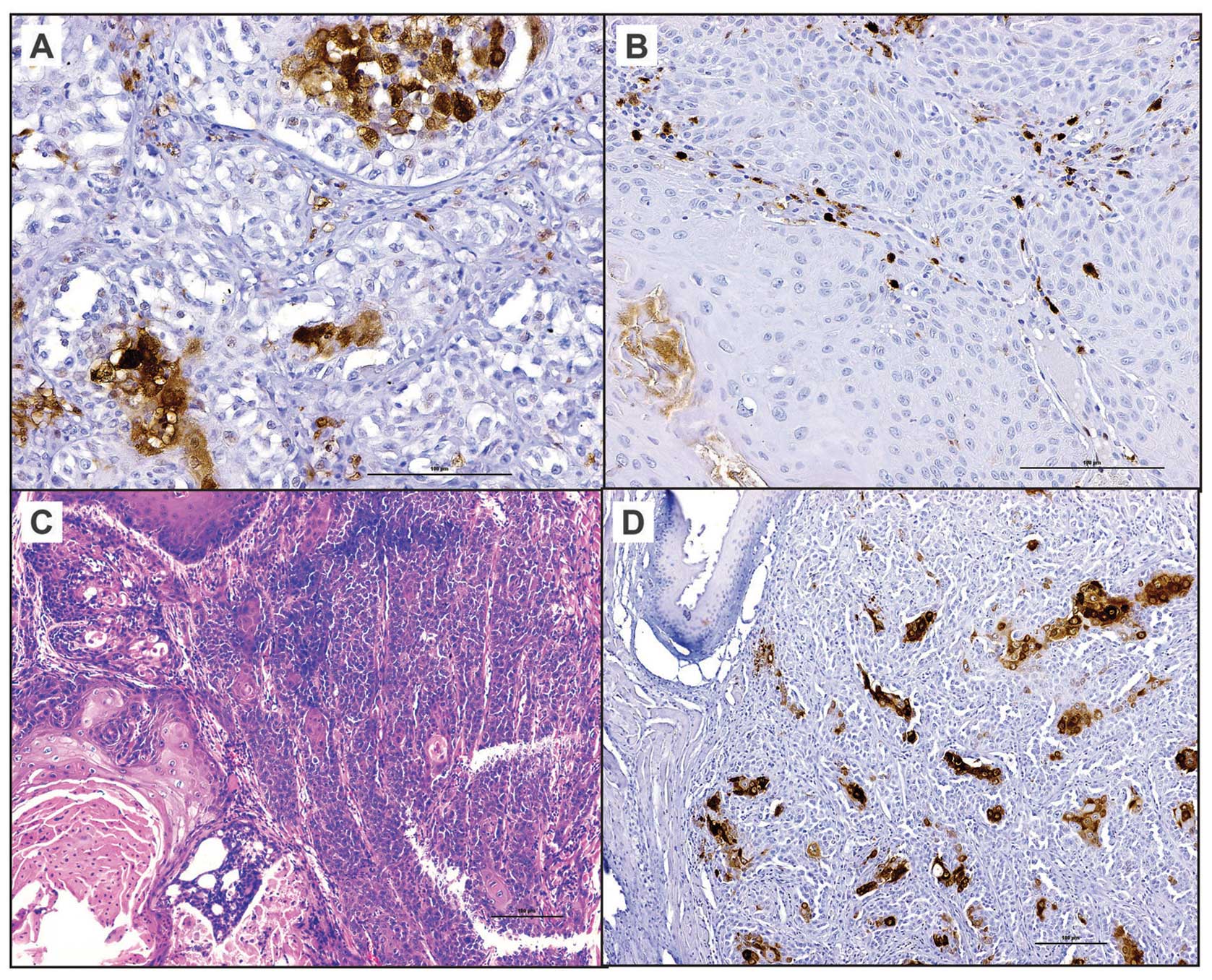
Expression of ALDH1 in FA and sporadic
HNSCC tumor samples
Tumor cells positive for ALDH1 were noted in three
of the five sporadic HNSCC tissue sections. Less than 10% of the
tumor cells stained positive for ALDH1 in sporadic HNSCC whereas
>25% of tumor cells are positive for ALDH1 in FA-HNSCC tissue
section (Fig. 4). Interestingly,
the FA-HNSCC tumor xenograft sections exhibited similar patterns of
ALDH1 expression as its parental tumor. Moreover, ALDH1-positivity
was noted in individual and small groups (3–5 cells/group) of tumor
cells in sporadic HNSCC tumor sections. In contrast, multiple large
foci of tumor cells express ALDH1 in FA-HNSCC sample and its
corresponding tumor xenograft (Fig.
4).
Discussion
The enzymatic activity of ALDH1, which can be
measured by Aldefluor assay, has been identified as a reliable
marker to identify normal and cancer stem cells (21). ALDH1 is widely used as a universal
functional marker to identify and isolate tumor cells, with
‘stemness’ phenotypes in several solid malignant tumors including
head and neck cancer (20). In
this study, we used Aldefluor assay to determine the presence and
size of the CSC fraction in a panel of cell lines derived from FA
and sporadic HNSCC. Our findings provide convincing evidence that
FA-HNSCC is highly enriched for CSC and confirm a previous study
reporting the presence of a subpopulation of tumor cells in
FA-HNSCC with stem cell-like properties (12). Evidence that ALDH1pos
cells in FA-HNSCC display a stem cell phenotype is further
supported by their expression of genes linked to stem cell
pluripotency and differentiation. We examined the expression of
mRNA transcripts of 14 genes linked to undifferentiated and
lineage-committed human ESC cells and iPS cells in
ALDHpos and ALDHneg cells using
gel-electrophoresis based RT-PCR. This qualitative method is
convenient and easily interpretable for the presence or absence of
the transcripts and a reliable alternative to microarray and
quantitative RT-PCR-based gene expression-profiling assays that
reveal the quantitative changes in genes that are differentially
expressed. Among the 14 genes examined, transcripts for Nanog,
Oct-3/4 and Stella are expressed only by ALDH1pos
FA-HNSCC cells and not by ALDH1neg cells. The
transcription factors Oct-3/4 and Nanog are crucial for the
maintenance of pluripotency and self-renewal of undifferentiated
ESC (26,27). Stella is a marker gene of
primordial germ cells and undifferentiated ESC but not expressed by
lineage-committed ESC (27).
Interestingly, SOX-2 is expressed by both ALDH1pos and
ALDH1neg cells albeit higher levels in
ALDH1pos cells which is in accordance with its function
as a transcription factor involved in both conferring pluripotency
to undifferentiated ESC and ectodermal lineage-committed ESC
(27). On the other hand, TP63, a
gene linked to ectodermal lineage-committed ESC, is also expressed
in both ALDH1pos and ALDH1neg cells but its
expression is more in ALDH1neg cells than
ALDH1pos cells. Genes linked to ectodermal (Nestin,
Otx2), endodermal (AFB, GATA-4, PDX-1, SOX17 and HNF-3β) and
mesodermal (Brachyury) lineage-committed ESC are not expressed
either by ALDH1pos or ALDH1neg cell
fractions. Our data suggest that CSC of FA-HNSCC express the
phenotype of undifferentiated ESC but not that of ectodermal
lineage-committed ESC.
CSC are clonogenic and undergo asymmetric cell
division resulting in self-renewal and multilineage differentiation
of their progenitors (14,28). Unlimited self-renewal potential and
formation of tumorspheres in vitro under low-attachment
condition are the hallmarks of CSC and defined their
tumor-initiating potential in vivo (29). On the contrary, cancer cells
lacking the stem cell phenotype are incapable of forming
tumorspheres when grown under similar culture conditions. The
ability of the tumor cells to form spheres in low-attachment and
serum-free culture conditions correlates with their ability to form
tumors in xenograft models (29).
Hence, tumorsphere-forming efficiency of cell lines correlates
positively with their respective fractions of CSC and in
vivo tumorigenicity (30). We
therefore compared the tumorsphere-forming capacity of FA (VU-1365)
and sporadic (UMSCC-22A) HNSCC cell lines. Our data clearly show
that VU-1365 poses a higher capacity of tumorsphere formation than
UMSCC-22A cells and confirms that FA-HNSCC cells contain a larger
fraction of tumor cells with CSC-phenotype compared to sporadic
HNSCC cells.
Next, we examined the ALDH1 expression in sporadic
and FA-HNSCC tumor tissue using immunohistochemistry. ALDH1
expression was noted in all HNSCC tumor sections examined. In
sporadic HNSCC tumors ALDH1pos cells were scattered as
single cells or small group of cells and represented a relatively
small population. By contrast, FA-HNSCC tumor tissue contained a
large fraction of ALDH1pos cells that were distributed
as sheets or islands of ALDH1pos cells among the tumor
tissue. Interestingly, CSC fraction and distribution pattern are
very similar in FA-HNSCC tumor and in the tumor xenograft derived
from the same tumor. Breast, lung, prostate and pancreatic
carcinomas with a high percentage of ALDH1pos cells
demonstrate an aggressive clinical course with poor prognosis
compared to the tumors with low ALDH1pos tumor cell
fraction (31–33). The relative abundance of
ALDH1pos CSC fraction may account for the biological
differences between the FA-HNSCC and sporadic HNSCC.
HNSCC represents the most frequently diagnosed solid
tumor in FA patients, developing at a significantly younger age
than the general population (3,5,6).
HNSCC in FA patients carries a poor prognosis and requires early
intervention and aggressive surgical treatment (3). Radiation and chemotherapy have
limited value in treating HNSCC in FA patients because their normal
cells are highly sensitive to DNA cross-linking agents (3). Moreover, FA-HNSCC patients have a
higher lifetime risk for developing multiple primary tumors than
the general population due to the presence of a widespread of
pre-malignant disease (3). This
phenomenon commonly known as ‘field cancerization’, is used to
describe multifocal pre-malignant disease, a higher than expected
prevalence of second primary tumors and the occurrence of
synchronous distant tumors within the upper aerodigestive tract
(34). Recent studies suggest that
genetic alterations that promote the development of CSC are
responsible for field cancerization (35).
CSC population is either derived directly from the
transformation of normal stem cells or from aberrant
de-differentiation of non-CSC acquiring the molecular armaments of
stem cells (14,28). In either case, mutations in the
fundamental self-renewal signaling cascades lead to the formation
of CSC, that are independent of regulatory pathways that govern
normal stem cells (36,37). We postulate that the defect in the
DNA repair pathways, a hallmark of FA, increases the risk of
mutation in the genes that guard self-renewal pathways of stem
cells and causes the enrichment of tumor cells with stemness in
FA-HNSCC. Recent evidence points to CSC as the key drivers of field
cancerization, resistance to chemotherapy and radiation therapy,
disease relapse and metastasis in solid malignancies (16). It is unlikely to be a coincidence
that CSC are found in abundance in FA-HNSCC with a high-risk for
field cancerization and aggressive biologic behavior compared to
sporadic HNSCC (3–5,38).
CSC survives the conventional chemoradiation therapy and maintains
their ability to re-populate to produce tumor recurrence and
metastasis (39,40). Hence, CSC-targeted therapy remains
an attractive approach for HNSCC in FA patients who currently have
very limited treatment options.
Acknowledgements
Grant support: NIH/NIDCR: 1RC2DE020785; CPRIT
RP101382; Pilot Award from ‘The Center for Clinical and
Translational Sciences (CCTS)’, UTHSC-Houston. We thank Drs Thomas
E. Carey, University of Michigan (UMSCC-22A), Ruud H. Brakenhoff,
Vrije University Medical Center, The Netherlands (VU-1131 and
VU-1365) and Laura Hayes, Oregon Health and Science University
(OHSU-974) for providing us the cell lines used in this study.
Abbreviations:
|
FA
|
Fanconi anemia
|
|
HNSCC
|
head and neck squamous cell
carcinoma
|
|
ALDH
|
aldehyde dehydrogenase
|
|
CSC
|
cancer stem cells
|
|
RT-PCR
|
reverse transcription-polymerase chain
reaction
|
|
FACS
|
fluorescence-assisted cell sorting
|
References
|
1
|
Levitus M, Joenje H and de Winter JP: The
Fanconi anemia pathway of genomic maintenance. Cell Oncol. 28:3–29.
2006.PubMed/NCBI
|
|
2
|
Kutler DI, Singh B, Satagopan J, et al: A
20-year perspective on the International Fanconi Anemia Registry
(IFAR). Blood. 101:1249–1256. 2003.PubMed/NCBI
|
|
3
|
Scheckenbach K, Wagenmann M, Freund M,
Schipper J and Hanenberg H: Squamous cell carcinomas of the head
and neck in Fanconi anemia: risk, prevention, therapy, and the need
for guidelines. Klin Padiatr. 224:132–138. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kutler DI, Auerbach AD, Satagopan J, et
al: High incidence of head and neck squamous cell carcinoma in
patients with Fanconi anemia. Arch Otolaryngol Head Neck Surg.
129:106–112. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Masserot C, Peffault de Latour R, Rocha V,
et al: Head and neck squamous cell carcinoma in 13 patients with
Fanconi anemia after hematopoietic stem cell transplantation.
Cancer. 113:3315–3322. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rosenberg PS, Socié G, Alter BP and
Gluckman E: Risk of head and neck squamous cell cancer and death in
patients with Fanconi anemia who did and did not receive
transplants. Blood. 105:67–73. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Birkeland AC, Auerbach AD, Sanborn E, et
al: Postoperative clinical radiosensitivity in patients with
fanconi anemia and head and neck squamous cell carcinoma. Arch
Otolaryngol Head Neck Surg. 137:930–934. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kutler DI, Wreesmann VB, Goberdhan A, et
al: Human papillomavirus DNA and p53 polymorphisms in squamous cell
carcinomas from Fanconi anemia patients. J Natl Cancer Inst.
95:1718–1721. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Alter BP, Giri N, Savage SA, Quint WG, de
Koning MN and Schiffman M: Squamous cell carcinomas in patients
with Fanconi anemia and dyskeratosis congenita: a search for human
papillomavirus. Int J Cancer. 133:1513–1515. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
van Zeeburg HJ, Snijders PJ, Pals G, et
al: Generation and molecular characterization of head and neck
squamous cell lines of fanconi anemia patients. Cancer Res.
65:1271–1276. 2005.PubMed/NCBI
|
|
11
|
Schlacher K, Wu H and Jasin M: A distinct
replication fork protection pathway connects Fanconi anemia tumor
suppressors to RAD51-BRCA1/2. Cancer Cell. 22:106–116. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gammon L, Biddle A, Fazil B, Harper L and
Mackenzie IC: Stem cell characteristics of cell sub-populations in
cell lines derived from head and neck cancers of Fanconi anemia
patients. J Oral Pathol Med. 40:143–152. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Alison MR, Islam S and Wright NA: Stem
cells in cancer: instigators and propagators? J Cell Sci.
123:2357–2368. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Beck B and Blanpain C: Unravelling cancer
stem cell potential. Nat Rev Cancer. 13:727–738. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Frank NY, Schatton T and Frank MH: The
therapeutic promise of the cancer stem cell concept. J Clin Invest.
120:41–50. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sampieri K and Fodde R: Cancer stem cells
and metastasis. Semin Cancer Biol. 22:187–193. 2012. View Article : Google Scholar
|
|
18
|
Noto Z, Yoshida T, Okabe M, et al: CD44
and SSEA-4 positive cells in an oral cancer cell line HSC-4 possess
cancer stem-like cell characteristics. Oral Oncol. 49:787–795.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Prince ME, Sivanandan R, Kaczorowski A, et
al: Identification of a subpopulation of cells with cancer stem
cell properties in head and neck squamous cell carcinoma. Proc Natl
Acad Sci USA. 104:973–978. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Clay MR, Tabor M, Owen JH, et al:
Single-marker identification of head and neck squamous cell
carcinoma cancer stem cells with aldehyde dehydrogenase. Head Neck.
32:1195–1201. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Marcato P, Dean CA, Giacomantonio CA and
Lee PW: Aldehyde dehydrogenase: its role as a cancer stem cell
marker comes down to the specific isoform. Cell Cycle.
10:1378–1384. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Deng S, Yang X, Lassus H, et al: Distinct
expression levels and patterns of stem cell marker, aldehyde
dehydrogenase isoform 1 (ALDH1), in human epithelial cancers. PLoS
One. 5:e102772010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim MP, Fleming JB, Wang H, et al: ALDH
activity selectively defines an enhanced tumor-initiating cell
population relative to CD133 expression in human pancreatic
adenocarcinoma. PLoS One. 6:e206362011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Landen CN Jr, Goodman B, Katre AA, et al:
Targeting aldehyde dehydrogenase cancer stem cells in ovarian
cancer. Mol Cancer Ther. 9:3186–3199. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Brenner JC, Graham MP, Kumar B, et al:
Genotyping of 73 UM-SCC head and neck squamous cell carcinoma cell
lines. Head Neck. 32:417–426. 2010.PubMed/NCBI
|
|
26
|
Loh YH, Wu Q, Chew JL, et al: The Oct4 and
Nanog transcription network regulates pluripotency in mouse
embryonic stem cells. Nat Genet. 38:431–440. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhao W, Ji X, Zhang F, Li L and Ma L:
Embryonic stem cell markers. Molecules. 17:6196–6236. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Clevers H: The cancer stem cell: premises,
promises and challenges. Nat Med. 17:313–319. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ponti D, Costa A, Zaffaroni N, et al:
Isolation and in vitro propagation of tumorigenic breast cancer
cells with stem/progenitor cell properties. Cancer Res.
65:5506–5511. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang S, Balch C, Chan MW, et al:
Identification and characterization of ovarian cancer-initiating
cells from primary human tumors. Cancer Res. 68:4311–4320. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Charafe-Jauffret E, Ginestier C, Iovino F,
et al: Aldehyde dehydrogenase 1-positive cancer stem cells mediate
metastasis and poor clinical outcome in inflammatory breast cancer.
Clin Cancer Res. 16:45–55. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jiang F, Qiu Q, Khanna A, et al: Aldehyde
dehydrogenase 1 is a tumor stem cell-associated marker in lung
cancer. Mol Cancer Res. 7:330–338. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li T, Su Y, Mei Y, et al: ALDH1A1 is a
marker for malignant prostate stem cells and predictor of prostate
cancer patients’ outcome. Lab Invest. 90:234–244. 2010.PubMed/NCBI
|
|
34
|
Ha PK and Califano JA: The molecular
biology of mucosal field cancerization of the head and neck. Crit
Rev Oral Biol Med. 14:363–369. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ha PK, Chang SS, Glazer CA, Califano JA
and Sidransky D: Molecular techniques and genetic alterations in
head and neck cancer. Oral Oncol. 45:335–339. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Blanpain C, Mohrin M, Sotiropoulou PA and
Passegué E: DNA-damage response in tissue-specific and cancer stem
cells. Cell Stem Cell. 8:16–29. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Rangwala F, Omenetti A and Diehl AM:
Cancer stem cells: repair gone awry? J Oncol. 2011:4653432011.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Taniguchi T and D’Andrea AD: Molecular
pathogenesis of Fanconi anemia: recent progress. Blood.
107:4223–4233. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Diehn M, Cho RW and Clarke MF: Therapeutic
implications of the cancer stem cell hypothesis. Semin Radiat
Oncol. 19:78–86. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ghiaur G, Gerber J and Jones RJ: Concise
review: cancer stem cells and minimal residual disease. Stem Cells.
30:89–93. 2012. View Article : Google Scholar : PubMed/NCBI
|