Introduction
Hepatocellular carcinoma (HCC) is the sixth most
common cancer and the third most common cause of cancer-related
mortality worldwide. HCC is also the leading cause of death among
cirrhotic patients (1,2). Most HCC cases occur in sub-Saharan
Africa and Eastern Asia; however, the incidence rate has been
increasing in some developed countries, including Japan, the United
Kingdom, France, and the United States (3,4). All
patients diagnosed with advanced stages of the disease exhibit very
limited survival, and to date, only treatment with the multikinase
inhibitor sorafenib has improved survival rates for these patients
(5,6).
In contrast to most other solid tumors, the
underlying cirrhotic liver disease found in HCC patients greatly
complicates the tumor-related prognosis, which presents a unique
situation in which accurate prognostic prediction is a relevant and
unmet need (2,7,8).
Chronic inflammation induced by chronic viral hepatitis, alcohol
consumption or aflatoxin and subsequent hepatocyte regeneration are
underlying causes of HCC (9).
Continuous inflammation occasionally damages DNA in the hepatocytes
of the regenerating liver and thereby increases the chance of
developing a gene alteration that may lead to carcinogenesis. The
molecular profiling of HCC has led to a better understanding of the
physiopathology of this neoplasm and has allowed the development of
novel therapeutic approaches (e.g., molecular targeted therapies)
for tumors previously considered to be therapy-refractory (10,11).
Integrative analyses of genetic and epigenetic information obtained
for the tumor and the surrounding tissue should be used to identify
novel biomarkers and therapeutic targets in HCC to improve existing
treatment algorithms and eventually design a more personalized
therapy for this devastating disease (12,13).
We have identified several HCC-related genes by
expression and epigenetic analyses (14,15).
From the exhaustive expression analysis obtained via our microarray
data, the B-cell translocation gene 1 (BTG1) was identified
as a candidate tumor suppressor gene for HCC. Human BTG1,
which is localized to chromosome 12q22, was originally identified
as a translocation partner of the c-Myc gene in a case of
B-cell chronic lymphocytic leukemia and belongs to a family of
anti-proliferative genes (16–18).
BTG1 is constitutively expressed in quiescent cells, and its
expression is downregulated as cells enter the growth cycle
(19,20). Experiments in which gene expression
was induced showed that BTG1 is a Bcl-2-regulated mediator
of apoptosis and that it negatively regulates cell proliferation in
breast and ovarian cancer (20,21).
However, the role of BTG1 in gastroenterological
malignancies including HCC remains unclear.
The aims of this study were to evaluate the clinical
significance of BTG1 expression, examine the regulatory
factors involved in BTG1 transcription, clarify the roles of
BTG1 in hepatocarcinogenesis and its subsequent progression,
and propose a potential diagnostic and therapeutic molecular target
for HCC.
Materials and methods
Ethics
This study conformed to the ethical guidelines of
the World Medical Association Declaration of Helsinki - Ethical
Principles for Medical Research Involving Human Subjects and has
been approved by the Institutional Review Board of Nagoya
University, Aichi, Japan (no. 2013–0295). Written informed consent
for usage of clinical samples and data, as required by the
Institutional Review Board, was obtained from all patients.
Sample collection
Nine HCC cell lines (Hep3B, HepG2, HLE, HLF, HuH1,
HuH2, HuH7, PLC/PRF/5 and SK-Hep1) were obtained from the American
Type Culture Collection (Manassas, VA, USA). Primary HCC tissues
and corresponding non-cancerous tissues were collected from 151
consecutive patients undergoing liver resection for HCC at Nagoya
University Hospital between January, 1998 and January, 2012.
Treatment after recurrence generally included the following
options: surgery, radiofrequency ablation, transcatheter arterial
chemoembolization, and chemotherapy according to tumor status and
liver function.
Tissue samples were collected, immediately flash
frozen in liquid nitrogen and stored at −80°C until RNA extraction
(28 days on average) was performed. Tumor samples ~5 mm2
in size that did not contain a necrotic component and were
confirmed to contain >80% tumor cells by definition were used
for RNA extraction. Corresponding non-cancerous liver tissue
samples were collected >2 cm away from the edge of the tumor,
were obtained from the same patient and did not contain any
regenerative or dysplastic nodules.
Microarray procedure
Sample collection, RNA extraction, and Affymetrix
HG-U133A and HG-U133B GeneChip (Affymetrix, Inc., Santa Clara, CA,
USA) gene expression arrays were performed as previously described
(22–24).
Quantitative real-time reverse
transcription polymerase chain reaction (RT-qPCR)
The BTG1 mRNA expression levels were analyzed
by RT-qPCR. Total RNA (10 μg per sample) was isolated and used to
generate complementary DNA. The primer sequences used in this study
are listed in Table I. RT-qPCR was
performed on nine HCC cell lines and 151 pairs of clinical samples
with a SYBR-Green PCR Core Reagents kit (Perkin-Elmer/Applied
Biosystems, Inc., Foster City, CA, USA) and included no-template
samples as a negative control. Real-time detection of the
SYBR-Green emission intensity was conducted with an ABI StepOnePlus
Real-Time PCR system (Perkin-Elmer/Applied Biosystems, Inc.). The
expression of glyceraldehyde-3-phosphate dehydrogenase
(GAPDH) mRNA was quantified in each sample for
standardization. For cell lines, biological replicates were tested
in triplicate. Technical replicates were performed in triplicate
for both cell lines and HCC tissues. The expression levels for each
sample are shown as the BTG1 value divided by the
GAPDH value. BTG1 mRNA expression was considered to
be downregulated in tumor tissues when its level was <40% of the
level in the corresponding non-cancerous tissues.
 | Table IPrimers and annealing
temperature. |
Table I
Primers and annealing
temperature.
| Gene | Experiment | Type | Sequence
(5′→3′) | Product size
(bp) | Annealing
temperature (°C) |
|---|
| BTG1 | RT-qPCR | Forward |
CTGCAGACTCAGCAGA | 104 | 60 |
| | Reverse |
CGATACAACGGTAACCCGA | | |
| Exon 1; HRM | Forwarvd |
CATCGCTCGTCTCTTCCTCT | 419 | 54 |
| sequencing | Reverse |
GACTCTGACAGATGTG | | |
| Exon 2; HRM | Forward |
CGATCCTAAGCGTTGTTTCTC | 497 | 56 |
| sequencing | Reverse |
TCATAATCCATCCCCAAGA | | |
| Bisulfite | Forward |
GTGGTATTATAAAGGGTGTTG | 118 | 62 |
| sequencing | Reverse | ACTCACTACTC | | |
| GAPDH | RT-qPCR | Forward |
GAGTGAGTCGGAGTC | 226 | 60 |
| | Probe | CAGCTCGTCTCAGC | | |
| | Reverse |
GAGATGGTGATGGGATTTC | | |
Mutational analysis
The BTG1 gene consists of two exons.
Mutational surveillance of HCC cell lines was performed in exon 1
and 2 of the BTG1 gene by high resolution melting (HRM)
analysis. HRM is known to be a reliable and concise technique for
the detection of genetic alterations (25–27).
Genomic DNA obtained from HCC cell lines was amplified with
specific primer pairs according to the manufacturer’s instructions
(Life Technologies, Carlsbad, CA, USA). All samples were tested in
triplicate. Eight wells of each PCR plate were allocated to
wild-type control DNA, and one well contained a non-template
control to validate the PCR. HRM was conducted using a StepOnePlus
instrument (Life Technologies) with a melting temperature range set
between 60 and 98°C. Scanning data were analyzed by HRM software
v3.0.1 (Life Technologies). The primers used for mutational
analysis are listed in Table
I.
Bisulfite sequencing analysis
The BTG1 gene contains a CpG island near the
promoter region; thus, we hypothesized that aberrant methylation is
responsible for regulating BTG1 transcription in HCC.
Genomic bisulfite-treated DNA from HCC cell lines was sequenced to
ascertain the levels of DNA methylation. The bisulfite treatment
and sequencing procedures were performed as previously reported
(28,29).
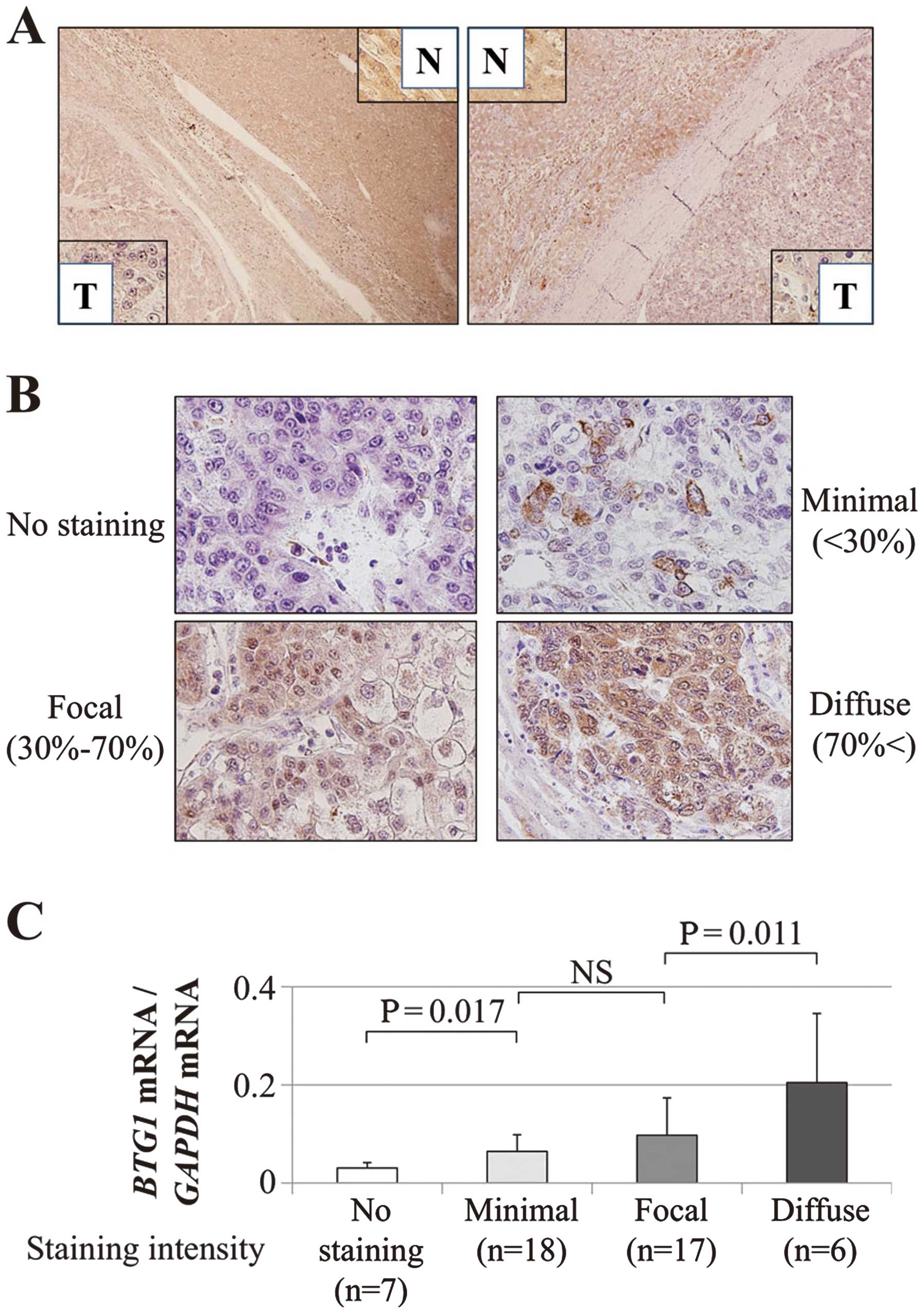
Immunohistochemistry (IHC)
IHC was performed to investigate BTG1 protein
localization in 48 representative sections of well-preserved HCC
tissue as described previously (30). Sections were incubated for 1 h at
room temperature with a rabbit antibody directed against
BTG1 (PA5-25035; Thermo Fisher Scientific, Inc., Rockford,
IL, USA) diluted 1:100 in Antibody Diluent (Dako, Carpinteria, CA,
USA) and then developed for 2 min using liquid
3,3′-diaminobenzidine as the substrate (Nichirei Corp., Tokyo,
Japan). The staining patterns were compared between HCC tissue and
the corresponding non-cancerous tissue. The intensity of
BTG1 protein expression was graded depending on the
percentage of stained cells as follows: no staining, minimal
(<30%); focal (30–70%); and diffuse (>70%) (31). To avoid subjectivity, specimens
were randomized and coded before analysis by two independent
observers blinded to the status of the samples. Each observer
evaluated all the specimens at least twice within a given time
interval to minimize intra-observer variation.
Statistical analysis
The values between the two groups were analyzed
using the Mann-Whitney U test. The χ2 test was used to
analyze the association between the expression status of
BTG1 and clinicopathological parameters. The strength of the
correlation between two variables was assessed by Spearman’s rank
correlation coefficient. Disease-specific and -free survival rates
were calculated using the Kaplan-Meier method, and the difference
in survival curves was analyzed using the log-rank test. We
performed multivariable regression analysis to detect prognostic
factors using the Cox proportional hazards model, and variables
with P<0.05 were entered into the final model. All statistical
analysis was performed using JMP 10 software (SAS Institute, Inc.,
Cary, NC, USA). P<0.05 was considered statistically
significant.
Results
Patient characteristics
The age of the 151 patients ranged from 34–84 years
(median 64 years), and the male-to-female ratio was 126:25.
Thirty-seven patients presented with hepatitis B infections, and 84
patients presented with hepatitis C infections. In terms of the
non-cancerous liver samples, the number of patients with normal
liver, chronic hepatitis, and cirrhosis were 10, 87, and 54,
respectively. When classified according to the 7th edition of the
UICC classification, 84, 39 and 18 patients were in stages I, II
and III, respectively.
Expression array
Gene expression that was reduced further in tumor
tissues than in the corresponding non-cancerous tissues was used to
identify new candidate tumor suppressors in HCC. BTG1
expression was reduced in HCC compared with normal tissue, with a
log2 ratio of −1.6 and −1.5 (Table II).
 | Table IIMicroarray results for BTG1
expression. |
Table II
Microarray results for BTG1
expression.
| Gene | Log2
ratio | Normal signal | Detection | Tumor signal | Detection | Probe ID | Chromosomal
location |
|---|
| BTG1 | −1.6 | 2544.9 | Positive | 757.1 | Positive | HU133p2_10370 | Chr 12q22 |
| −1.5 | 1389 | Positive | 418.8 | Positive | HU133p2_10369 | Chr 12q22 |
BTG1 mRNA expression and regulatory
mechanisms in HCC cell lines
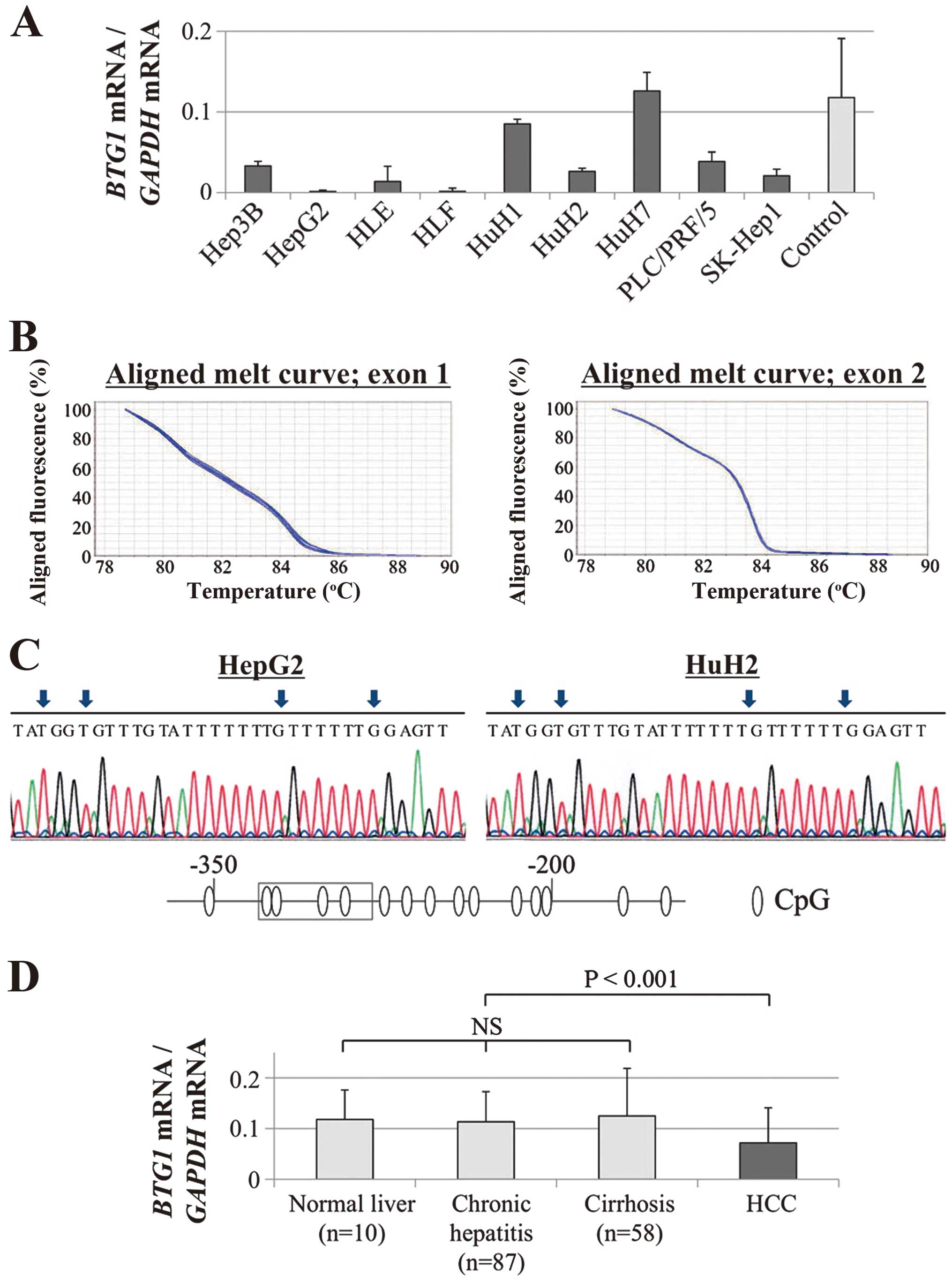
Decreases in BTG1 mRNA were confirmed in
eight (89%) of the nine HCC cell lines compared with the median
expression level in non-cancerous liver tissues; these results
demonstrate the heterogeneity of BTG1 expression in HCC cell
lines (Fig. 1A). No mutations were
detected by the HRM analysis of BTG1 exons 1 and 2 (Fig. 1B). Direct nucleotide sequence
analysis of bisulfite-treated GC cell lines showed absence of
hypermethylation of BTG1 promoter region in all GC cell
lines (Fig. 1C).
Expression status of BTG1 in 151 clinical
HCC samples
The BTG1 mRNA expression levels of
non-cancerous tissue samples were categorized pathologically into
normal liver (n=10), chronic hepatitis (n=87), and cirrhosis
(n=58). Upon evaluation of these samples, no significant
differences were found, suggesting that the expression of
BTG1 mRNA in non-cancerous liver was not affected by
background liver fibrosis (Fig.
1D). In 129 (85%) of 151 patients, the expression level of
BTG1 mRNA was lower in the HCC tissues than in the
corresponding normal tissues. The HCC tissues exhibited
significantly lower expression levels of BTG1 mRNA than the
corresponding normal tissues (P<0.001, Fig. 1D).
Expression patterns of the BTG1 protein were
evaluated by IHC. Representative cases with downregulated
BTG1 mRNA expression in HCC tissues exhibited reduced
expression of the BTG1 protein in the cytoplasm of the
cancerous tissues compared with the adjacent non-cancerous tissues
(Fig. 2A). Overall, the staining
intensity (shown in Fig. 2B) of
the BTG1 protein in 48 patients was consistent with the
RT-qPCR data (Fig. 2C).
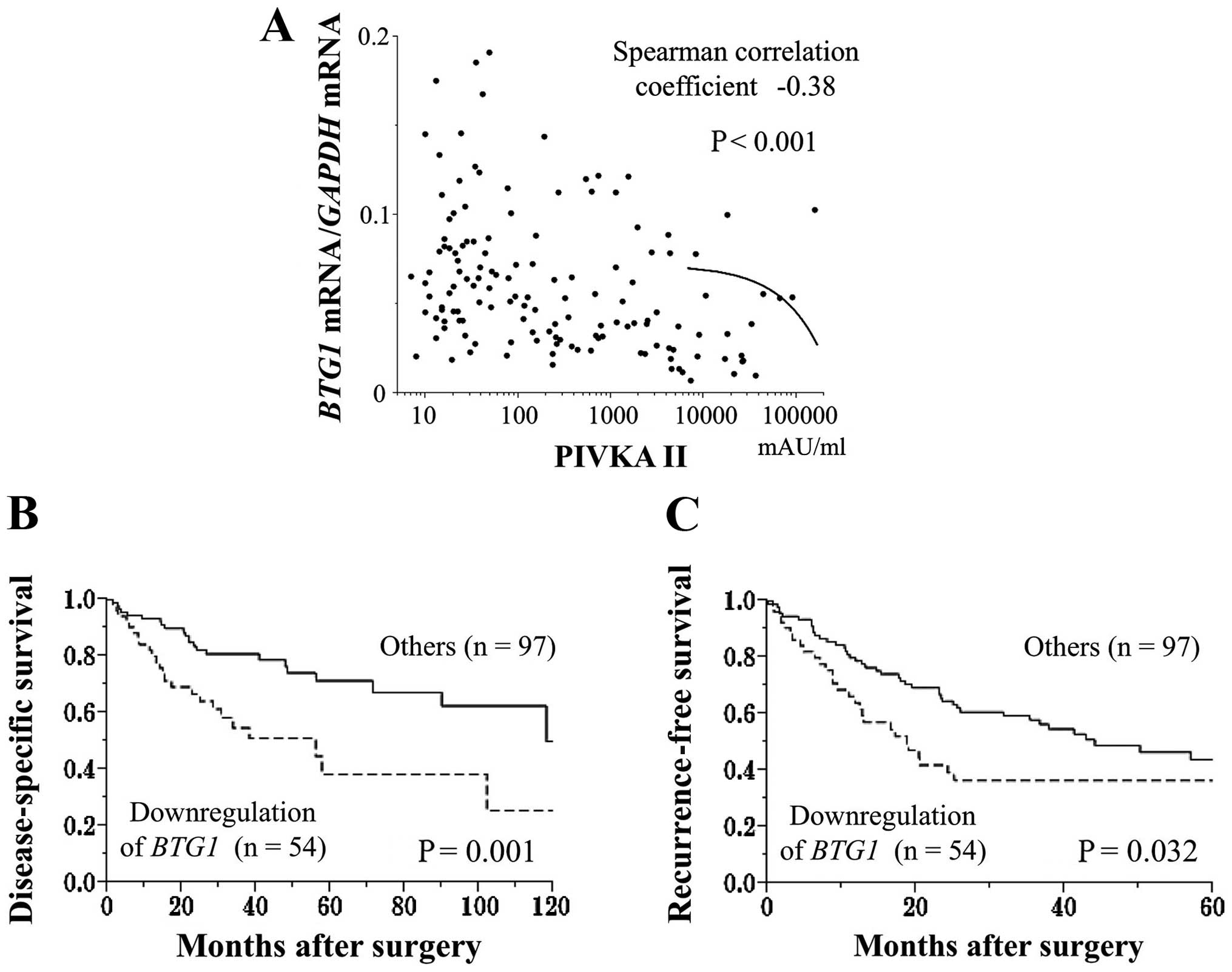
Prognostic values of the expression
status of BTG1
Fifty-four of 151 HCC patients showed substantial
downregulation (<40%) of BTG1 mRNA in HCC tissues
compared with non-cancerous tissues. The downregulation of
BTG1 mRNA in the HCC samples was significantly associated
with male gender, protein induced by vitamin K antagonists (PIVKA)
II >40 mAU/ml, tumor size ≥3 cm, tumor differentiation (poorly
to moderately differentiated), serosal infiltration, vascular
invasion, advanced UICC stage, and extra-hepatic recurrence
(Table III). The BTG1
mRNA expression levels in HCC tissues were inversely correlated
with preoperative PIVKA II levels (Fig. 3A). Patients exhibiting a
downregulation of BTG1 mRNA expression in the HCC samples
had a significantly shorter disease-specific survival rate than the
other patients (2-year survival rates, 67 and 82%, respectively,
Fig. 3B). Multivariate analysis
identified the downregulation of BTG1 mRNA as an independent
prognostic factor for HCC (hazard ratio 2.12, 95% confidence
interval 1.12–4.04, P=0.022, Table
IV). In terms of recurrence-free survival rates, patients with
a substantial downregulation of BTG1 mRNA in the HCC samples
had significantly earlier recurrence rates after surgery than the
other patients (2-year recurrence-free survival rates: 39 and 65%,
respectively, P=0.032, Fig.
3C).
 | Table IIIAssociation between expression status
of BTG1 mRNA and clinicopathological parameters in 151 HCC
patients. |
Table III
Association between expression status
of BTG1 mRNA and clinicopathological parameters in 151 HCC
patients.
| Clinicopathological
parameters | Downregulation of
BTG1 mRNA (n) | Others (n) | P-value |
|---|
| Age |
| <65 year | 19 | 48 | 0.088 |
| ≥65 year | 35 | 49 | |
| Gender |
| Male | 51 | 75 | 0.004a |
| Female | 3 | 22 | |
| Background
liver |
| Normal liver | 4 | 6 | 0.497 |
| Chronic
hepatitis | 34 | 53 | |
| Cirrhosis | 16 | 38 | |
| Pugh-Childs
classification |
| A | 48 | 92 | 0.187 |
| B | 6 | 5 | |
| Hepatitis
virus |
| Absent | 14 | 16 | 0.387 |
| HBV | 12 | 25 | |
| HCV | 28 | 56 | |
| AFP (ng/ml) |
| ≤20 | 24 | 57 | 0.091 |
| >20 | 30 | 40 | |
| PIVKA II
(mAU/ml) |
| ≤40 | 9 | 49 | <0.001a |
| >40 | 45 | 48 | |
| Tumor
multiplicity |
| Solitary | 39 | 78 | 0.253 |
| Multiple | 15 | 19 | |
| Tumor size |
| <3.0 cm | 10 | 37 | 0.010a |
| ≥3.0 cm | 44 | 60 | |
|
Differentiation |
| Well | 7 | 28 | 0.022a |
| Moderate to
poor | 47 | 69 | |
| Growth type |
| Expansive
growth | 45 | 82 | 0.847 |
| Invasive
growth | 9 | 15 | |
| Serosal
infiltration |
| Absent | 34 | 80 | 0.009a |
| Present | 20 | 17 | |
| Formation of
capsule |
| Absent | 14 | 33 | 0.299 |
| Present | 40 | 64 | |
| Infiltration to
capsule |
| Absent | 22 | 46 | 0.428 |
| Present | 32 | 51 | |
| Septum
formation |
| Absent | 14 | 39 | 0.074 |
| Present | 40 | 58 | |
| Vascular
invasion |
| Absent | 35 | 79 | 0.025a |
| Present | 19 | 18 | |
| Margin status |
| Negative | 43 | 80 | 0.668 |
| Positive | 11 | 17 | |
| UIC pathological
stage |
| I | 26 | 68 | 0.014a |
| II | 17 | 22 | |
| III | 11 | 7 | |
| Extra-hepatic
recurrence |
| Absent | 33 | 79 | 0.007a |
| Present | 21 | 18 | |
 | Table IVPrognostic factors in 151 patients
with hepatocellular carcinoma. |
Table IV
Prognostic factors in 151 patients
with hepatocellular carcinoma.
| | Univariate | Multivariable |
|---|
| |
|
|
|---|
| Variable | n | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age (≥65) | 84 | 1.92 | 1.07–3.57 | 0.030a | 1.77 | 0.96–3.38 | 0.069 |
| Gender (male) | 126 | 1.27 | 0.60–3.13 | 0.553 | | | |
| Background liver
(cirrhosis) | 54 | 1.58 | 0.88–2.81 | 0.123 | | | |
| Pugh-Childs
classification (B) | 11 | 0.93 | 0.28–2.32 | 0.889 | | | |
| AFP (>20
ng/ml) | 70 | 1.90 | 1.07–3.42 | 0.029a | 1.55 | 0.81–2.96 | 0.181 |
| PIVKA II (>40
mAU/ml) | 93 | 2.10 | 1.14–4.07 | 0.016a | 1.16 | 0.56–2.51 | 0.695 |
| Tumor multiplicity
(multiple) | 34 | 2.09 | 1.11–3.76 | 0.023a | 1.77 | 0.91–3.32 | 0.092 |
| Tumor size (≥3.0
cm) | 104 | 2.20 | 1.13–4.71 | 0.020a | 1.11 | 0.50–2.62 | 0.810 |
| Tumor
differentiation (well) | 35 | 0.55 | 0.25–1.10 | 0.095 | | | |
| Growth type
(invasive growth) | 24 | 1.44 | 0.69–2.76 | 0.318 | | | |
| Serosal
infiltration | 37 | 2.51 | 1.32–4.61 | 0.006a | 1.12 | 0.55–2.24 | 0.742 |
| Formation of
capsule | 104 | 1.05 | 0.57–2.02 | 0.884 | | | |
| Infiltration to
capsule | 83 | 1.20 | 0.67–2.18 | 0.537 | | | |
| Septum
formation | 98 | 0.87 | 0.49–1.60 | 0.651 | | | |
| Vascular
invasion | 37 | 3.40 | 1.87–6.07 | <0.001a | 2.26 | 1.13–4.55 | 0.022a |
| Margin status
(positive) | 28 | 2.64 | 1.42–4.73 | 0.003a | 2.30 | 1.21–4.23 | 0.012a |
| Downregulation of
BTG1 mRNA | 54 | 2.55 | 1.43–4.57 | 0.002a | 2.12 | 1.12–4.04 | 0.022a |
Discussion
BTG is a nuclear protein that is imported into the
nucleus through a nuclear localization signal; its
nucleocytoplasmic translocation depends on the stage of cell growth
and is mediated by a nuclear export signal (32–34).
Accordingly, the BTG family, which is thought to play an intimate
role in the proliferation of cancer cells, has attracted attention
in recent years (34,35). BTG1 has been shown to
enhance homeobox B9-mediated transcription in transfected cells and
mediate its antiproliferative function. As shown by DNA
fragmentation and nuclear condensation, BTG1 localizes to
specific macrophage-rich regions in human lesions and apoptotic
cells (36). BTG1 mRNA is
abundantly expressed in quiescent endothelial cells and is
decreased upon the addition of angiogenic growth factors (19).
In this study, the expression status and regulatory
mechanisms of BTG1 were investigated in HCC. Following the
confirmation that BTG1 mRNA expression is remarkably
suppressed in most HCC cell lines, the somatic mutation and DNA
methylation statuses were evaluated as possible mechanisms of
suppression. No mutations were detected in any of the HCC cell
lines examined by the HRM. In addition, bisulfite sequencing showed
absence of hypermethylation in the BTG1 promoter in all GC
cell lines. These findings were consistent with those in acute
lymphoblastic leukemia (37), and
further study will be needed to elucidate the alterative underlying
molecular pathway suppressing BTG1 transcription.
Interestingly, the expression analysis of clinical
samples demonstrated important clinical implications for the
expression of BTG1. BTG1 was downregulated in most
HCC tissues, and the strong suppression of BTG1 was an
independent prognostic factor associated with early recurrence.
These results indicate that BTG1 is a putative tumor
suppressor gene that affects both carcinogenesis and the subsequent
progression of HCC. The potential use of BTG1 expression as
a prognostic biomarker is supported by the finding that BTG1
expression in HCC tissues was inversely correlated with the serum
levels of PIVKA II, an important HCC tumor marker.
In clinical practice, aggressive pre- and
post-operative systemic therapy could be considered for patients
exhibiting strong downregulation of BTG1 identified in
biopsies or surgical specimens in anticipation of early recurrence
and an adverse prognosis. BTG1 interacts with and regulates
the activity of protein arginine methyl transferase (PRMT)1
(38,39). Members of this enzyme family,
including PRMT1, are considered global regulators of gene
expression that act as transcriptional coregulators of the arginine
methylation of histone tails and are critical regulators of
transcription (40). Based on our
results, the forced expression or artificial modification of
interacting molecules (including PRMT1) of BTG1 may be used
as novel therapeutic approaches for the treatment of HCC. For
future consideration, external validation is necessary, and
functional analysis of the BTG1 gene could help to further
clarify the role that BTG1 plays in the progression of
HCC.
In summary, the reduced expression of BTG1
mRNA was associated with early recurrence rates and subsequent poor
prognoses in patients with HCC. Our results indicate that altered
BTG1 expression might affect hepatocarcinogenesis and may
represent a novel biomarker for the initiation of carcinogenesis
and the progression of HCC.
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Llovet JM, Burroughs A and Bruix J:
Hepatocellular carcinoma. Lancet. 362:1907–1917. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
El-Serag HB: Hepatocellular carcinoma:
recent trends in the United States. Gastroenterology. 127(Suppl 1):
S27–S34. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Minguez B and Lachenmayer A: Diagnostic
and prognostic molecular markers in hepatocellular carcinoma. Dis
Markers. 31:181–190. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Llovet JM, Ricci S, Mazzaferro V, et al:
Sorafenib in advanced hepatocellular carcinoma. N Engl J Med.
359:378–390. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Miki D, Ochi H, Hayes CN, Aikata H and
Chayama K: Hepatocellular carcinoma: towards personalized medicine.
Cancer Sci. 103:846–850. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Herath NI, Leggett BA and MacDonald GA:
Review of genetic and epigenetic alterations in
hepatocarcinogenesis (Review). J Gastroenterol Hepatol. 21:15–21.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Villanueva A, Newell P, Chiang DY,
Friedman SL and Llovet JM: Genomics and signaling pathways in
hepatocellular carcinoma. Semin Liver Dis. 27:55–76. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shiraha H, Yamamoto K and Namba M: Human
hepatocyte carcinogenesis (Review). Int J Oncol. 42:1133–1138.
2013.PubMed/NCBI
|
|
10
|
Bird A: Perceptions of epigenetics.
Nature. 447:396–398. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Khare S, Zhang Q and Ibdah JA: Epigenetics
of hepatocellular carcinoma: role of microRNA. World J
Gastroenterol. 19:5439–5445. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kanda M, Nomoto S, Okamura Y, et al:
Detection of metallothionein 1G as a methylated tumor suppressor
gene in human hepatocellular carcinoma using a novel method of
double combination array analysis. Int J Oncol. 35:477–483.
2009.PubMed/NCBI
|
|
13
|
Hernandez-Gea V, Toffanin S, Friedman SL
and Llovet JM: Role of the microenvironment in the pathogenesis and
treatment of hepatocellular carcinoma. Gastroenterology.
144:512–527. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kanda M, Nomoto S, Nishikawa Y, et al:
Correlations of the expression of vascular endothelial growth
factor B and its isoforms in hepatocellular carcinoma with
clinico-pathological parameters. J Surg Oncol. 98:190–196. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Takami H, Kanda M, Oya H, et al:
Evaluation of MAGE-D4 expression in hepatocellular carcinoma in
Japanese patients. J Surg Oncol. 108:557–562. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cho JW, Kim JJ, Park SG, et al:
Identification of B-cell translocation gene 1 as a biomarker for
monitoring the remission of acute myeloid leukemia. Proteomics.
4:3456–3463. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lin WJ, Gary JD, Yang MC, Clarke S and
Herschman HR: The mammalian immediate-early TIS21 protein and the
leukemia-associated BTG1 protein interact with a protein-arginine
N-methyltransferase. J Biol Chem. 271:15034–15044. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rouault JP, Rimokh R, Tessa C, et al:
BTG1, a member of a new family of antiproliferative genes. EMBO J.
11:1663–1670. 1992.PubMed/NCBI
|
|
19
|
Prévôt D, Voeltzel T, Birot AM, et al: The
leukemia-associated protein Btg1 and the p53-regulated protein Btg2
interact with the homeoprotein Hoxb9 and enhance its
transcriptional activation. J Biol Chem. 275:147–153. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhao Y, Gou WF, Chen S, Takano Y, Xiu YL
and Zheng HC: BTG1 expression correlates with the pathogenesis and
progression of ovarian carcinomas. Int J Mol Sci. 14:19670–19680.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sheng SH, Zhao CM and Sun GG: BTG1
expression correlates with the pathogenesis and progression of
breast carcinomas. Tumour Biol. 35:3317–3326. 2014. View Article : Google Scholar
|
|
22
|
Kanda M, Nomoto S, Okamura Y, et al:
Promoter hypermethylation of fibulin 1 gene is associated with
tumor progression in hepatocellular carcinoma. Mol Carcinog.
50:571–579. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nomoto S, Kanda M, Okamura Y, et al:
Epidermal growth factor-containing fibulin-like extracellular
matrix protein 1, EFEMP1, a novel tumor-suppressor gene detected in
hepatocellular carcinoma using double combination array analysis.
Ann Surg Oncol. 17:923–932. 2010. View Article : Google Scholar
|
|
24
|
Okamura Y, Nomoto S, Kanda M, et al:
Reduced expression of reelin (RELN) gene is associated with high
recurrence rate of hepatocellular carcinoma. Ann Surg Oncol.
18:572–579. 2011. View Article : Google Scholar
|
|
25
|
Kanda M, Knight S, Topazian M, et al:
Mutant GNAS detected in duodenal collections of secretin-stimulated
pancreatic juice indicates the presence or emergence of pancreatic
cysts. Gut. 62:1024–1033. 2013. View Article : Google Scholar
|
|
26
|
Kanda M, Matthaei H, Wu J, et al: Presence
of somatic mutations in most early-stage pancreatic intraepithelial
neoplasia. Gastroenterology. 142:730–733. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kanda M, Sadakari Y, Borges M, et al:
Mutant TP53 in duodenal samples of pancreatic juice from patients
with pancreatic cancer or high-grade dysplasia. Clin Gastroenterol
Hepatol. 11:719–730. 2013. View Article : Google Scholar :
|
|
28
|
Hibino S, Kanda M, Oya H, et al: Reduced
expression of DENND2D through promoter hypermethylation is an
adverse prognostic factor in squamous cell carcinoma of the
esophagus. Oncol Rep. 31:693–700. 2014.
|
|
29
|
Shimizu D, Kanda M, Nomoto S, et al:
Identification of intragenic methylation in the TUSC1 gene as a
novel prognostic marker of hepatocellular carcinoma. Oncol Rep.
31:1305–1313. 2014.
|
|
30
|
Kanda M, Nomoto S, Oya H, et al:
Downregulation of DENND2D by promoter hypermethylation is
associated with early recurrence of hepatocellular carcinoma. Int J
Oncol. 44:44–52. 2014.
|
|
31
|
Kanda M, Shimizu D, Nomoto S, et al:
Prognostic impact of expression and methylation status of DENN/MADD
domain-containing protein 2D in gastric cancer. Gastric Cancer. Apr
3–2014.(Epub ahead of print). View Article : Google Scholar
|
|
32
|
Kawamura-Tsuzuku J, Suzuki T, Yoshida Y
and Yamamoto T: Nuclear localization of Tob is important for
regulation of its antiproliferative activity. Oncogene.
23:6630–6638. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rodier A, Rochard P, Berthet C, et al:
Identification of functional domains involved in BTG1 cell
localization. Oncogene. 20:2691–2703. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Winkler GS: The mammalian
anti-proliferative BTG/Tob protein family. J Cell Physiol.
222:66–72. 2010. View Article : Google Scholar
|
|
35
|
Buitenkamp TD, Pieters R, Zimmermann M, et
al: BTG1 deletions do not predict outcome in Down syndrome acute
lymphoblastic leukemia. Leukemia. 27:251–252. 2013. View Article : Google Scholar
|
|
36
|
Lee H, Cha S, Lee MS, Cho GJ, Choi WS and
Suk K: Role of antiproliferative B cell translocation gene-1 as an
apoptotic sensitizer in activation-induced cell death of brain
microglia. J Immunol. 171:5802–5811. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Waanders E, Scheijen B, van der Meer LT,
et al: The origin and nature of tightly clustered BTG1 deletions in
precursor B-cell acute lymphoblastic leukemia support a model of
multiclonal evolution. PLoS Genet. 8:e10025332012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Berthet C, Guéhenneux F, Revol V, et al:
Interaction of PRMT1 with BTG/TOB proteins in cell signalling:
molecular analysis and functional aspects. Genes Cells. 7:29–39.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
van Galen JC, Kuiper RP, van Emst L, et
al: BTG1 regulates glucocorticoid receptor autoinduction in acute
lymphoblastic leukemia. Blood. 115:4810–4819. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pal S and Sif S: Interplay between
chromatin remodelers and protein arginine methyltransferases. J
Cell Physiol. 213:306–315. 2007. View Article : Google Scholar : PubMed/NCBI
|