Introduction
Lung cancer is the leading cause of cancer-related
mortality both globally and in China. Statistics indicate that
884,500 new lung cancer cases were diagnosed and 778,000
individuals died from lung cancer in developing countries in 2008
alone (1). Specifically, non-small
cell lung cancer (NSCLC) accounts for ~80–85% of lung cancer cases
(2). However, the lack of
effective early detection methods and limited options for combined
therapies both contribute to the dismal statistics associated with
lung cancer. Since the initial identification of the lethal-7
(let-7) gene as a regulator in Caenorhabditis elegans
development, members of the let-7 family have frequently
been deleted in lung cancer, cell lines, mouse NSCLC xenografts and
NSCLC patients (3–6). Subsequent studies have also
identified additional microRNAs (miRNAs) and implicated several
dysregulated miRNAs in lung cancer.
miRNAs are short endogenous non-protein-coding RNAs
that are ~22 nucleotides in length and play important roles in
regulating human gene expression. By base pairing to the 3′
untranslated region (3′ UTR) of the target messenger RNA (mRNA),
miRNAs mediate post-transcriptional gene silencing by inhibiting
mRNA translation or cleaving the target mRNA (7,8).
Various miRNAs are involved in diverse cellular processes,
including cell proliferation, apoptosis, development and
differentiation (6,9). Accumulating evidence shows that
miRNAs function as oncogenes or tumor suppressors and can be
grossly dysregulated in various cancers including lung cancer
(10).
miR-124, a brain-specific miRNA, was first reported
to be abundantly expressed in neuronal cells (11). Recent studies have suggested that
miR-124 is significantly downregulated in several human cancers,
such as gastric, cervical, bladder and breast cancer. These studies
also suggest that this downregulation may be caused by DNA
hypermethylation (12–14). In addition, several studies have
shown that miR-124 exhibits oncogenic activity by directly
suppressing signal transducer and activator of transcription 3
(STAT3) expression (15,16). STAT3, which might be an important
target of miR-124, upregulates the expression of cyclin D1,
survivin, Bcl-xL and vascular endothelial growth factor (VEGF),
which are responsible for suppressing cell apoptosis and inducing
tumor growth (17–20). To date, increasing evidence has
indicated that miR-124 may function as a tumor suppressive factor
by downregulating STAT3 in various human cancers. However, the
relationship between miR-124 function and lung cancer remains
unclear.
In the present study, we explored the role of
miR-124 in lung cancer and searched for a possible relationship
between miR-124 and STAT3 using TargetScan software. The findings
were confirmed using a luciferase assay. In addition, we
retrospectively examined the expression of miR-124 and its target
gene STAT3 in postoperative NSCLC samples and adjacent normal lung
tissues. Furthermore, we investigated the relationship between
miR-124 and clinicopathological features and survival in NSCLC
patients, the results of which suggested that miR-124 might be a
clinically useful biomarker for the prognosis of NSCLC.
Materials and methods
Patients and tissue specimens
All of the patients included in the study underwent
surgical resection procedures at the Thoracic Surgery Department of
the Affiliated Hospital of Qingdao University, Qingdao, China. One
hundred and sixty-four eligible patients diagnosed with a primary
tumor in stages I, II or III were enrolled into the present study
between February 2009 and May 2013. None of the patients had
received any treatment for lung cancer prior to surgery, and all
patients had a preoperative computed tomography scan. Patients with
diffuse lobar or multifocal bronchioalveolar carcinoma and patients
who had malignancy within the previous 5 years were ineligible. The
NSCLC stage was based on the American Joint Committee on Cancer
(AJCC) TNM System. Tumors were graded as well, moderately or poorly
differentiated. After the surgical specimens were obtained,
individual-matched normal lung tissues adjacent to the proximal
excision margin were harvested. A total of 164 pairs of specimens
were used to generate paraffin-embedded tissue microarrays. The
remaining specimens were immediately snap-frozen in liquid nitrogen
and stored at −80°C until further analysis.
Cell culture
The A549 human lung cancer cell line, NCL-H460 cells
and BEAS-2E normal lung epithelial cells were obtained from the
Cell Resource Center of Shanghai Institutes for Biological
Sciences. The cell lines were routinely cultured in DMEM-1640
medium (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal
bovine serum (FBS; Gibco), 100 U/ml penicillin-G, and 100 μg/ml
streptomycin, and were cultivated in a humidified incubator with 5%
CO2 at 37°C.
Cell transfection
The miR-124 mimics (miR-124) and negative control
mimics (miR-NC) were all purchased from Beyotime Institute of
Biotechnology (Co., Ltd., Shanghai, China). The cells were seeded
in 12-well plates and grown to 40% confluence before transfection.
The oligonucleotides were transfected at a final concentration of
50 nM using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) in
accordance with the manufacturer’s instructions. The cells were
divided into three groups according to the treatment and were named
as follows: i) miR-124 (transfected with miR-124-mimics); ii)
miR-NC (transfected with negative control mimics); and iii)
untransfected control. RNA or protein was isolated 48 h after
transfection and used for qRT-PCR or western blotting,
respectively. The transfections were repeated in triplicate a
minimum of 3 times.
Cell viability analysis
Cell proliferation was determined using methyl
thiazolyl tetrazolium (MTT) assays. Briefly, A549 cells
(5×103 cells/well) were plated in 96-well plates with
DMEM containing 10% FBS. After 24 h of culturing, the A549 cells
were transfected with miR-124 or miR-NC mimics as described above.
The MTT assay was completed 0, 12, 24, 48 and 72 h after gene
transfection. The absorbance in each well was measured with a
microplate reader set at 450 and 630 nM. All experiments were
performed in triplicate and repeated three times. For the apoptosis
analysis, cultured cells were harvested by trypsinization and
washed twice with ice-cold PBS. Forty-eight hours after
transfection, the cells (5×105 cells/ml) from each
sample were stained with Annexin V-FITC and PI using the protocol
included in the Apoptosis Detection kit I (BD Biosciences, San
Jose, CA, USA). A fluorescence-activated cell sorting FACSCalibur
instrument (BD Biosciences) was used for this assay, and the data
generated were subsequently analyzed with CellQuest software (BD
Biosciences). The experiments were performed in triplicate and
repeated three times.
Luciferase assay
The 3′ UTR of STAT3 containing wild-type or mutant
miR-124 target sites was amplified by PCR and cloned into the
XhoI/NotI site of the pmiR-RB-REPORT vector. The
XhoI/NotI site was downstream of the Renilla
reporter gene. The mutant constructs were created by mutating the
seed regions of the miR-124 sites (5′-GUGCCU-3′). The primers for
the wild-type STAT3 3′ UTR were
5′-CCCTCGAGGGAGCTGAGAACGGAAGCTGCA-3′ (forward) and
5′-ATTTGCGGCCGCAGCTGTTCTGCCTCACCTGTGGG-3′ (reverse), while the
mutagenesis primers were 5′-CCCTGATATCACATCCACAGAAACAACCT-3′
(reverse) and 5′-AGGTTGTTTCTGTGGATGTGATATCAGGG-3′ (forward). All of
the constructs were confirmed using restriction digests and DNA
sequencing. For the luciferase reporter assay, the A549 cell line
was co-transfected with luciferase constructs containing the 3′ UTR
of STAT3 (with wild-type or mutant miR-124 binding sites) and
miR-124 or miR-NC mimics. All of the transfections were conducted
in triplicate with Lipofectamine 2000. Luciferase activity was
measured 48 h after transfection using a dual luciferase reporter
system (Promega, Madison, WI, USA) according to the manufacturer’s
protocol and normalized to the corresponding Renilla
luciferase activity.
Quantitative real-time reverse
transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted from fresh tissues or cells
using TRIzol reagent (Gibco). The concentration and purity of the
RNA was determined using a NanoDrop® ND-1000
spectrophotometer (Thermo Fisher Scientific, Inc., Wilmington, DE,
USA). miR-124 expression was determined using a TaqMan microRNA
assay kit (Applied Biosystems, Foster City, CA, USA). To determine
the expression of STAT3 and glyceraldehyde-3-phosphate
dehydrogenase (GAPDH), 1 μg of total RNA was subjected to
first-strand cDNA synthesis (Takara, Shiga, Japan) and examined
using a PrimeScript RT reagent kit (Takara). Real-time PCR was
performed using SYBR-Green PCR Master Mix (Takara) on the ABI
7500HT System. All of the primers used in the present study were
designed by Sangon Biotech Co., Ltd. (Shanghai, China). The
expression of U6 and GAPDH was used for normalization. The relative
expression of mRNA was determined using the 2−ΔΔCt
method (21). All of the real-time
PCR reactions were carried out in triplicate. A list of primers
used in the reactions is presented in Table I.
 | Table IA list of PCR primers used in the
reactions. |
Table I
A list of PCR primers used in the
reactions.
| Gene | Primer
sequences |
|---|
| miR-124 | F:
5′-GGACTTTCTTCATTCACACCG-3′
R: 5′-GACCACTGAGGTTAGAGCCA-3′ |
| U6 | F:
5′-CTCGCTTCGGCAGCACATATACT-3′
R: 5′-ACGCTTCACGAATTTGCGTGTC-3′ |
| STAT3 | F:
5′-GAAGAATCCAACAACGGC-3′
R: 5′-TCACAATCAGGGAAGCAT-3′ |
| GAPDH | F:
5′-CAACGACCACTTTGTCAAGCTCA-3′
R: 5′-GCTGGTGGTCCAGGGGTCTTACT-3′ |
Western blotting
Tissues were lysed using RIPA buffer (Sigma, St.
Louis, MO, USA). The soluble protein concentration was determined
by the Bradford method. Total protein (10 μg) was run on a 10–15%
SDS-PAGE gel and transferred to polyvinylidene difluoride (PVDF)
membranes. The membranes were blocked with 1% Tween 20-PBS at 4°C
overnight before being washed and incubated with primary antibodies
against STAT3 and GAPDH (Bioworld Co., Dublin, OH, USA; diluted
1:500 in TBS-A) for 1 h at room temperature. After washing, the
membranes were incubated with the secondary antibodies (Abcam Co.,
Cambridge, UK; diluted 1:1,000 in TBS-A) for 1 h at room
temperature. The immunoblots were developed using an
electrochemiluminescence kit and imaged with the Vilber Fusion FX5
automatic gel imaging analysis system (Vilber, Marne La Vallée,
France).
Immunohistochemistry (IHC)
Immunohistochemistry was performed using standard
techniques. Briefly, 4-μm thick paraffin-embedded specimens were
dewaxed in xylene and rehydrated in graded alcohols. Endogenous
peroxidase activity was blocked using 3% hydrogen peroxide. Antigen
retrieval was accomplished in citrate buffer (pH 6.0) using a
microwave. A polyclonal rabbit anti-human STAT3 antibody (1:200
dilution; Santa Cruz Biotechnology, Santa Cruz, CA, USA) was added
and the samples were incubated overnight at 4°C. After washing, the
sections were incubated with a secondary antibody for 40 min at
37°C and subsequently incubated with streptavidin-conjugated
horseradish peroxidase (HRP) and diaminobenzidine (DAB) and
counterstained with hematoxylin. The primary antibody was replaced
by an isotope IgG in the negative control. Representative viable
tissue sections were scored semi-quantitatively using light
microscopy. The proportion of positively stained cells was graded
as follows: score 0, completely negative samples; score 1, samples
with up to 10% positive cells; score 2, samples with 11–50%
positive cells; and score 3, samples with 50% positive cells. The
staining intensity in tumor cells was scored as 0, no staining, 1,
weak staining, 2, moderate staining or 3, strong staining. The
staining index was calculated as the product of the staining
intensity score and the proportion score, which had a value of 0,
1, 2, 3, 4, 6 or 9. The final score was achieved by comparing the
scores obtained by different observers, and any discrepancies were
resolved by consensus. In the case of a disagreement, the slides
were reexamined and the observers reached a consensus. A staining
index ≥4 was defined as high expression, and an index <4 was
defined as low expression.
Follow-up
Follow-up was carried out by telephone or written
correspondence, while clinical examinations and chest radiography
were performed every 3 months for 3 years and every 6 months
thereafter. Disease-free survival (DFS) was evaluated for the
period from the date of operation to the appearance of tumor
progression. Overall survival (OS) was evaluated for the period
from the date of surgery to the date of death (or last follow-up).
The study concluded on December 31, 2013. The overall follow-up
rate was 100%, and the median follow-up time was 31 months with a
range from 2 months to 58 months.
Ethics statement
All of the specimens were obtained from the Central
Laboratory of the Affiliated Hospital of Qingdao University, and
all of the patients provided written informed consent for the use
of their tissues. The present study was approved by the Ethics
Committee of the Affiliated Hospital of Qingdao University.
Statistical analysis
All of the analyses were performed using SPSS 19.0
software (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism 5.0
(GraphPad Software, San Diego, CA, USA). All of the numerical data
were expressed as the mean ± SD. P-values <0.05 were defined as
statistically significant. The results were analyzed using the
Student’s t-test or one-way analysis of variance (ANOVA). The
Chi-square test was used to analyze the relationship between
miR-124 or STAT3 expression and clinicopathological symptoms.
Bivariate correlations between the study variables were calculated
using Spearman’s rank-order correlation coefficients. The survival
curves were analyzed using Kaplan-Meier methodology, and the
difference in distribution was evaluated with the log-rank test.
The influence of each variable on survival was examined by
univariate and multivariate Cox regression analysis.
Results
Patient characteristics
A total of 164 patients were enrolled in the present
study (Table II). The patients
consisted of 115 men and 49 women with a median age of 59 years
(range, 22–76 years). The NSCLC tumors consisted of 106 (64.6%)
adenocarcinomas (AC), 52 (31.7%) squamous cell carcinomas (SCC) and
6 (3.7%) carcinomas of other cell types. The post-surgical
pathologic stage was determined by the TNM classification. The
number of patients with disease stages I, II and III was 47
(28.6%), 29 (17.7%) and 88 (53.7%), respectively.
 | Table IIClinicopathological features of
patients and miR-124 and STAT3 expression levels in NSCLC. |
Table II
Clinicopathological features of
patients and miR-124 and STAT3 expression levels in NSCLC.
| | miR-124 expression
level | | STAT3 expression
level | |
|---|
| |
| |
| |
|---|
|
Characteristics | Cases 164 | Low expression
n=116 (%) | High expression
n=48 (%) | P-value | Low expression n=57
(%) | High expression
n=107 (%) | P-value |
|---|
| Gender | | | | 0.105 | | | 0.290 |
| Male | 115 | 85 (81.34) | 30 (33.66) | | 38 (39.97) | 77 (75.03) | |
| Female | 49 | 31 (34.66) | 18 (14.34) | | 19 (17.03) | 30 (31.97) | |
| Age (years) | | | | 0.378 | | | 0.316 |
| ≥60 | 75 | 50 (53.05) | 25 (21.95) | | 34 (26.07) | 41 (48.93) | |
| <60 | 89 | 66 (62.95) | 23 (26.05) | | 23 (30.93) | 66 (58.07) | |
| Smoking index
(SI) | | | | 0.565 | | | 0.349 |
| ≥400 | 86 | 62 (60.83) | 24 (25.17) | | 28 (29.89) | 58 (56.11) | |
| <400 | 78 | 54 (55.17) | 24 (22.83) | | 29 (27.11) | 49 (50.89) | |
| Tumor site | | | | 0.208 | | | 0.436 |
| Left lobe | 64 | 44 (45.27) | 20 (18.73) | | 25 (22.24) | 39 (41.76) | |
| Right lobe | 100 | 72 (70.73) | 28 (29.27) | | 32 (34.76) | 68 (65.24) | |
| Pleural
invasion | | | | 0.190 | | | 0.815 |
| Absent | 45 | 36 (31.83) | 9 (13.17) | | 15 (15.64) | 30 (29.36) | |
| Present | 119 | 80 (84.17) | 39 (34.83) | | 42 (41.36) | 77 (77.64) | |
| Lymph node
metastasis | | | | 0.017 | | | 0.008 |
| Absent | 61 | 37 (43.15) | 24 (17.85) | | 30 (21.20) | 31 (39.80) | |
| Present | 103 | 79 (72.85) | 24 (30.15) | | 27 (35.80) | 76 (67.20) | |
| TNM stage | | | | 0.006 | | | 0.007 |
| IA | 5 | 2 (3.54) | 3 (1.46) | | 4 (1.74) | 1 (3.26) | |
| IB | 42 | 25 (29.71) | 17 (12.29) | | 19 (14.60) | 23 (27.40) | |
| IIA | 22 | 15 (15.56) | 7 (6.44) | | 7 (7.65) | 15 (14.35) | |
| IIB | 7 | 6 (4.95) | 1 (2.05) | | 4 (2.43) | 3 (4.57) | |
| IIIA | 57 | 43 (40.32) | 14 (16.68) | | 16 (19.81) | 41 (37.19) | |
| IIIB | 31 | 25 (21.93) | 6 (9.07) | | 7 (10.77) | 24 (20.23) | |
|
Differentiation | | | | 0.001 | | | <0.001 |
| Well | 13 | 5 (9.20) | 8 (3.80) | | 9 (4.52) | 4 (8.48) | |
| Moderately | 63 | 38 (44.56) | 25 (18.44) | | 33 (21.90) | 30 (41.10) | |
| Poorly | 86 | 71 (60.83) | 15 (25.17) | | 15 (29.89) | 71 (56.11) | |
| Pathological
type | | | | 0.578 | | | 0.838 |
| AC | 106 | 74 (74.98) | 32 (31.02) | | 37 (36.84) | 70 (69.16) | |
| SCC | 52 | 37 (36.78) | 15 (15.22) | | 18 (18.07) | 34 (33.93) | |
| Other types | 6 | 5 (4.24) | 1 (1.76) | | 2 (2.09) | 4 (3.91) | |
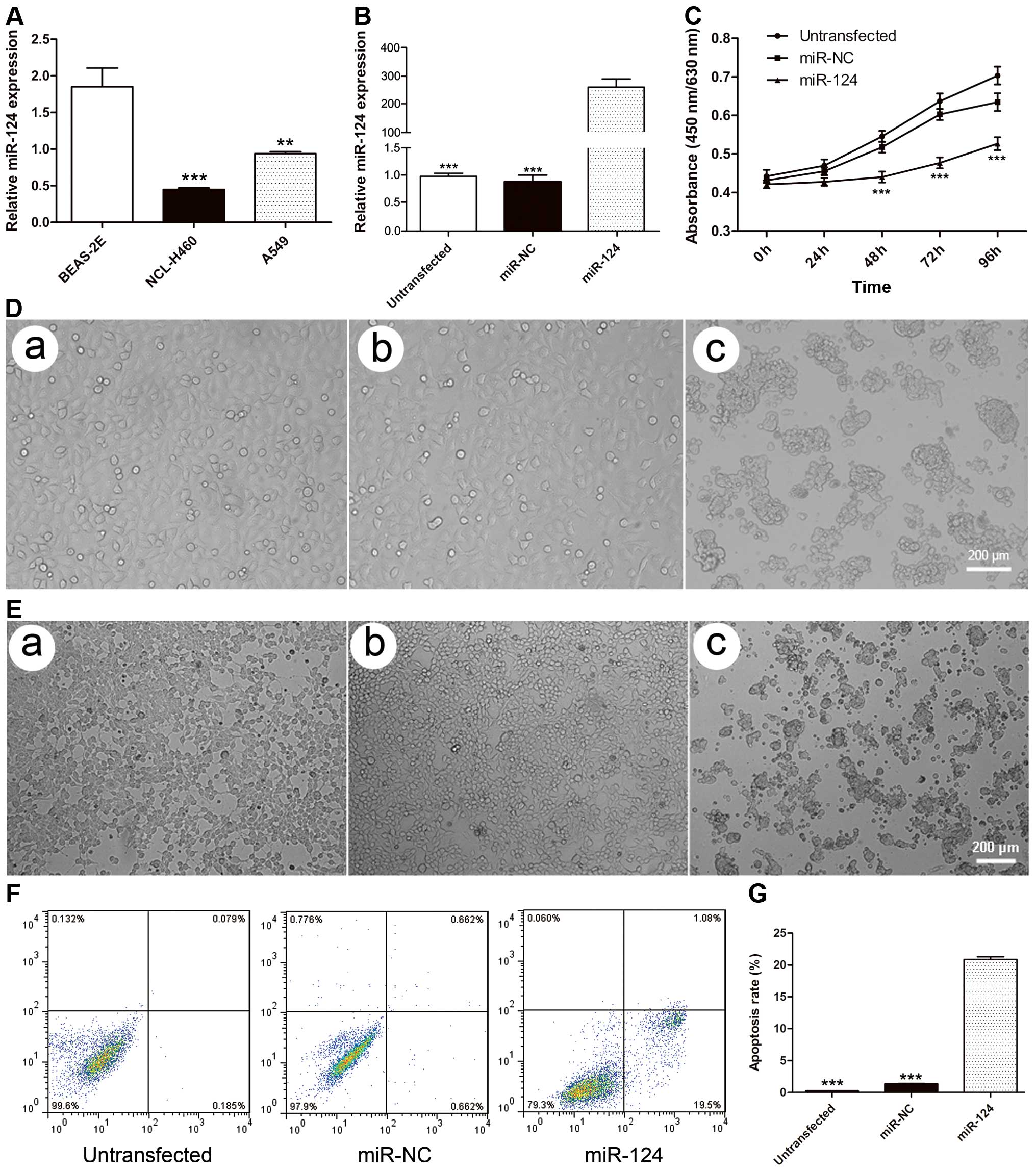
miR-124 is downregulated in human NSCLC
cells
We performed real-time PCR in the A549 and NCL-H460
NSCLC cell lines and the BEAS-2E normal lung epithelial line to
confirm the expression level of miR-124. The results showed that
miR-124 expression in both A549 and NCL-H460 cells was
significantly lower than in normal lung epithelial BEAS-2E cells
(P=0.0015 and P<0.001, respectively) (Fig. 1A), confirming the downregulation of
miR-124 in NSCLC cells.
miR-124 inhibits cell proliferation and
induces apoptosis in A549 cells
To explore the expression and significance of
miR-124 in lung cancer, we transfected miR-124 mimics or negative
control mimics into both A549 and NCL-H460 cells. The expression
level of miR-124 was detected using qRT-PCR 48 h after
transfection. The cells transfected with miR-124 mimics showed a
significant, 300-fold increase in miR-124 expression compared with
miR-NC cells or untransfected cells (P<0.001) (Fig. 1B). We further investigated the
effect of miR-124 on cell survival. The cell lines transfected with
miR-124 mimics were significantly less viable 24, 48 and 72 h
post-transfection compared with miR-NC cells or untransfected cells
(Fig. 1C). The cells
overexpressing miR-124 began to shrink in size and display vague
membranes and an irregular shape 48 h after transfection (Fig. 1D and E). Few early or late
apoptotic cells were detected in untransfected and miR-NC cells
(0.2608±0.019 vs. 1.358±0.301%; P=0.059), whereas the cells
transfected with miR-124 mimics exhibited a significantly higher
percentage of early and late apoptotic cells (20.88±2.84;
P<0.001) (Fig. 1F and G).
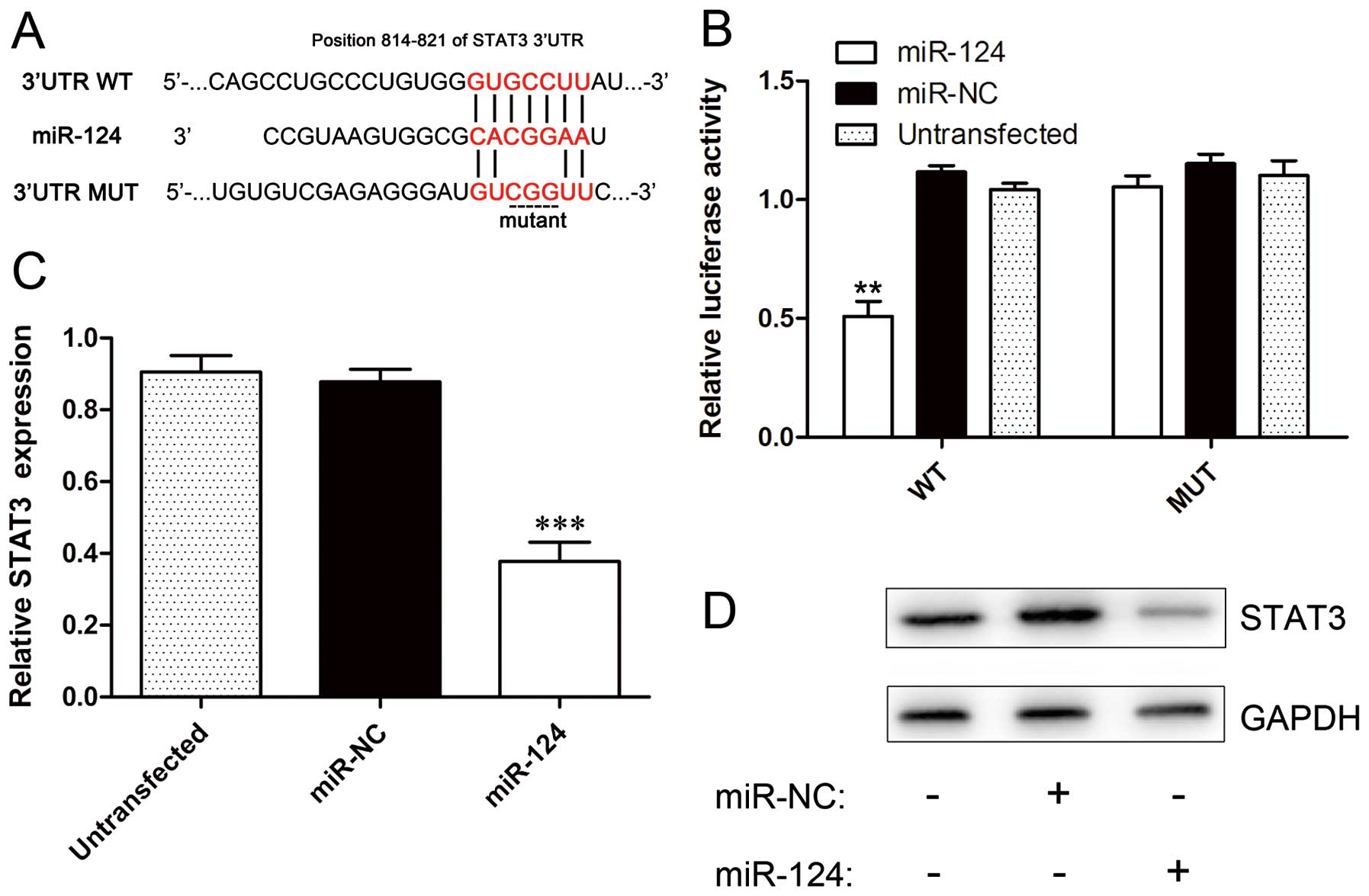
miR-124 directly targets STAT3 in NSCLC
cells
STAT3 has been reported as a potential target of
miR-124 during T cell-mediated immune clearance of glioma (16). However, the relationship between
miR-124 and STAT3 has not been validated in NSCLC. We used
TargetScan target prediction software (http://www.targetscan.org/) to analyze the potential
interaction between miR-124 and STAT3, which is a potential target
of miR-124 and contains putative miR-124 target sites in the 3′ UTR
(Fig. 2A). To confirm that STAT3
is a direct target of miR-124, we cloned the 3′ UTR of STAT3 into
the pGL3-control vector and performed reporter assays. The result
showed that the relative luciferase activity of the wild-type STAT3
3′ UTR reporter construct was notably decreased in A549-miR-124
cells compared to A549-miR-NC (P=0.0024) or untransfected cells
(P=0.0032). However, the relative luciferase activity of the mutant
STAT3 3′ UTR reporter construct was not significantly different
from the luciferase activity of the A549-miR-NC cells and failed to
respond to miR-124 (Fig. 2B). To
further prove that STAT3 is a target of miR-124, we evaluated the
expression levels of STAT3 in the three cell lines 48 h after
transfection using real-time PCR and western blotting. The results
from these assays show that STAT3 was significantly downregulated
in A549-miR-124 cells compared to A549-miR-NC or untransfected
cells (P<0.001) (Fig. 2C and
D).
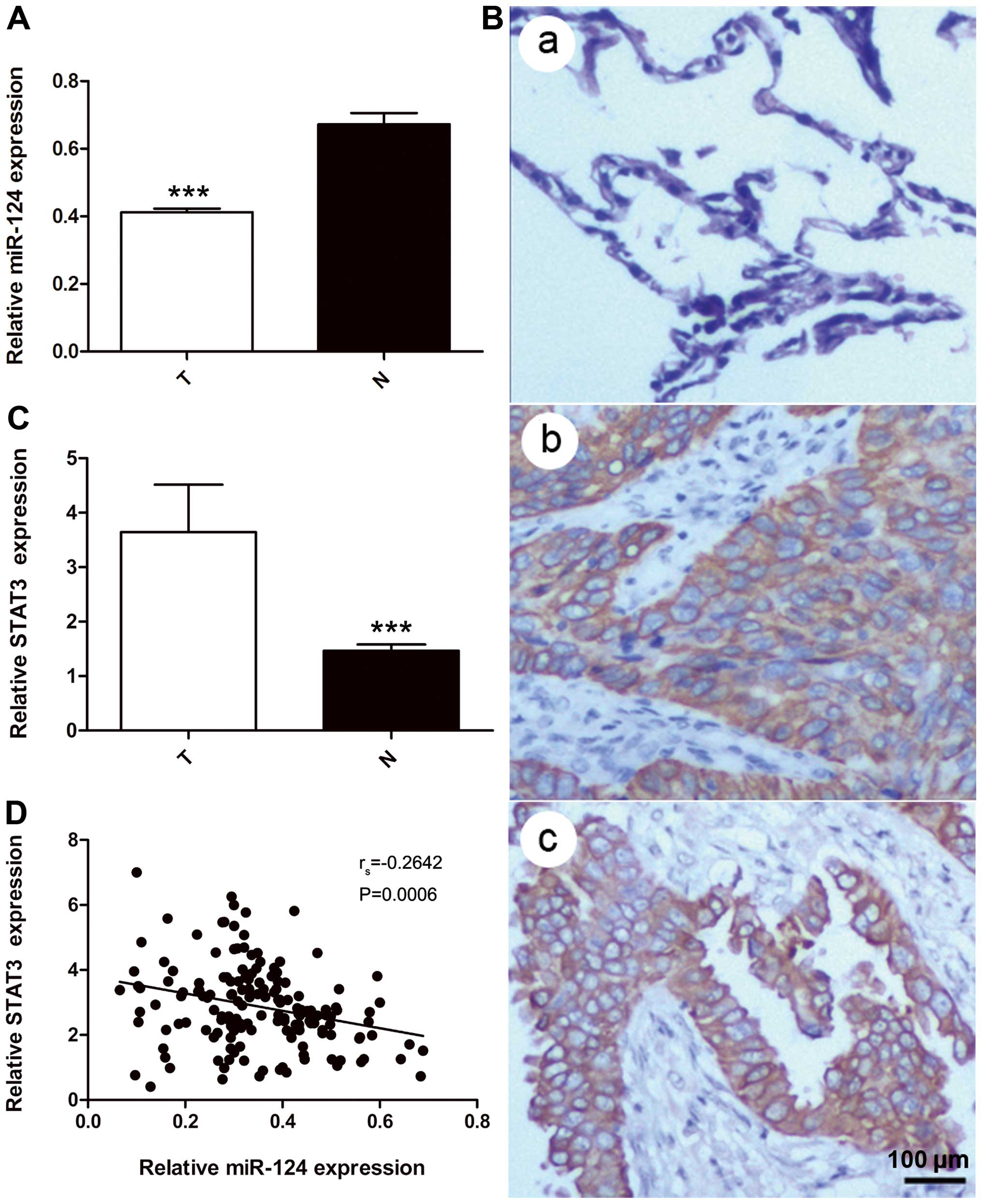
Expression of miR-124 and STAT3 in NSCLC
tissues
miR-124 and STAT3 levels were detected in all 164
pairs of NSCLC tissues and adjacent normal lung tissue by real-time
PCR. U6 snRNA, which are not deregulated in lung cancer, was used
for normalization (22). These
results indicated that the expression of miR-124 was significantly
lower in NSCLC tissues (0.351±0.129) than in adjacent normal lung
tissues (0.661±0.304; P=0.0002) (Fig.
3A). The IHC analysis showed that STAT3 localized to the
cytoplasm of cells (Fig. 3B). The
relative expression of STAT3 was significantly higher in NSCLC
tissues than adjacent normal lung tissues (2.858±1.264 vs.
1.476±0.672; P<0.001) (Fig.
3C). In addition, Spearman’s rank-order correlation analysis
revealed that STAT3 expression in NSCLC tissues was inversely
correlated with the expression of miR-124 (rs=−0.2642;
P=0.0006) (Fig. 3D).
The correlation between miR-124 and STAT3
expression and clinicopathological features of NSCLC
The relationship between miR-124 or STAT3 expression
and clinicopathological NSCLC characteristics is shown in Table II. The miR-124 expression levels
in NSCLC tissues were closely associated with the differentiation
grade (P=0.001), lymph node metastasis (P=0.017) and TNM stage
(P=0.006). However, a statistical analysis revealed no significant
correlations between miR-124 expression and gender, age, SI,
location, pleura invasion or pathological type. As shown in
Table II, STAT3 expression
correlated significantly with tumor differentiation, lymph node
metastasis and TNM stage (P<0.001, P=0.007 and 0.008,
respectively). In contrast, STAT3 expression did not correlate with
gender, age, SI, location, pleural invasion or pathological
type.
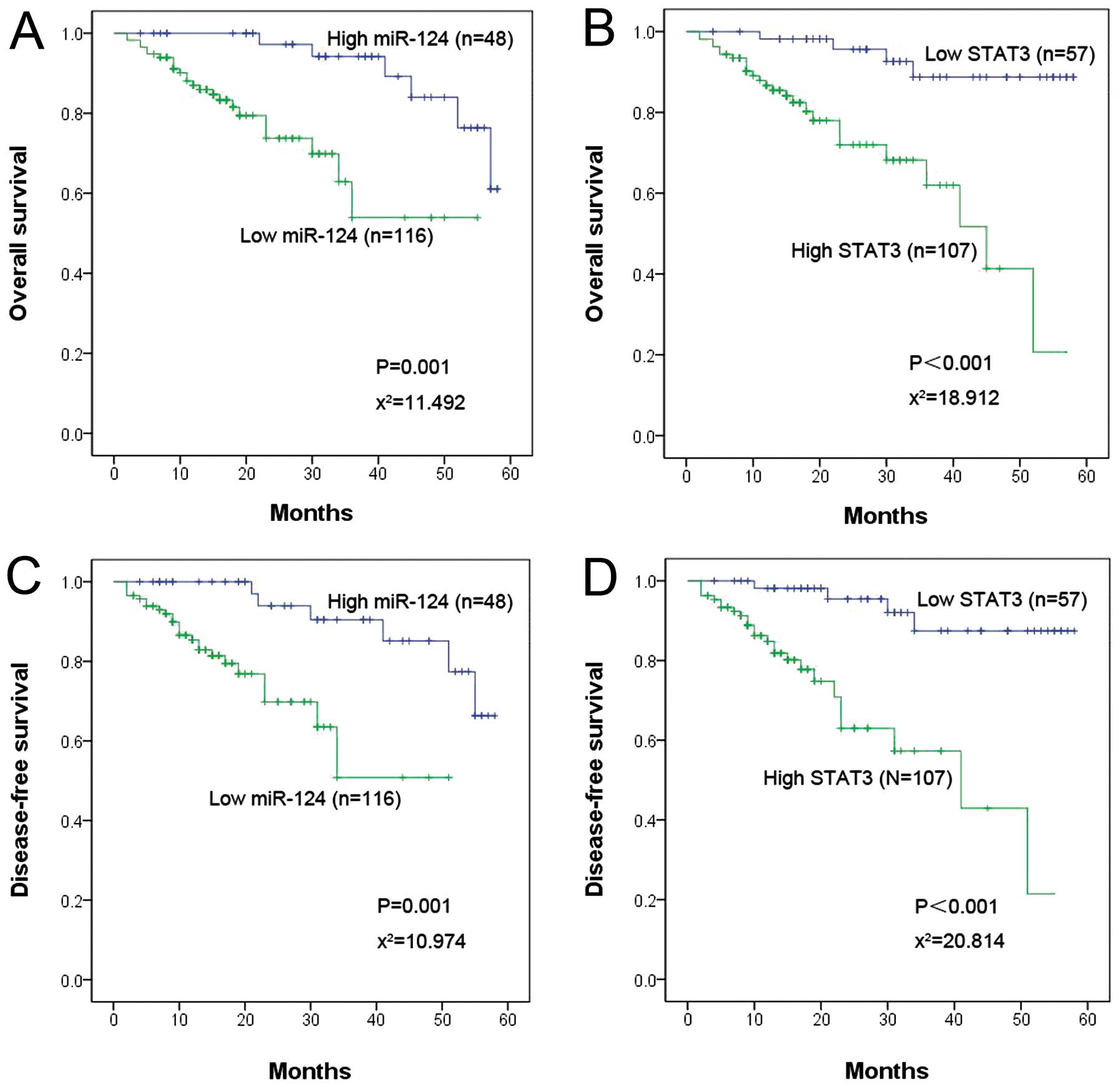
The expression of miR-124 and STAT3 and
NSCLC patient prognosis
Thirty of the patients in the study died of NSCLC
cancer during the follow-up period. The 1-year overall survival and
DFS rates in the miR-124 upregulated group were 87.5 and 81.2%,
respectively, but these rates were only 68.1 and 57.8% in the
miR-124 downregulated group. A Kaplan-Meier survival analysis was
performed to further analyze the association between miR-124 and
STAT3 expression and patient prognosis. The results of this
analysis suggested that patients with low miR-124 or high STAT3
expression had poor OS and DFS (Fig.
4). Previously it was suggested that patients with positive
lymph nodes are more prone to recurrence and metastasis compared
with negative lymph node patients (23). Therefore, an additional analysis of
the relationship between miR-124 expression and patient survival
was performed in positive lymph node and negative lymph node
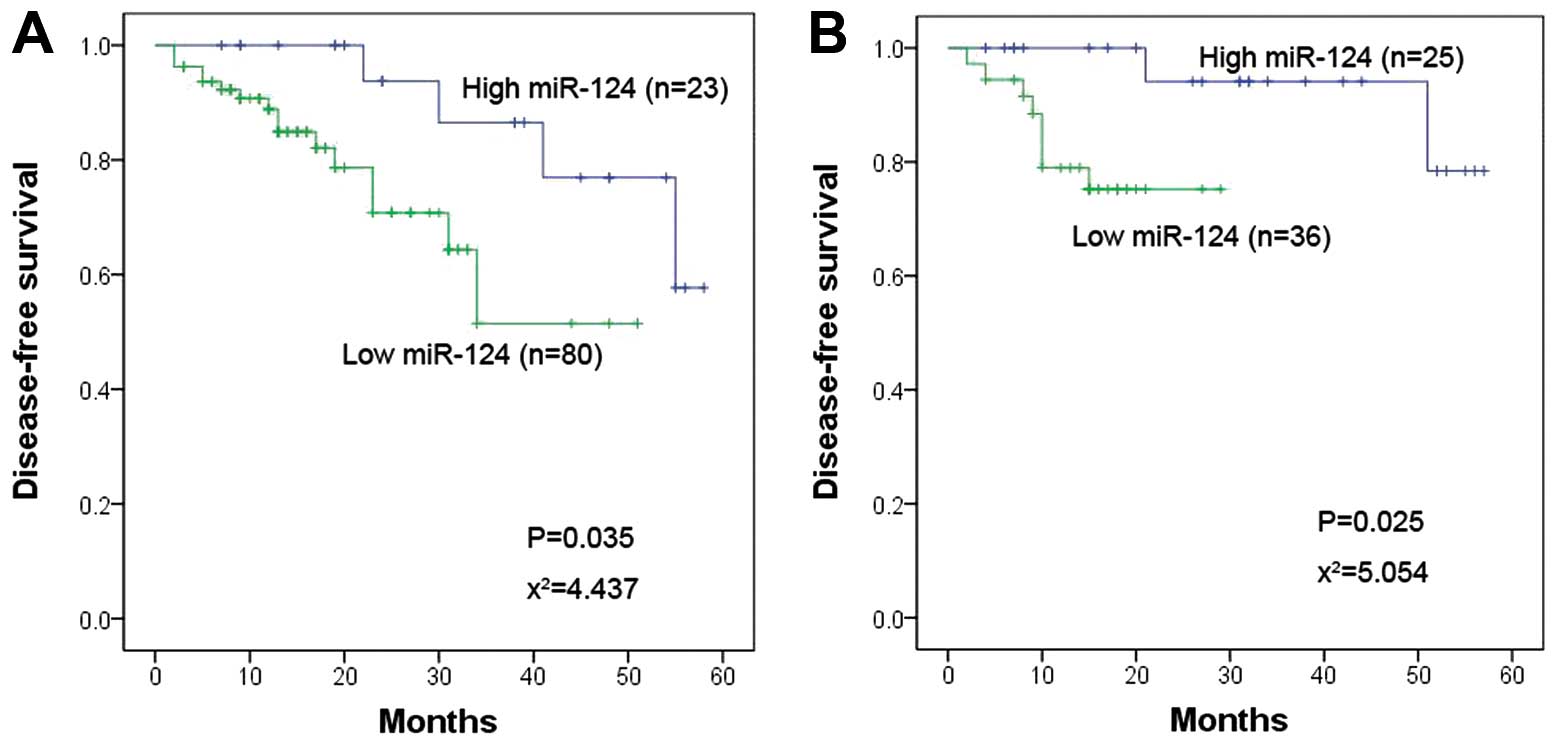
patients. The results suggested that miR-124 expression was
significantly associated with DFS in both positive and negative
lymph node groups (P=0.035 vs. 0.025, respectively) (Fig. 5A and B). Furthermore, a Cox
multivariate analysis showed that miR-124 expression (positive
lymph node group, P=0.004; negative lymph node group, P=0.009),
lymph node metastasis (positive lymph node group, P=0.037; negative
lymph node group, P=0.048) and STAT3 expression (positive lymph
node group, P=0.007; negative lymph node group, P=0.004) were
significantly correlated with OS and DFS (Tables III and IV).
 | Table IIICox multivariate regression analysis
of miR-124 expression, STAT3 expression, gender, age, SI, tumor
site, pleural invasion, pathological type, grade of differentiation
and stage in relation to OS in patients with NSCLC. |
Table III
Cox multivariate regression analysis
of miR-124 expression, STAT3 expression, gender, age, SI, tumor
site, pleural invasion, pathological type, grade of differentiation
and stage in relation to OS in patients with NSCLC.
| Variables |
Unfavorable/favorable | β | SE | HR | 95% CI | P-value |
|---|
| Gender | Male/female | 0.779 | 0.584 | 2.180 | 0.694–6.853 | 0.182 |
| Age (years) | ≥60/<60 | −0.325 | 0.407 | 0.723 | 0.325–1.606 | 0.426 |
| SI | ≥400/<400 | −0.265 | 0.539 | 0.767 | 0.267–2.205 | 0.623 |
| Tumor site | Left/right
lobe | 0.393 | 0.445 | 1.481 | 0.619–3.544 | 0.378 |
| Pleural
invasion | Present/absent | −0.463 | 0.590 | 0.502 | 0.158–1.595 | 0.242 |
| Lymph node
metastasis | Present/absent | 1.559 | 0.758 | 4.753 | 1.093–20.677 | 0.037 |
| Pathological
type | AC/SCC/others | −0.469 | 1.142 | 0.625 | 0.067–5.862 | 0.979 |
|
Differentiation |
Poor/moderate/well | 0.097 | 0.836 | 1.102 | 0.214–5.670 | 0.928 |
| Stage |
IIIB/IIIA/IIB/IIA/IB/IA | 1.136 | 0.657 | 3.115 | 0.860–11.285 | 0.161 |
| miR-124
expression |
Downregulated/upregulated | −1.998 | 0.699 | 0.136 | 0.034–0.534 | 0.004 |
| STAT3
expression |
Downregulated/upregulated | −1.737 | 0.639 | 0.176 | 0.050–0.616 | 0.007 |
 | Table IVCox multivariate regression analysis
of miR-124 expression, STAT3 expression, gender, age, SI, tumor
site, pleural invasion, pathological type, grade of differentiation
and stage in relation to DFS in patients with NSCLC. |
Table IV
Cox multivariate regression analysis
of miR-124 expression, STAT3 expression, gender, age, SI, tumor
site, pleural invasion, pathological type, grade of differentiation
and stage in relation to DFS in patients with NSCLC.
| Variables |
Unfavorable/favorable | β | SE | HR | 95% CI | P-value |
|---|
| Gender | Male/female | 0.706 | 0.628 | 2.026 | 0.592–6.936 | 0.261 |
| Age (years) | ≥60/<60 | −0.265 | 0.402 | 0.767 | 0.349–1.728 | 0.547 |
| SI | ≥400/<400 | −0.332 | 0.611 | 0.718 | 0.217–2.374 | 0.587 |
| Tumor site | Left/right
lobe | 0.492 | 0.439 | 1.635 | 0.691–3.868 | 0.263 |
| Pleural
invasion | Present/absent | −0.385 | 0.606 | 0.680 | 0.207–2.231 | 0.525 |
| Lymph node
metastasis | Present/absent | 1.535 | 0.775 | 4.640 | 1.016–21.195 | 0.048 |
| Pathological
type | AC/SCC/others | −0.182 | 1.158 | 0.833 | 0.086–8.061 | 0.978 |
|
Differentiation |
Poor/moderate/well | 0.295 | 0.836 | 1.343 | 0.261–6.913 | 0.925 |
| Stage |
IIIB/IIIA/IIB/IIA/IB/IA | 1.034 | 0.671 | 2.813 | 0.756–10.471 | 0.196 |
| miR-124
expression |
Downregulated/upregulated | −1.785 | 0.686 | 0.168 | 0.044–0.644 | 0.009 |
| STAT3
expression |
Downregulated/upregulated | −1.872 | 0.648 | 0.154 | 0.043–0.548 | 0.004 |
Discussion
Current research has indicated that abnormal
expression of miRNAs plays a critical role in tumor formation and
development (24,25). miR-124 is predominantly expressed
in normal brain and plays an important role in neural processes
from normal neural cell function to the development of central
nervous system tumors, supporting its potential role in cancer
(26,27). SLC16A1, the target of miR-124,
regulates lactic acid export during aerobic glycolysis in
medulloblastoma (28). miR-124
also inhibits the expression of SOS1 in glioblastoma (29). Recently, a number of experiments
have shown that ectopic expression of miR-124 is involved in the
development of other non-central nervous system tumors. For
example, in hepatocellular carcinoma (HCC), cell aggressiveness is
modulated by miR-124, which represses the expression of ROCK2 and
EZH2 (30). In addition, in mouse
models of HCC, miR-124 suppresses the expression of IL-6R and
reduces STAT3 activation in transformed cells (31). In breast cancer, miR-124 inhibits
the invasive and metastatic potential of cells by targeting the
CD151 gene (32). Moreover,
miR-124 promotes tumor progression and metastasis by downregulating
Rac1 in pancreatic cancer and participates in the progression of
glioblastoma by targeting PPP1R13L (33,34).
Taken together, these findings indicate that miR-124 is a promising
tumor suppressor gene that negatively regulates oncogenes or genes
that control the growth, proliferation, invasion and apoptosis of
certain tumor cells by inhibiting the expression of some
transcription repressors.
The results of the present study indicate that
miR-124 inhibits the tumorigenic potential of lung cancer cells by
downregulating STAT3. This conclusion is based on the following
observations. First, overexpression of miR-124 suppressed cell
proliferation and promoted apoptosis in NSCLC cells. Second, STAT3
was downregulated in NSCLC cells overexpressing miR-124 following
transfection with miR-124 mimics. Third, TargetScan and luciferase
activity data showed that the 3′ UTR of STAT3 was directly targeted
by miR-124. STAT3, a member of the signal transduction and
activation of transcription (STAT) family, has been shown to
participate in the formation of various cancers by promoting cell
proliferation, angiogenesis, and invasion or by inhibiting
apoptosis (35,36). In colorectal cancer, overexpression
of miR-124 suppresses the growth of colorectal cancer cells by
directly binding to the 3′ UTR of STAT3, similar to the effects
observed following knocking down of STAT3 by specific siRNA
(15). As the putative primary
target of miR-124, STAT3 has been studied in various cancers
(15,37,38).
The data presented herein shed light on the relationship between
miR-124 and STAT3 in lung cancer cells. In addition, we further
investigated the expression of miR-124 and STAT3 at the tissue
level. Our findings demonstrated that the expression of miR-124 was
lower while STAT3 was higher in NSCLC tissues compared to adjacent
normal lung tissues. Moreover, Spearman’s rank-order correlation
analysis indicated an inverse correlation between STAT3 and
miR-124, further confirming that the downregulation of miR-124
resulted in STAT3 overexpression.
Accumulating studies have shown that downregulation
of miR-124 is associated with a worse survival in patients with
solid tumors, including pancreatic duct adenocarcinoma (33), colorectal (39) and renal cell cancer (40). The clinical data obtained in the
present study revealed that a decrease in the expression of miR-124
or an increase in the expression of STAT3 correlated closely with
the TNM stage, differentiation grade and lymph node metastasis.
miR-124 was downregulated in patients with poor differentiation or
lymph node metastasis, which suggested that its downregulation
might be acquired during the process of tumor progression and
perhaps even during the acquisition of metastatic potential.
Berghmans et al (41)
constructed a prognostic score for OS using a linear combination of
miR CT values that were weighted using Cox’s regression
coefficients in advanced NSCLC patients treated with first-line
chemotherapy. The results showed that miR-124 had no predictive
role in terms of the response to chemotherapy but contributed to
the prognostic outcome in OS (41). Moreover, miRNA profiling, which
commonly included miR-124, revealed an association of miR-124 with
the prognosis of stage I NSCLC after resection (42). All of these findings supported the
potential prognosis value of miR-124 in NSCLC. Therefore, we
further explored the relationship between the expression of miR-124
and survival in postoperative NSCLC patients. We found that a low
level of miR-124 expression was associated with poor survival in
terms of both DFS and OS, and a Cox proportional hazard regression
analysis revealed the prognostic importance of the expression level
of STAT3. Based on the above results, we concluded that miR-124
expression correlated closely with the survival of patients with
NSCLC and might be a useful prognostic marker in this disease.
In conclusion, the results of the present study
support the hypothesis that miR-124 is a tumor-suppressing miRNA
that regulates STAT3 activity and functions as a useful biomarker
for the prognosis of postoperative NSCLC patients. However, the
conclusions of the present study are limited because we did not
explore the detailed relationship between miR-124 and the STAT3
pathway in lung cancer cells. In addition, we did not analyze the
relationship between the expression of miR-124 and adjuvant
therapy. The prognostic significance of miR-124 expression in NSCLC
patients receiving adjuvant therapy requires an additional
prospective controlled study. Finally, additional studies in
vitro and larger-scale statistical analyses of this clinical
study have yet to be performed.
Acknowledgements
The present study was supported by the Jieping Wu
Foundation of China (nos. 320.6753.1219 and 320.6750.13210), and
the Department of Science and Technology of Shandong province
(Contract nos. 2012YD18042, 2011YD18004 and 2010GWZ20260).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Herbst RS, Heymach JV and Lippman SM: Lung
cancer. N Engl J Med. 359:1367–1380. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Calin GA, Sevignani C, Dumitru CD, et al:
Human microRNA genes are frequently located at fragile sites and
genomic regions involved in cancers. Proc Natl Acad Sci USA.
101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Johnson CD, Esquela-Kerscher A, Stefani G,
et al: The let-7 microRNA represses cell proliferation pathways in
human cells. Cancer Res. 67:7713–7722. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kumar MS, Erkeland SJ, Pester RE, et al:
Suppression of non-small cell lung tumor development by the let-7
microRNA family. Proc Natl Acad Sci USA. 105:3903–3908. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Takamizawa J, Konishi H, Yanagisawa K, et
al: Reduced expression of the let-7 microRNAs in human lung cancers
in association with shortened postoperative survival. Cancer Res.
64:3753–3756. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lin PY, Yu SL and Yang PC: MicroRNA in
lung cancer. Br J Cancer. 103:1144–1148. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu J, Carmell MA, Rivas FV, et al:
Argonaute2 is the catalytic engine of mammalian RNAi. Science.
305:1437–1441. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu X, Sempere LF, Galimberti F, et al:
Uncovering growth-suppressive MicroRNAs in lung cancer. Clin Cancer
Res. 15:1177–1183. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ando T, Yoshida T, Enomoto S, et al: DNA
methylation of microRNA genes in gastric mucosae of gastric cancer
patients: its possible involvement in the formation of epigenetic
field defect. Int J Cancer. 124:2367–2374. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shimizu T, Suzuki H, Nojima M, et al:
Methylation of a panel of microRNA genes is a novel biomarker for
detection of bladder cancer. Eur Urol. 63:1091–1100. 2013.
View Article : Google Scholar
|
|
14
|
Lv XB, Jiao Y, Qing Y, et al: miR-124
suppresses multiple steps of breast cancer metastasis by targeting
a cohort of pro-metastatic genes in vitro. Chin J Cancer.
30:821–830. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang J, Lu Y, Yue X, et al: MiR-124
suppresses growth of human colorectal cancer by inhibiting STAT3.
PLoS One. 8:e703002013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wei J, Wang F, Kong LY, et al: miR-124
inhibits STAT3 signaling to enhance T cell-mediated immune
clearance of glioma. Cancer Res. 73:3913–3926. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Grivennikov SI and Karin M: Dangerous
liaisons: STAT3 and NF-kappaB collaboration and crosstalk in
cancer. Cytokine Growth Factor Rev. 21:11–19. 2010. View Article : Google Scholar :
|
|
18
|
Zhang X, Zhang J, Wang L, Wei H and Tian
Z: Therapeutic effects of STAT3 decoy oligodeoxynucleotide on human
lung cancer in xenograft mice. BMC Cancer. 7:1492007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kanda N, Seno H, Konda Y, et al: STAT3 is
constitutively activated and supports cell survival in association
with survivin expression in gastric cancer cells. Oncogene.
23:4921–4929. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang G, Huang C, Cao J, Huang KJ, Jiang T
and Qiu ZJ: Lentivirus-mediated shRNA interference targeting STAT3
inhibits human pancreatic cancer cell invasion. World J
Gastroenterol. 15:3757–3766. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ikari A, Sato T, Watanabe R, Yamazaki Y
and Sugatani J: Increase in claudin-2 expression by an
EGFR/MEK/ERK/c-Fos pathway in lung adenocarcinoma A549 cells.
Biochim Biophys Acta. 1823:1110–1118. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Peltier HJ and Latham GJ: Normalization of
microRNA expression levels in quantitative RT-PCR assays:
identification of suitable reference RNA targets in normal and
cancerous human solid tissues. RNA. 14:844–852. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Howington JA, Blum MG, Chang AC, Balekian
AA and Murthy SC: Treatment of stage I and II non-small cell lung
cancer: diagnosis and management of lung cancer, 3rd ed: American
College of Chest Physicians evidence-based clinical practice
guidelines. Chest. 143:e278S–313S. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Raveche ES, Salerno E, Scaglione BJ, et
al: Abnormal microRNA-16 locus with synteny to human 13q14 linked
to CLL in NZB mice. Blood. 109:5079–5086. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hatley ME, Patrick DM, Garcia MR, et al:
Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21.
Cancer Cell. 18:282–293. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Clark AM, Goldstein LD, Tevlin M, Tavare
S, Shaham S and Miska EA: The microRNA miR-124 controls gene
expression in the sensory nervous system of Caenorhabditis elegans.
Nucleic Acids Res. 38:3780–3793. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fowler A, Thomson D, Giles K, et al:
miR-124a is frequently down-regulated in glioblastoma and is
involved in migration and invasion. Eur J Cancer. 47:953–963. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li KK, Pang JC, Ching AK, et al: miR-124
is frequently downregulated in medulloblastoma and is a negative
regulator of SLC16A1. Hum Pathol. 40:1234–1243. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lv Z and Yang L: MiR-124 inhibits the
growth of glioblastoma through the downregulation of SOS1. Mol Med
Rep. 8:345–349. 2013.PubMed/NCBI
|
|
30
|
Zheng F, Liao YJ, Cai MY, et al: The
putative tumour suppressor microRNA-124 modulates hepatocellular
carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut.
61:278–289. 2012. View Article : Google Scholar
|
|
31
|
Hatziapostolou M, Polytarchou C, Aggelidou
E, et al: An HNF4alpha-miRNA inflammatory feedback circuit
regulates hepatocellular oncogenesis. Cell. 147:1233–1247. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Han ZB, Yang Z, Chi Y, et al: MicroRNA-124
suppresses breast cancer cell growth and motility by targeting
CD151. Cell Physiol Biochem. 31:823–832. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang P, Chen L, Zhang J, et al:
Methylation-mediated silencing of the miR-124 genes facilitates
pancreatic cancer progression and metastasis by targeting Rac1.
Oncogene. 33:514–524. 2014. View Article : Google Scholar
|
|
34
|
Zhao WH, Wu SQ and Zhang YD:
Downregulation of miR-124 promotes the growth and invasiveness of
glioblastoma cells involving upregulation of PPP1R13L. Int J Mol
Med. 32:101–107. 2013.PubMed/NCBI
|
|
35
|
Haura EB, Turkson J and Jove R: Mechanisms
of disease: insights into the emerging role of signal transducers
and activators of transcription in cancer. Nat Clin Pract Oncol.
2:315–324. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yu H, Pardoll D and Jove R: STATs in
cancer inflammation and immunity: a leading role for STAT3. Nat Rev
Cancer. 9:798–809. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lu Y, Yue X, Cui Y, Zhang J and Wang K:
MicroRNA-124 suppresses growth of human hepatocellular carcinoma by
targeting STAT3. Biochem Biophys Res Commun. 441:873–879. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li Y, Zhang Z, Liu X, et al: miR-124
functions as a tumor suppressor in the endometrial carcinoma cell
line HEC-1B partly by suppressing STAT3. Mol Cell Biochem.
388:219–231. 2014. View Article : Google Scholar
|
|
39
|
Wang MJ, Li Y, Wang R, et al:
Downregulation of microRNA-124 is an independent prognostic factor
in patients with colorectal cancer. Int J Colorectal Dis.
28:183–189. 2013. View Article : Google Scholar
|
|
40
|
Gebauer K, Peters I, Dubrowinskaja N, et
al: Hsa-mir-124–3 CpG island methylation is associated with
advanced tumours and disease recurrence of patients with clear cell
renal cell carcinoma. Br J Cancer. 108:131–138. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Berghmans T, Ameye L, Willems L, et al:
Identification of microRNA-based signatures for response and
survival for non-small cell lung cancer treated with
cisplatin-vinorelbine A ELCWP prospective study. Lung Cancer.
82:340–345. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Patnaik SK, Kannisto E, Knudsen S and
Yendamuri S: Evaluation of microRNA expression profiles that may
predict recurrence of localized stage I non-small cell lung cancer
after surgical resection. Cancer Res. 70:36–45. 2010. View Article : Google Scholar
|