Introduction
MicroRNAs (miRNAs) constitute a class of small
non-coding RNAs of approximately 19–25 nucleotides that
post-transcriptionally regulate gene expression. These small RNA
molecules regulate or tune finely protein-coding gene expression in
animals and plants. miRNAs usually bind to the 3′-untranslated
region (3′-UTR) of target mRNAs, leading to mRNA degradation and/or
translational repression (1).
Therefore, they play significant roles in cell proliferation, cell
death, as well as many other important cellular processes (2,3).
Consequently, miRNAs have emerged as significant molecules that are
implicated in carcinogenesis and metastasis of many different
cancer cell types. miRNAs in carcinogenesis can target transcripts
of tumor-suppressor genes and/or proto-oncogenes.
Colorectal cancer (CRC) is one of the most frequent
cancers and a serious cause of cancer-related deaths all over the
world (4). About 5–10% of all
colorectal cancers develop as a defined hereditary cancer syndrome.
The two main forms of hereditary cancer syndromes are hereditary
non-polyposis colorectal cancer (HNPCC) and familial adenomatous
polyposis (FAP) (5). Recent
studies have pointed out the connection between CRC and the
expression levels of specific miRNAs such as miR-143 and miR-145,
in an attempt to clarify their role in carcinogenesis in colonic
mucosae (6).
miR-224 expression is deregulated in several human
malignancies, including prostate (7,8),
breast (9), ovarian (10) and cervical cancer (11), pancreatic ductal adenocarcinoma
(12), hepatocellular carcinoma
(13–15), colorectal cancer (16–18),
thyroid cancer (19), glioma
(20), as well as acute myeloid
leukemia (21), strongly
suggesting that miR-224 expression plays a role in the general
process of carcinogenesis. With regard to prostate cancer, miR-224
is expressed by perineural cancer cells and is likely to be
involved in perineural invasion, which constitutes the dominant
pathway for local invasion in this cancer (7). Overexpression of miR-224 plays a
major role in Hep G2 cells, as it is considered to regulate the
migration and invasion of these hepatocellular carcinoma cells
(15). Moreover, high miR-224
levels in highly invasive and metastatic pancreatic ductal
adenocarcinoma account for downregulation of CD40 protein
expression at the cell surface (12).
miR-224 targets protein-coding genes, modulates the
levels of the respective proteins and is thus involved in the
pathogenesis of several diseases. The apoptosis inhibitor 5 (API5)
(13), whose expression prevents
apoptosis after growth factor deprivation (22) is a target of miR-224. By
downregulating API5 expression, miR-224 increases apoptotic cell
death of Hep G2 cells; however, miR-224 overexpression in these
cells was also shown to increase cell proliferation (13). API5 depletion sensitizes cancer
cells to chemotherapy (22).
Significantly augmented API5 expression has been detected in
biopsies of lung and colorectal tumors, compared to biopsies from
adjacent normal tissues. Therefore, API5 protein is upregulated in
tumor epithelial cells and has been proposed as a potential
prognostic marker in colorectal cancer (23).
Furthermore, in breast cancer cells miR-224
compromises the expression of RAF1 kinase inhibitor protein (RKIP),
a tumor suppressor that protects against metastasis and genomic
instability (9). A recent study
showed that RKIP expression is diffused in normal colorectal mucosa
and progressively lost towards tumor center and front. Loss of RKIP
expression predicts features of epithelial-mesenchymal transition
(EMT) and is also associated with distant metastasis; however, its
prognostic significance is restricted to the tumor center (24). The CD40 gene constitutes
another confirmed target of miR-224 (12). CD40, a member of the tumor necrosis
factor receptor (TNFR) superfamily, is a co-stimulatory protein
found on antigen presenting cells and is required for their
activation (25). CD40 is highly
expressed in various established human CRC cell lines in culture as
well as in CRC specimens. Activation of this receptor by
membrane-bound CD40L, but not by soluble agonists, has been shown
to trigger programmed cell death in CD40-positive CRC cells and to
induce secretion of proinflammatory cytokines (26).
The aim of this study was to investigate the
prognostic potential of miR-224 expression and its putative
clinical application in colorectal adenocarcinoma prognosis. We
developed a highly sensitive quantitative real-time PCR methodology
and used it to quantify miR-224 levels in malignant colorectal
tumors and adjacent non-cancerous colorectal mucosae.
Materials and methods
Tissue samples
We collected 115 cancerous and 66 paired
non-cancerous colorectal tissue specimens from patients who
underwent surgical treatment for primary colorectal adenocarcinoma
at the University General Hospital ‘Attikon’, between 2000 and
2010. Tumor tissues were histologically characterized by a
pathologist and frozen in liquid nitrogen immediately after
resection. Informed consent was obtained from all patients
participating in the study. The study was approved by the
institutional Ethics Committee of the University General Hospital
‘Attikon’ (Athens, Greece) and performed in accordance with the
ethical standards of the 1964 Declaration of Helsinki and its later
amendments.
Patients’ clinicopathological parameters included
the tumor size, histological grade and stage of the disease
according to the TNM classification. The TNM staging system updated
in May, 2011 (27), combines tumor
invasion (T), regional lymph node status (N), and presence or
absence of distant metastases (M). Patients’ clinical and biologic
characteristics are summarized in Table I. Follow-up information included
disease status (disease-free or recurrence) and survival status
(alive or deceased), along with the dates of the events and the
cause of death. The median disease-free survival (DFS) was 32.0
months (range, 3.0–120.0) and the median overall survival (OS) was
31.0 months (range, 1.0–120.0). Patient age ranged from 37.0 to
93.0 years with a mean (±SE) of 67.8 (±1.1) (Table II).
 | Table IClinical and biological
characteristics of colorectal adenocarcinoma patients. |
Table I
Clinical and biological
characteristics of colorectal adenocarcinoma patients.
| Variable | No. of patients
(%) |
|---|
| Total | 115 |
| Sex |
| Male | 60 |
| Female | 55 |
| Histological
grade |
| I | 13 (11.3%) |
| II | 79 (68.7%) |
| III | 23 (20.0%) |
| T |
| T1 | 2 (1.8%) |
| T2 | 12 (10.4%) |
| T3 | 68 (59.1%) |
| T4 | 33 (28.7%) |
| N |
| N0 | 67 (58.3%) |
| N1 | 30 (26.1%) |
| N2 | 18 (15.6%) |
| M |
| M0 | 102 (88.7%) |
| M1 | 13 (11.3%) |
| TNM stage |
| I | 13 (11.3%) |
| II | 52 (45.2%) |
| III | 37 (32.2%) |
| IV | 13 (11.3%) |
 | Table IIDistribution of the numerical
variables of the study in the cohort of colorectal adenocarcinoma
patients. |
Table II
Distribution of the numerical
variables of the study in the cohort of colorectal adenocarcinoma
patients.
| | | Percentile |
|---|
| | |
|
|---|
| Variable | Mean ± SE | Range | 25th | 50th (median) | 75th |
|---|
| miR-224 expression
(RQU) |
| in tumors
(n=115) | 47.39±4.02 | 1.81–187.75 | 16.14 | 34.27 | 64.30 |
| in non-cancerous
specimens (n=66) | 15.84±1.74 | 2.16–63.25 | 6.10 | 11.34 | 21.25 |
| Patient age
(years) | 67.8±1.1 | 37.0–93.0 | 59.0 | 69.0 | 77.0 |
| Tumor size
(cm2) | 23.2±1.8 | 0.8–132.0 | 10.5 | 18.2 | 30.0 |
| DFS (months) | 39.9±3.2 | 3.0–120.0 | 17.0 | 32.0 | 52.0 |
| OS (months) | 40.5±2.9 | 1.0–120.0 | 18.0 | 31.0 | 53.0 |
Human CRC cell line culture
Human colorectal adenocarcinoma Caco-2 cells were
subcultured in DMEM medium, adjusted to contain 10% fetal bovine
serum (FBS), 100 kU/l penicillin, 0.1 g/l streptomycin and 2 mM
L-glutamine. Cells were seeded at a concentration of
0.5×105 cells/ml and incubated for 48 h at 37°C, in a
humidified atmosphere containing 5% CO2, before being
collected for further use.
Total RNA extraction, polyadenylation and
reverse transcription
Tissue specimens were pulverized and then dissolved
in TRI Reagent® (Molecular Research Center, Inc.
Cincinnati, OH, USA). Following the manufacturer’s instructions,
total RNA was extracted from homogenized tumors and Caco-2 cells,
diluted in RNA Storage Solution (Life Technologies Ltd., Carlsbad,
CA, USA), and stored at −80°C until use. The concentration and
purity of total RNA were assessed spectrophotometrically at 260 and
280 nm. Next, polyadenylation of total RNA and reverse
transcription into first-strand cDNA was performed, as previously
described (8). In brief, 2 μg of
total RNA were polyadenylated using recombinant poly(A) polymerase
(New England Biolabs Ltd., Whitby, ON, Canada) in the presence of
ATP (80 μM). Next, first-strand cDNA was synthesized from 2 μg of
polyadenylated total RNA, using M-MLV Reverse Transcriptase (Life
Technologies Ltd.) and an oligo-dT-adapter sequence
(5′-GCGAGCACAGAATTAA TACGACTCACTATAGGTTTTTTTTTTTTVN-3′, where V=G,
A, C and N=G, A, T, C) as primer. The final reaction volume was 20
μl.
Quantitative real-time PCR
Quantitative real-time PCR (qPCR) was performed
using the SYBR-Green chemistry in a 7500 Fast Real-Time PCR System
(Applied Biosystems, Foster City, CA, USA). Based on the published
sequences of mature miR-224 and SNORD48 (small nucleolar RNA, C/D
box 48; also known as U48 or RNU48) with GenBank accession numbers
NR_029638.1 and NR_002745.1, respectively, two miRNA gene-specific
primers were designed and used in combination with a common reverse
primer to generate two respective amplicons. The sequence of the
miR-224 forward primer was 5′-CAAGTCACTAGTGGTTCCGTTAA-3′ and that
of the SNORD48 forward primer was 5′-TGATGATGAC
CCCAGGTAACTCT-3′. The sequence of the common reverse primer,
complementary to the oligo-dT-adapter, was 5′-GCGA
GCACAGAATTAATACGAC-3′. The resulting PCR amplicons for miR-224 and
SNORD48 were 65- and 105-bp long, respectively. The reaction
mixture contained 1 μl of 10-fold diluted cDNA, 5 μl KAPA™
SYBR® FAST qPCR Kits (2X) (Kapa Biosystems Inc., Woburn,
MA, USA), and 2 μl of gene-specific primers (final concentration:
200 nM each), in a final reaction volume of 10 μl. The cycling
conditions were as follows: a denaturation step at 95°C for 3 min,
followed by 40 cycles of 95°C for 3 sec, for denaturation of the
PCR products, and 60°C for 30 sec, for primer annealing and
extension. Each real-time PCR reaction was performed in duplicate
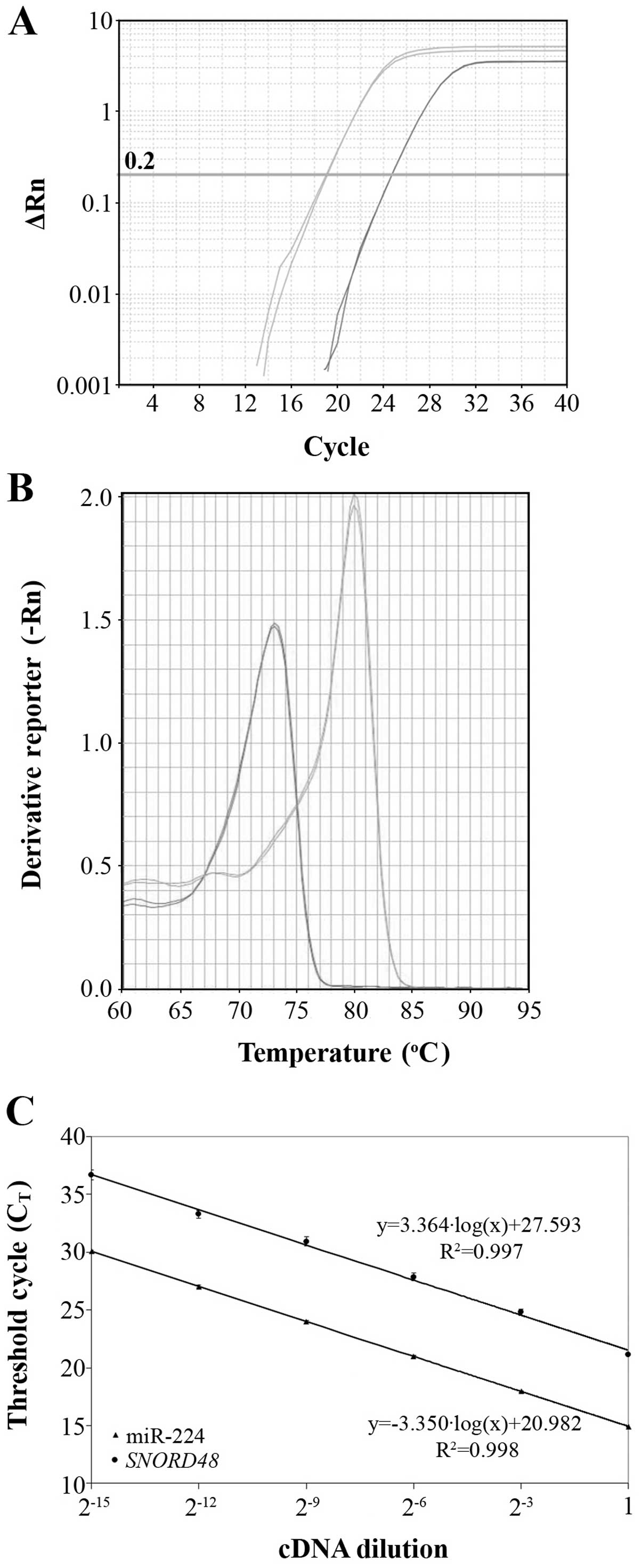
to evaluate the reproducibility of data (Fig. 1A).
In order to distinguish between the specific PCR
products and primer-dimers or other non-specific products, melting
curves of the PCR products were generated after amplification, by
heating the reaction mixtures from 60 to 95°C with a heating rate
of 0.3°C/sec and continuously acquiring fluorescence emission data.
Primer-dimers and/or other non-specific products are characterized
by a lower Tm than those of the miR-224 and SNORD48
amplicons (Fig. 1B).
Calculations and validation of the
comparative CT (2−ΔΔCT) method for miR-224
quantification
Calculations were made using the comparative
CT (2−ΔΔCT) method. The prerequisites for the
application of the comparative CT (2−ΔΔCT)
method (28) were checked in a
validation experiment, in which CT values of miR-224 and
SNORD48 were measured in a dilution series of Caco-2 cDNA.
Real-time PCR efficiency (E) for amplification of each gene
was calculated using the following formula: E =
−1+10(−1/_), where α is the slope of the corresponding
amplification plot. As illustrated in Fig. 1C, the slopes of miR-224 and
SNORD48 amplification plots are very similar (−3.364 and
−3.350, respectively), which clearly indicates similar efficiencies
for the corresponding amplicons (98.3 and 98.8%, respectively).
In the current study, SNORD48 was used as an
endogenous control gene so as to normalize PCRs for the RNA amount
added to the reverse transcription reactions, while the colorectal
adenocarcinoma cell line Caco-2 was used as a calibrator in order
to make PCRs from distinct runs comparable (29).
The normalized result of each sample was calculated
as the ratio of miR-224 copies to SNORD48 copies divided by
the same ratio that had been previously calculated for Caco-2
cells. The normalized (2−ΔΔCT) amounts of tissue sample
miR-224 levels were then multiplied with the average ratio of
miR-224 copies to SNORD48 copies of Caco-2 cells (2–6.611),
calculated by the difference between the y-intercepts of the
regression lines shown in Fig. 1C.
This procedure produced comparable results that do not depend on
the miR-224 expression levels of Caco-2 cells. Finally, normalized
results were multiplied by 1,000 and designated as relative
quantification units (RQU), standing for miR-224 copies/1,000
SNORD48 copies.
Statistical analysis
As the distribution of the expression levels of
miR-224 in our cohort of patients was not Gaussian, analysis of the
differences in the two groups of specimens (colorectal
adenocarcinomas vs. non-cancerous mucosae) was performed with the
non-parametric Mann-Whitney U test. Moreover, miR-224 levels
between paired tissue samples were compared using the Wilcoxon
signed-rank test.
So as to determine the optimal cut-off point for
categorization of patients into miR-224-positive and
miR-224-negative as there are no established cut-off points, we
used the X-tile software, an algorithm that facilitated the
determination of an optimal cut-off point by correcting for the use
of minimum p-value statistics (30). This cut-off point was 37.79 RQU,
equal to the 56th percentile. According to this cut-off value,
miR-224 expression in each specimen was categorized as negative or
positive.
We also constructed receiver operating
characteristic (ROC) curves for miR-224 expression levels, by
plotting sensitivity versus (1-specificity) and the areas under the
ROC curves (AUC) were analyzed by Hanley and McNeil method. To
further examine the discriminatory value of miR-224 expression in
colorectal adenocarcinoma, we performed univariate binary logistic
regression analysis.
Associations between miR-224 expression status and
survival of the patients were assessed by Kaplan-Meier DFS and OS
curves. The differences between the curves were evaluated by the
log-rank (Mantel-Cox) test. We also developed Cox proportional
hazard regression models, so as to assess the association between
the prognostic markers and the relative risks for relapse and death
of patients. Multivariate Cox regression models were adjusted for
the aforementioned established clinicopathological parameters.
Finally, Kaplan-Meier survival analysis was carried out to evaluate
the prognostic potential of miR-224 expression with regard to DFS
and OS in groups of patients, stratified according to distinct
clinicopathological features. The level of statistical significance
was defined at a probability value of less than 0.05
(p<0.05).
Results
miR-224 expression levels in colorectal
adenocarcinoma tissue specimens and adjacent non-cancerous
colorectal mucosae
miR-224 expression was significantly higher in
colorectal adenocarcinoma tissues than in non-cancerous mucosae,
ranging from 1.81 to 187.75 RQU with a mean (±SE) of 47.39 (±4.02)
in the former, while varying between 2.16 and 63.25 RQU with a mean
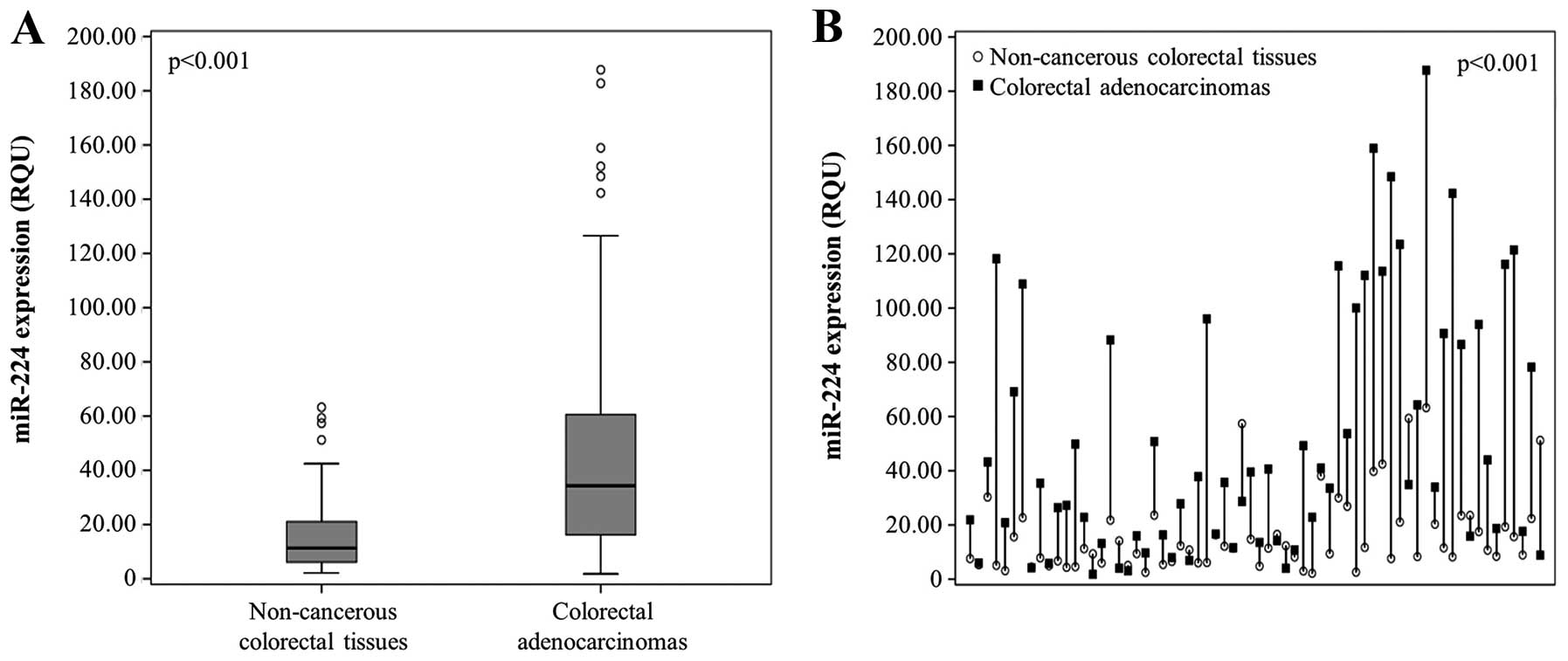
(±SE) of 15.84 (±1.74) in the latter (Table II and Fig. 2A). Comparison of miR-224 levels
between colorectal tumors and their adjacent non-cancerous mucosae
uncovered the profound overexpression of this molecule in the
malignant colorectal tumors [55 out of 66 (83.3%) examined tissue
pairs, p<0.001; Fig. 2B].
miR-224 expression of each colorectal adenocarcinoma
was then categorized into one of two groups (positive or negative),
as described in Materials and methods; thus, 51 (44.3%) cases were
classified as miR-224-positive and 64 (55.7%) as
miR-224-negative.
Discriminatory value of miR-224
expression in colorectal adenocarcinoma
So as to evaluate the potential of miR-224
expression as a predictive biomarker for the discrimination between
colorectal adenocarcinoma and non-cancerous colorectal tissues, we
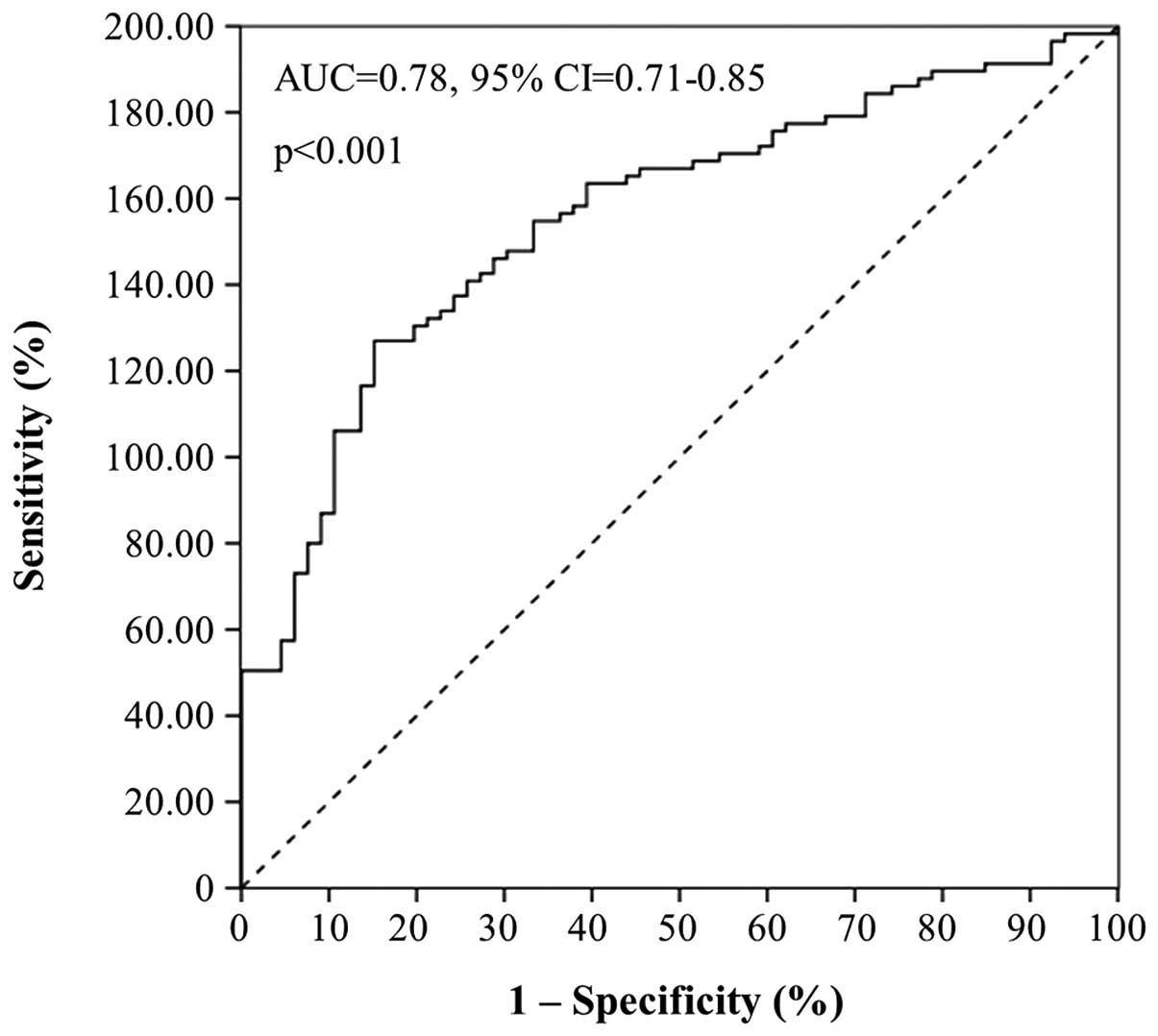
performed ROC and logistic regression analyses. As illustrated by
the ROC curve in Fig. 3, miR-224
expression was found to distinguish very efficiently colorectal
adenocarcinoma from healthy colorectal mucosae [area under the
curve (AUC)=0.78, 95% confidence interval (95% CI)=0.71–0.85,
p<0.001].
Univariate logistic regression analysis revealed
that high miR-224 levels constitute a predictor of the presence of
colorectal adenocarcinoma. Analysis of miR-224 expression as a
dichotomous variable showed that miR-224 positivity in colorectal
mucosae predicts a 7-fold higher risk for adenocarcinoma (crude
odds ratio = 6.72, 95% CI = 2.83–15.96, p<0.001).
miR-224 expression predicts short-term
relapse in colorectal adenocarcinoma patients
Follow-up information was available for 104
patients; however, 13 patients were diagnosed with distant
metastases before or at the time of surgery and were excluded from
DFS analysis. Out of the remaining 91 patients, 15 (16.5%) relapsed
during the respective follow-up periods. In Cox univariate
regression analysis (Table III),
a 3.5-fold higher risk of recurrence was predicted for colorectal
adenocarcinoma patients bearing tumors with positive miR-224
expression status [hazard ratio (HR)=3.52, 95% CI=1.20–10.33,
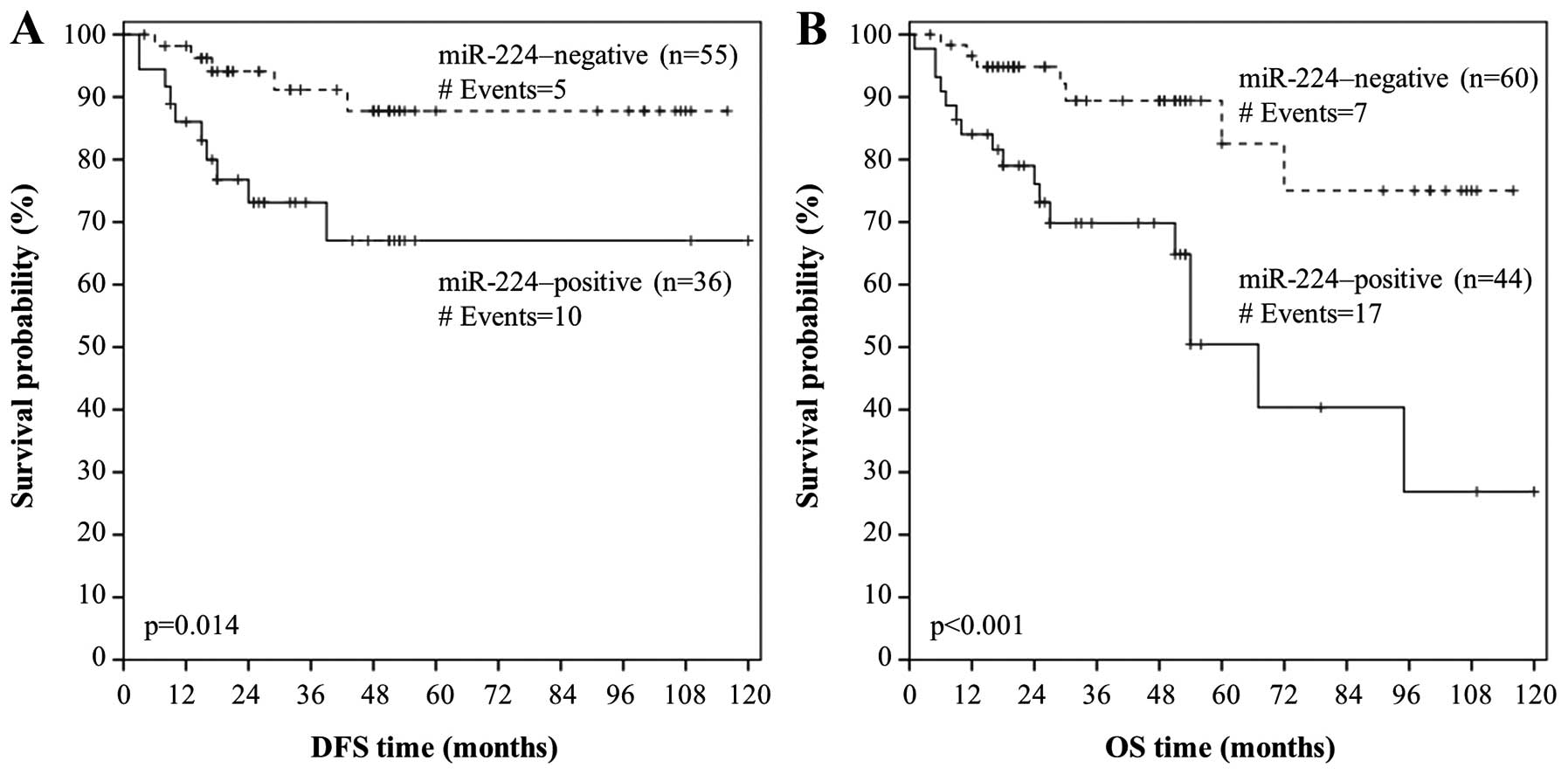
p=0.022). We also performed Kaplan-Meier survival analysis so as to
evaluate miR-224 expression in terms of predicting DFS. In
agreement with the aforementioned results, Kaplan-Meier DFS curves
illustrated that colorectal adenocarcinoma patients with
miR-224-positive tumors had significantly shorter DFS (p=0.014),
compared to those who had a miR-224-negative colorectal
adenocarcinoma (Fig. 4A).
 | Table IIImiR-224 expression and DFS of
colorectal adenocarcinoma patients. |
Table III
miR-224 expression and DFS of
colorectal adenocarcinoma patients.
| Disease-free
survival |
|---|
| Univariate analysis
(n=91) | Multivariate
analysisd (n=91) |
|---|
|
|
|
|---|
| Covariate | HRb | 95% CIc | p-value | HRb | 95% CIc | p-value |
|---|
| miR-224 expression
statusa |
| Negative | 1.00 | | | 1.00 | | |
| Positive | 3.52 | 1.20–10.33 | 0.022 | 4.61 | 1.41–15.09 | 0.012 |
| Tumor size | 0.96 | 0.92–1.01 | 0.10 | 0.93 | 0.87–1.00 | 0.061 |
| Histological grade
(ordinal) | 2.33 | 0.91–5.96 | 0.078 | 6.30 | 1.61–24.65 | 0.008 |
| T (ordinal) | 1.36 | 0.59–3.17 | 0.47 | 1.74 | 0.72–4.24 | 0.22 |
| Positive nodal
status | 1.01 | 0.48–2.13 | 0.98 | 0.43 | 0.16–1.15 | 0.093 |
In the multivariate Cox regression analysis
(Table III), miR-224 expression
predicted a significantly unfavorable prognostic outcome (HR=4.61,
95% CI=1.41–15.09, p=0.012), which was independent of tumor size,
histological grade, tumor invasion, and regional lymph node
status.
miR-224 expression as an independent
prognosticator of poor OS in colorectal adenocarcinoma
Regarding OS, out of 104 colorectal adenocarcinoma
patients for whom followup data were available, 24 patients (23.1%)
died during the accrual follow-up period. Cox univariate regression
analysis (Table IV) demonstrated
that patients with miR-224-positive colorectal adenocarcinoma were
at higher risk of death (HR=4.08, 95% CI=1.68–9.88, p=0.002),
compared to patients whose colorectal adenocarcinoma was
miR-224-negative. Hence, enhanced miR-224 expression seems also to
constitute a strong unfavorable predictor of OS. Histological grade
of the tumor as well as TNM stage, which is a resultant of tumor
invasion, regional lymph node status and presence of distant
metastases, were significant prognosticators of OS (p=0.014 and
p<0.001, respectively), as expected. In accordance with these
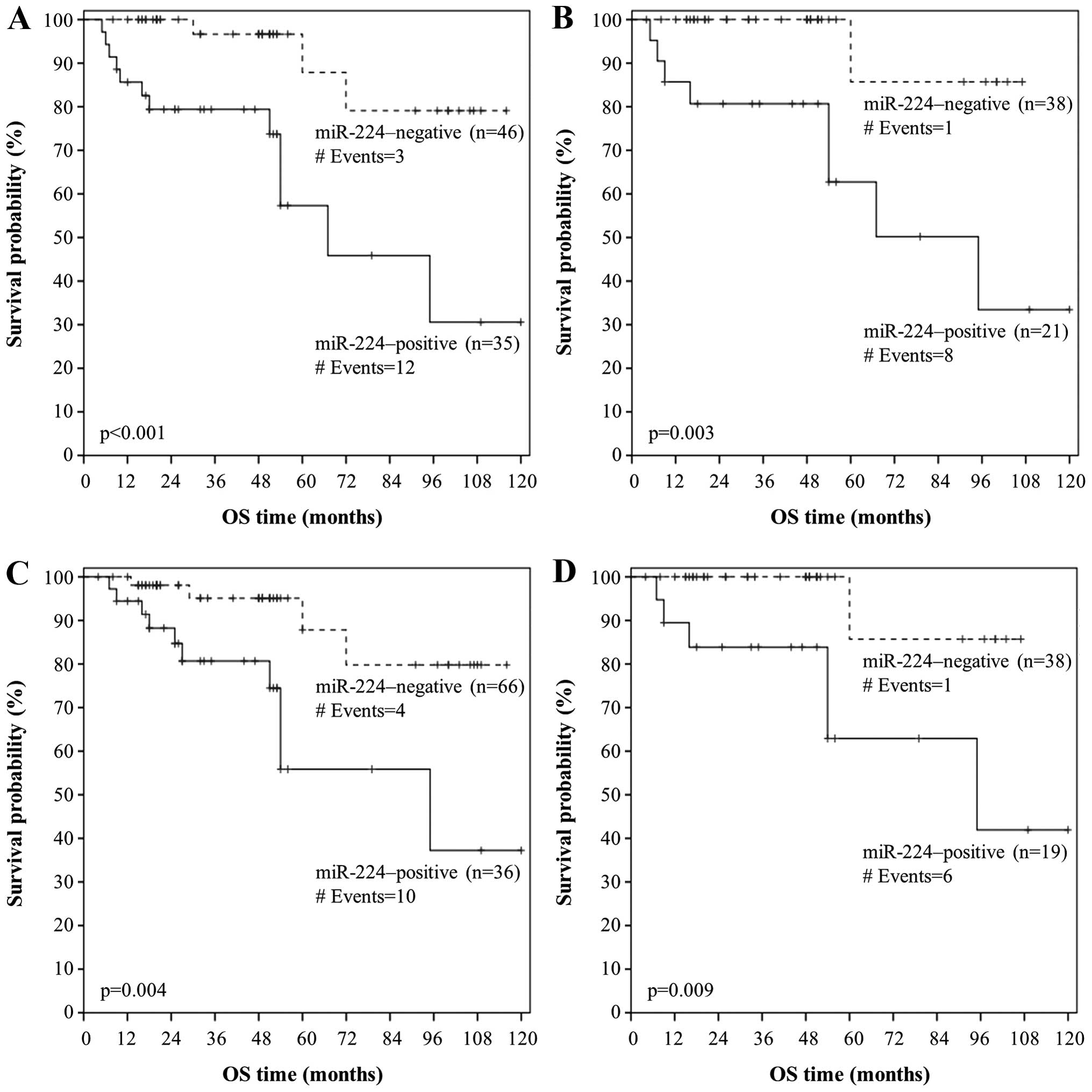
results, Kaplan-Meier OS analysis revealed that patients with
miR-224-positive colorectal adenocarcinoma were more likely to
succumb to their disease later than patients with a
miR-224-negative malignancy (p<0.001; Fig. 4B).
 | Table IVmiR-224 expression and OS of
colorectal adenocarcinoma patients. |
Table IV
miR-224 expression and OS of
colorectal adenocarcinoma patients.
| OS |
|---|
|
|
|---|
| Variable | HRb | 95% CIc | p-value |
|---|
| Univariate
analysis (n=104) |
| miR-224 expression
statusa |
| Negative | 1.00 | | |
| Positive | 4.08 | 1.68–9.88 | 0.002 |
| Tumor size | 1.01 | 0.99–1.03 | 0.47 |
| Histological grade
(ordinal) | 2.58 | 1.21–5.50 | 0.014 |
| T (ordinal) | 3.76 | 1.79–7.91 | <0.001 |
| Positive nodal
status | 2.02 | 1.22–3.33 | 0.006 |
| Distant
metastasis | 10.90 | 4.65–25.55 | <0.001 |
| TNM stage
(ordinal) | 3.77 | 2.18–6.53 | <0.001 |
|
| Multivariate
analysis (n=104) |
| miR-224 expression
statusa,d |
| Negative | 1.00 | | |
| Positive | 4.41 | 1.72–11.34 | 0.002 |
| Tumor size | 0.97 | 0.94–1.01 | 0.13 |
| Histological grade
(ordinal) | 2.60 | 0.97–6.98 | 0.057 |
| T (ordinal) | 3.12 | 1.33–7.35 | 0.009 |
| Positive nodal
status | 1.43 | 0.77–2.66 | 0.26 |
| Distant
metastasis | 6.06 | 2.14–17.15 | <0.001 |
|
| miR-224 expression
statusa,e |
| Negative | 1.00 | | |
| Positive | 3.74 | 1.48–9.44 | 0.005 |
| Tumor size | 0.99 | 0.96–1.02 | 0.57 |
| Histological grade
(ordinal) | 1.71 | 0.74–3.98 | 0.21 |
| TNM stage
(ordinal) | 3.49 | 1.87–6.51 | <0.001 |
In the multivariate Cox regression analysis
(Table IV), miR-224 positivity
remained a statistically significant indicator of poor OS in
colorectal adenocarcinoma, independent of tumor size, histological
grade, invasion, nodal status, and presence of distant metastases
(HR=4.41, 95% CI=1.72–11.34, p=0.002). More importantly, miR-224
expression retained its independent prognostic significance in
colorectal adenocarcinoma (HR=3.74, 95% CI=1.48–9.44, p=0.005) even
when the multivariate Cox regression model was adjusted for tumor
size, histological grade and TNM stage.
Prognostic value of miR-224 expression in
colorectal adenocarcinoma patients, stratified according to tumor
histological grade, nodal status and disease stage
Owing to the fact that patients with
well-differentiated tumors as well as those without regional nodal
metastases or, at least, without distant metastases are
substantially different from patients with poorly-differentiated
colorectal adenocarcinoma or advanced-stage patients with
metastases, respectively, in terms of their prognosis and
postoperative treatment, Kaplan-Meier survival analysis was carried
out to assess the prognostic value of miR-224 expression regarding
DFS and OS for each group of patients, stratified according to the
aforementioned clinicopathological features. As depicted in
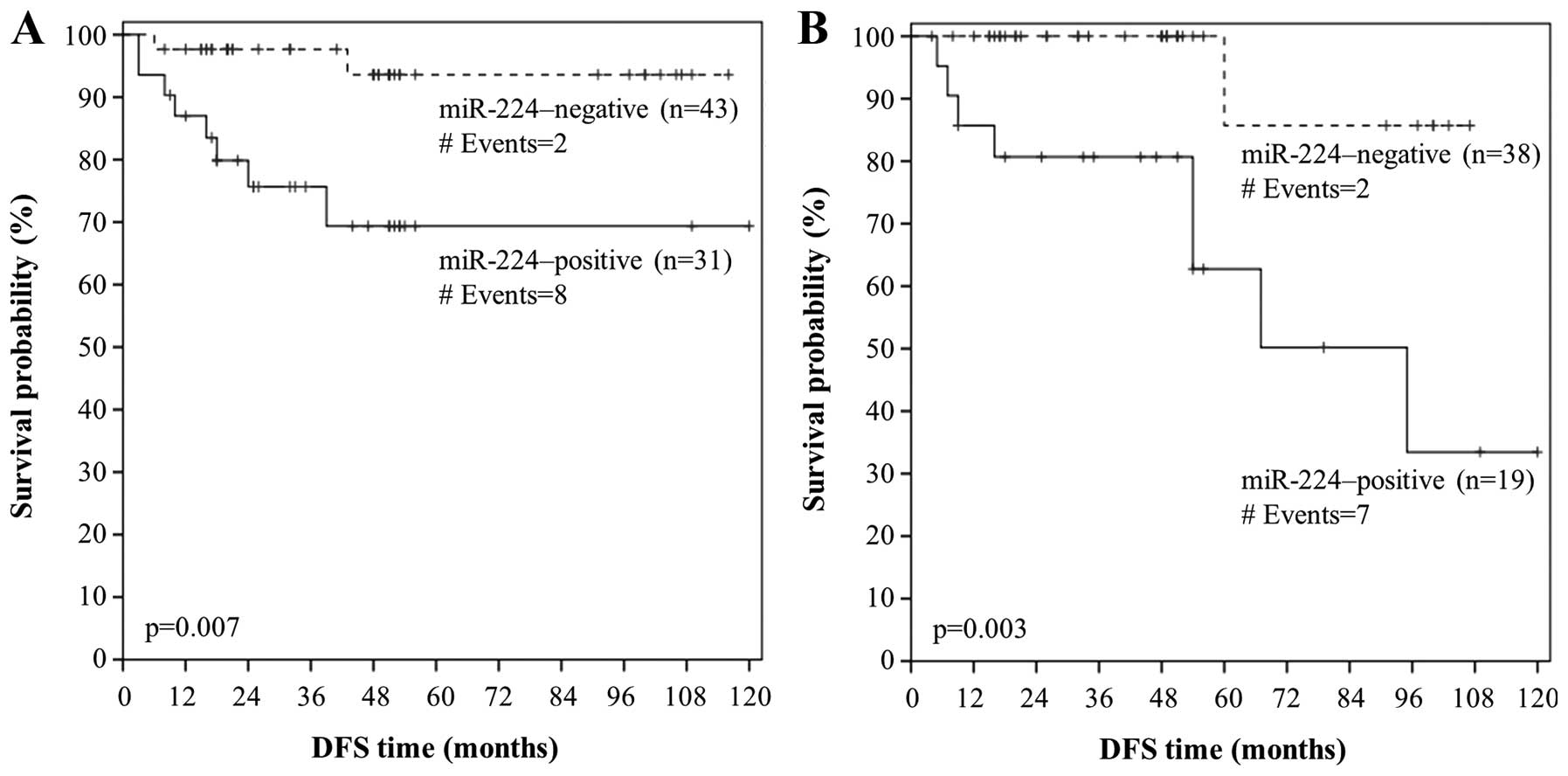
Figs. 5A and 6A, patients with well-(grade I) or
moderately-differentiated (grade II) colorectal adenocarcinoma
strongly expressing miR-224 had lower DFS and OS probabilities than
patients with miR-224-negative grade I/II tumors (p=0.007 and
p<0.001, respectively). Furthermore, node-negative (N0) patients
with colorectal adenocarcinoma that was positive for miR-224 tended
to relapse or succumb to their disease earlier than node-negative
patients with miR-224-negative carcinomas (p=0.003 in both cases),
as shown by the respective Kaplan-Meier DFS and OS curves (Figs. 5B and 6B). Interestingly, miR-224 positivity
predicts an unfavorable outcome also in metastasis-free patients,
in terms of OS (p=0.004; Fig. 6C).
More importantly, patients with miR-224-positive early-stage (TNM
stage I/II) colorectal adenocarcinoma had lower OS rates than did
patients bearing miR-224-negative early-stage tumors (p=0.009;
Fig. 6D).
Discussion
Colorectal adenocarcinoma, which constitutes by far
the most common type of colorectal cancer, is a multistep process
involving mutations in proto-oncogenes and tumor-suppressor genes
with deleterious effects on the respective proteins, defective
apoptosis, chromosomal or microsatellite instability, and aberrant
expression of several cancer-related genes, including those being
transcribed into long non-coding RNAs that are usually subjected to
further splicing, resulting in the generation of tiny RNA molecules
such as miRNAs (31,32). These small regulatory RNAs play a
key role in gene expression regulation by targeting specific
complementary regions of mRNAs, mainly located in the
3′-untranslated region (3′-UTR) of the latter. Thus, miRNAs
regulate post-transcriptionally the protein levels of their targets
by impeding the translation of the targeted mRNAs, but can also
trigger target degradation (33).
Occasionally, they can also affect histone modification and DNA
methylation on gene promoter sites, again resulting in modulations
of expression of their targets (34,35).
miRNA functional studies in CRC have uncovered their implication in
critical pathways such as EGFR (36), p53 (37), NFκB (38), Wnt/β-catenin (39,40)
and β-catenin/APC pathways (41),
as well as in the regulation of EMT (38) and cancer stem cell maintenance
(38,42). It is therefore evident that
alterations of specific miRNAs can significant contribute to
colorectal carcinogenesis.
Besides having a functional role in the initiation
and progression of colorectal cancer, miRNAs are about to introduce
a new era of diagnostic, prognostic and therapeutic modalities for
this human malignancy (43–45).
These small molecules are being intensively evaluated as biomarkers
and therapeutic targets for colorectal cancer (46), as their expression patterns are
subjected to remarkable alterations in tumor tissues (45), blood (plasma) (47,48)
and feces (49–51). In addition to miRNA expression
signatures (48), miRNA
methylation signatures may provide candidate biomarkers for the
detection of malignant colonocytes, since miRNAs are epigenetically
modified in CRC (52,53). Polymorphisms in miRNAs or
miRNA-binding sites have also particular interest, as they may
modify ones risk of developing cancer (46,54).
Thus, miRNA transcriptome constitutes a rich pool of novel and
emerging tumor biomarkers.
The oncogenic properties of miR-224 have been
demonstrated by several studies using cell line models originating
from different cancer types. miR-224 overexpression can affect
crucial cellular processes such as apoptosis (13), cell proliferation (13,15,55),
cell migration and invasion (15,55,56).
With regard to miR-224 targets, many genes are predicted to be
downregulated by this oncomiR, yet only a handful of those have
been validated. The most important miR-224 targets include SMAD
family member 4 (SMAD4) (55),
API5 (13) and RKIP (9). In human colorectal adenocarcinoma HCT
116 cells, the common signaling transducer SMAD4 is directly
downregulated by miR-224 (57),
which hence attenuates the transcriptional response of TGF-β
signaling transduction to promote cell proliferation (58). Downregulation of RKIP expression is
also very important in colorectal carcinogenesis, as this inhibitor
of RAF1 kinase is a master modulator of many critical intracellular
signaling cascades that control cellular growth, motility,
apoptosis, genomic integrity and therapeutic resistance (59). Finally, as aforementioned,
elimination of API5 sensitizes cancer cells to chemotherapy
(22), whereas miR-224 depletion
and, probably, subsequent API5 overexpression render human
colorectal adenocarcinoma HT-29 cells resistant to methotrexate
(60). Nonetheless, the in
vivo function and the clinical significance of miR-224
overexpression in CRC remain unclear.
Our study investigated the potential diagnostic and
prognostic value of miR-224 expression in colorectal
adenocarcinoma. Since previous studies failed to detect circulating
miR-224 (17), we used tumor
tissue and adjacent non-cancerous colorectal mucosae. In accordance
with previous findings (16–18),
our study showed that miR-224 expression is significantly
upregulated in colorectal adenocarcinoma in comparison with
non-cancerous adjacent mucosae (p<0.001). Taking this step
further, we also showed that miR-224 expression levels can
discriminate colorectal adenocarcinoma patients from normal
population, as indicated by ROC analysis (p<0.001) and
univariate binary logistic regression models (p<0.001). Recent
studies examining the diagnostic potential of several other miRNAs,
including miR-106a (61), miR-141
(62), miR-17-3p (63), miR-21 (61,62),
miR-29a (64) and miR-92 (62–64),
have shown that miRNAs can provide important diagnostic information
with an enhanced AUC. Therefore, it would be useful to analyze
miR-224 expression in combination with other miRNAs, in order to
create a multiparametric panel of markers for colorectal
adenocarcinoma diagnosis with relatively good accuracy.
Undoubtedly, this model will need to be validated within a larger
cohort and may be further improved by addition of other tumor
markers.
To the best of our knowledge, this is the first
study examining the prognostic significance of miR-224 expression
in colorectal adenocarcinoma, in terms of DFS. Cox univariate
regression analysis showed that high miR-224 expression in
colorectal adenocarcinoma predicts an increased risk of relapse,
and Kaplan-Meier survival analysis demonstrated significantly lower
DFS rates for miR-224-positive patients. These findings seem to
partially contradict the results of a previous study demonstrating
that miR-224 along with miR-221* promote colorectal
tumor growth and metastasis in mice (65). However, there are two major
differences that should be taken into account: first, that study
compared miR-224 between metastatic and non-metastatic samples,
while it did not examine the prognostic potential of miR-224;
second, the human material included in that study is restricted
(only 20 human CRC samples) compared to our material (115 human
colorectal adenocarcinoma samples).
Interestingly, the unfavorable prognostic value of
miR-224 with regard to DFS is independent of established
clinicopathological parameters such as histological
differentiation, tumor invasion, and regional lymph node
metastasis, as revealed by the multivariate Cox regression
analysis. Moreover, miR-224 overexpression retained its unfavorable
prognostic significance in the subgroups of colorectal
adenocarcinoma patients with grade I/II tumors or negative nodal
status. This is especially important when considering that about
30% of CRC patients with histopathology-negative regional lymph
nodes (N0) die from metastatic disease, reflected by microscopic
lymph node metastases that are overlooked by currently used
techniques (66).
miR-224 expression appears to constitute also an
independent unfavorable prognosticator of OS in colorectal
adenocarcinoma. As demonstrated by both Kaplan-Meier and Cox
regression analysis, miR-224 overexpression is an adverse
prognostic factor in colorectal adenocarcinoma, as it predicts a
quite 4-fold risk of death for patients, independently of the
clinicopathological features of the TNM stage. This finding not
only confirms the results of Liao et al, according to which
miR-224 expression predicts poor outcome in CRC patients
independently of tumor invasion and regional lymph node status
(67), but takes it a step further
by showing that miR-224 positivity, defined as expression over the
optimal cut-off point (not by arbitrary choice of the median as
cut-off value), is also independent of the presence (or absence) of
distant metastases. In addition to the above findings, we show that
high miR-224 expression constitutes a predictor of poor survival
even among patients with apparently similar OS probabilities, such
as those with negative regional lymph nodes, without distant
metastasis, or at an early TNM stage (I or II), thus suggesting its
putative future exploitation by multiparametric prognostic models
composed of a gamut of molecular biomarkers with prognostic value
in colorectal adenocarcinoma.
In conclusion, we demonstrated that miR-224 is
upregulated in colorectal adenocarcinoma compared with adjacent
non-cancerous mucosae, that miR-244 expression could be used to
discriminate malignant from normal colorectal tissue, and that
miR-224 positivity is an independent prognosticator of short-term
relapse and poor OS in colorectal adenocarcinoma. Additional
studies are hence necessitated to thoroughly evaluate the potential
of miR-224 as a prognostic biomarker in colorectal
adenocarcinoma.
Acknowledgements
This study was financially supported by the
Commission of the European Community through the INsPiRE project
(EU-FP7-REGPOT-2011-1, proposal 284460).
References
|
1
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xu P, Vernooy SY, Guo M and Hay BA: The
Drosophila microRNA Mir-14 suppresses cell death and is required
for normal fat metabolism. Curr Biol. 13:790–795. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brennecke J, Hipfner DR, Stark A, Russell
RB and Cohen SM: bantam encodes a developmentally regulated
microRNA that controls cell proliferation and regulates the
proapoptotic gene hid in Drosophila. Cell. 113:25–36. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Weitz J, Koch M, Debus J, Hohler T, Galle
PR and Buchler MW: Colorectal cancer. Lancet. 365:153–165. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lynch HT and de la Chapelle A: Hereditary
colorectal cancer. N Engl J Med. 348:919–932. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pagliuca A, Valvo C, Fabrizi E, et al:
Analysis of the combined action of miR-143 and miR-145 on oncogenic
pathways in colorectal cancer cells reveals a coordinate program of
gene repression. Oncogene. 32:4806–4813. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Prueitt RL, Yi M, Hudson RS, et al:
Expression of microRNAs and protein-coding genes associated with
perineural invasion in prostate cancer. Prostate. 68:1152–1164.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mavridis K, Stravodimos K and Scorilas A:
Downregulation and prognostic performance of microRNA 224
expression in prostate cancer. Clin Chem. 59:261–269. 2013.
View Article : Google Scholar
|
|
9
|
Huang L, Dai T, Lin X, et al: MicroRNA-224
targets RKIP to control cell invasion and expression of metastasis
genes in human breast cancer cells. Biochem Biophys Res Commun.
425:127–133. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
White NM, Chow TF, Mejia-Guerrero S, et
al: Three dysregulated miRNAs control kallikrein 10 expression and
cell proliferation in ovarian cancer. Br J Cancer. 102:1244–1253.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shen SN, Wang LF, Jia YF, Hao YQ, Zhang L
and Wang H: Upregulation of microRNA-224 is associated with
aggressive progression and poor prognosis in human cervical cancer.
Diagn Pathol. 8:692013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mees ST, Mardin WA, Sielker S, et al:
Involvement of CD40 targeting miR-224 and miR-486 on the
progression of pancreatic ductal adenocarcinomas. Ann Surg Oncol.
16:2339–2350. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Y, Lee AT, Ma JZ, et al: Profiling
microRNA expression in hepatocellular carcinoma reveals
microRNA-224 up-regulation and apoptosis inhibitor-5 as a
microRNA-224-specific target. J Biol Chem. 283:13205–13215. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ladeiro Y, Couchy G, Balabaud C, et al:
MicroRNA profiling in hepatocellular tumors is associated with
clinical features and oncogene/tumor suppressor gene mutations.
Hepatology. 47:1955–1963. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li Q, Wang G, Shan JL, et al: MicroRNA-224
is upregulated in HepG2 cells and involved in cellular migration
and invasion. J Gastroenterol Hepatol. 25:164–171. 2010. View Article : Google Scholar
|
|
16
|
Arndt GM, Dossey L, Cullen LM, et al:
Characterization of global microRNA expression reveals oncogenic
potential of miR-145 in metastatic colorectal cancer. BMC Cancer.
9:3742009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vega AB, Pericay C, Moya I, et al:
microRNA expression profile in stage III colorectal cancer:
Circulating miR-18a and miR-29a as promising biomarkers. Oncol Rep.
30:320–326. 2013.
|
|
18
|
Wang YX, Zhang XY, Zhang BF, Yang CQ, Chen
XM and Gao HJ: Initial study of microRNA expression profiles of
colonic cancer without lymph node metastasis. J Dig Dis. 11:50–54.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nikiforova MN, Tseng GC, Steward D, Diorio
D and Nikiforov YE: MicroRNA expression profiling of thyroid
tumors: biological significance and diagnostic utility. J Clin
Endocrinol Metab. 93:1600–1608. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lu S, Wang S, Geng S, Ma S, Liang Z and
Jiao B: Upregulation of microRNA-224 confers a poor prognosis in
glioma patients. Clin Transl Oncol. 15:569–574. 2013. View Article : Google Scholar
|
|
21
|
Li Z, Lu J, Sun M, et al: Distinct
microRNA expression profiles in acute myeloid leukemia with common
translocations. Proc Natl Acad Sci USA. 105:15535–15540. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rigou P, Piddubnyak V, Faye A, et al: The
antiapoptotic protein AAC-11 interacts with and regulates
Acinus-mediated DNA fragmentation. EMBO J. 28:1576–1588. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Koci L, Chlebova K, Hyzdalova M, et al:
Apoptosis inhibitor 5 (API-5; AAC-11; FIF) is upregulated in human
carcinomas in vivo. Oncol Lett. 3:913–916. 2012.PubMed/NCBI
|
|
24
|
Koelzer VH, Karamitopoulou E, Dawson H,
Kondi-Pafiti A, Zlobec I and Lugli A: Geographic analysis of RKIP
expression and its clinical relevance in colorectal cancer. Br J
Cancer. 108:2088–2096. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chatzigeorgiou A, Lyberi M, Chatzilymperis
G, Nezos A and Kamper E: CD40/CD40L signaling and its implication
in health and disease. Biofactors. 35:474–483. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Georgopoulos NT, Merrick A, Scott N, Selby
PJ, Melcher A and Trejdosiewicz LK: CD40-mediated death and
cytokine secretion in colorectal cancer: a potential target for
inflammatory tumour cell killing. Int J Cancer. 121:1373–1381.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nagtegaal ID, Quirke P and Schmoll HJ: Has
the new TNM classification for colorectal cancer improved care?
Nature reviews Clin Oncol. 9:119–123. 2012. View Article : Google Scholar
|
|
28
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
30
|
Camp RL, Dolled-Filhart M and Rimm DL:
X-tile: a new bio-informatics tool for biomarker assessment and
outcome-based cut-point optimization. Clin Cancer Res.
10:7252–7259. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chung DC: The genetic basis of colorectal
cancer: insights into critical pathways of tumorigenesis.
Gastroenterology. 119:854–865. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Colussi D, Brandi G, Bazzoli F and
Ricciardiello L: Molecular pathways involved in colorectal cancer:
implications for disease behavior and prevention. Int J Mol Sci.
14:16365–16385. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huntzinger E and Izaurralde E: Gene
silencing by microRNAs: contributions of translational repression
and mRNA decay. Nat Rev Genet. 12:99–110. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tan Y, Zhang B, Wu T, et al:
Transcriptional inhibiton of Hoxd4 expression by miRNA-10a in human
breast cancer cells. BMC Mol Biol. 10:122009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hawkins PG and Morris KV: RNA and
transcriptional modulation of gene expression. Cell Cycle.
7:602–607. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mlcochova J, Faltejskova P, Nemecek R,
Svoboda M and Slaby O: MicroRNAs targeting EGFR signalling pathway
in colorectal cancer. J Cancer Res Clin Oncol. 139:1615–1624. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ma Q, Wang X, Li Z, et al: microRNA-16
represses colorectal cancer cell growth in vitro by regulating the
p53/survivin signaling pathway. Oncol Rep. 29:1652–1658.
2013.PubMed/NCBI
|
|
38
|
Ma Y, Li W and Wang H: Roles of miRNA in
the initiation and development of colorectal carcinoma. Curr Pharm
Des. 19:1253–1261. 2013.
|
|
39
|
Yamada N, Noguchi S, Mori T, Naoe T, Maruo
K and Akao Y: Tumor-suppressive microRNA-145 targets catenin
delta-1 to regulate Wnt/beta-catenin signaling in human colon
cancer cells. Cancer Lett. 335:332–342. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fu J, Tang W, Du P, et al: Identifying
microRNA-mRNA regulatory network in colorectal cancer by a
combination of expression profile and bioinformatics analysis. BMC
Syst Biol. 6:682012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Nagel R, le Sage C, Diosdado B, et al:
Regulation of the adenomatous polyposis coli gene by the miR-135
family in colorectal cancer. Cancer Res. 68:5795–5802. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yu Y, Kanwar SS, Patel BB, et al:
MicroRNA-21 induces stemness by downregulating transforming growth
factor beta receptor 2 (TGFbetaR2) in colon cancer cells.
Carcinogenesis. 33:68–76. 2012. View Article : Google Scholar :
|
|
43
|
Dong Y, Wu WK, Wu CW, Sung JJ, Yu J and Ng
SS: MicroRNA dysregulation in colorectal cancer: a clinical
perspective. Br J Cancer. 104:893–898. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhai H and Ju J: Implications of microRNAs
in colorectal cancer development, diagnosis, prognosis, and
therapeutics. Front Genet. 2:pii00078. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Menendez P, Villarejo P, Padilla D,
Menendez JM and Rodriguez-Montes JA: Implications of the
histological determination of microRNAs in the screening, diagnosis
and prognosis of colorectal cancer. J Surg Oncol. 108:70–73. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Schetter AJ and Harris CC: Alterations of
microRNAs contribute to colon carcinogenesis. Semin Oncol.
38:734–742. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Madhavan D, Cuk K, Burwinkel B and Yang R:
Cancer diagnosis and prognosis decoded by blood-based circulating
microRNA signatures. Front Genet. 4:1162013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Bartley AN, Yao H, Barkoh BA, et al:
Complex patterns of altered MicroRNA expression during the
adenoma-adenocarcinoma sequence for microsatellite-stable
colorectal cancer. Clin Cancer Res. 17:7283–7293. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kalimutho M, Del Vecchio Blanco G, Di
Cecilia S, et al: Differential expression of miR-144* as
a novel fecal-based diagnostic marker for colorectal cancer. J
Gastroenterol. 46:1391–1402. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Xiao YF, Yong X, Fan YH, Lu MH, Yang SM
and Hu CJ: microRNA detection in feces, sputum, pleural effusion
and urine: Novel tools for cancer screening (Review). Oncol Rep.
30:535–544. 2013.PubMed/NCBI
|
|
51
|
Wu CW, Ng SS, Dong YJ, et al: Detection of
miR-92a and miR-21 in stool samples as potential screening
biomarkers for colorectal cancer and polyps. Gut. 61:739–745. 2012.
View Article : Google Scholar
|
|
52
|
Cho WC: Epigenetic alteration of microRNAs
in feces of colorectal cancer and its clinical significance. Expert
Rev Mol Diagn. 11:691–694. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kalimutho M, Di Cecilia S, Del Vecchio
Blanco G, et al: Epigenetically silenced miR-34b/c as a novel
faecal-based screening marker for colorectal cancer. Br J Cancer.
104:1770–1778. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Srivastava K and Srivastava A:
Comprehensive review of genetic association studies and
meta-analyses on miRNA polymorphisms and cancer risk. PLoS One.
7:e509662012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yao G, Yin M, Lian J, et al: MicroRNA-224
is involved in transforming growth factor-beta-mediated mouse
granulosa cell proliferation and granulosa cell function by
targeting Smad4. Mol Endocrinol. 24:540–551. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Li X, Shen Y, Ichikawa H, Antes T and
Goldberg GS: Regulation of miRNA expression by Src and contact
normalization: effects on nonanchored cell growth and migration.
Oncogene. 28:4272–4283. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wang Y, Ren J, Gao Y, et al: MicroRNA-224
targets SMAD family member 4 to promote cell proliferation and
negatively influence patient survival. PLoS One. 8:e687442013.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Miyaki M and Kuroki T: Role of Smad4
(DPC4) inactivation in human cancer. Biochem Biophys Res Commun.
306:799–804. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Al-Mulla F, Bitar MS, Taqi Z and Yeung KC:
RKIP: much more than Raf kinase inhibitory protein. J Cell Physiol.
228:1688–1702. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Mencia N, Selga E, Noe V and Ciudad CJ:
Underexpression of miR-224 in methotrexate resistant human colon
cancer cells. Biochem Pharmacol. 82:1572–1582. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Link A, Balaguer F, Shen Y, et al: Fecal
MicroRNAs as novel biomarkers for colon cancer screening. Cancer
Epidemiol Biomarkers Prev. 19:1766–1774. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Cheng H, Zhang L, Cogdell DE, et al:
Circulating plasma MiR-141 is a novel biomarker for metastatic
colon cancer and predicts poor prognosis. PLoS One. 6:e177452011.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ng EK, Chong WW, Jin H, et al:
Differential expression of microRNAs in plasma of patients with
colorectal cancer: a potential marker for colorectal cancer
screening. Gut. 58:1375–1381. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Huang Z, Huang D, Ni S, Peng Z, Sheng W
and Du X: Plasma microRNAs are promising novel biomarkers for early
detection of colorectal cancer. Int J Cancer. 127:118–126. 2010.
View Article : Google Scholar
|
|
65
|
Yuan K, Xie K, Fox J, et al: Decreased
levels of miR-224 and the passenger strand of miR-221 increase
MBD2, suppressing maspin and promoting colorectal tumor growth and
metastasis in mice. Gastroenterology. 145:853–864. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Hyslop T and Waldman SA: Molecular staging
of node negative patients with colorectal cancer. J Cancer.
4:193–199. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Liao WT, Li TT, Wang ZG, et al:
microRNA-224 promotes cell proliferation and tumor growth in human
colorectal cancer by repressing PHLPP1 and PHLPP2. Clin Cancer Res.
19:4662–4672. 2013. View Article : Google Scholar : PubMed/NCBI
|