Introduction
Although global statistics show that gastric cancer
is the fourth most common cancer in men and the fifth most common
cancer in women, the mortality from gastric cancer ranks third to
lung and liver cancer (1).
Advances in diagnosis and therapeutics have improved long-term
survival for early gastric cancer; however, the prognosis of
advanced and metastatic gastric cancer is still not satisfactory. A
better understanding of the molecular pathologies of metastatic
gastric cancer will lead to progress in both diagnostics and
treatment.
Metastasis is a critical determinant of cancer
morbidity. Tumor cell metastasis is a multiphasic process including
cell transformation, growth, angiogenesis, invasion, dissemination
and survival in the circulation and cell growth at distant sites
from the primary tumor location. Each step requires distinct
molecular program regulating the interaction between the tumor
cells, the extracellular matrix (ECM) and the cells of the stroma.
Molecules participating in these steps can also be considered as
potential prognostic factors. Diverse molecular regulators,
including adhesion receptor families, receptor tyrosine kinase,
cytoskeleton protein, adapters and signaling molecules, are
involved in the regulation of migration and invasion to tumor cells
(2).
The carboxyl terminus of Hsc-70-interacting protein
(CHIP), encoded by the STUB1 gene, was first cloned and
characterized in human heart (3).
CHIP contains two important domains. The N-terminal domain of CHIP
consists of three tetratricopeptide repeat (TPR) domains mediating
protein-protein interactions, and a U-box domain at its C-terminus
displaying E3 ubiquitin ligase activity (4). CHIP functions in maintaining the
protein homeostasis in the cytoplasm by promoting the
ubiquitination and depredation of chaperone-bound and misfolded
proteins including glucocorticoid receptor (GR), cystic fibrosis
transmembrane conductance regulator (CFTR) and mutated superoxide
dismutase 1 (SOD1) (5–7). CHIP is also involved in the
degradation of multiple oncogenic proteins.
Though the exact function of CHIP in tumorigenesis
has not been uncovered, accumulated evidence indicates that CHIP
predominantly functions as a tumor suppressor. Several studies
indicate that CHIP expression can be considered as an independent
prognostic factor for cancer patients including gastric, breast and
colorectal cancers (8–10). In human colorectal cancer cells,
CHIP inhibits the malignancy possibly through negatively regulating
the NF-κB activity (10). In
breast cancer cells, CHIP is implicated in the modulation the
stability of estrogen receptor (ESRI) and Her-2/neu (ERBB2). CHIP
silencing in breast cancer cells results in rapid tumor growth and
other malignant phenotypes (11).
CHIP overexpression in the MDA-MB-231 breast cancer cells promotes
the ubiquitination of TNF receptor-associated factor 2 (TRAF2) and
subsequently inactivates the TRAF2-NF-κB signaling (12). In gastric cancer cells, it has been
reported that CHIP can degrade the classical NF-κB member RelA/p65
and inhibit the IL-8-induced angiogenesis (13).
However, the role of CHIP in the metastasis of
gastric cancer is still poorly understood. In this study, we
investigated the functional importance and potential mechanism of
CHIP in the migration and invasion in gastric cancer.
Materials and methods
Clinical samples
Gastric cancer tissues were obtained, with written
consent, from 164 gastric cancer patients at the Department of
General Surgery, the First Affiliated Hospital of Soochow
University, from 2008 to 2013. All patients had an accurate
pathology diagnosis and underwent surgery without chemotherapy or
radiotherapy before operation.
Tissue culture
The human gastric cancer cell line AGS was purchased
from American Type Culture Collection (ATCC). All cell lines were
cultured in F-12 (Hyclone, HAM’S/F-12) supplemented with 10% fetal
bovine serum (FBS), 100 U/ml penicillin, 100 μg/ml streptomycin and
2 mM glutamine. The cells were cultured at 37°C in a humidified
atmosphere of 5% CO2.
Transfection
CHIP overexpression vector was constructed based on
the MSCV vector containing GFP. Full-length CDS of CHIP was
amplified by PCR from human breast tissue and cloned into MSCV-GFP
vector (MSCV-GFP-hCHIP). Primer sequences of CHIP are as follows:
5′-AAAAAGATCTGGCGGCATGAAGGGCAAGG-3′ and
5′-GGGAACCTCAGTAGTCCTCCACCC-3′. Phoenix A packaging cells were
transfected with MSCV-GFP-hCHIP or control vector (MSCV-GFP) by
FuGENE HD (Roche, China). Lentivirus supernatants were used to
infect the target cells.
Quantitative real-time PCR (qRT-PCR)
Equivalent amounts of RNA (2 μg) were
reverse-transcribed with Superscript M-MLV (Promega, China).
Triplicates were performed for all qRT-PCR reactions with a
LightCycler 480 System (Roche). Primers for qRT-PCR were designed
using Primer-BLAST (PubMed). Primers were synthesized from
Invitrogen (China). The housekeeping gene (β-actin) and
target genes were reverse transcribed together in a single run. The
reaction with 2X LC480 SYBR Green I Master Mix (Roche) was
according to the manufacturer’s protocol. For data analysis, a
target gene transcript was quantified in comparison to the house
keeping gene (β-actin) as a reference.
Western blot analysis
Whole-cell extracts were prepared with RIPA buffer
according to standard procedures. Proteins were fractionated on an
SDS-PAGE gel and transferred to PVDF membranes. After blocking,
membranes were probed with different antibodies (Abs). Membranes
were then washed and incubated with an appropriate secondary
antibody. Proteins were detected and scanned with the Gel Imaging
system (PeiQing, China). Band density was normalized to the
α-tubulin, actin or GAPDH reference. Abs against Bcl-2 (sc-7382),
Cyclin D1 (sc-20044), TRAF-2 (sc-876), RelA (sc-372), p50
(sc-7178), p100/p52 (sc-3017), RelB (sc-226) and c-Rel (sc-70) were
purchased from Santa Cruz Biotechnology. Abs against Bim (#2819),
AKT (#4691), p-AKT (Ser473) (#4060), p-AKT (Thr308) (#2965), TRAF-3
(#4729), ITGB1 (#9699), MMP-2 (#4022), MMP-9 (#2270) and CHIP
(#2080) were obtained from Cell Signaling Technology. α-tubulin
(AJ1034a), actin (AT0001) and GAPDH (#5174) were purchased from
Abgent.
Cell growth assay
Cell growth was performed with the xCelligence RTCA
instrument (Roche). In this assay, impedance for indicated times
was continuously monitored by the system and the value was
indicated as ‘Cell index’ which was determined by the number of
cells seeded, the overall size and morphology of the cells and the
degree to which the cells interact with the sensor surface. Cell
Index was continuously monitored by the system and data were
collected and analyzed by RTCA software 1.2.
Cell cycle analysis
The cells were detected using flow cytometry. After
48-h continuous culture, cells were harvested and fixed by 70%
ethanol for 24 h at 4°C. Then single cell suspensions were prepared
to stain DNA using PI staining based on the manufacturer’s
instructions. Cell cycle was measured by FACSCalibur (BD
Biosciences, China) with at least three independent experiments
performed.
Ki-67 assay
Cells were cultured in F-12 medium supplemented with
10% fetal bovine serum on cover slides. After continuous culture
for 24, 48 and 72 h, the slides were fixed in cold 4%
paraformaldehyde for 30 min and treated with 1% Triton PBS solution
for 10 min. Then 1X PBS with 10% FBS was used to block the cells
for 45 min at room temperature. Cells were incubated with a Ki-67
antibody (1:100, Santa Cruz Biotechnology, USA). Following 1X PBS
washing, sections were incubated for 30 min using the secondary
antibody (rabbit anti-mouse IgA-B, GK500705, GTVision III).
Finally, 3,3-diaminobenzine (DAB) was used to visualize the
immunoreactive products. Results were evaluated by the System
Microscope IX71 (Olympus, Japan).
Terminal nucleotidyl transferase-mediated
nick end labeling (TUNEL) assay
Cells were cultured on cover slides for 24, 48 and
72 h in a humidified incubator at 37°C and 5% CO2.
According to the manufacturer’s instructions of TUNEL system kit
(Roche), the slides were fixed by cold 4% paraformaldehyde for 30
min. Following 1X PBS washing, 3% H2O2
methanol solution was used to block the slides for 10 min at 20°C.
Then the slides were treated using 1% Triton PBS solution for 2 min
on ice after PBS washing. Avoiding light, 50 μl of TUNEL reaction
solution was applied to incubate the cells on slides for 60 min at
37°C. Following PBS washing, the signals of TUNEL were converted
using peroxidase (POD) for 30 min at 37°C and the sections were
treated with DAB for 3 min at room temperature. Results were
examined by the light System Microscope IX71 (Olympus, Japan).
Cell migration assay
Cell migration was also performed with the
xCelligence RTCA instrument (Roche). In this assay, CIM-plate with
upper chamber and lower chamber was used. F-12 (180 μl)
supplemented with 10% FBS was added in each wells on the lower
chamber. Cells were suspended in F-12 FBS-free media, 60,000
cells/well were added in the upper chamber. After attachment, cell
migration towards lower chamber containing 10% FBS media was
continuously monitored by the system and data were collected and
analyzed by RTCA 1.2 software.
Scratch healing assay
For the wound healing assays, cells were treated
with 10 mg/ml mitomycin C (Sigma, USA) for 3 h. Then, cells were
wounded using a pipette tip, and F-12 with 10% FBS was added. The
wound closure was observed for 48 h, with the light System
Microscope IX71. The wound healing ability was calculated, compared
to the width of wound closure for 0 h.
Cell invasion assay
Cell invasion was also performed with the
xCelligence RTCA instrument (Roche). In this assay, CIM-plate with
two chambers were used. F-12 (180 μl) supplemented with 10% FBS was
added to each well in the lower chamber. Cells were suspended in
F-12 FBS-free media, then 60,000 cells/well were added to the
uppper chamber. After attachment, cells invading through Matrigel
(cat. no. 356234, BD Biosecience, China) towards lower chamber
containing 10% FBS media were continuously monitored by the system
and data were collected and analyzed by RTCA 1.2 software.
Gelatinase zymography
For the assessment of MMP-2 and −9, gelatin
zymography assays were used. Gelatinase zymography was performed in
an 8% SDS-PAGE gel in the presence of 0.1% gelatin under
non-reducing conditions. Culture media with sample buffer were
loaded for SDS-PAGE with Tris-glycine SDS buffer. Samples were not
boiled before electrophoresis. Following electrophoresis, the gels
were washed twice in 2.5% Triton X-100 for 30 min at room
temperature to remove SDS. The gels were then incubated at 37°C
overnight in substrate buffer containing 50 mM Tris-HCl and 10 mM
CaCl2 at pH 8.0 and stained with 0.5% Coomassie Blue
R250 in 50% methanol and 10% glacial acetic acid for 30 min and
destained. Upon renaturation of the enzyme, the gelatinases
digested the gelatin to produce clear bands against an intensely
stained background.
Immunohistochemistry (IHC)
Tissue IHC was performed using a standard
peroxidase-based staining method. Tissue sections (4 μm) were
incubated in a dry oven at 60°C for 1 h and then dewaxed in xylene
for 3×10 min, rehydrated with graded ethanol in 100, 100, 95, 90,
80 and 70% ethanol for 5 min each. Then antigen retrieval was
performed by pretreatment of the slides in 0.01 M citrate buffer
(pH 6.0) using a microwave oven. Subsequently, the sections were
treated with 3% hydrogen peroxide (H2O2) for
10 min in order to block endogenous peroxidase. The sections were
washed with 1X phosphate buffered saline (PBS) (pH 7.4) and were
incubated with rabbit anti-CHIP antibody (dilution 1:200; CST)
overnight at 4°C. The sections were then washed with 1X PBS and
incubated with biotinylated goat anti-rabbit IgG. For each sample,
the omission of primary antibody was used as a negative control.
Finally, 3, 3-diaminobenzine (DAB) was used to visualize the
immunoreactive products. The results were evaluated by the System
Microscope IX71. The staining index was expressed as the proportion
of positive staining cells (<25%, +; 25–50%, ++; 50–75%, +++;
>75%, ++++).
Statistical analysis
All experiments were repeated at least three times.
The data were expressed as the mean ± standard deviation from
experiments in replicate. All statistical analysis was performed by
Graphpad software. The differences between groups were valued using
Student’s t-test. A P-value of ≤0.05 was considered significant and
a P-value of ≤0.01 as highly significant.
Results
Establishing a CHIP overexpressing cell
line
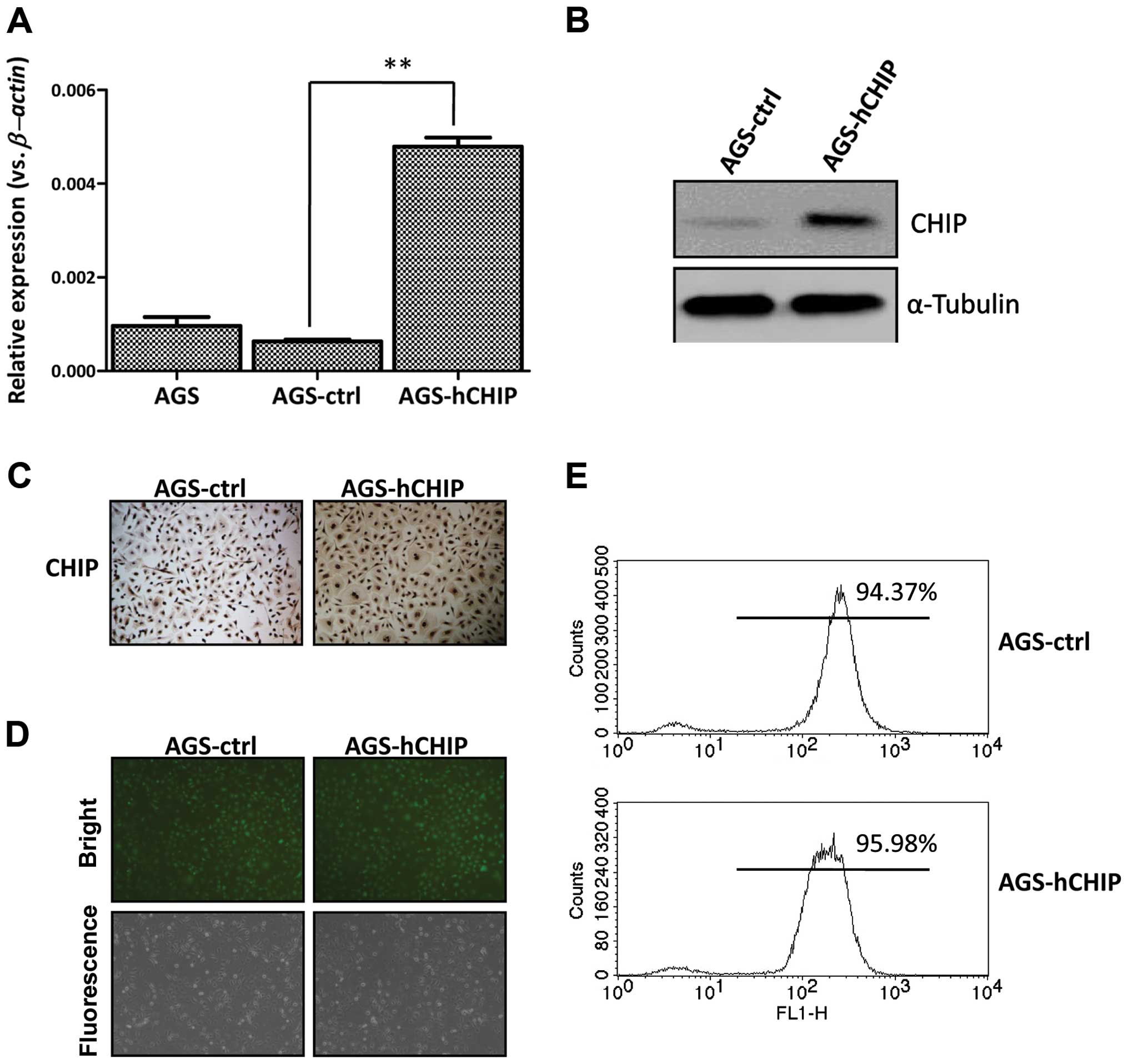
To establish a CHIP overexpressing cell line, the
reconstructed plasmid carrying both green fluorescent protein (GFP)
and human CHIP cDNA were transfected into the AGS human
gastric cancer cells. The individual monoclones were subsequently
selected by puromycin with a final concentration of 5 ng/μl for two
weeks. In parallel, an AGS cell line expressing GFP only was
established as a control. The selected monoclones were further
expanded and examined for the CHIP expression by qRT-PCR. As
shown in Fig. 1A, the CHIP
expression at mRNA level in a single clone, which was transfected
with the CHIP-expressing plasmid (AGS-CHIP), was significantly
higher than that in the control cells (AGS-control). The CHIP
expression at protein level was also markedly increased in the
AGS-CHIP cells compared to that in the AGS-control cells by western
blotting (Fig. 1B). The induced
expression of CHIP was further confirmed by an IHC assay (Fig. 1C). When observed under fluorescence
microscopy, both the established AGS-CHIP and the AGS-control cells
presented strong GFP signals (Fig.
1D). The purities of the two established cell lines were 94.37
and 95.98%, respectively, which were examined by flow cytometry
(Fig. 1E). This indicated that the
AGS gastric cell line overexpressing the human CHIP was established
successfully.
CHIP overexpression inhibits AGS cell
growth
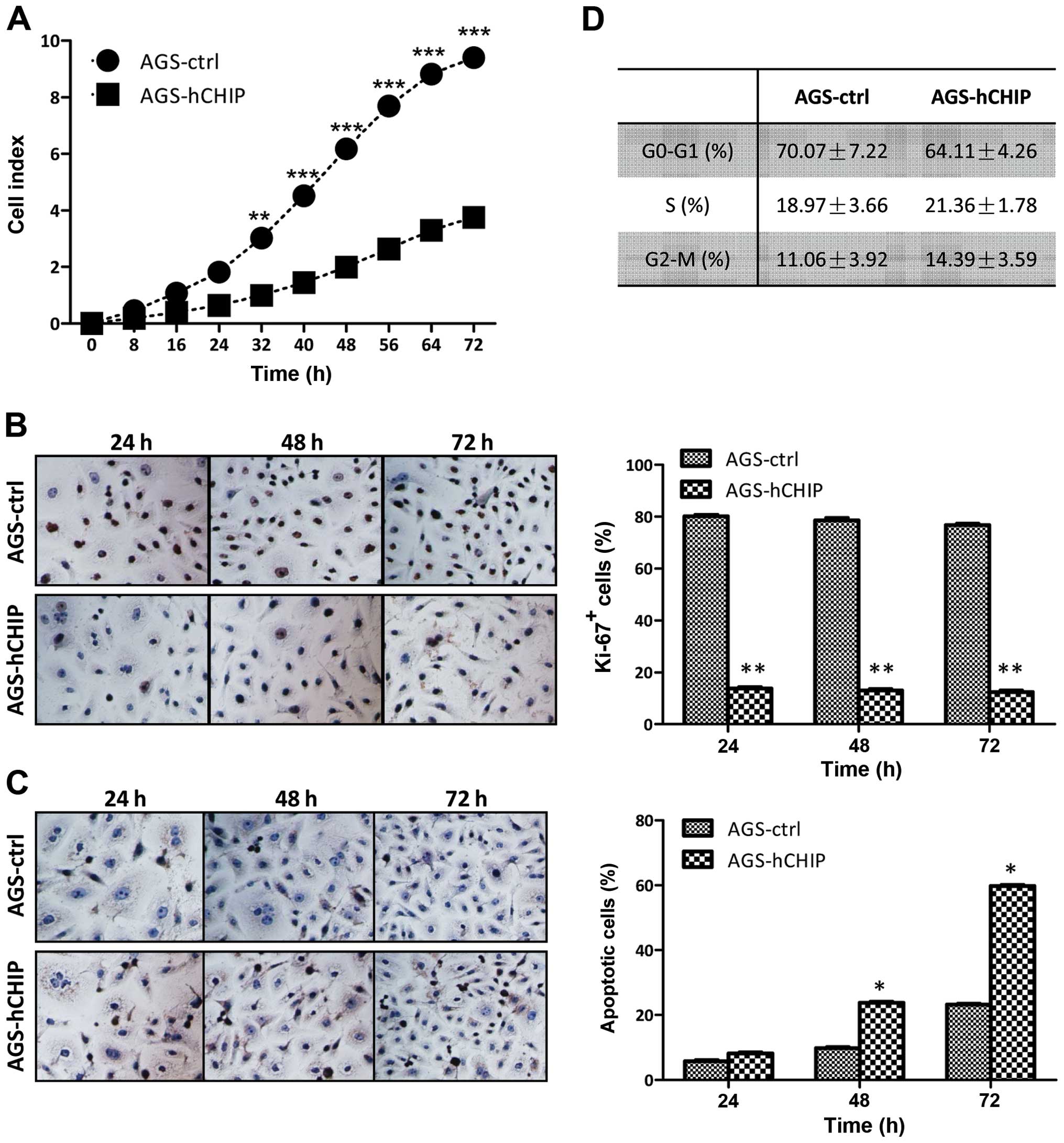
The cell growth affected by the CHIP overexpression
was detected by a real-time xCelligence system using E-plates. The
AGS cells overexpressing CHIP grew much slower than that of the
AGS-control cells and there was a statistically significant
difference between the two established cell lines during the 72-h
continuous monitoring (Fig. 2A).
The cell cycle analysis and cellular DNA content measurement were
examined by flow cytometry and no obvious differences were observed
between the AGS-hCHIP and the AGS-control cells in any of the three
phases (G0–G1, S and G2-M)
(Fig. 2B). To investigate whether
CHIP overexpression affects the proliferation capability of the AGS
cells, a Ki-67 cell proliferation assay was performed. The
frequencies of Ki-67-positive cells in the AGS-hCHIP group were
13.78±0.50, 13.11±0.50 and 12.45±0.70%; while those of
Ki-67-positive cells in the AGS-control group were 80.11±0.7,
78.56±1.00 and 76.78±0.50% at 24, 48 and 72 h, respectively
(Fig. 2C). The TUNEL assay was
carried out to quantitatively analyze the apoptotic cells. As
showed in Fig. 2D, both the
AGS-hCHIP and the AGS-control cells underwent apoptosis in a
time-dependent manner. The percentages of apoptotic cells in the
AGS-hCHIP group were 8.22±0.50, 23.78±1.50 and 59.78±3.50%; while
those of apoptotic cells in the AGS-control cells were 5.78±0.50,
9.78±0.80 and 23.22±1.50% at 24, 48 and 72 h, respectively. Taken
together, it is shown that CHIP overexpression in AGS cells led to
increased apoptosis and decreased cellular proliferation.
Therefore, CHIP plays a pivotal role in cell growth of the AGS
gastric cancer cells due to regulation of apoptosis and cellular
proliferation.
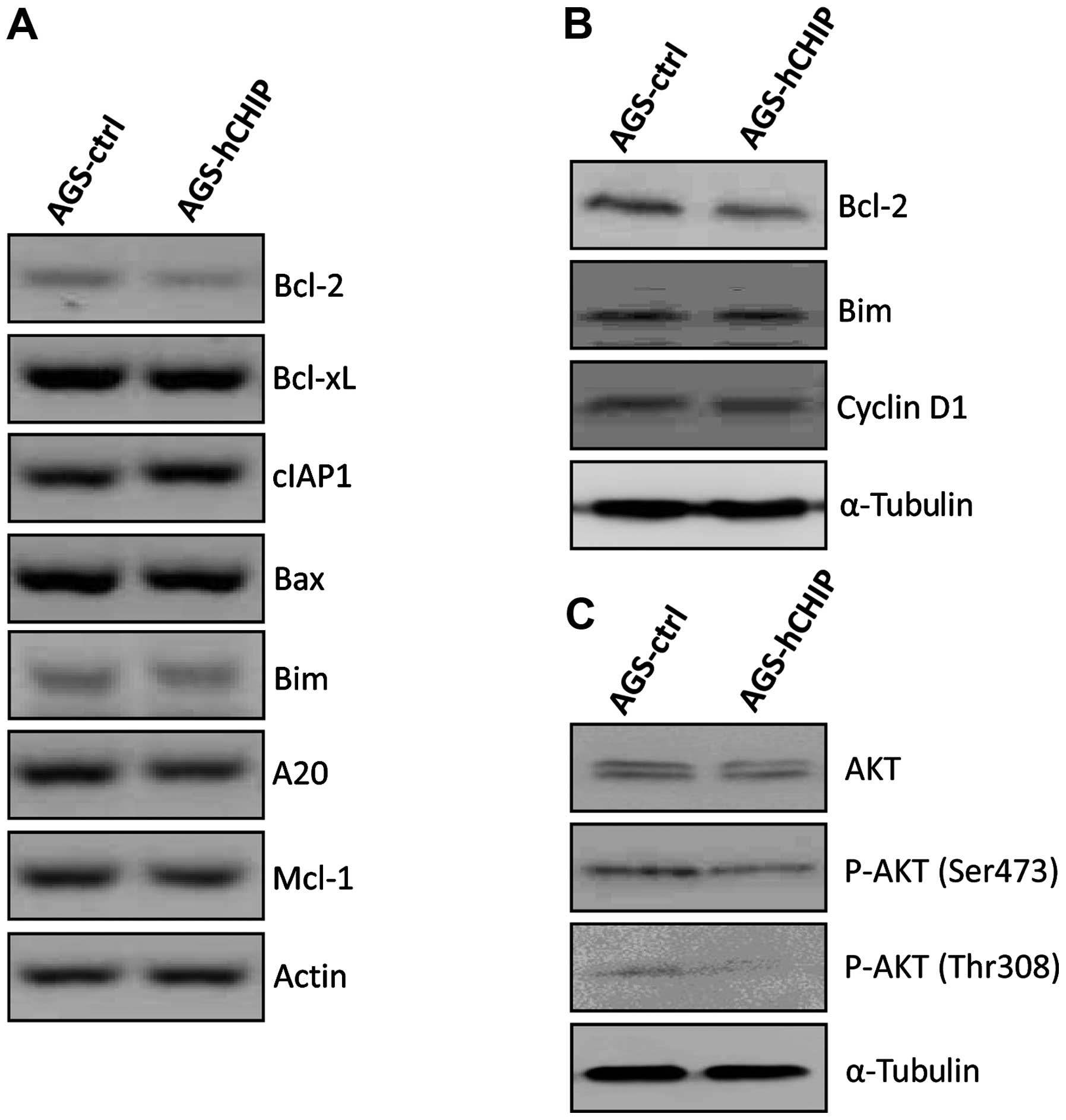
CHIP overexpression regulates Bcl-2
expression
The mRNA expression of the pro-apoptotic genes
including Bax, Bim and Mcl-1, as well as the
anti-apoptotic genes including Bcl-2, Bcl-xl, cIAP1 and
A20, were measured by qRT-PCR. As shown in Fig. 3A, the expression of the
Bcl-2 gene at the mRNA level was markedly decreased in the
AGS-hCHIP cells compared to that of the AGS-control cells. The
reduced Bcl-2 expression was also detected at the protein level by
western blotting in the AGS-hCHIP cells (Fig. 3B). No significant changes were
observed in other apoptosis-associated genes including Bax, Bim,
Mcl-1, Bcl-xl, cIAP1 and A20 at mRNA level. The
expression of Cyclin D1 was also comparable between the AGS-control
and AGS-hCHIP cells (Fig. 3B).
The AKT signaling molecules were examined to
investigate whether the AKT signaling pathway is involved in the
cellular proliferation affected by the CHIP overexpression. The
expression level of AKT was similar in the whole-cell extracts of
the AGS-control and AGS-hCHIP cells. However, CHIP overexpression
led to a clear reduction in the expression levels of p-AKT (Ser473)
and p-AKT (Thr 308) (Fig. 3C).
Taken together, these results indicated that the increased
apoptosis in the CHIP-overexpressing AGS cells is likely attributed
to the inhibition of the Bcl-2 expression. The suppressed
phosphorylation of AKT due to CHIP overexpression is involved in
the reduced cellular proliferation.
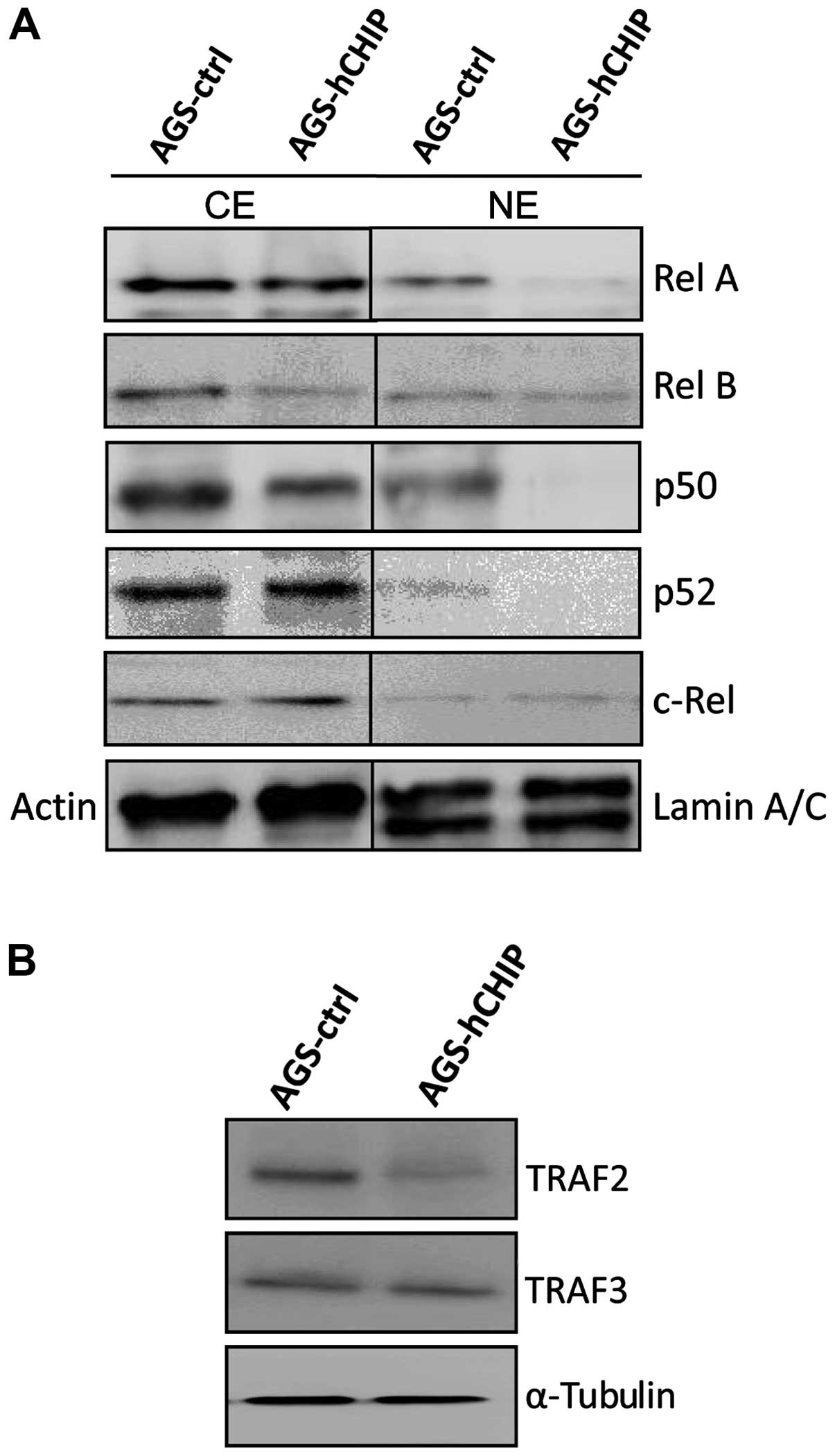
CHIP negatively regulates the NF-κB
signaling
The expression of RelA/p65 and p50, representing the
canonical NF-κB activities, was clearly decreased in both the
cytoplasmic (CE) and nuclear fraction (NE) in the AGS-hCHIP cells
compared to that in the AGS-control cells. Moreover, the expression
of RelB, representing the non-canonical NF-κB activity, was clearly
reduced in the cytoplasmic fraction but not in nuclear fraction of
the AGS-hCHIP cells. However, no obvious changes in the expression
of p52 and c-Rel were observed between the AGS-hCHIP cells and the
AGS-control cells (Fig. 4A).
TRAF2, which positively regulates both the canonical and the
non-canonical NF-κB activity, was also slightly decreased in the
AGS-hCHIP cells. TRAF3, which negatively regulates the
non-canonical NF-κB activity, was not affected by the
overexpression of CHIP (Fig. 4B).
Taken together, it was indicated that CHIP overexpression
suppressed the expression of TRAF2, which further inhibited not
only the classical NF-κB activity but also the alternative NF-κB
activity in the AGS gastric cancer cells.
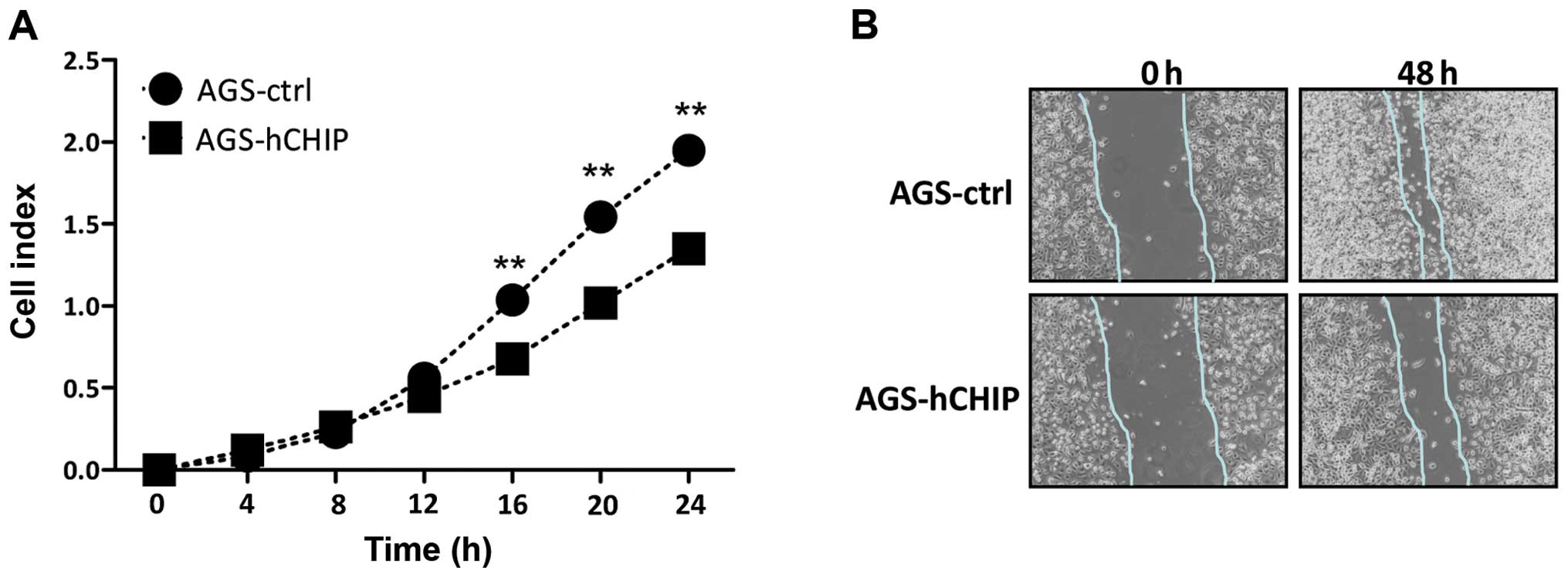
CHIP overexpression attenuates the
migration ability of AGS gastric cancer cells
The migration ability affected by the overexpression
of CHIP was investigated by a real-time xCelligence system using
CIM-plates. The AGS cells over-expressing CHIP migrated markedly
slower than that of the AGS-control cells during the 24-h
continuous monitoring and there was a statistically significantly
difference in the migration assay between the two established cell
lines (Fig. 5A). The in
vitro scratch assay was also performed to measure
quantitatively the migration ability of the cells. A scratched cell
monolayer was created in both cell lines and the images were
captured at the beginning and after 48 h. At 48 h it was shown that
the AGS-hCHIP cells migrated from the edge towards the scratch
center much slower than that of the AGS-control cells (Fig. 5B). Thus, the overexpression of CHIP
clearly inhibited the migration ability of the AGS cells.
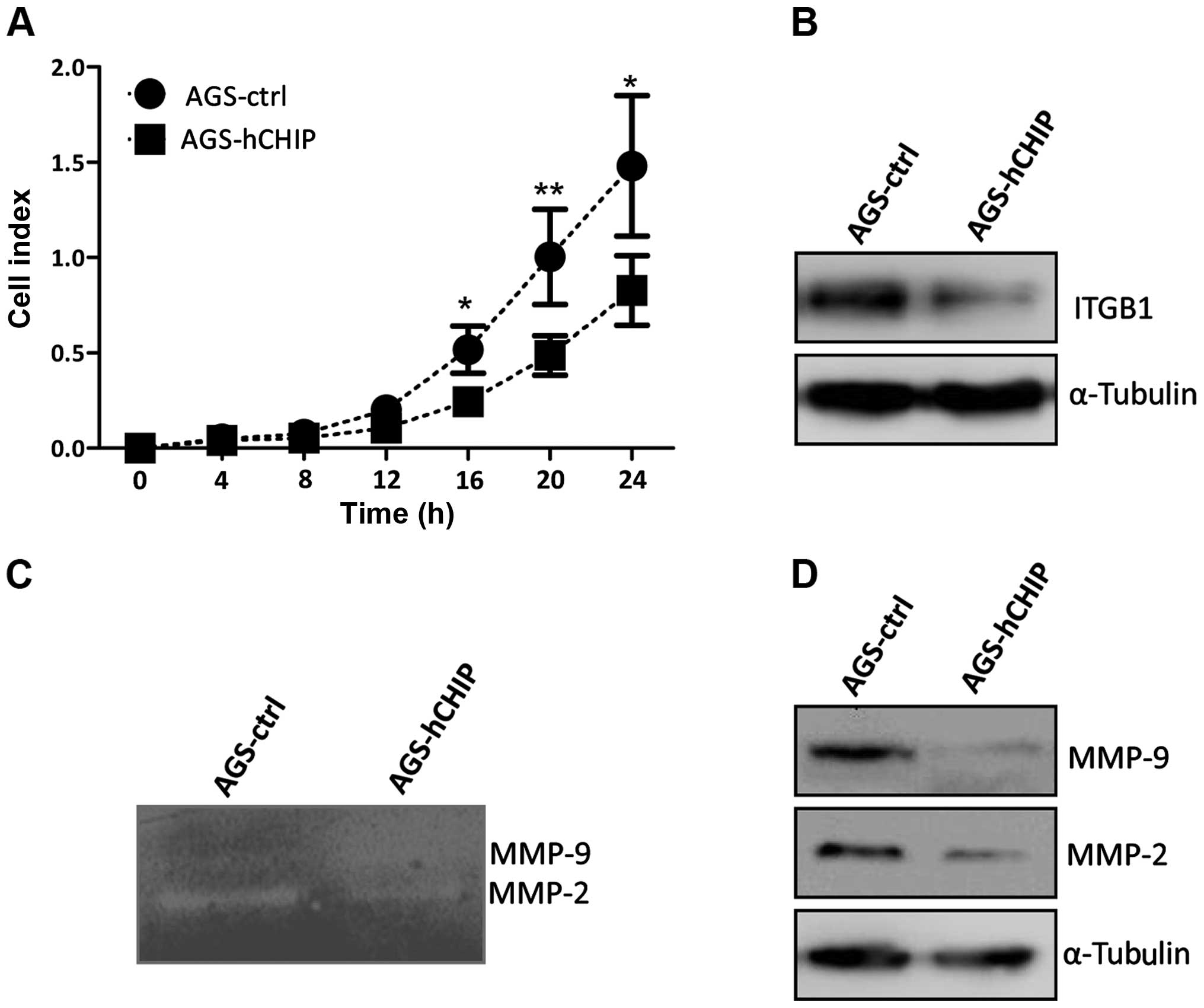
CHIP overexpression affects the invasion
ability of AGS gastric cancer cells
The invasion ability affected by the overexpression
of CHIP was also measured by a real-time xCelligence system using
Matrigel (dilution at 1:40)-coated CIM-plates. The AGS cells
overexpressing CHIP invaded through Matrigel markedly slower than
that of the AGS-control cells and there was a statistically
significant difference in the invasion ability between the
established cell lines during the 24-h continuous monitoring
(Fig. 6A). The integrin β-1
expression at the protein level was also dramatically decreased in
the AGS-hCHIP cells compared to that of the AGS-control cells
(Fig. 6B). The gelatin zymography
experiment was further performed to explore the relative amounts of
active and inactive gelatinase (MMP-2 or MMP-9). As showed in
Fig. 5B, both the MMP-2 and −9
activities were decreased in the AGS-hCHIP cells compared to the
AGS-control cells (Fig. 6C).
Moreover, the expression of MMP-2 and −9 in the AGS-hCHIP cells at
protein level was also significantly lower than the AGS-control
cells (Fig. 6D). Thus, the results
here indicated that overexpression of CHIP suppressed the migration
and invasion ability of the AGS gastric cancer cells, which was
correlated with the decreased activity of MMP-2 and −9 and the
downregulated integrin β-1 expression.
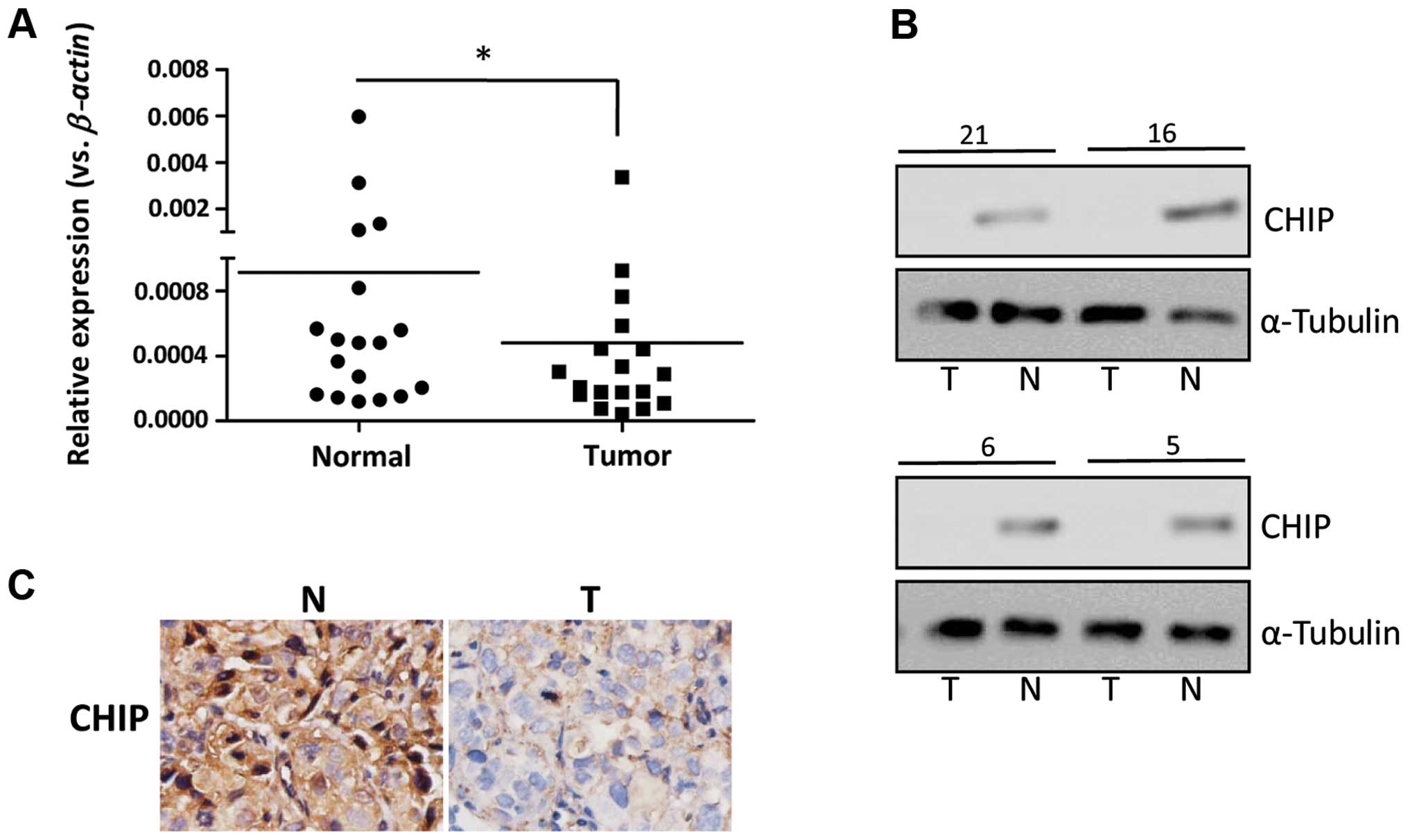
CHIP expression in human gastric cancer
tissues
The expression of CHIP was examined in individual
human gastric cancer tissues. As shown in Fig. 7A, the average mRNA level of the
CHIP gene detected in 18 gastric cancer tissues was clearly
lower than that of the paired normal gastric mucosa by qRT-PCR. The
western blotting was performed to examine the expression of CHIP in
gastric tumor tissues at the protein level. Representative results
are presented in Fig. 7B, showing
that the expression of CHIP was detected in the normal tissues
while the expression of CHIP was decreased in most of the gastric
cancer samples or was hardly detected. The expression of CHIP was
detected in both the nucleus and the cytoplasm of the normal
tissues by IHC assay, while the expression of CHIP was
predominantly localized in cytoplasm of the gastric tumor cells.
Moreover, a strong staining of CHIP was detected in the gastric
normal mucosa, while a relatively weak staining of CHIP was found
in the gastric tumor cells (Fig.
7C). A total of 164 gastric cancer samples were assayed for the
CHIP expression by IHC and the correlation between the expression
of CHIP and the clinical features was investigated. As shown in
Table I, the expression level of
CHIP in gastric cancer tissue was negatively correlated with the
differentiation status and the TNM stages of the gastric cancer
patients, but was not correlated with age, gender and tumor
diameters. These results indicated that CHIP expression was
frequently decreased in gastric cancer tissues and the decreased
expression of CHIP was an indicator of an unfavorable
prognosis.
 | Table IAssociation of CHIP expression with
clinicopatho-logic features of 164 gastric cancers patients. |
Table I
Association of CHIP expression with
clinicopatho-logic features of 164 gastric cancers patients.
|
Characteristics | No. of patients
(total: 164) | CHIP expression
+-++ | CHIP expression
+++-++++ | P-value |
|---|
| Age (years) | | | | |
| ≤60 | 45 | 22 | 23 | 0.834 |
| >60 | 119 | 56 | 63 | |
| Gender | | | | |
| Male | 66 | 30 | 36 | 0.279 |
| Female | 88 | 33 | 55 | |
| Tumor diameter
(cm) | | | | |
| ≤5 | 87 | 36 | 51 | 0.306 |
| >5 | 77 | 38 | 39 | |
|
Differentiation | | | | |
|
Well-moderately | 96 | 78 | 18 | <0.001 |
| Poorly | 68 | 6 | 62 | |
| TNM stage | | | | |
| I–II | 70 | 23 | 47 | 0.002 |
| III–IV | 84 | 49 | 35 | |
Discussion
In this study, we show that CHIP overexpression
clearly affected the ability of migration and invasion of the AGS
gastric cancer cells, which was associated with the decreased
activities of MMP-2 and −9 and the downregulated integrin β-1. Not
only the canonical NF-κB but also the non-canonical NF-κB activity
was influenced by the presence of CHIP overexpression. TRAF2, which
positively regulates both NF-κB signaling, was also clearly
reduced. In addition, we show that CHIP played a critical role in
cell growth of the AGS gastric cancer cells via regulating cellular
proliferation and survival.
In the CHIP-overexpressing AGS gastric cancer cells,
the reduction of TRAF2 as well as NF-κB subunits was observed. A
previous report suggested that CHIP affects NF-κB-mediated cell
invasion via regulating TRAF2 expression in the breast cancer cells
(12). Particularly, RelA/p65,
representing the classical NF-κB activity was found downregulated
in that report. In mammals, the transcription factor family of
NF-κB includes RelA/p65, NF-κB1/p50, RelB, NF-κB2/p52 and c-Rel.
NF-κB subunits play critical roles in cell survival, inflammation,
proliferation, apoptosis and tumorigenesis (14). The activation of NF-κB subunits
triggers the canonical signaling pathway which is characterized by
the activation of RelA-p50 heterodimers and the non-canonical
signaling pathway that activates RelB-p52 heterodimers (15). The non-canonical NF-κB-triggering
receptors contain a TRAF-binding motif, recruiting different TRAF
members, particularly TRAF2 and TRAF3 (16,17).
TRAF2 is important for activation of the canonical NF-κB pathway
but also mediates their stimulatory effects on the c-Jun N-terminal
kinase pathway (18). In addition,
TRAF2 is involved in the degradation of the NF-κB inducing kinase
(NIK) that triggers the non-canonical NF-κB signaling pathway
(19,20). In general, TRAF2 are considered as
a positive regulator of both canonical and non-canonical NF-κB
activation. In this study, the reduction of RelA and p50 as well as
RelB was observed in the presence of CHIP overexpression. Growing
attention is paid to understanding the role of RelB in the
pathogenesis of diverse malignancies, such as prostate cancer,
breast cancer and lymphoid malignancy. Targeting RelB has also been
recommended to be a valuable strategy in overcoming radiation
resistance in the prostate and breast cancer (21,22).
The affects of CHIP on RelB would further support the idea that
CHIP functions as a tumor suppressor in gastric cancer. Other NF-κB
subunits, such as c-Rel and p52, stayed unchanged in the
CHIP-overexpressing AGS gastric cancer cells. The expression of
TRAF3, a negative regulator of non-canonical NF-κB activation, was
not affected by CHIP overexpression. These results suggest that in
the gastric cancer cells, CHIP negatively regulates TRAF2
expression, which subsequently inhibits both canonical and
non-canonical NF-κB expression. The potential mechanism by which
CHIP negatively regulates the expression of TRAF2 remains unclear.
Ubiquitination and proteasome-mediated degradation of TRAF2 by CHIP
might be one of the explanations.
Gastric cancer is a very aggressive malignant tumor
due to its invasive nature and early metastatic ability.
Degradation of the ECM and basement membrane barriers are essential
steps in the pathogenesis of gastric cancer. Many studies have
confirmed the critical roles of matrix metalloproteinases (MMPs)
and their inhibitors in the growth and progression of gastric
cancer. The human MMPs family consists of at least 26 proteases,
which are subdivided into collagenases, gelatinases, stromelysins
and matrilysins (23). MMP-2 and
−9 are well-characterized gelatinases and are closely associated
with cancer invasion and metastasis due to their strong proteolytic
activity of ECM. While MMP-2 promotes cleavage of ECM proteins,
MMP-9 modulates permeability of the vascular endothelium (24). The mRNA expression of MMP-2 and −9
in gastric cancer tissue is significantly higher than that in
normal tissue (25–26). The expression of MMP-2 and −9 is
also correlated with clinicopathological features of tumor
patients. Overexpression of MMP-2 and −9 indicates worse survival
of gastric cancer patients (27–29).
NF-κB and AP-1 are important transcription factors in regulating
the activity of MMP-2 and −9, as the gene promoter contains NF-κB
and AP-1 binding sites (30). In
gastric cancer cells, MMP-2 and −9 could be also upregulated in
response to IL-1β stimulation, which is p38-mediated and
AP-1-dependent (31). In this
study, we found that CHIP overexpression in the AGS gastric cancer
cells inhibited the production and the activity of MMP-2 and −9,
indicating by the western blotting and gelatinase zymography assay.
The migration and invasion abilities of the AGS cells were largely
affected by CHIP overexpression. The data reported here is in line
with the previous results that CHIP overexpression suppresses the
invasion ability and inhibits the expression of MMP-9 in breast
cancer cells (12). However, the
expression of urokinase plasminogen activator (uPA) was not
affected by the overexpression of CHIP in the AGS cells (data not
shown); unlike that in the MDA-MB-231 breast cancer cells.
In this study, we also observed that the expression
of integrin β-1 was downregulated in the CHIP-overexpressing
gastric cancer cells. Integrin β-1, encoded by the ITGB1
gene, belongs to the family of heterodimeric transmembrane cell
surface receptors that contain 18α and 8β subunits. Integrin β-1,
as a direct target of mir-29c, positively regulates gastric cancer
cell adhesion, invasion and migration (32). Overexpression of integrin β-1 has
been found in various epithelial malignancies including breast
cancer and glioblastoma, during invasion, angiogenesis and
metastasis. High levels of integrin β-1 expression have been
detected in gastric cancer and shown to be associated with poor
prognosis and recurrence in patients with gastric cancer (33,34).
Several studies have also shown that expression of integrin β-1 is
associated with peritoneal metastasis of gastric cancer and
functional blocking of integrin β-1 significantly reduces the
peritoneal metastasis of gastric cancer in vivo (35,36).
A typical binding site for NF-κB is located in the promoter of the
human ITGB1 gene (37). In
a small cell lung cancer cell line (SCLC), blocking RelB expression
can prevent the increase of integrin β-1 (38). Here, we found in gastric cancer
cells, that CHIP negatively regulated the integrin β-1 expression.
The RelB reduction would be one of the reasons for repressed
integrin β-1 expression. The inhibited expression of RelA/p65 in
the presence of CHIP was involved in the downregulation of MMP-2
and −9 in the gastric cancer cells. The decreased activities of
MMP-2 and −9, together with the downregulated integrin β-1
contributed to the diminished migration and invasion abilities of
gastric cancer cells.
CHIP is generally considered as a tumor suppressor.
However, a few studies have also suggested that CHIP may have
opposing roles in certain cancers, such as gliomas and esophageal
squamous cell carcinoma (39,40).
In this study, we found that CHIP expression level was often
decreased in gastric cancer patients, either in mRNA or protein
levels. We also found that the level of CHIP expression was
significantly associated with the differentiation status and the
TNM staging, suggesting that the decreased expression of CHIP might
indicate poor prognosis. These findings are in line with previous
findings (13,41).
Increased apoptosis and decreased cellular
proliferation, which lead to slower cell growth, were observed in
the AGS cells transfected with CHIP. Clear reduction of Bcl-2, a
target gene of RelB (42), would
be one of the reasons for increased apoptosis. The decreased
phosphoration of AKT and NF-κB signaling contributed to the
decreased cellular proliferation. In contrast, it is reported that
in normal breast epithelial MCF10A and breast cancer MCF7 cells,
CHIP activates PI3K/AKT survival factors and induce apoptosis
(43).
Taken together, we found that CHIP overexpression in
gastric cancer cells affected several aspects of malignant
phenotypes, including cell growth, migration and invasion. The
defected TRAF2-NF-κB signaling downregulated several important
molecules, including apoptosis genes, MMPs and integrin β-1,
involved in these processes. Particularly, RelB, representing the
non-canonical NF-κB, was reduced in the presence of CHIP in the
gastric cancer cells. The negative correlation of CHIP expression
with clinical features of gastric cancer patients support the idea
that CHIP is a tumor suppressor in gastric cancer.
Acknowledgements
This study was supported by Jiangsu Provincial
Natural Science Foundation of China (F.G., grant no. BK2011306) and
National Natural Science Foundation of China (F.G., grant no.
81172433 and W.C.C., grant no. 81272737).
References
|
1
|
Ferlay J, Soerjomataram II, Dikshit R, et
al: Cancer incidence and mortality worldwide: sources, methods and
major patterns in GLOBOCAN 2012. Int J Cancer. Sep 13–2014.(Epub
ahead of print). PubMed/NCBI
|
|
2
|
Bozzuto G, Ruggieri P and Molinari A:
Molecular aspects of tumor cell migration and invasion. Ann Ist
Super Sanita. 46:66–80. 2010.PubMed/NCBI
|
|
3
|
Ballinger CA, Connell P, Wu Y, et al:
Identification of CHIP, a novel tetratricopeptide repeat-containing
protein that interacts with heat shock proteins and negatively
regulates chaperone functions. Mol Cell Biol. 19:4535–4545.
1999.PubMed/NCBI
|
|
4
|
McDonough H and Patterson C: CHIP: a link
between the chaperone and proteasome systems. Cell Stress
Chaperones. 8:303–308. 2003. View Article : Google Scholar
|
|
5
|
Connell P, Ballinger CA, Jiang J, et al:
The co-chaperone CHIP regulates protein triage decisions mediated
by heat-shock proteins. Nat Cell Biol. 3:93–96. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Meacham GC, Patterson C, Zhang W, Younger
JM and Cyr DM: The Hsc70 co-chaperone CHIP targets immature CFTR
for proteasomal degradation. Nat Cell Biol. 3:100–105. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Urushitani M, Kurisu J, Tateno M, et al:
CHIP promotes proteasomal degradation of familial ALS-linked mutant
SOD1 by ubiquitinating Hsp/Hsc70. J Neurochem. 90:231–244. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Patani N, Jiang W, Newbold R and Mokbel K:
Prognostic implications of carboxyl-terminus of Hsc70 interacting
protein and lysyl-oxidase expression in human breast cancer. J
Carcinog. 9:92010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun C, Li HL, Shi ML, Liu QH, Bai J and
Zheng JN: Diverse roles of C-terminal Hsp70-interacting protein
(CHIP) in tumorigenesis. J Cancer Res Clin Oncol. 140:189–197.
2014. View Article : Google Scholar
|
|
10
|
Wang Y, Ren F, Feng Y, et al: CHIP/Stub1
functions as a tumor suppressor and represses NF-kappaB-mediated
signaling in colorectal cancer. Carcinogenesis. 35:983–991. 2014.
View Article : Google Scholar
|
|
11
|
Kajiro M, Hirota R, Nakajima Y, et al: The
ubiquitin ligase CHIP acts as an upstream regulator of oncogenic
pathways. Nat Cell Biol. 11:312–319. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jang KW, Lee KH, Kim SH, et al: Ubiquitin
ligase CHIP induces TRAF2 proteasomal degradation and NF-kappaB
inactivation to regulate breast cancer cell invasion. J Cell
Biochem. 112:3612–3620. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang S, Wu X, Zhang J, et al: CHIP
functions as a novel suppressor of tumour angiogenesis with
prognostic significance in human gastric cancer. Gut. 62:496–508.
2013. View Article : Google Scholar
|
|
14
|
Xu J, Zhou P, Wang W, Sun A and Guo F:
RelB, together with RelA, sustains cell survival and confers
proteasome inhibitor sensitivity of chronic lymphocytic leukemia
cells from bone marrow. J Mol Med (Berl). 92:77–92. 2014.
View Article : Google Scholar
|
|
15
|
Vallabhapurap US and Karin M: Regulation
and function of NF-kappaB transcription factors in the immune
system. Annu Rev Immunol. 27:693–733. 2009. View Article : Google Scholar
|
|
16
|
Shih VF, Tsui R, Caldwell A and Hoffmann
A: A single NFkappaB system for both canonical and non-canonical
signaling. Cell Res. 21:86–102. 2011. View Article : Google Scholar
|
|
17
|
Sun SC: Non-canonical NF-kappaB signaling
pathway. Cell Res. 21:71–85. 2011. View Article : Google Scholar
|
|
18
|
Shu HB, Takeuchi M and Goeddel DV: The
tumor necrosis factor receptor 2 signal transducers TRAF2 and
c-IAP1 are components of the tumor necrosis factor receptor 1
signaling complex. Proc Natl Acad Sci USA. 93:13973–13978. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Karl I, Jossberger-Werner M, Schmidt N, et
al: TRAF2 inhibits TRAIL- and CD95L-induced apoptosis and
necroptosis. Cell Death Dis. 5:e14442014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hayden MS and Ghosh S: NF-kappaB, the
first quarter-century: remarkable progress and outstanding
questions. Genes Dev. 26:203–234. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Holley AK, Xu Y, St Clair DK and St Clair
WH: RelB regulates manganese superoxide dismutase gene and
resistance to ionizing radiation of prostate cancer cells. Ann NY
Acad Sci. 1201:129–136. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang X, Belguise K, O’Neill CF, et al:
RelB NF-kappaB represses estrogen receptor alpha expression via
induction of the zinc finger protein Blimp1. Mol Cell Biol.
29:3832–3844. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lukaszewicz-Zajac M, Mroczko B and
Szmitkowski M: Gastric cancer - the role of matrix
metalloproteinases in tumor progression. Clin Chim Acta.
412:1725–1730. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Galis ZS, Muszynski M, Sukhova GK, et al:
Cytokine-stimulated human vascular smooth muscle cells synthesize a
complement of enzymes required for extracellular matrix digestion.
Circ Res. 75:181–189. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chu D, Zhang Z, Li Y, et al: Matrix
metalloproteinase-9 is associated with disease-free survival and
overall survival in patients with gastric cancer. Int J Cancer.
129:887–895. 2011. View Article : Google Scholar
|
|
26
|
Zhang M, Zhu GY, Gao HY, Zhao SP and Xue
Y: Expression of tissue levels of matrix metalloproteinases and
tissue inhibitors of metalloproteinases in gastric adenocarcinoma.
J Surg Oncol. 103:243–247. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Alakus H, Grass G, Hennecken JK, et al:
Clinicopathological significance of MMP-2 and its specific
inhibitor TIMP-2 in gastric cancer. Histol Histopathol. 23:917–923.
2008.PubMed/NCBI
|
|
28
|
Gao ZL, Zhang C, Du GY and Lu ZJ: Clinical
significance of changes in tumor markers, extracellular matrix,
MMP-9 and VEGF in patients with gastric carcinoma.
Hepatogastroenterology. 54:1591–1595. 2007.PubMed/NCBI
|
|
29
|
Zhou Y, Li G, Wu J, et al:
Clinicopathological significance of E-cadherin, VEGF, and MMPs in
gastric cancer. Tumour Biol. 31:549–558. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Eberhardt W, Huwiler A, Beck KF, Walpen S
and Pfeilschifter J: Amplification of IL-1 beta-induced matrix
metalloproteinase-9 expression by superoxide in rat glomerular
mesangial cells is mediated by increased activities of NF-kappa B
and activating protein-1 and involves activation of the
mitogen-activated protein kinase pathways. J Immunol.
165:5788–5797. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang Q, Lan F, Wang X, et al:
IL-1beta-induced activation of p38 promotes metastasis in gastric
adenocarcinoma via upregulation of AP-1/c-fos, MMP2 and MMP9. Mol
Cancer. 13:182014. View Article : Google Scholar
|
|
32
|
Han TS, Hur K, Xu G, et al: MicroRNA-29c
mediates initiation of gastric carcinogenesis by directly targeting
ITGB1. Gut. May 28–2014.(Epub ahead of print). pii:
gutjnl-2013306640. PubMed/NCBI
|
|
33
|
Zhao ZS, Li L, Wang HJ and Wang YY:
Expression and prognostic significance of CEACAM6, ITGB1, and CYR61
in peripheral blood of patients with gastric cancer. J Surg Oncol.
104:525–529. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xu ZY, Chen JS and Shu YQ: Gene expression
profile towards the prediction of patient survival of gastric
cancer. Biomed Pharmacother. 64:133–139. 2010. View Article : Google Scholar
|
|
35
|
Lin MT, Chang CC, Lin BR, et al: Elevated
expression of Cyr61 enhances peritoneal dissemination of gastric
cancer cells through integrin alpha2beta1. J Biol Chem.
282:34594–34604. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ura H, Denno R, Hirata K, Yamaguchi K and
Yasoshima T: Separate functions of alpha2beta1 and alpha3beta1
integrins in the metastatic process of human gastric carcinoma.
Surg Today. 28:1001–1006. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ahmed KM, Zhang H and Park CC: NF-kappaB
regulates radioresistance mediated by beta1-integrin in
three-dimensional culture of breast cancer cells. Cancer Res.
73:3737–3748. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Saito T, Sasaki CY, Rezanka LJ, Ghosh P
and Longo DL: p52-independent nuclear translocation of RelB
promotes LPS-induced attachment. Biochem Biophys Res Commun.
391:235–241. 2010. View Article : Google Scholar :
|
|
39
|
Xu T, Zhou Q, Zhou J, et al: Carboxyl
terminus of Hsp70-interacting protein (CHIP) contributes to human
glioma oncogenesis. Cancer Sci. 102:959–966. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wen J, Luo KJ, Hu Y, Yang H and Fu JH:
Metastatic lymph node CHIP expression is a potential prognostic
marker for resected esophageal squamous cell carcinoma patients.
Ann Surg Oncol. 20:1668–1675. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gan L, Liu DB, Lu HF, et al: Decreased
expression of the carboxyl terminus of heat shock cognate 70
interacting protein in human gastric cancer and its clinical
significance. Oncol Rep. 28:1392–1398. 2012.PubMed/NCBI
|
|
42
|
Wang X, Belguise K, Kersual N, et al:
Oestrogen signalling inhibits invasive phenotype by repressing RelB
and its target BCL2. Nat Cell Biol. 9:470–478. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lv Y, Song S, Zhang K, Gao H and Ma R:
CHIP regulates AKT/FoxO/Bim signaling in MCF7 and MCF10A cells.
PLoS One. 8:e833122013. View Article : Google Scholar :
|