Introduction
Ubiquitination is an important post-translational
modification in cells, maintaining homeostasis through numerous
cellular processes such as cell growth, proliferation, apoptosis,
DNA damage response, and immune response (1,2).
Ubiquitin is composed of 76 amino acids and is well conserved in
the eukaryotic genome. In addition, ubiquitin regulates its
targeted proteins for proteasomal degradation or signal
transduction in an ATP-dependent manner through making a covalent
bond of lysine sites on its binding proteins (3). Within the cells, polyubiquitination
has several different roles according to the position of the
attachment site on target proteins (4). Ubiquitin has seven lysine residues
(K6, K11, K27, K29, K33, K48, and K63). On these residues, other
ubiquitins are attached as mono- or polyubiquitin chain (1). K48-linked polyubiquitin chains
regulate the proteasomal degradation of target proteins (5), and K63-linked and linear
polyubiquitin chains control protein kinase activation and cell
signaling (6,7).
Deubiquitination is a reversible reaction of
ubiquitination and this process requires for deubiquitinating
enzymes (DUBs). DUBs can induce deconjugation of the ubiquitin from
target proteins and lead to regulation of stability or downstream
signaling for targeting proteins. Deubiquitination, in common with
ubiquitination, regulates diverse biochemical pathways in cells.
The importance of many DUBs has been verified in the receptor
tyrosine kinase signaling, signal transduction, gene transcription,
DNA repair, proliferation, and mitosis (8). The human genome encodes ~100 DUBs; in
general, DUBs are divided into at least six subfamilies, as
follows: ubiquitin-specific processing proteases (USPs/UBPs),
ubiquitin carboxy-terminal hydrolases (UCHs),
Jad1/Pad/MPN-domain-containing metallo-enzymes (JAMMs), Otu-domain
ubiquitin-aldehyde-binding proteins (OTUs), Ataxin-3/Josephin, and
monocyte chemotactic protein-induced proteases (MCPIPs) (9–12).
These six subfamilies are classified in accordance with the
ubiquitin-binding domain. They have specific regions including Cys,
Asp/Asn and His boxes in the domain of catalytic activity, and JAMM
subfamily members are zinc-dependent metallo-proteases (13).
The pyruvate kinase isoform M2 (PKM2) is regarded as
a major protein regulating metabolism signaling in cancer. In
normal conditions, PKM2 acts as a pyruvate kinase with a tetramer
form. In cancer cells, however, PKM2 plays a role as a protein
kinase with a dimer form. Expression of the PKM2 dimer impedes the
glycolytic metabolism, thereby inducing the Warburg effect. This
effect is the main process through which most cancer cells
predominantly produce energy, involving a high rate of glycolysis
(14). Increased conversion of
glucose to regulate lactate is a key feature of many cancer cells
with rapid growth. Increased PKM2 expression enables regulation of
lactate production in cancer cells. In hypoxic condition, PKM2
leads to the accumulation of upstream glycolytic metabolites, which
stimulate the pentose phosphate pathway, thereby promoting
macromolecule biosynthesis and inhibiting intracellular reactive
oxygen species (ROS) generation (15). In addition, PKM2 dimers are
translocated into the nucleus and stimulate the transcription
factors such as hypoxia-inducible factor (HIF)-1, HIF-2, signal
transducer and activator of transcription 4 (STAT4), and
octamer-binding transcription factor 4 (Oct-4). Therefore, cell
proliferation and cell growth are increased (16).
USP20 is one of the DUBs belonging to the USP
subfamily. It has been identified as a regulator of HIF-1α
(17), tumor necrosis factor (TNF)
receptor-associated factor 6 (TRAF6), and Tax (18). A recent study suggested that USP20
is also involved in homologous recombination (HR), one of the DNA
repair pathways, by stabilizing Rad17 as a DNA damage response
protein (19). A previous study
revealed that the over-expression of USP20 increased the expression
level of HIF-1α by regulating degradation signal (17). Moreover, PKM2 stimulates the
binding of HIF-1α at the promoter of target genes such as the
recruitment of coactivators, histone acetylation, and gene
transcription (20). The
expression of HIF-1α is increased in the majority of human cancers
and acts as a major regulator of O2 homeostasis
(21). In this study, we
identified PKM2 as a putative substrate of USP20 with proteomic
analysis tools. We also demonstrated that PKM2 can be a binding
partner for USP20, which downregulates the ubiquitination level by
DUB activity.
Material and methods
Cell culture and transfection
HeLa cells were cultured in Dulbecco’s modified
Eagle’s medium (Gibco BRL, Rockville, MD, USA) contained with 10%
fetal bovine serum (Gibco BRL) and 1% penicillin/streptomycin
(Gibco BRL). Cells were transfected using 10 mM polyethlylenimine
(PEI, Polysciences, Warrington, WA, USA) with 6 μg plasmid DNA in
100-mm culture dish.
Two-dimensional electrophoresis (2-DE)
and matrix assisted laser desorption/ionization time-of-flight mass
spectrometry (MALDI-TOF/MS)
2-DE was conducted as previously described (22). Cells were lysed with 2-DE lysis
buffer (7 M CO(NH2)2, 2 M
SC(NH2)2, 4.5% CHAPS, 100 mM DTE, 40 mM Tris,
pH 8.8), and samples were applied to pH 3–10 nonlinear gradient
strips (Amersham Biosciences, Uppsala, Sweden). IEF was performed
at 80,000 Vh. After IFR, strips were loaded on 9–16% linear
gradient polyacrylamide gels and the gels were stained with
Coomassie Brilliant Blue (CBB) G-250 for 12 h. Stained gels were
scanned with a Bio-Rad GS710 densitometer (Bio-Rad Laboratories,
Richmond, CA, USA) and analyzed using Image Master Platinum 5.0
image analysis program (Amersham Biosciences, Glattbrugg,
Switzerland).
For MALDI-TOF/MS analysis, each peptide was
concentrated by a POROS R2, Oligo R3 column (Applied Biosystems,
Foster City, CA, USA). After washing the column with 70%
CH3CN, 100% CH3CN and then 50 mM
(NH4)HCO3, samples were applied to the R2, R3
column and eluted with cyano-4-hydroxycinamic acid (CHCA)
(Sigma-Aldrich, St. Louis, MO, USA). Samples were dissolved in 70%
CH3CN and 2% HCO2H onto the MALDI plate
(Opti-TOF™ 384-well Insert, Applied Biosystems) (23).
MALDI-TOF/MS was performed on 4800 MALDI-TOF/TOF™
Analyzer (Applied Biosystems) equipped with a 355-nm Nd:YAG laser.
The pressure in the TOF analyzer is approximately 7.6e-07 Torr. The
mass spectra were obtained in the reflection mode with an
accelerating voltage of 20 kV and sum from either 500 laser pulses
and calibrated using the 4700 calibration mixture (Applied
Biosystems). NCBInr database (downloaded May 12, 2014, # of entries
= 7,051,549 sequence) searching was performed with the MASCOT
search engine (http://www.matrixscience.com). The peak
list-generating software Data Explorer version 4.4 (PerSeptive
Biosystems, Wiesbaden, Germany) was used. Database search criteria
were, taxonomy, Homo sapiens, fixed modification;
carboxyamidomethylated (+57) at cysteine residue; variable
modification; oxidized (+16) at methionine residues, maximum
allowed missed cleavage, 1, MS tolerance, 100 ppm. Only peptides
resulting from trypsin digests were considered, and trypsin peak
(842.5090, 2211.1040) was used for internal calibration.
Reverse transcription PCR (RT-PCR)
Total RNAs were extracted from HeLa cells after 24 h
of transfection using TRIzol™ reagent (Invitrogen, Carlsbad, CA,
USA). cDNA of each sample was constructed by the SuperScript III
system (Invitrogen). PCR conditions were 20 sec denaturation at
95°C, 30 sec annealing at 55°C, and 40 sec extension at 72°C for
all genes. All primer sets were assessed by gel electrophoresis to
confirm amplification from extracted cDNA of a single band of the
expected size.
Immunoprecipitation
After 24 h of incubation from transfection, cells
were harvested. Phosphate buffered saline (PBS) was used as a
washing solution. Harvested cells were lysed with a lysis buffer
(150 mM NaCl, 1% Triton X-100, 20 mM Tris-HCl at pH 8.0, 1 mM EDTA,
100 mM PMSF, protease inhibitor cocktail). A 1.5 mg aliquot of cell
lysates was incubated with mouse anti-Flag (Sigma-Aldrich), mouse
anti-PKM2, and mouse anti-Myc (Santa Cruz Biotechnology, Santa
Cruz, CA, USA) antibodies at 4°C. Protein A/G PLUS-Agarose beads
(Santa Cruz Biotechnology) was added to the lysates and incubated
at 4°C. Immunoprecipitated proteins were separated on 8%
polyacrylamide gels.
Western blotting
The protein concentration was identified using the
Bio-Rad Protein Assay reagent (Bio-Rad Laboratories, Hercules, CA,
USA). Cell lysates were boiled with 2X SDS sample buffer and loaded
on 8% polyacrylamide gels. The separated proteins on SDS-PAGE gel
were transferred to polyvinylidene difluoride (PVDF) membranes.
After transfer to the membranes, they were incubated in 3% skim
milk and primary antibody mixture (1:1000) overnight at 4°C.
Membranes were incubated with the secondary peroxidase labeled
anti-mouse IgG antibody (1:10000) (Santa Cruz Biotechnology). Then,
the bands were detected using X-ray developer and exposure to
film.
Immunofluorescence
At 24 h after transfection, cells were seeded onto
12-mm flame sterilized coverslips. After incubation, cells were
fixed with 4% paraformaldehyde (Sigma-Aldrich). The cells were
incubated with primary goat anti-USP20 and mouse anti-PKM2
antibodies (Santa Cruz Biotechnology). Then, the cells were
incubated with 1:200 diluted fluorescein-isothiocyanate-labeled
anti-goat or anti-mouse IgG (Molecular Probes, Carlsbad, CA, USA)
as a secondary antibody. Cells were stained with 4′
6-diamindino-2-phenylindole (DAPI; Molecular Probes) for nuclei
staining. Afterwards, coverslips were mounted onto glass slides
using 90% glycerol/100 mM Tris (pH 8.0) and cell images were
captured by a confocal microscope (Leica TCS SP8, Leica, Wetzlar,
Germany).
DNA constructs and mutagenesis
A catalytically inactive form of USP20 was
constructed by site-directed mutagenesis using QuikChange™
Site-Directed Mutagenesis Kit (Statagene, La Jolla, CA, USA). The
forward primer (5′-CCT CGG GAA CTC CAG CTA CAT GAA CGC-3′) and
reverse primer (5′-GCG TTC ATG TAG CTG GAG TTC CCG AGG-3′) were
used for replacing cysteine to serine at position 154 of USP20. The
HA-ubiquitin (K48R) and HA-ubiquitin (K63R)
constructs were as previously described (24).
Results
Investigation of putative substrates for
USP20
It has been demonstrated that USP20 is involved in
intracellular regulatory mechanisms such as proliferation of cancer
cells, DNA repair, and the immune system (17–19).
TRAF6, HIF-1α, and Rad17 were identified as USP20 associated
proteins. When the USP20 is overexpressed, the expression of TRAF6
and HIF-1α is stabilized (17–19).
USP20 has important roles in tumorigenesis, the immune system, and
DNA repair, other additional roles of USP20 are not yet fully
understood. In order to investigate other binding partners of
USP20, we performed 2-DE and MALDI-TOF/MS analysis using HeLa cells
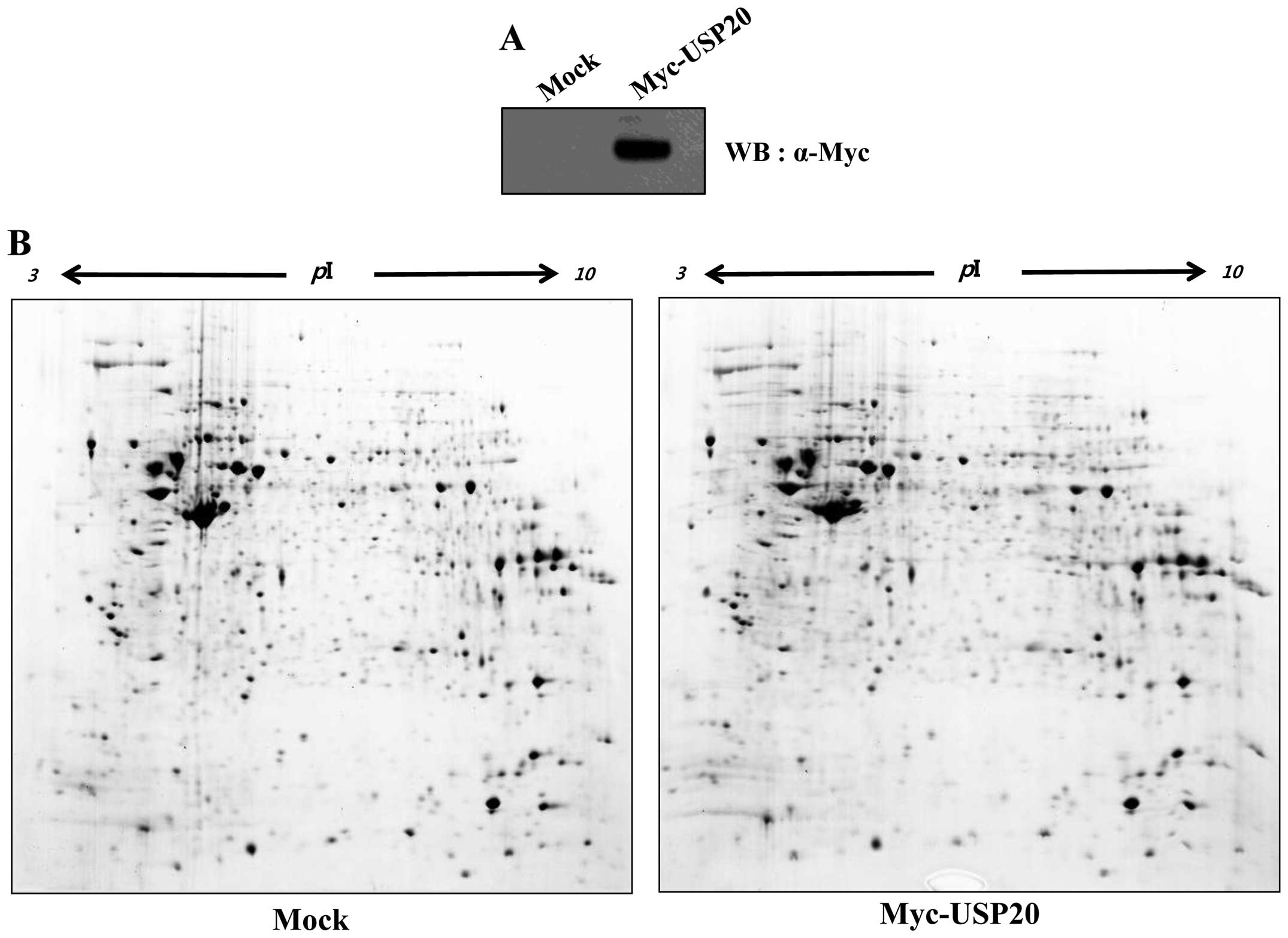
with overexpression of USP20. Protein spots were detected in the
acrylamide gel (Fig. 1). Among
them, 11 upregulated spots were detected. Before the proteomic
analysis, we verified the expression of USP20 in HeLa cells
(Fig. 1A), and performed 2-DE
analysis to identify putative substrates of USP20 (Fig. 1B). In USP20-transfected samples, 7
were downregulated, and 4 were upregulated (Fig. 1C and D). Isolated spots were
analyzed by mass spectrometry (Table
I).
 | Table IProteins identified by
MALDI-TOF/MS. |
Table I
Proteins identified by
MALDI-TOF/MS.
| Place | Spot ID | Accession
number | Protein name | Score | Matched peptides
number | Sequence coverage
(%) | Theoretical
MW/PI |
|---|
| Myc | 1538 | gi|48928058 | Small
ubiquitin-related modifier 3 (SUMO3) precursor | 70 | 7/50 (14%) | 46 | 11630/5.32 |
| 1551 | gi|5410451 | Interferon-induced
protein p78 | 118 | 18/60 (30%) | 37 | 75886/5.60 |
| 1577 | gi|3063997 |
CMP-N-acetylneuraminate-β-galactosamide-α-2,3-sialyltransferase
(ST3GAL1) | 92 | 11/60 (18%) | 41 | 30462/9.50 |
| 1752 | gi|348239 | Unnamed | 78 | 15/68 (22%) | 34 | 58083/7.60 |
| 1694 | gi|228008398 | Synaptotagmin
binding, cytoplasmic RNA interacting protein isoform 3
(SYNCRIP) | 65 | 11/60 (18%) | 30 | 58927/7.18 |
| 1696 | gi|12804225 | Unknown | 117 | 21/60 (35%) | 42 | 59887/5.45 |
| 2680 | gi|16306550 | Selenium-binding
protein 1 (SELENBP1) | 73 | 10/40 (25%) | 33 | 52358/5.93 |
| USP20 | 1642 | gi|193788722 | Hypothetical
protein LOC51507 | 67 | 10/56 (17%) | 45 | 33865/8.86 |
| 1731 | gi|33286418 | Pyruvate kinase
isozymes M1/M2 isoform M2 (PKM2) | 119 | 18/60 (30%) | 34 | 58470/7.96 |
| 1796 | gi|1208427 | ER-60 protease | 117 | 16/60 (26%) | 35 | 57160/5.98 |
| 2608 | gi|111550164 | BORIS transcription
factor transcript variant A5 (CTCFL) | 76 | 14/43 (32%) | 27 | 71158/8.51 |
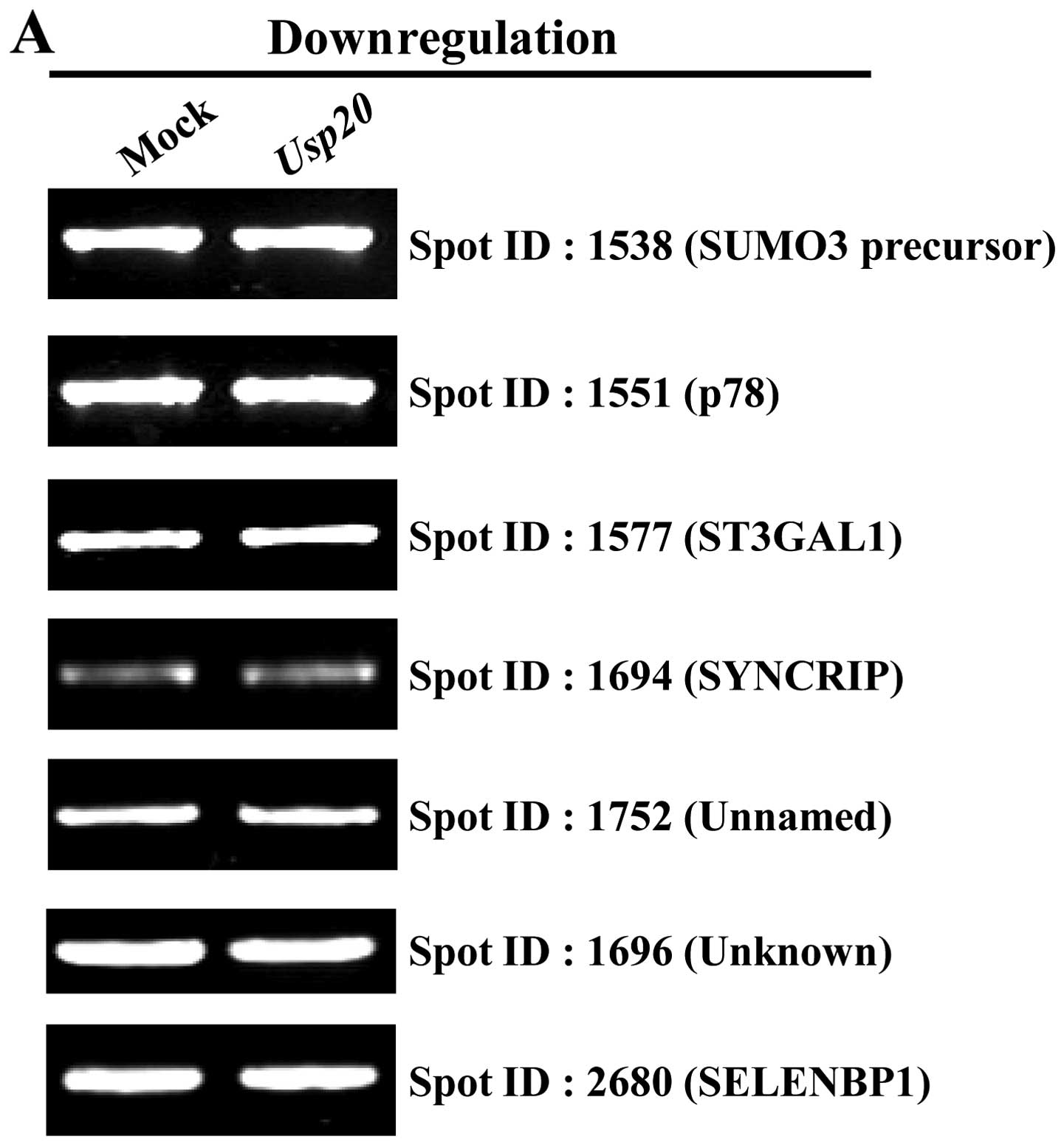
To investigate whether the screened substrates are
regulated by the ubiquitin-proteasome pathway, we performed RT-PCR
to determine the effect of USP20 overexpression on the
transcription (Fig. 2, Table II). As expected, the results
indicate that USP20 has no effect on the transcriptional level of
genes encoding proteins identified from proteomics analysis
(Fig. 2). From the data, we
selected one of the proteins, pyruvate kinase isozymes M1/M2
isoform M2 (PKM2) for further analysis. A previous study showed
that PKM2 functions as a co-activator with HIF-1α (21). In addition, it has been
demonstrated that HIF-1α is stabilized by USP20 (17). Our proteomic analysis with USP20
overexpression showed that PKM2 has a high identity score and
exhibits two times higher expression than that of a control.
Furthermore, the mRNA level was not different in the
USP20-transfected condition (Fig.
2B). Collectively, these results indicate that PKM2 can be a
genuine protein for one of the UPS20 substrates.
 | Table IIPrimers used for RT-PCR analysis. |
Table II
Primers used for RT-PCR analysis.
| Name | Sequences |
|---|
| Small
ubiquitin-related modifier 3 precursor | FP: 5′-CCA AGG AGG
GTG TGA AGA CA-3′
RP: 5′-GTG TCC TCG TCC TCC ATC TC-3′ |
| Interferon-induced
protein p78 | FP: 5′-GGG AAG GAA
TGG GAA TCA GT-3′
RP: 5′-ATG CTG AGA GCC TCT GTG GT-3′ |
|
CMP-N-acetylneuraminate-β-galactosamide-α-2,3-sialyltransferase | FP: 5′-GGG AGA TGC
CAT CAA CAA GT-3′
RP: 5′-TGT TTC CAG AAA CCC TTT CG-3′ |
| Unnamed | FP: 5′-ATA TGC CAC
TCC GTG GAA AG-3′
RP: 5′-GAA GGA GCC TTC ACT GCA TC-3′ |
| Synaptotagmin
binding, cytoplasmic RNA interacting protein isoform 3 | FP: 5′-TGT GGG AAA
GAT CCC AAG AG-3′
RP: 5′-TTG GCA ACT GAG ATG CAG AC-3′ |
| Unknown | FP: 5′-GAT GGC TGA
GAT TGC TGT GA-3′
RP: 5′-TTG GTT TGG GTG GTT CAA AT-3′ |
| Selenium-binding
protein 1 | FP: 5′-ATC ACC GAC
ATC CTG CTC TC-3′
RP: 5′-CCG TTT TCC CTT GAC CAC TA-3′ |
| Hypothetical
protein LOC51507 | FP: 5′-GAG AGC GAA
GCT GGA AAA GA-3′
RP: 5′-ACT GTC AGC GAT GGA CCT CT-3′ |
| Pyruvate kinase
isozymes M1/M2 isoform M2 | FP: 5′-ATG AGT ACC
ATG CGG AGA CC-3′
RP: 5′-AGT CCA GCC ACA GGA TGT TC-3′ |
| ER-60 protease | FP: 5′-ATG GGC CTG
TGA AGG TAG TG-3′
RP: 5′-GTG GCA TCC ATC TTG GCT AT-3′ |
| BORIS transcription
factor transcript variant A5 | FP: 5′-CTA CAA GCT
GAA ACG CCA CA-3′
RP: 5′-AGC AGA ACA GTA GCG GCA TT-3′ |
| GAPDH | FP: 5′-ACC ACA GTC
CAT GCC ATC AC-3′
RP: 5′-TCC ACC ACC CTG TTG CTG TA-3 |
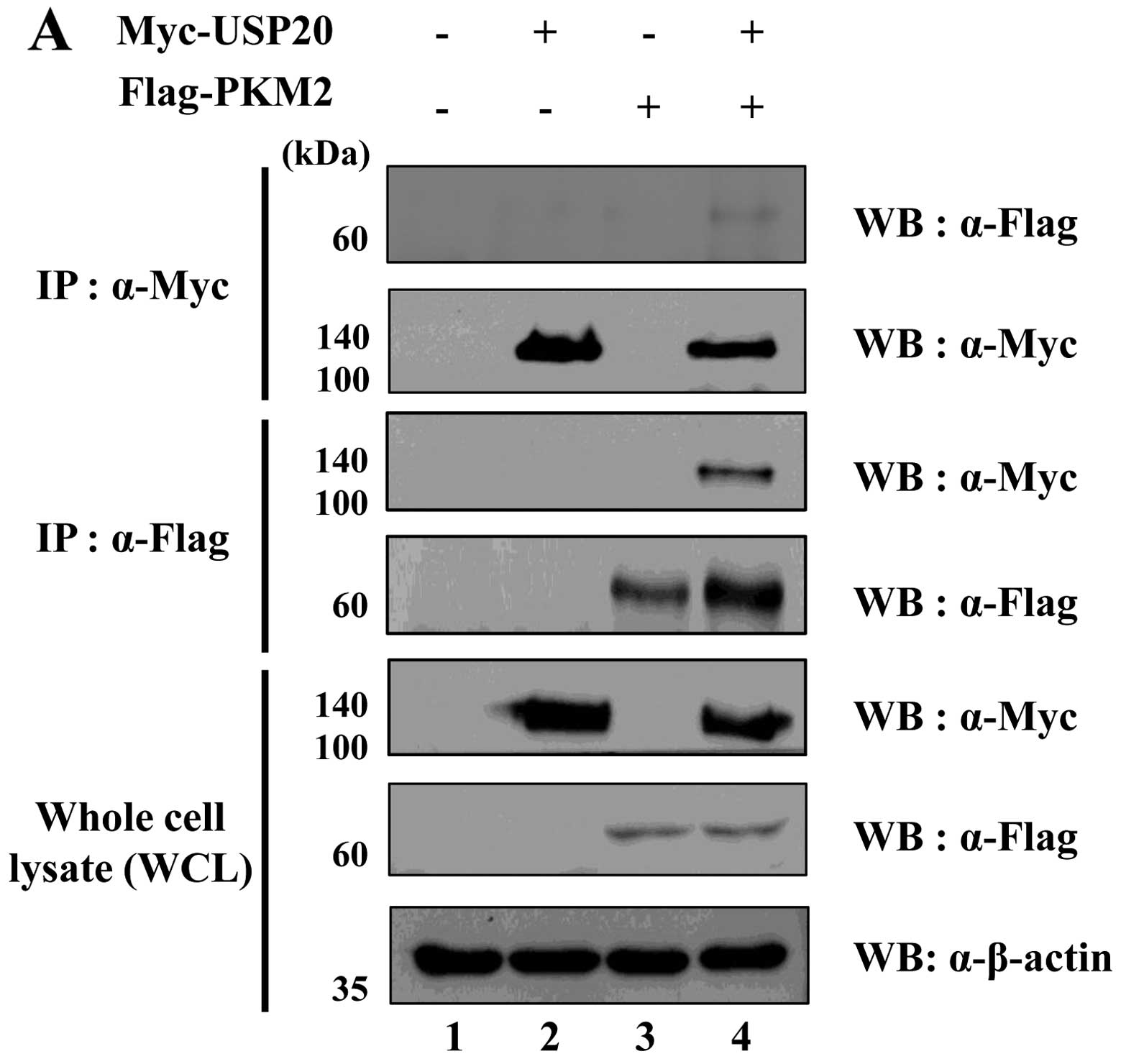
USP20 interacts with PKM2
To demonstrate the binding between USP20 and PKM2,
we performed immunoprecipitation analysis using transfected samples
with Myc- or Flag-tagged USP20 and PKM2 expressing
HeLa cells. Interaction of these two proteins was confirmed by
immunoprecipitation assay using an anti-Myc or an anti-Flag
antibody. As expected, the result showed that USP20 binds with PKM2
exogenously (Fig. 3A). We next
performed immunofluorescence (IF) assay using HeLa cells to check
the localization of these two proteins in HeLa cells (Fig. 3B). The results showed that both
USP20 and PKM2 were co-localized in the cytosol and nucleus. Taken
together, our biochemical assay revealed that USP20 was
specifically bound with PKM2.
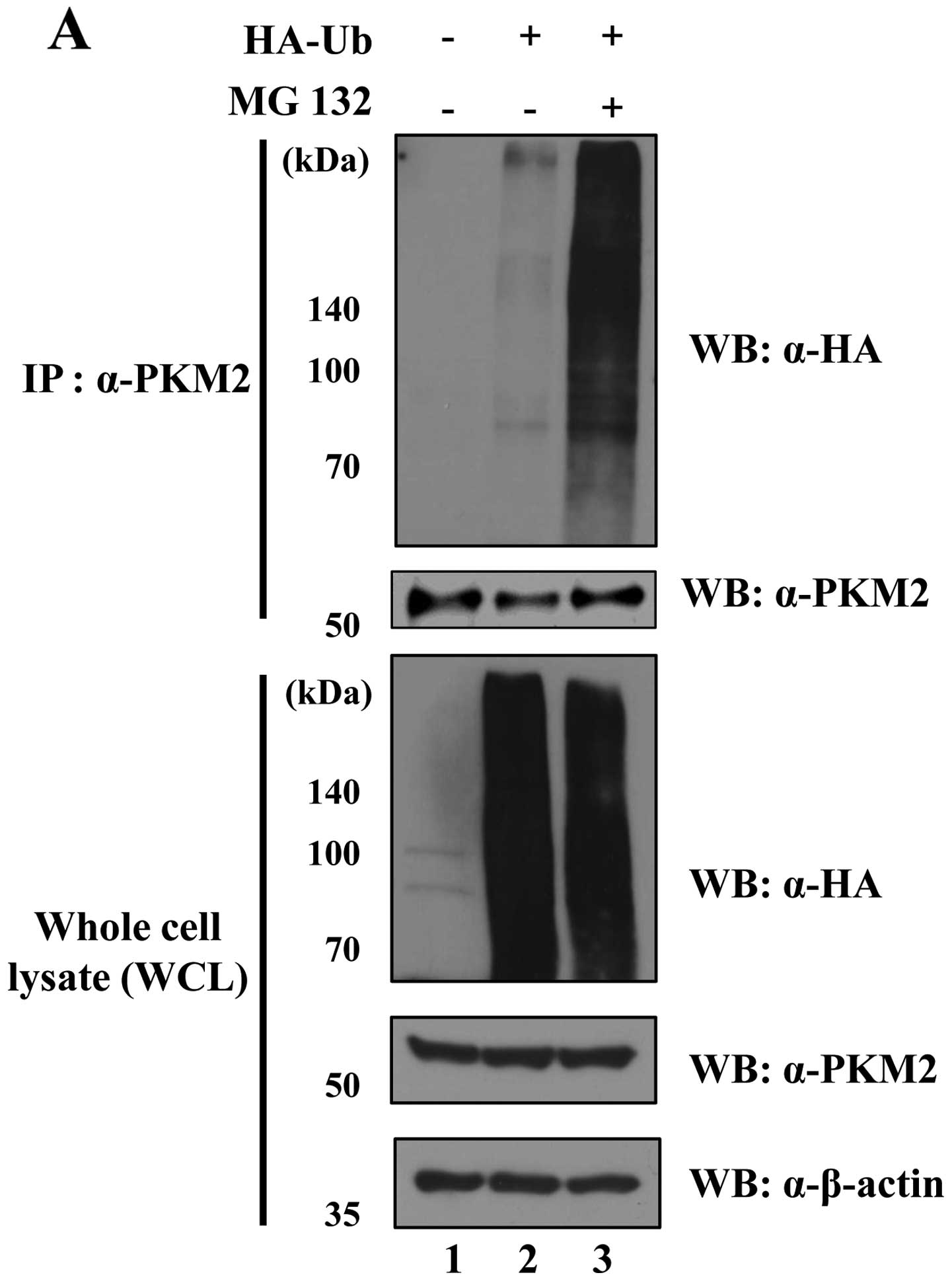
PKM2 is regulated by the
ubiquitin-proteasome system
A previous study revealed the posttranslational
modification of PKM2 (25). For
example, extracellular signal-regulated kinase (ERK) phosphorylates
PKM2, leading to the induction of c-Myc expression as a
transcription factor with β-catenin as a co-transcription factor in
the nucleus (25). However,
evidence of PKM2 that undergoes the proteasomal degradation pathway
has not yet been found. Since we demonstrated the interaction
between USP20 and PKM2 (Fig. 3),
we next determined whether PKM2 is degraded by the 26S proteasome.
The level of ubiquitination for PKM2 was dramatically increased
with the treatment of MG132, a proteasome inhibitor (Fig. 4A). Polyubiquitination has different
roles in cells depending on the lysine site. While Lys48-linked
polyubiquitination is involved in the degradation of proteins
through the 26S proteasome, Lys63-linked polyubiquitination is
related to intracellular signaling pathways (26). In order to analyze the
ubiquitination pattern of PKM2, we designed mutant constructs of
ubiquitin; HA-ubiquitin (K48R) and HA-ubiquitin
(K63R). Polyubiquitination has different roles in cells
depending on the lysine site. K48R and K63R constructs were only
Lys48 or Lys63 changed to Arg. On transfection of HA-ubiquitin
(K48R), we found a dramatically increased ubiquitination level
of PKM2. In contrast, the K63-mutated ubiquitin did not increase
PKM2 ubiquitination (Fig. 4B).
These results suggest that PKM2 can build polyubiquitin chains on
both K48 and K63, and be regulated by the ubiquitin proteasome
pathway.
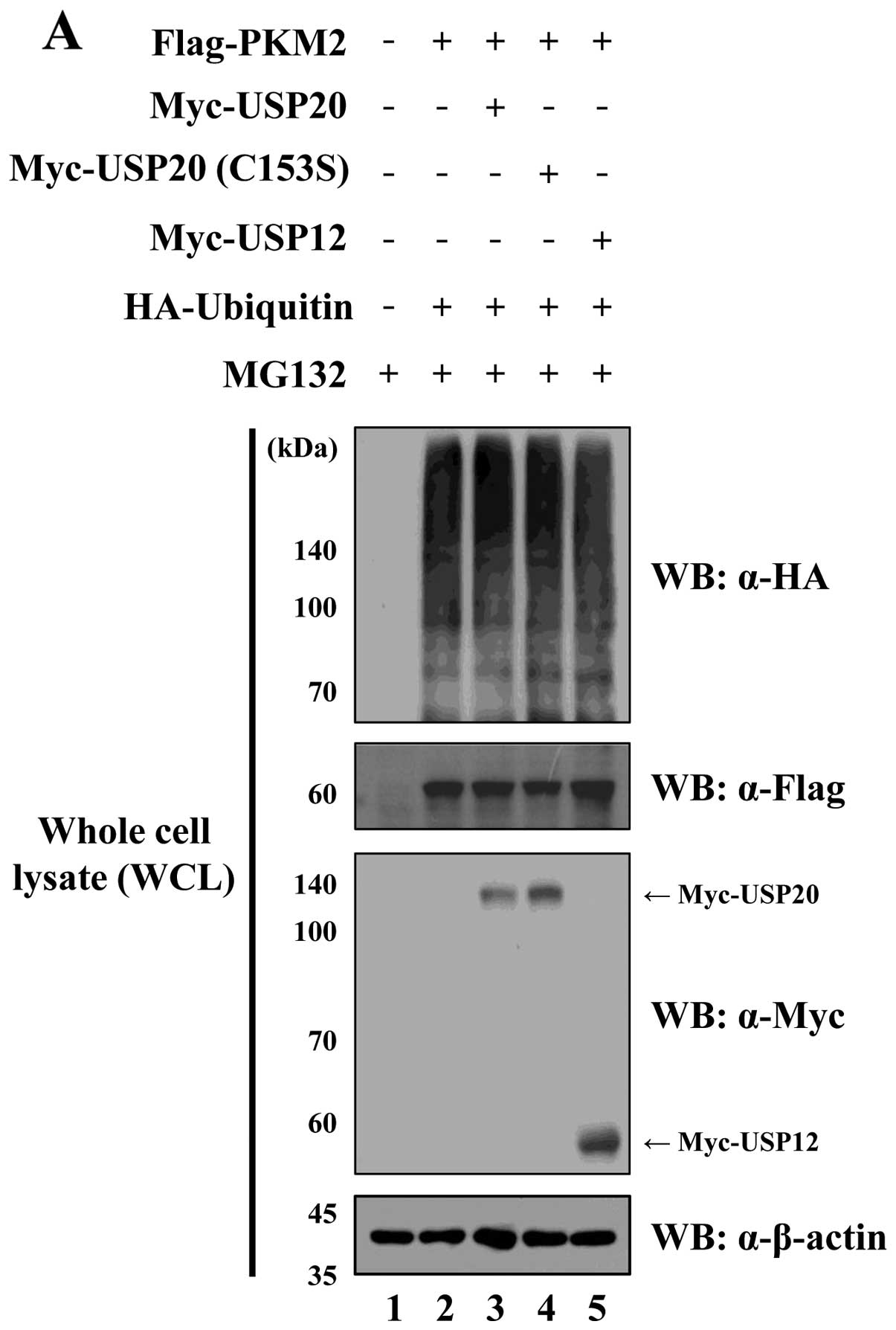
USP20 regulates PKM2 by its DUB
activity
To identify the molecular mechanism by which USP20
engages in DUB activity for PKM2 regulation, Usp20 and
Pkm2 were expressed with HA-ubiquitin in HeLa cells.
Additionally, USP20 (C153S) lacking catalytic activity was used as
a negative control for catalytic activity for USP20. When Flag-PKM2
was immunoprecipitated by an anti-Flag antibody, the wild-type
USP20 dramatically reduced the ubiquitination level of PKM2.
However, the catalytically inactive form of USP20 and USP12 as a
negative control could not affect the ubiquitination level of PKM2
(Fig. 5). These data confirm that
USP20 is a specific DUB for PKM2.
To determine whether USP20 has effect on the
regulation of PKM2 by DUB activity, we expressed USP20 protein in a
dose-dependent manner in HeLa cells. The result indicated that PKM2
expression did not increase in the USP20 transfected condition
(Fig. 5C). Taken together, these
data indicate that USP20 can be a specific DUB for PKM2, and it
would regulate the signaling progress of PKM2.
Discussion
Ubiquitination is a crucial mechanism for regulating
a number of cellular processes (27). DUBs mediate the reversible reaction
of ubiquitination by depolymerization and removal of ubiquitin from
target proteins. These enzymes are very important in cellular
homeostasis and cancer (28,29).
Many studies have demonstrated the importance of DUBs and their
functions in cancers and other cellular functions. In the mechanism
of carcinogenesis, DUBs not only play a tumor suppression role in
cancer cells, but also have oncogenic function by stabilizing some
oncogenes (30). Therefore,
specific inhibitors of DUBs have been proposed as putative
therapeutic targets (31).
Herein, we investigated USP20, as an important
enzyme in diverse cellular mechanisms, regulating TRAF6, HIF1-α,
β2AR, and Rad17 (18,32).
However, the detailed mechanisms of USP20 activity, and its diverse
functions are not fully understood compared to the other DUBs.
In this study, we performed 2-DE analysis and
identified PKM2 as one of the target proteins for USP20. PKM2 is a
main protein in tumor metabolism. In 1924, Otto Warburg identified
a specific metabolism in cancer cells. In cancer cells, glucose was
converted to lactate in the normoxia, that is known as the Warburg
effect, or aerobic glycolysis (33). In this effect, PKM2 accumulates
pyruvate converting it to lactate. As a result, the pentose
phosphate pathway (PPP) is promoted and macromolecules are
synthesized for use in the proliferation of cancer cells (34). When PKM2 is localized in the
nucleus, it interacts with HIF-1α and promotes the transactivation
of encoding cell proliferation proteins in cancer cells. In the
prolyl hydroxylase 3 (PHD3)-stimulated condition, PHD3 enhances the
binding with PKM2 and HIF-1α. The binding of PKM2 and HIF-1α
induces the expression of HIF-1α-targeted proteins, such as GLUT1,
LDHA, PDK1, and VEGF, GLUT1, LDHA, and PDK1 inducing the glycolytic
pathway in cancer cells, while VEGF is an angiogenesis protein
(20).
We confirmed the interaction between USP20 and PKM2,
using a co-immunoprecipitation assay and immunofluorescence
analysis, and the result showed that USP20 was bound with PKM2 and
they are co-localized in the cytosol (Fig. 3). We also investigated the patterns
of ubiquitination for PKM2. We generated mutant constructs of
ubiquitin, and found that PKM2 is ubiquitinated by Lys48- and
Lys63-polyubiquitin chains.
In summary, we found PKM2 to be a target of USP20
and demonstrated the specific deubiquitination of PKM2 by USP20.
Through these results, it is feasible to elucidate the cellular
cancer mechanism and identify a new target for cancer therapy. In
order to investigate the diverse regulation of USP20 and PKM2,
further functional studies should be performed.
Acknowledgements
This work was supported by Basic Science Research
Program through the National Research Foundation of Korea (NRF)
funded by the Ministry of Education, Science and Technology
(2013-0141), and by Exchange Professor Program of LG Yonam
Foundation (2012).
References
|
1
|
Yang WL, Zhang X and Lin HK: Emerging role
of Lys-63 ubiquitination in protein kinase and phosphatase
activation and cancer development. Oncogene. 29:4493–4503. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cole AJ, Clifton-Bligh RJ and Marsh DJ:
Ubiquitination and cancer: Histone H2B monoubiquitination: roles to
play in human malignancy. Endocr Relat Cancer. 22:T19–T33. 2015.
View Article : Google Scholar
|
|
3
|
Tu Y, Chen C, Pan J, Xu J, Zhou ZG and
Wang CY: The Ubiquitin Proteasome Pathway (UPP) in the regulation
of cell cycle control and DNA damage repair and its implication in
tumorigenesis. Int J Clin Exp Pathol. 5:726–738. 2012.PubMed/NCBI
|
|
4
|
Weissman AM, Shabek N and Ciechanover A:
The predator becomes the prey: Regulating the ubiquitin system by
ubiquitylation and degradation. Nat Rev Mol Cell Biol. 12:605–620.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hershko A and Ciechanover A: The ubiquitin
system for protein degradation. Annu Rev Biochem. 61:761–807. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kawadler H and Yang X: Lys63-linked
polyubiquitin chains: Linking more than just ubiquitin. Cancer Biol
Ther. 5:1273–1274. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rieser E, Cordier SM and Walczak H: Linear
ubiquitination: A newly discovered regulator of cell signalling.
Trends Biochem Sci. 38:94–102. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hussain S, Zhang Y and Galardy PJ: DUBs
and cancer: The role of deubiquitinating enzymes as oncogenes,
non-oncogenes and tumor suppressors. Cell Cycle. 8:1688–1697. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Todi SV and Paulson HL: Balancing act:
Deubiquitinating enzymes in the nervous system. Trends Neurosci.
34:370–382. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lim KH, Ramakrishna S and Baek KH:
Molecular mechanisms and functions of cytokine-inducible
deubiquitinating enzymes. Cytokine Growth Factor Rev. 24:427–431.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kouranti I, McLean JR, Feoktistova A,
Liang P, Johnson AE, Roberts-Galbraith RH and Gould KL: A global
census of fission yeast deubiquitinating enzyme localization and
interaction networks reveals distinct compartmentalization profiles
and overlapping functions in endocytosis and polarity. PLoS Biol.
8:e10004712010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bhattacharya S and Ghosh MK: Cell death
and deubiquitinases: perspectives in cancer. Biomed Res Int.
2014:4351972014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
MacGurn JA, Hsu PC and Emr SD: Ubiquitin
and membrane protein turnover: From cradle to grave. Annu Rev
Biochem. 81:231–259. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ferreira LM: Cancer metabolism: The
Warburg effect today. Exp Mol Pathol. 89:372–380. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Grüning NM, Rinnerthaler M, Bluemlein K,
Mülleder M, Wamelink MM, Lehrach H, Jakobs C, Breitenbach M and
Ralser M: Pyruvate kinase triggers a metabolic feedback loop that
controls redox metabolism in respiring cells. Cell Metab.
14:415–427. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gao X, Wang H, Yang JJ, Liu X and Liu ZR:
Pyruvate kinase M2 regulates gene transcription by acting as a
protein kinase. Mol Cell. 45:598–609. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li Z, Wang D, Messing EM and Wu G: VHL
protein-interacting deubiquitinating enzyme 2 deubiquitinates and
stabilizes HIF-1alpha. EMBO Rep. 6:373–378. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yasunaga J, Lin FC, Lu X and Jeang KT:
Ubiquitin-specific peptidase 20 targets TRAF6 and human T cell
leukemia virus type 1 tax to negatively regulate NF-kappaB
signaling. J Virol. 85:6212–6219. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shanmugam I, Abbas M, Ayoub F, Mirabal S,
Bsaili M, Caulder EK, Weinstock DM, Tomkinson AE, Hromas R and
Shaheen M: Ubiquitin-specific peptidase 20 regulates Rad17
stability, checkpoint kinase 1 phosphorylation and DNA repair by
homologous recombination. J Biol Chem. 289:22739–22748. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Luo W, Hu H, Chang R, Zhong J, Knabel M,
O’Meally R, Cole RN, Pandey A and Semenza GL: Pyruvate kinase M2 is
a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell.
145:732–744. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Semenza GL: Regulation of metabolism by
hypoxia-inducible factor 1. Cold Spring Harb Symp Quant Biol.
76:347–353. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim YS, Kim MS, Lee SH, Choi BC, Lim JM,
Cha KY and Baek KH: Proteomic analysis of recurrent spontaneous
abortion: Identification of an inadequately expressed set of
proteins in human follicular fluid. Proteomics. 6:3445–3454. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Choi BK, Cho YM, Bae SH, Zoubaulis CC and
Paik YK: Single-step perfusion chromatography with a throughput
potential for enhanced peptide detection by matrix-assisted laser
desorption/ionization-mass spectrometry. Proteomics. 3:1955–1961.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ramakrishna S, Suresh B, Lee EJ, Lee HJ,
Ahn WS and Baek KH: Lys-63-specific deubiquitination of SDS3 by
USP17 regulates HDAC activity. J Biol Chem. 286:10505–10514. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo
F, Lyssiotis CA, Aldape K, Cantley LC and Lu Z: ERK1/2-dependent
phosphorylation and nuclear translocation of PKM2 promotes the
Warburg effect. Nat Cell Biol. 14:1295–1304. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Perrett CA, Lin DY and Zhou D:
Interactions of bacterial proteins with host eukaryotic ubiquitin
pathways. Front Microbiol. 2:1432011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ramakrishna S, Suresh B and Baek KH: The
role of deubiquitinating enzymes in apoptosis. Cell Mol Life Sci.
68:15–26. 2011. View Article : Google Scholar
|
|
28
|
D’Arcy P, Brnjic S, Olofsson MH, et al:
Inhibition of proteasome deubiquitinating activity as a new cancer
therapy. Nat Med. 17:1636–1640. 2011. View
Article : Google Scholar
|
|
29
|
Vucic D, Dixit VM and Wertz IE:
Ubiquitylation in apoptosis: A post-translational modification at
the edge of life and death. Nat Rev Mol Cell Biol. 12:439–452.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fraile JM, Quesada V, Rodríguez D, Freije
JM and López-Otín C: Deubiquitinases in cancer: New functions and
therapeutic options. Oncogene. 31:2373–2388. 2012. View Article : Google Scholar
|
|
31
|
Mattern MR, Wu J and Nicholson B:
Ubiquitin-based anticancer therapy: Carpet bombing with proteasome
inhibitors vs surgical strikes with E1, E2, E3, or DUB inhibitors.
Biochim Biophys Acta. 1823:2014–2021. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Berthouze M, Venkataramanan V, Li Y and
Shenoy SK: The deubiquitinases USP33 and USP20 coordinate beta2
adrenergic receptor recycling and resensitization. EMBO J.
28:1684–1696. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Semenza GL: HIF-1: Upstream and downstream
of cancer metabolism. Curr Opin Genet Dev. 20:51–56. 2010.
View Article : Google Scholar :
|