Introduction
Endometrial cancer is one of the most common
malignant tumors of the reproductive tract among women in developed
counties. In Poland in 2010, over five thousand women were
diagnosed with endometrial cancer of whom over a thousand died
(http://epid.coi.waw.pl/krn/). The
average age at diagnosis is 65, and mortality rate increases with
the age of patient (1,2). Many potential risk factors have been
characterized, such as obesity, menopausal status, unopposed
estrogens, low parity and nullparity and history of cancer in
family (1–3) however, the mechanisms by which they
work are not fully understood.
Moreover, conditions such as diabetes mellitus and
polycystic ovarian syndrome increase the risk of endometrial cancer
development (4). Most endometrial
cancers are sporadic, and can be divided into two general groups
based on histopathological features and molecular profiles:
endometroid endometrial cancers (EECs) and non-endometroid
endometrial cancers (NEECs) (1–3).
Type I, or endometrioid endometrial cancers (EEC), are more common
and characterized by estrogen-dependence, microsatellite
instability, loss of heterozygosity of the PTEN tumor
suppressor gene and genetic alterations in the RAS and WNT
pathways. Type II, non-endometrioid endometrial cancers (NEECs),
are characterized by estrogen-independence with an abnormal
accumulation of p53 protein, inactivation of the p16 gene,
overexpression of the HER2/neu gene and decreased expression
of the CDH1 gene (5).
However, it is important to note that the molecular pathways of the
two cancer types are not fully understood.
The epithelial-mesenchymal transition (EMT) is a
complex process which results in transformation of epithelial to
mesenchymal cells. Cells of the epithelium are characterized by
polarity and non-motility, while mesenchymal cells are loosely
organized and able to migrate (6–8).
These features are a consequence of changes in gene expression
profile i.e., increases in expression of mesenchymal markers such
as vimentin and N-cadherin, and donwregulation of epithelial
markers such as E-cadherin and cytokeratins, which act as thigh and
adherence junction proteins (9,10).
Although EMT is associated with embryogenesis and organ development
under physiological conditions, it has also been associated with
cancer progression and metastasis (10–12).
A number of transcription factors, i.e., SNAI1, SNAI2, TWIST and
ZEB1/2, which are known as the E-cadherin repressors, are the key
regulators triggering EMT progression (9,11,13).
Changes in EMT marker expression are important in endometrial
cancer, as cancer cells invade the myometrium, and this invasion is
considered one of the most important prognostic factors for types I
and II EC (14).
The WW domain-containing oxidoreductase gene
(WWOX), also known as FOR or WOX1, is a tumor
suppressor gene which spans more than one million base pairs mapped
at locus 16q23.3-q24 in the common chromosomal fragile site FRA16D.
The WWOX gene encodes a chain of 414 amino acids
constituting a 46-kDa protein containing two WW domains in the
NH2-terminal region with a short-chain dehydrogenase/reductase
domain, which suggests that it may interact with the receptors
regulating steroid hormones. The first WW domain interacts with
many partners such as ERBB 4 (15,16),
transcription factor AP2γ (17,18),
YAP (15), Jun (19), HIF1α (20) and others (21). A high level of WWOX
expression has been observed in hormone-dependent tissues such as
those of the ovary, mammary gland or testis, which together with
the enzymatic specificity of the SDR domain, indicates its role in
the metabolism of steroid hormones (22,23).
Loss of WWOX expression has been observed during the
development of various types of cancer. Moreover, numerous studies
have demonstrated correlations between the decrease of WWOX
expression and the presence of adverse prognostic factors,
including shorter survival and a higher degree of malignancy in
various types of carcinoma, especially such hormone-dependent ones
as those of the breast (5,16,24),
prostate (25), ovary (26) and others (27–31).
The suppressive character of the WWOX gene has been
confirmed in many in vitro studies, where WWOX
overexpression resulted in inhibition of cell proliferation,
increased apoptosis and inhibition of anchorage-independent growth
(32–34). Moreover, mice with a knocked out
WWOX gene resulted in a greater incidence of numerous
disorders such as bone metabolic defects, improper glandular
development and disruption of steroidogenesis (23). However, the experiments performed
in our laboratory on variety of tumor types, indicated that
WWOX functioning in the cell appears to be more complex and
is probably not only limited to well-known tumor suppressor
properties.
We redirected our interest into WWOX
regulation of cell adhesion and motility properties, as Gourley
et al reported that overexpression of WWOX in an
ovarian cancer cell line resulted in decreased adhesion to
fibronectin (35), and WWOX
ectopic overexpression in breast cancer and colon cancer cell lines
has also been observed to result in increased potential of invasion
with the modulation of cell adhesive properties (36,37).
Moreover, in our previous study on endometrial cancer, we observed
that WWOX negatively correlated with CDH1 expression,
a well known epithelial marker downregulated during EMT (38). The aim of the study, as a
continuation of previous observations, was to determine its role in
the EMT process using both an in vitro model and endometrial
tumor samples.
The overexpression of WWOX in an ECC1
well-differentiated steroid-responsive endometrial cell line was
induced via stable retroviral transduction. The changes in global
gene expression profile after WWOX induction were assessed
by microarray and the biological effects connected with EMT process
were assessed by migration through a basement membrane,
anchorage-independent growth, adhesion to ECM proteins and MMPs
activity. RT-qPCR analysis of chosen EMT markers was also performed
on 164 tumor samples.
Materials and methods
Tumor samples
The study included 164 endometrioid endometrial
carcinomas (EECs, type I) obtained from the Department of
Gynecological Oncology, Medical University of Lodz. The samples
were classified according to FIGO classification system and also
classified into risk of recurrence category according to Pecorelli
(39). The mean age was 62 years
(median 61; range 31–83 years). Tumor samples were stored at −80°C
in RNA Later buffer (Ambion®; Thermo Fisher Scientific,
Waltham, MA, USA).
The study was performed after receiving the consent
of the patients, and was approved by the the Ethics Committee of
the Medical University of Lodz (RNN/208/11/KE).
Cell lines and reagents
The ECC-1 cell line was obtained from the American
Type Culture Collection (ATCC). This endometrial cancer cell line
is hormone-responsive, as well as estrogen (α and β), androgen and
progesterone receptor-positive (40). Cells were cultured in advanced
RPMI-1640 medium supplemented with heat-inactivated fetal bovine
serum (Invitrogen Life Technologies, Carlsbad, CA, USA),
L-glutamine (Invitrogen Life Technologies), HEPES buffer
(Invitrogen Life Technologies) and antibiotics (penicillin 50 U/ml;
streptomycin 50 μg/ml; neomycin 100 μg/ml) (Invitrogen Life
Technologies). The cells were incubated at 37°C in a humidified
incubator with 5% CO2.
Transduction
The process was performed with retroviral vector
pLNCX2, either with or without WWOX cDNA. To increase the
effectiveness of transduction, the infectious medium was
supplemented with 8 μg/ml polybrene (Sigma-Aldrich, MO, USA). After
24 h, the medium was removed and replaced with advanced RPMI-1640
(Invitrogen Life Technologies) with standard supplements. For
selection, the cells were cultured in the presence of 400 mg/ml
G418 (Sigma-Aldrich) for 14 days. The stable transductants were
analyzed by RT-qPCR and western blot assay.
RT-qPCR
Total RNA from cells and tumor tissues was isolated
using TRIzol reagent (Invitrogen Life Technologies), and 10 μg of
RNA was reverse-transcribed with ImProm RT-II reverse transcriptase
(Promega Corp., Madison, WI, USA).
RT-qPCR was performed with LightCycler 480 II (Roche
Diagnostics GmbH, Mannheim, Germany). PCR products were detected
with SYB R Green I and qPCR Core kit for SYBR Green I (Eurogentec,
Southampton, UK). Reactions were performed in duplicate. The
relative expression of WWOX, CDH1, and VIM were
analysed, as were two CDH1 repressors: ZEB1 and
SNAI1. The relative expression of all investigated genes was
normalized to three reference genes: RPLPO, RPS17 and
H3F3A. All information regarding primer sequences, PCR
reaction conditions and the length of received products is
available upon request. The relative expression level of all
investigated genes was calculated using the Roche algorithm
(41) with the Universal Human
Reference RNA (Stratagene, La Jolla, CA, USA) as a calibrator.
Microarray assay
The microarrays were performed using Human OneArray
Whole Genome Microarray v 5.1 (Phalanx Biotech, USA) in four
replicates (dual labelling protocol). A ULS™ Labeling kit (Kreatech
Diagnostics, The Netherlands) was used to alternately/cross label
single-stranded cDNA of two variants (ECC1/vector, ECC1/WWOX) with
Cy3 and Cy5 as well as Universal Human Reference RNA
(Stratagene).
The slide was prepared for hybridization and
pre-hybridization according to the manufacturer’s protocol. The
hybridization process was performed in a 2X SSPE humidity chamber
at 42°C for 16–18 h. The following buffers were used for
post-hybridization washes: 1X SSPE/0.03% SDS (2 min, 42°C), 1X SSPE
(2 min, RT), 0.1X SSPE (rinsed several times, RT) (Phalanx
Biotech).
ProScanArray (Perkin-Elmer, USA) was used for
scanning and ScanArray Express for preliminary normalization.
Lowess and statistical analysis were performed with TM4 software
suite provided by The Institute for Genomic Research (http://www.tm4.org/). After modified t-test analysis,
the fold change was calculated and the Pantherdb online ontology
application (www.pantherdb.org) was used to classify
the received genes according to ontological terms. The results were
regarded as significant at p<0.05.
Protein extraction and western
blotting
The endometrial cancer cells were lysed in RIPA
protein extraction buffer supplemented with proteases, phosphatase
inhibitor cocktail and PMSF (Sigma-Aldrich). The protein
concentration was measured using the Bradford method (Bio-Rad
Laboratories, CA, USA), run on 10% SDS-PAGE gel electrophoresis and
transferred to a PVDF membrane (Sigma-Aldrich). The membranes were
blocked in a buffer of 5% non-fat milk in TBST for 1 h at room
temperature and then incubated for 19 h at 4°C with primary
antibodies to goat anti-human WWOX, 1:100; mouse anti-E-cadherin,
1:100; mouse anti-vimentin, 1:100 (sc:20529, sc:8426, sc:51721,
Santa Cruz Biotechnology Inc., Dallas, TX, USA). Following
incubation, the membranes were washed three times with TBST and
incubated with secondary antibodies conjugated with alkaline
phosphatase (Sigma-Aldrich) for 1 h. The membranes were washed
three times in TBST and developed using Novex® AP
Chromogenic Substrate (Invitrogen Life Technologies).
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) at a dilution of
1:1,000 was used as the reference protein (sc-59540, Santa Cruz
Biotechnology Inc.). All biological tests were performed in a
minimum of three replicates.
Adhesion assay
Cell adhesion was evaluated using Cell Biolabs
CytoSelect™ Cell Adhesion assay kit (Cell BioLabs, San Diego, CA,
USA) according to the manufacturer’s instructions. This test
provides a rapid and quantitative method for accessing the cell
adhesion potential. The ECC1 cells were seeded at a density of
1.5×105 cells in a total volume of 150 μl serum-free
media to each well. The cells were incubated for 1 h at 37°C. The
absorbance was measured at 560 nm with a BioTek plate reader
(BioTek US, USA).
Cell invasion assay
The cell invasiveness potential was determined using
a Colorimetric CytoSelect™ Cell Invasion Assay Kit Cell (Cell
BioLabs) according to the manufacturer’s protocol. The cell
suspension was incubated at a density of 3×105/well in
serum-free media for 48 h. The absorbance was measured at 560 nm on
a BioTek reading plate (BioTek US). Each variant was tested in
quadruplicate.
Zymography
ECC1 cells were seeded in 6-well plates at a density
of 1.5×106/well. Twenty-four hours before collection,
the medium was replaced with serum-free RPMI medium.
Qubit® Protein assay kits (Invitrogen Life Technologies)
were used to quantify protein content. Samples containing 15 μg of
protein were separated in 10% SDS-page gel containing 2 mg/ml
gelatin. After electrophoresis, the gel was washed twice for 30 min
in 2.5% Triton X-100 wash buffer and then incubated at 37°C for 18
h in a solution containing 50 mM Tris (pH 7.5) (Sigma-Aldrich), 10
mM CaCl2 (Sigma-Aldrich) and 200 mM NaCl
(Sigma-Aldrich). Following this, the gel was stained with 0.125%
Coomassie brilliant blue R-250 (Sigma-Aldrich) and de-stained with
solution (30% methanol, 10% acetic acid, 60% distilled water)
(Sigma-Aldrich). A protein marker was used to measure the molecular
weights of the analysed enzymes. Densitometric analysis with ImageJ
Analysis 1.34s software (Wayane Rasband, National Institutes of
Health, USA, http://rsb.info.nih.gov/ij/) was used to quantify MMP
activities.
Soft agar
A soft agar assay was performed to analyze the
differences in anchorage-independent growth between ECC1/WWOX and
ECC1/vec. Six-well plates were coated with 2 ml 0.9% low melting
agarose (AppliChem, MO, USA) in RPMI-1640 with 10% FBS. Both
variants of ECC1 cells were suspended in 0.3% low melting agarose
at a concentration of 104 cells per well. The cells were
incubated at 37°C in a humidified incubator for 14 days. The plates
were stained with 1 ml of 0.005% crystal violet (Sigma-Aldrich) in
water and colonies were counted using ImageJ Analysis 1.34 s
software (Wayane Rasband, National Institutes of Health, USA,
http://rsb.info.nih.gov/ij/).
Statistical analysis
The correlation of expression was performed using
the non-parametric Spearman’s rank correlation test. The
Mann-Whitney t-test was used to determine the differences between
the levels of expression of the investigated genes and clinical
factors in patients with endometrial cancer. The Aspin-Welsch
t-test was used to determine the statistical significance of the
difference observed between cell variants in all biological
tests.
Results
Confirmation of WWOX gene transduction in
ECC1
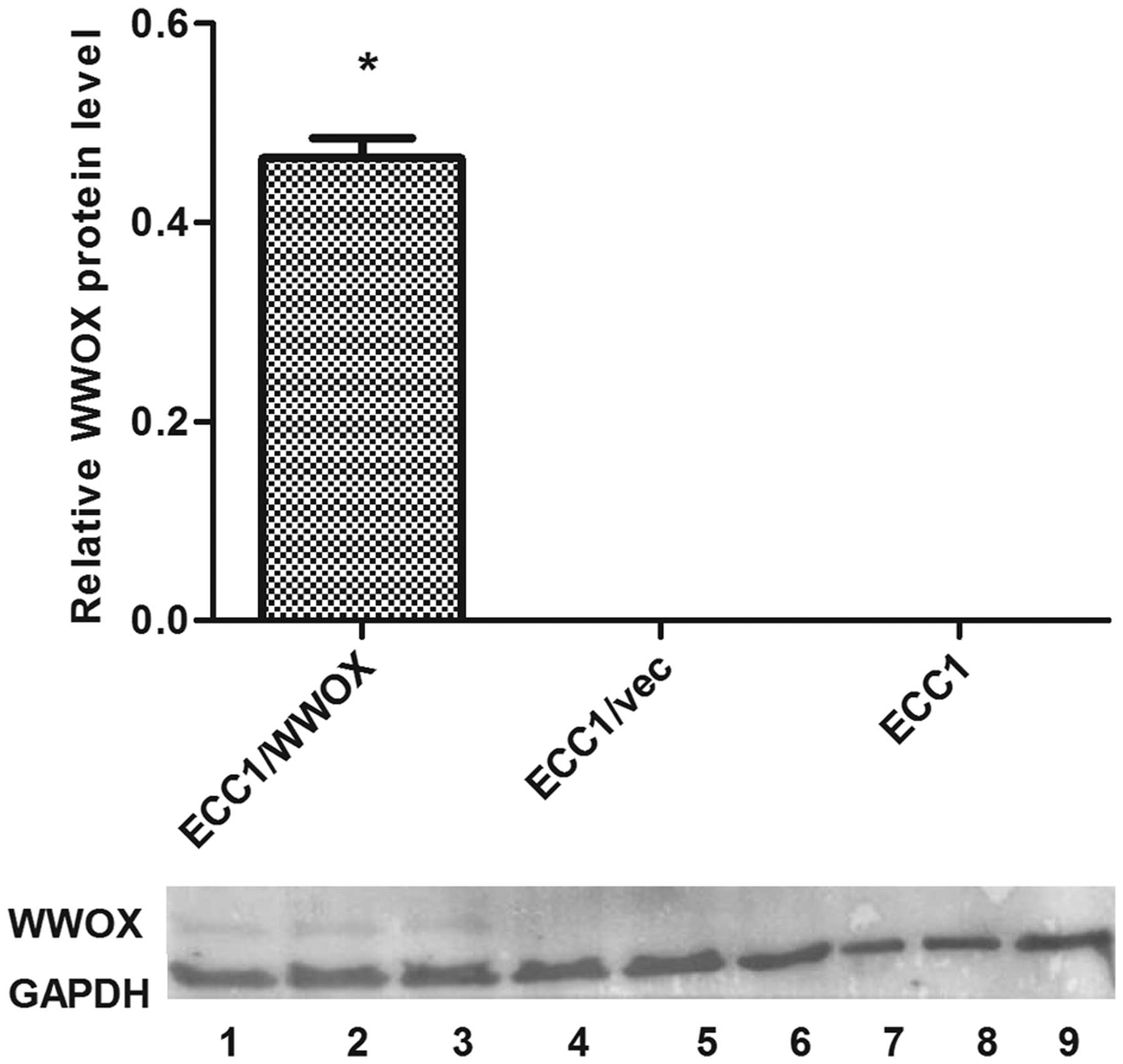
The transduction resulted in the increase of WWOX
expression both on the mRNA and protein levels. A >180-fold
change of WWOX gene expression was observed on the mRNA
level: the mean relative expression values being 0.019 and 3.34 for
ECC1/vector and ECC1/WWOX respectively. The change on the protein
level is shown (Fig. 1).
Microarray analysis
Our microarray analysis revealed more than 800 genes
with changed expression as the result of WWOX ectopic
overexpression (p<0.05). The identified genes belong to several
signalling pathways associated with regulation of cell
differentiation, apoptosis and cellular communication/tissue
architecture (Table I). The
predominant ones being the Wnt signalling pathway (13 genes),
integrin signalling pathway (10 genes), inflammation mediated by
chemokine and cytokine signalling pathway (6 genes),
gonadotropin-releasing hormone receptor pathway (11 genes),
angiogenesis (6 genes), TGF-β signalling pathways (8 genes),
apoptosis (5 genes), and the cadherin signaling pathway (6 genes).
The ontology classification (by cellular pathways) by WWOX
overexpression is included in Table
II. The two main EMT markers found in the cadherin signalling
pathway were vimentin and E-cadherin.
 | Table IOntological analysis of genes in
biological process regulated by WWOX gene in ECC1 cell
line. |
Table I
Ontological analysis of genes in
biological process regulated by WWOX gene in ECC1 cell
line.
| Biological
process | No. of genes |
|---|
| Metabolic
process | 368 |
| Primary metabolic
process | 299 |
| Unclassified | 266 |
| Cellular
process | 262 |
| Cell
communication | 147 |
| Biological
regulation | 138 |
|
Nucleobase-containing compound metabolic
process | 138 |
| Protein metabolic
process | 136 |
| Localization | 124 |
| Transport | 120 |
| Developmental
process | 117 |
| RNA metabolic
process | 102 |
| Regulation of
biological process | 87 |
| Transcription,
DNA-dependent | 82 |
| Transcription from
RNA polymerase II promoter | 80 |
| Multicellular
organismal process | 70 |
|
Single-multicellular organism process | 70 |
| Regulation of
nucleobase-containing compound metabolic process | 69 |
| System
development | 69 |
| Cell cycle | 68 |
| Response to
stimulus | 65 |
| Immune system
process | 62 |
| Regulation of
transcription from RNA polymerase II promoter | 62 |
| Cellular protein
modification process | 61 |
| Protein
transport | 60 |
| Intracellular
protein transport | 60 |
| System process | 60 |
| Regulation of
catalytic activity | 51 |
| Regulation of
molecular function | 51 |
| Neurological system
process | 48 |
| Cellular component
organization or biogenesis | 46 |
| Cellular component
organization | 43 |
| Mesoderm
development | 43 |
| Nervous system
development | 42 |
| Ectoderm
development | 42 |
| Cell adhesion | 41 |
| Biological
adhesion | 41 |
| Cell-cell
signaling | 41 |
| Anatomical
structure morphogenesis | 28 |
| Cell-cell
adhesion | 27 |
| Cellular component
morphogenesis | 26 |
| Death | 26 |
| Cell death | 26 |
| Apoptotic
process | 26 |
| Mitosis | 26 |
 | Table IIThe number of genes in pathways
regulated by WWOX gene in ECC1 cell line. |
Table II
The number of genes in pathways
regulated by WWOX gene in ECC1 cell line.
| Pathway | No. of genes |
|---|
| Unclassified | 667 |
| Wnt signalling
pathway | 13 |
|
Gonadotropin-releasing hormone receptor
pathway | 11 |
| Integrin signalling
pathway | 10 |
| TGF-β signalling
pathway | 8 |
| Alzheimer
disease-presenilin pathway | 7 |
| Angiogenesis | 6 |
| Inflammation
mediated by chemokine and cytokine signalling pathway | 6 |
| Huntington
disease | 6 |
| Cadherin signalling
pathway | 6 |
| Apoptosis
signalling pathway | 5 |
| Heterotrimeric
G-protein signalling pathway-Gq α and Go α mediated pathway | 5 |
A 1.6-fold increase in expression of the CDH1
gene and a 1.9-fold decrease in expression of VIM mRNA were
observed in the ECC1 cell line overexpressing WWOX.
Microarray data were submitted to the GEO Database (reference
series: GSE61041).
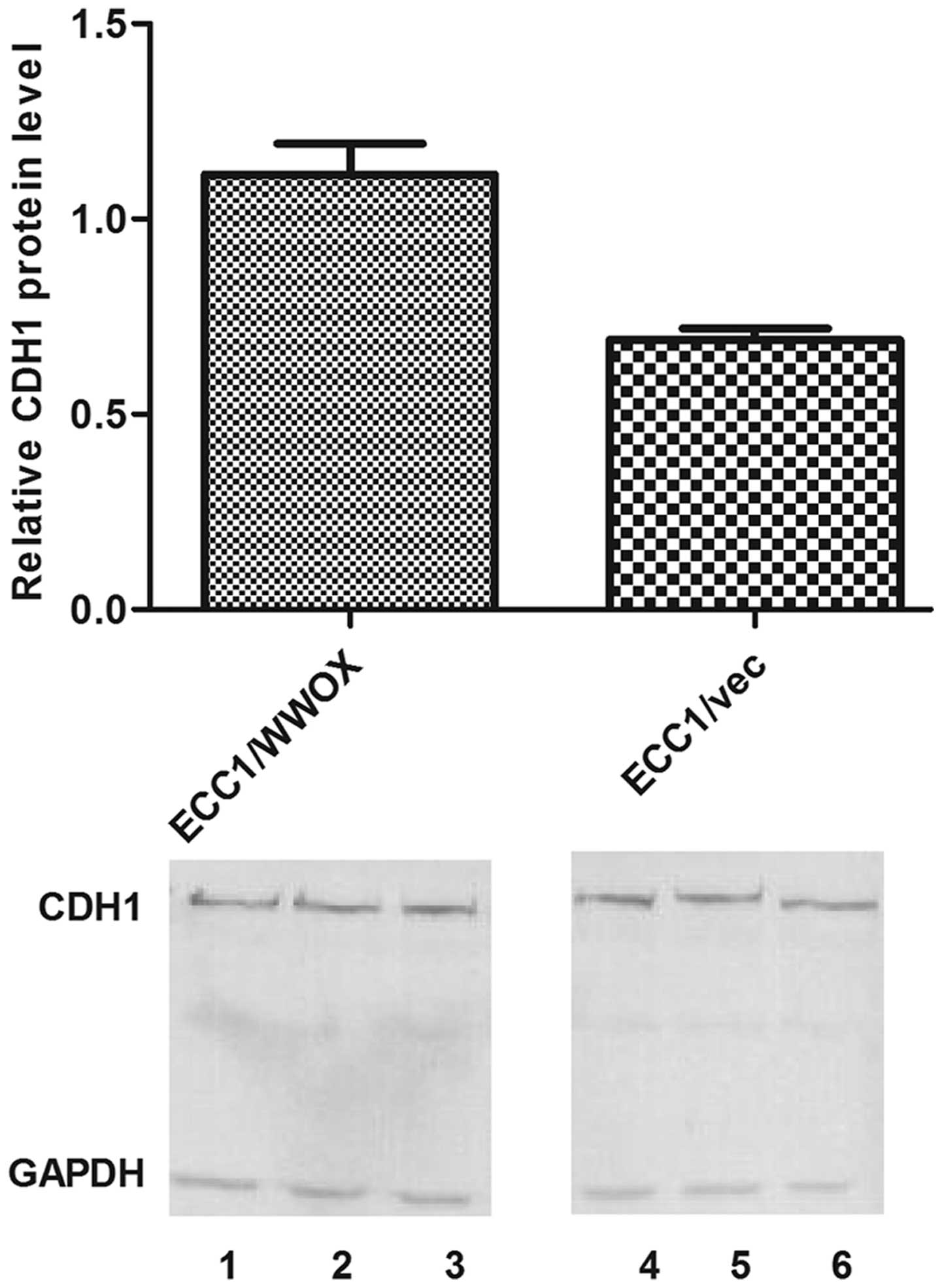
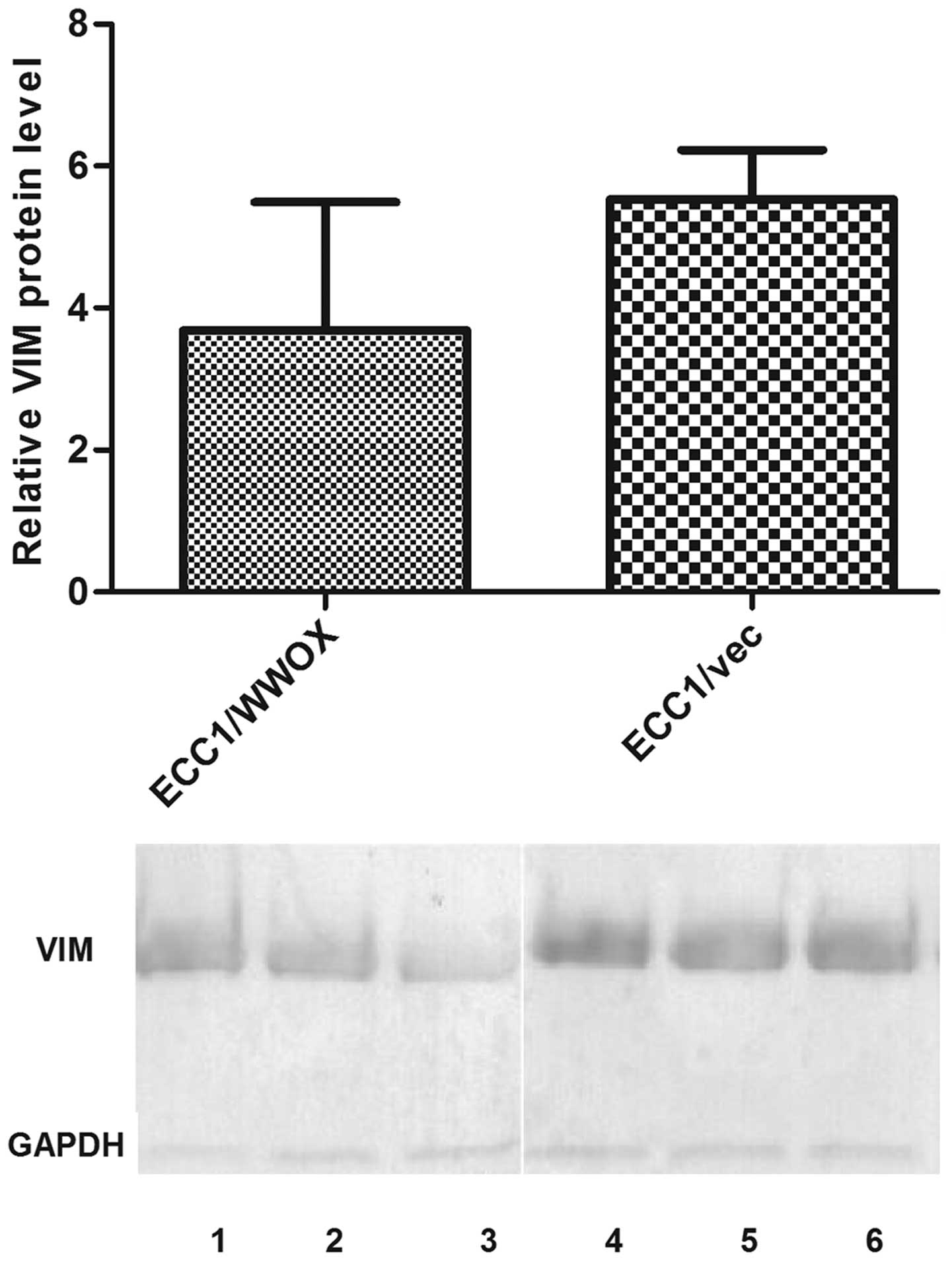
Microarray validation
The observed change in expression of CDH1 and
VIM was confirmed with RT-qPCR: a 3.5-fold increase of
CDH1 (mean relative expression 0.48 vs 1.67 for ECC1/vector
and ECC1/WWOX respectively); 2-fold decrease for VIM
(0.00035 vs 0.00019 for ECC1/vector and ECC1/WWOX respectively).
Similar changes were also noted on protein level (Figs. 2 and 3).
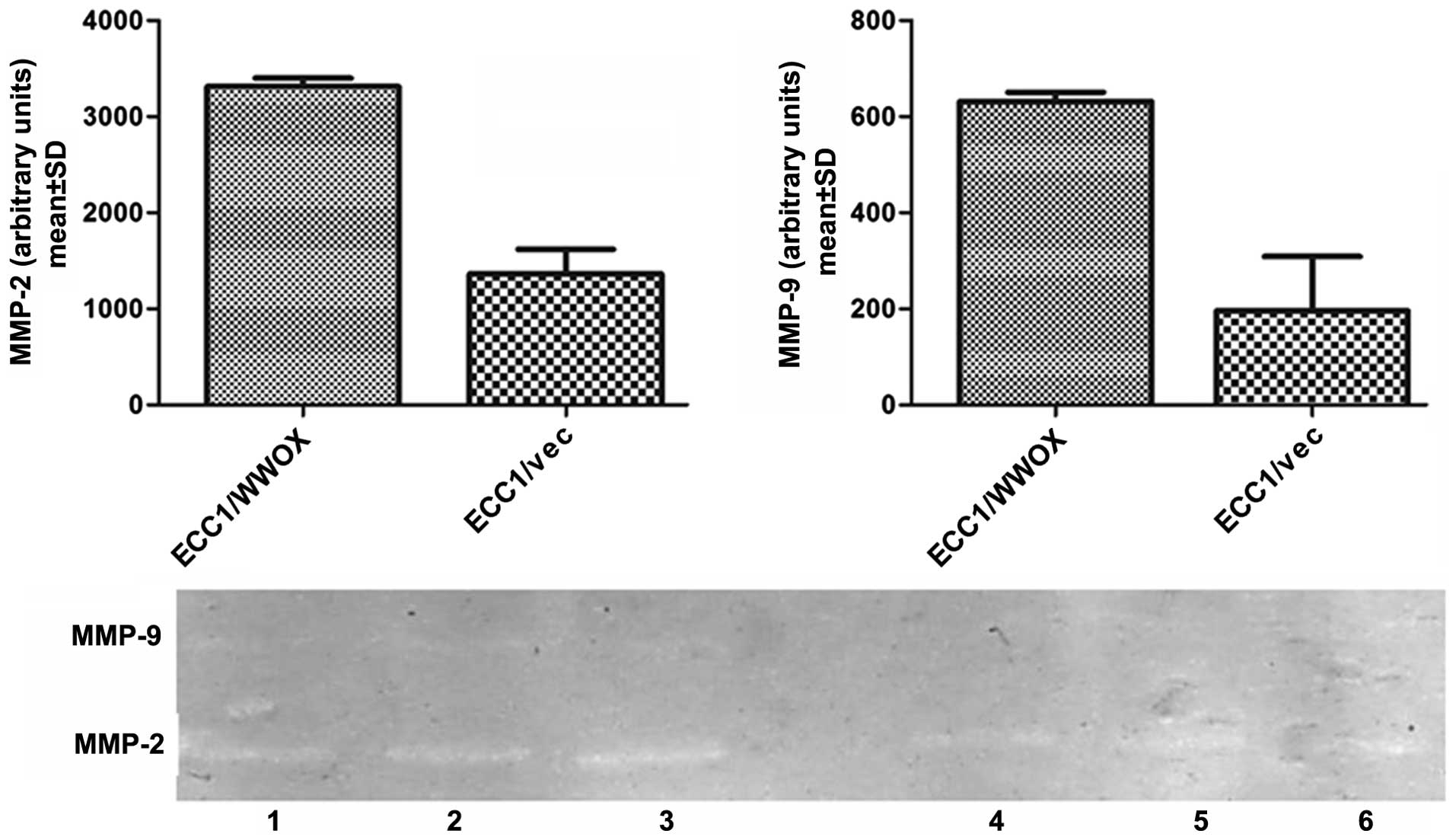
The influence of WWOX on invasiveness,
adhesion potential and cell growth in suspension
Overexpression of the WWOX gene resulted in a
1.3-fold increase in migration in comparison to the ECC1/vector
(p<0.05) (Fig. 4) and greater
induction of metalloproteinase activity i.e., MMP-2 and -9
(Fig. 5). These results suggest
that increased expression of WWOX influences the motility
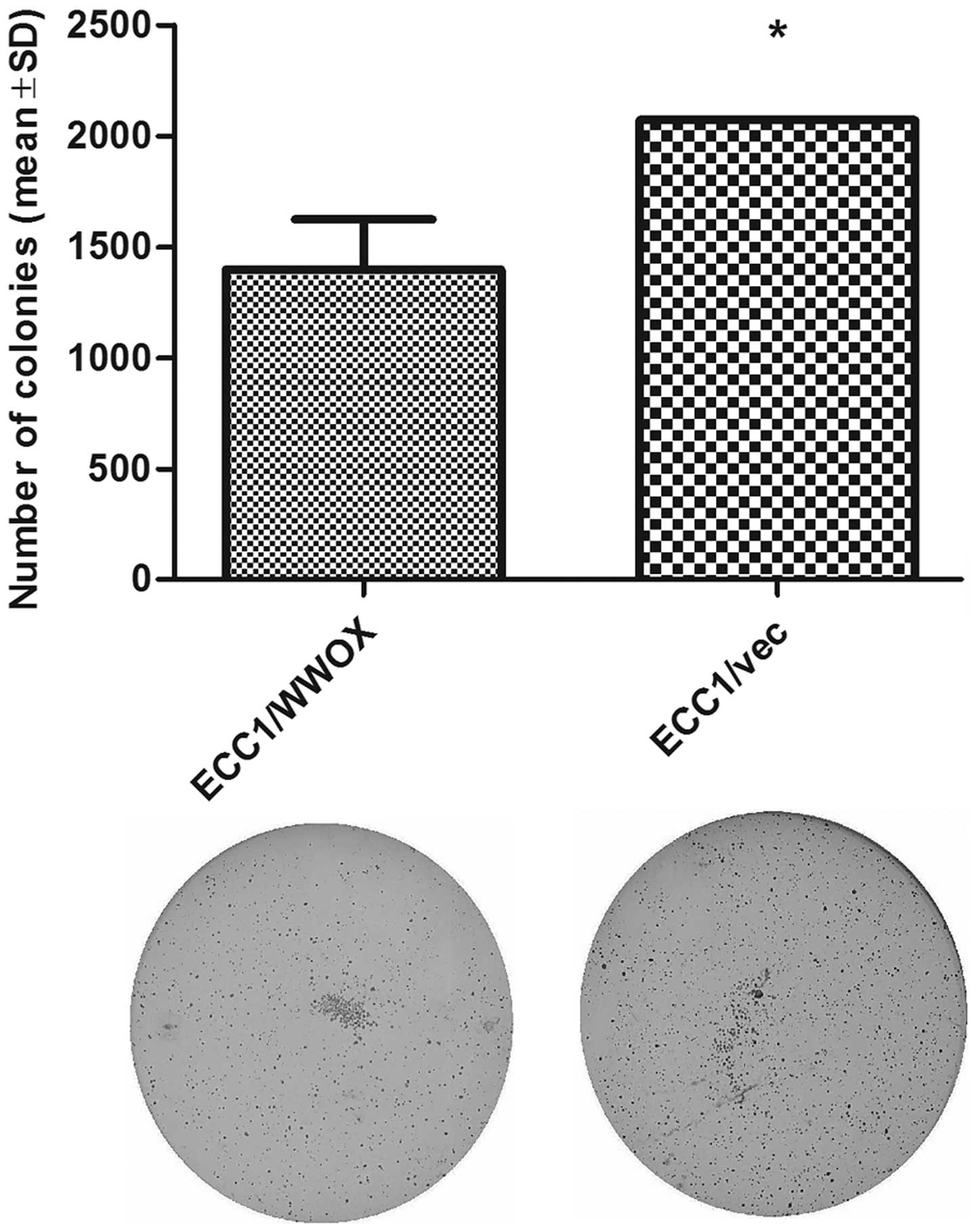
and invasive potential of the cells. However, the soft agar assay,
which measures the ability of cells to grow in suspension, in a
similar way to cancer cells being able to survive after detachment,
revealed a reduced colony number for the ECC1/WWOX cell line
variant (Fig. 6, p<0.05).
The invasion and growth in soft agar assay results
indicate that even though ECC1/WWOX cells demonstrated a higher
basic membrane transition ability, they were unable to survive
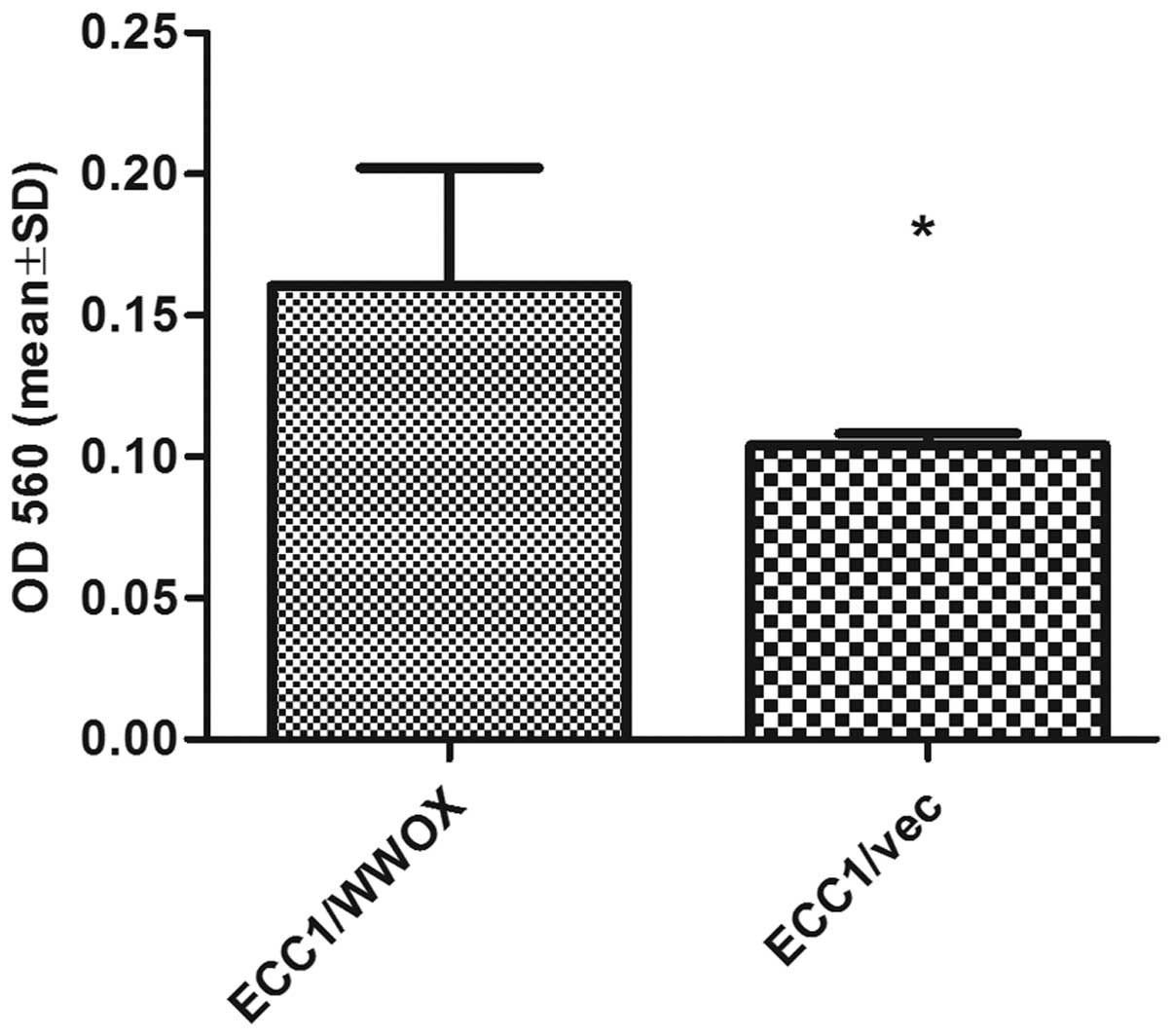
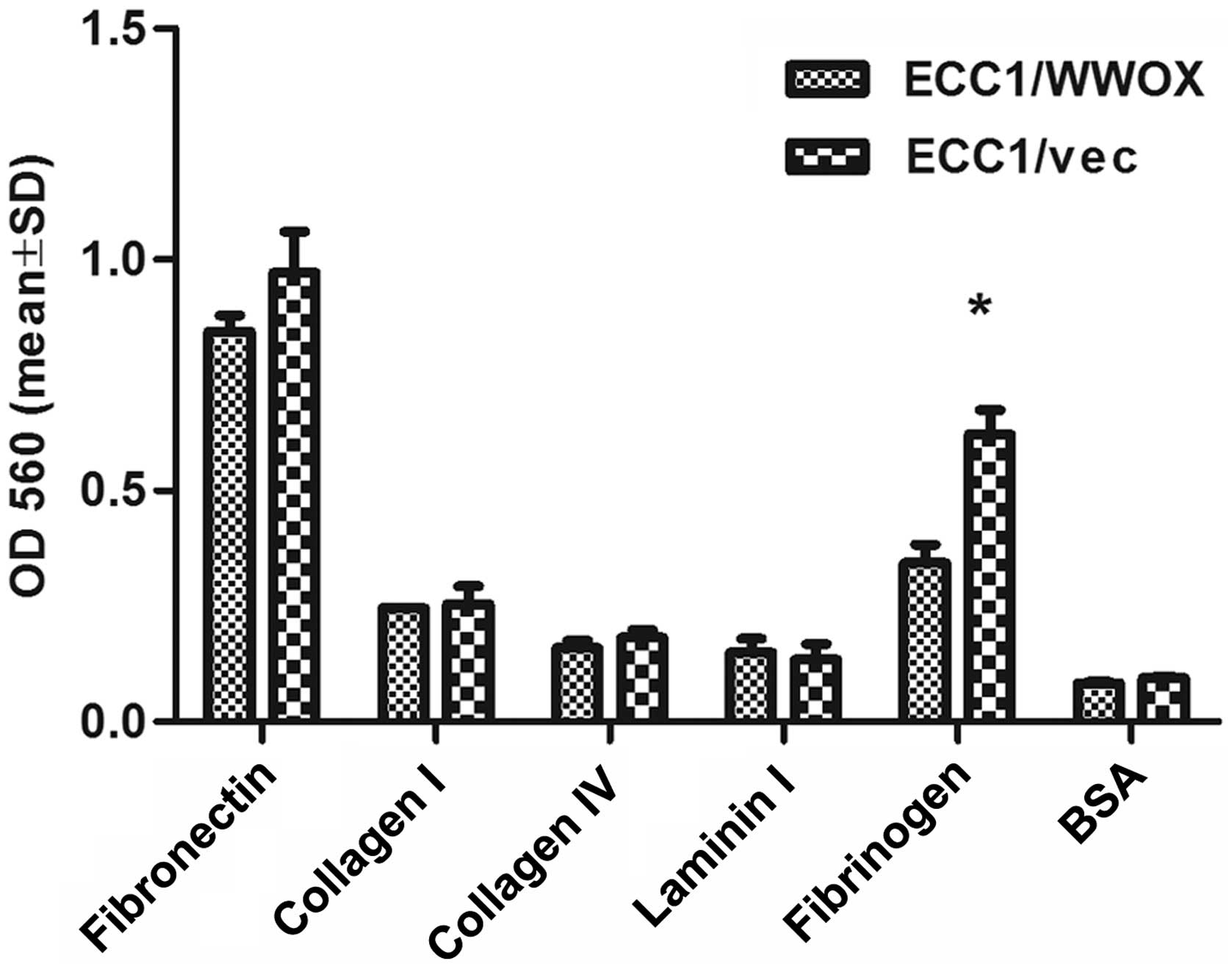
after detachment. The cells overexpressing WWOX demonstrated
a significantly lower ability to adhere to fibrinogen (1.8-fold
change; p<0.05) (Fig. 7).
Expression analysis in tumor samples
In the tumor samples, the EMT markers were also
analyzed in relation to WWOX. ZEB1 and SNAI1
were included in the EMT marker gene panel. A positive correlation
was observed between WWOX and ZEB1
(Rs=0.26; p=0.0004), and a negative correlation between
WWOX and VIM (Rs=-0.268, p=0.0005). A
negative correlation is known to exist between WWOX and
CDH1 (38) and this was
confirmed by the increased number of samples. The correlation with
SNAI1 was statistically insignificant (Rs=0.029;
p>0.05). The remaining correlations are presented in Table III. Similar to other tumor types,
the expression of WWOX in endometrial cancer inversely
correlated with the risk of cancer recurrence, i.e., tumors with
low risk demonstrated higher WWOX expression than those with
medium and high risk. A statistically significant difference was
found between the low (n=38, median expression 2.33) and medium
(n=83, median expression 1.12) risks of disease recurrence
(p=0.023). More detailed information of all correlations with
clinical factors is presented in Table IV.
 | Table IIIThe correlation analysis between the
levels of expression of WWOX and other EMT tumor-related
genes in endometrial carcinoma patients. |
Table III
The correlation analysis between the
levels of expression of WWOX and other EMT tumor-related
genes in endometrial carcinoma patients.
| Gene | Spearman’s rank
correlation | P-value |
|---|
|
WWOX/VIM | −0.268 | 0.0005 |
|
WWOX/ZEB1 | 0.260 | 0.0004 |
|
WWOX/CDH1 | −0.512 | 0.0001 |
|
WWOX/SNAI1 | 0.029 | 0.7114 (NS) |
 | Table IVThe correlation analysis between
WWOX, ZEB1, VIM, CDH1 genes and clinical factors in
endometrial carcinoma patients. |
Table IV
The correlation analysis between
WWOX, ZEB1, VIM, CDH1 genes and clinical factors in
endometrial carcinoma patients.
| Clinical
factor | n | Median of
WWOX mRNA level (range) | P-value | Median of
ZEB1 mRNA level (range) | P-value | Median of
VIM mRNA level (range) | P-value | Median of
CDH1 mRNA level (range) | P-value |
|---|
| Risk of
reccurence | | | | | | | | | |
| Low | 38 | 2.33
(1.24–3.40) | 0.012
(low/medium) | 0.35
(0.18–0.76) | 0.06
(low/medium) | 0.06
(0.02–0.17) | 0.44
(low/medium) | 0.34
(0.05–0.69) | 0.023
(low/medium) |
| Medium | 83 | 1.12
(0.75–1.43) | 0.19
(medium/high) | 0.15
(0.10–0.23) | 0.13
(high-medium) | 0.05
(0.01–0.10) | 0.68
(medium/high) | 0.69
(0.51–0.94) | 0.15
(medium/high) |
| High | 25 | 1.24
(0.86–2.77) | 0.44
(low/high) | 0.25
(0.11–0.51) | 0.58
(low/high) | 0.04
(0.01–0.12) | 0.23
(low/high) | 0.34
(0.10–0.95) | 0.69
(low/high) |
| NS | 18 | | | | | | | | |
| Grade | | | | | | | | | |
| 1 | 48 | 1.86
(1.05–2.57) | 0.05 (1 vs 2) | 0.15
(0.08–0.45) | 0.42 (1 vs 2) | 0.03
(0.01–0.08) | 0.06 (1 vs 2) | 0.26
(0.07–0.70) | 0.048 (1 vs
2) |
| 2 | 79 | 1.03
(0.75–1.37) | 0.14 (2 vs 3) | 0.30
(0.18–0.47) | 0.06 (2 vs 3) | 0.09
(0.04–0.14) | 0.14 (2 vs 3) | 0.79
(0.53–1.03) | 0.1 (2 vs 3) |
| 3 | 30 | 1.59
(0.83–4.21) | 0.86 (1 vs 3) | 0.13
(0.09–0.21) | 0.44 (1 vs 3) | 0.05
(0.01–0.09) | 0.83 (1 vs 3) | 0.39
(0.16–0.77) | 0.86 (1vs 3) |
| NS | 7 | | | | | | | | |
| FIGO stage | | | | | | | | | |
| I | 98 | 1.30
(0.90–1.72) | 0.87 (I vs II) | 0.19
(0.12–0.34) | 0.70 (I vs II) | 0.06
(0.03–0.11) | 0.48 (I vs II) | 0.64
(0.47–0.79) | 0.35 (I vs II) |
| II | 25 | 1.36
(0.64–2.86) | 0.44 (II vs
III) | 0.19
(0.09–0.43) | 0.38 (II vs
III) | 0.03
(0.03–0.19) | 0.96 (II vs
III) | 0.94
(0.4–1.28) | 0.11 (II vs
III) |
| III | 26 | 1.34
(0.86–4.27) | 0.43 (I vs
III) | 0.23
(0.11–0.51) | 0.60 (I vs
III) | 0.05
(0.01–0.12) | 0.34 (I vs
III) | 0.29
(0.10–0.95) | 0.33 (I vs
III) |
| NS | 15 | | | | | | | | |
| Myometrium
invasion | | | | | | | | | |
| <1/2 | 83 | 1.61
(1.24–2.12) | 0.12 | 0.20
(0.12–0.34) | 0.96 | 0.07
(0.04–0.11) | 0.18 | 0.53
(0.32–0.70) | 0.30 |
| >1/2 | 69 | 0.97
(0.81–1.36) | | 0.19
(0.12–0.31) | | 0.04
(0.01–0.08) | | 0.65
(0.24–1.15) | |
| Without | 4 | 0.72 | | 0.18 | | 0.13 | | 0.33 | |
| NS | 8 | | | | | | | | |
| Lymph node
metastasis | | | | | | | | | |
| Negative | 125 | 1.34
(0.97–1.66) | 0.42 | 0.17
(0.11–0.27) | 0.13 | 0.05
(0.02–0.08) | 0.65
(0.49–0.83) | 0.17 | |
| Positive | 24 | 1.34
(0.86–2.65) | | 0.27
(0.11–0.63) | | 0.05
(0.01–0.13) | 0.29
(0.10–0.64) | | |
| NS | 15 | | | | | | | | |
Discussion
The process of epithelial to mesenchymal transition
and its regulation seems to be important in endometrial
carcinogenesis and disease progression. A review of the literature
indicates that this is the first report to investigate the
influence of the WWOX gene on EMT in endometrial tumor
samples and in an endometrial cancer cell line model.
The present study is a continuation of previous
observations that WWOX inversely correlates with CDH1
expression, which suggests that alterations in the expression of
this tumor suppressor gene probably influence the process of cell
adhesion (38).
WWOX overexpression was induced in a
well-differentiated endometrial cancer cell line and its effect on
EMT progression was analysed on microarrays and by multiple
biological assays. The microarray analysis revealed significant
changes (p<0.05) in the expression of two main EMT markers,
oppositely regulated in relation to WWOX expression level
i.e., CDH1 was upregulated and VIM was downregulated.
This result was confirmed on both the protein and mRNA levels.
This observed upregulation of CDH1 in the
cancer cell line stands in contrast to our previous report, which
revealed a negative correlation between CDH1 and
WWOX. However, in the present study, the larger, tumor
sample population also demonstrated a negative correlation. This
discrepancy between the cell line model and the tumor samples in
the present study may be explained by comparing the complexity of
tumor architecture between the ECM matrix and the cell line in a 2D
in vitro culture. The well-differentiated nature of the cell
line within the experiment may also have an impact on WWOX
functioning, as shown in previous studies on colon cancer cell
lines (36). However, the
relationship between CDH1 and WWOX implies that the
WWOX gene has an influence on the regulation of the adhesion
process. The loss of E-cadherin expression is a hallmark of EMT,
and its reduced transcription in endometrial cancer was found to
correlate with the depth of myometrial invasion (14,42).
In the increased number of tumor samples used in the
present study, loss of CDH1 expression tended to be observed
in grade 3 tumors and those with the highest risk of recurrence
(p>0.05). However, interestingly, grade 2 tumors and those with
medium risk of recurrence demonstrated significantly higher
CDH1 expression on the mRNA level in comparison to grade 1
tumors and those with a low risk of recurrence.
WWOX mRNA expression was found to have an
inverse association with risk of recurrence and tumor grade. Tumors
with low risk of recurrence and grade 1 demonstrated significantly
higher WWOX expression, which confirms that loss of
WWOX is connected with tumor progression, as seen in other
tumor types (30,43,44).
The inverse correlation with WWOX and CDH1 seen in
tumor samples may also suggest WWOX regulatory role in the
process of differentiation, since reduced CDH1 protein level is
associated with diminished differentiation (42).
Even though the overexpression of WWOX
resulted in the increase of CDH1 expression in the in
vitro model, the result of the invasion assay revealed that the
ECC1/WWOX cell line variant had increased invasion capacity and
migratory potential, as overexpression of WWOX resulted in
increased expression of metalloproteinases MMP2/MMP9. This
observation is consistent with previous findings in breast and
colon cancer cell lines ectopically overexpressing WWOX protein
(36,37), but also implies that the
interaction between WWOX and CDH1 may be modulated by other
EMT-related genes. Hence, WWOX expression was compared with
ZEB1 and SNAIL1 expression on the mRNA level. Of
those two genes, a positive correlation was only noticed between
WWOX and ZEB1.
The observed positive correlation between
WWOX and ZEB1 may further confirm the idea that
WWOX may play a important role in EMT induction in terms of
cell remodelling, with suppressed acquisition of the mesenchymal
phenotype; this negative correlation was previously noted with
vimentin, a mesenchymal marker, both in an in vitro model
and in tumor samples.
In a normal human uterus, ZEB1 expression is
regulated by steroid hormones. It is higher in both stroma and the
myometrium in the secretory phase than in the proliferative phase.
While the ZEB1 gene is silenced in the normal endometrial
epithelium (45), it is expressed
in the tumor-associated stroma of endometrial cancers, as well as
in epithelial-derived cancer cells, with its level correlating with
disease staging (45,46). Thus, activation of ZEB1
expression in epithelial cancer cells causes EMT, through
repression of CDH1 and induction of vimentin (46). In the present study, especially in
tumor samples, it seems that the negative correlation between
WWOX and CDH1 may be a result of ZEB1
interplay in the functional axis of WWOX-CDH1, and is not present
in the stable microenvironment of cell culture.
However, ZEB1-regulated acquisition of mesenchymal
phenotype was found to be incompleted in both the in vitro
model and tumor samples. High WWOX expression resulted in
decreased VIM expression. This negative correlation was
confirmed on the protein level in the in vitro ECC1 cultures
and has recently also been observed for ovarian cancer (47). The possibility that mesenchymal
progression was incomplete as a result of VIM downregulation
is confirmed by the observation that the WWOX-transducted
cells were not able to form colonies in suspension, suggesting that
they have lost some of their aggressiveness. This observation is
consistent with previous findings on ovarian and cervical cancer
cell lines (32,33) and confirms the tumor suppression
properties of WWOX. Moreover, our findings also support
those of Gourley et al that WWOX may be involved in
tissue architecture organization through modulation of the adhesion
potential of cells to different ECM proteins (35). Significantly reduced adhesion to
fibrinogen and tendency to fibrinonectin was demonstrated by ECC1
cells with high ectopic expression of WWOX.
The received tendency for decreased adhesion to
fibronectin is consistent with previous findings in ovarian cancer
(35), but this is the first study
to reveal reduced adhesion to fibrinogen after WWOX
overexpression. Importantly, it has been hypothesized that
fibrinogen is able to facilitate metastasis by enhancing adhesion,
thus promoting the survival of tumor cells circulating in blood
vessels. Elevated fibrinogen expression was associated with
metastasis in lung, colon, ovarian and breast cancers (48,49).
Moreover, in breast cancer, fibrinogen-mediated migration was
observed only in such malignant tumor cell lines as MDA-MB-231 and
MCF-7, whereas this ability was decreased in the non-malignant
MCF-10A cell line (49). Hence,
our observations on the endometrial cancer cell line seems to
confirm that WWOX overexpression mediates the loss of
aggressiveness and that this gene is involved in the regulation of
adhesion potential.
In conclusion, this study investigated the role of
the WWOX gene in the process of EMT in endometrial cancer.
On both tumor and cell line models, statistically significant
correlations were noted with CDH1 and vimentin, which are the main
markers of EMT. Our observations indicate that WWOX may be
involved in the initiation of EMT, leading to changes in cell
adhesion and motility. They also suggest that WWOX plays a
suppressive role in the process of mesenchymal phenotype
acquisition, resulting in reduction of cell features associated
with aggressiveness. However, further research in this direction is
required.
Acknowledgements
We acknowledge Dr hab Agnieszka
Piastowska-Ciesielska for access to BioTek reading plate and
methodological support to western blot assay. This study was
supported by the National Center of Sciences N N407 168940.
References
|
1
|
Amant F, Moerman P, Neven P, Timmerman D,
Van LE and Vergote I: Endometrial cancer. Lancet. 366:491–505.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Leslie KK, Thiel KW, Goodheart MJ, De
Geest K, Jia Y and Yang S: Endometrial cancer. Obstet Gynecol Clin
North Am. 39:255–268. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Merritt MA and Cramer DW: Molecular
pathogenesis of endometrial and ovarian cancer. Cancer Biomark.
9:287–305. 2010.
|
|
4
|
Troisi R, Potischman N, Hoover RN, Siiteri
P and Brinton LA: Insulin and endometrial cancer. Am J Epidemiol.
146:476–482. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bednarek AK, Laflin KJ, Daniel RL, Liao Q,
Hawkins KA and Aldaz CM: WWOX, a novel WW domain-containing protein
mapping to human chromosome 16q23.3–24.1, a region frequently
affected in breast cancer. Cancer Res. 60:2140–2145.
2000.PubMed/NCBI
|
|
6
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Garg M: Epithelial-mesenchymal transition
- activating transcription factors - multifunctional regulators in
cancer. World J Stem Cells. 5:188–195. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tanaka Y, Terai Y, Kawaguchi H, Fujiwara
S, Yoo S, Tsunetoh S, Takai M, Kanemura M, Tanabe A and Ohmichi M:
Prognostic impact of EMT
(epithelial-mesenchymal-transition)-related protein expression in
endometrial cancer. Cancer Biol Ther. 14:13–19. 2013. View Article : Google Scholar :
|
|
10
|
Huber MA, Kraut N and Beug H: Molecular
requirements for epithelial-mesenchymal transition during tumor
progression. Curr Opin Cell Biol. 17:548–558. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Thiery JP: Epithelial-mesenchymal
transitions in development and pathologies. Curr Opin Cell Biol.
15:740–746. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Grünert S, Jechlinger M and Beug H:
Diverse cellular and molecular mechanisms contribute to epithelial
plasticity and metastasis. Nat Rev Mol Cell Biol. 4:657–665. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Montserrat N, Mozos A, Llobet D, Dolcet X,
Pons C, de Herreros AG, Matias-Guiu X and Prat J: Epithelial to
mesenchymal transition in early stage endometrioid endometrial
carcinoma. Hum Pathol. 43:632–643. 2012. View Article : Google Scholar
|
|
14
|
Abal M, Llauradó M, Doll A, Monge M, Colas
E, González M, Rigau M, Alazzouzi H, Demajo S, Castellví J, et al:
Molecular determinants of invasion in endometrial cancer. Clin
Transl Oncol. 9:272–277. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Aqeilan RI, Donati V, Palamarchuk A,
Trapasso F, Kaou M, Pekarsky Y, Sudol M and Croce CM: WW
domain-containing proteins, WWOX and YAP, compete for interaction
with ErbB-4 and modulate its transcriptional function. Cancer Res.
65:6764–6772. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Aqeilan RI, Donati V, Gaudio E, Nicoloso
MS, Sundvall M, Korhonen A, Lundin J, Isola J, Sudol M, Joensuu H,
et al: Association of Wwox with ErbB4 in breast cancer. Cancer Res.
67:9330–9336. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Aqeilan RI, Palamarchuk A, Weigel RJ,
Herrero JJ, Pekarsky Y and Croce CM: Physical and functional
interactions between the Wwox tumor suppressor protein and the
AP-2gamma transcription factor. Cancer Res. 64:8256–8261. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guler G, Huebner K, Himmetoglu C, Jimenez
RE, Costinean S, Volinia S, Pilarski RT, Hayran M and Shapiro CL:
Fragile histidine triad protein, WW domain-containing
oxidoreductase protein Wwox, and activator protein 2gamma
expression levels correlate with basal phenotype In breast cancer.
Cancer. 115:899–908. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gaudio E, Palamarchuk A, Palumbo T,
Trapasso F, Pekarsky Y, Croce CM and Aqeilan RI: Physical
association with WWOX suppresses c-Jun transcriptional activity.
Cancer Res. 66:11585–11589. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Abu-Remaileh M and Aqeilan RI: Tumor
suppressor WWOX regulates glucose metabolism via HIF1α modulation.
Cell Death Differ. 21:1805–1814. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Abu-Odeh M, Bar-Mag T, Huang H, Kim T,
Salah Z, Abdeen SK, Sudol M, Reichmann D, Sidhu S, Kim PM, et al:
Characterizing WW domain interactions of tumor suppressor WWOX
reveals its association with multiprotein networks. J Biol Chem.
289:8865–8880. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sałuda-Gorgul A, Seta K, Nowakowska M and
Bednarek AK: WWOX oxidoreductase - substrate and enzymatic
characterization. Z Naturforsch C. 66:73–82. 2011. View Article : Google Scholar
|
|
23
|
Aqeilan RI, Hagan JP, de Bruin A, Rawahneh
M, Salah Z, Gaudio E, Siddiqui H, Volinia S, Alder H, Lian JB, et
al: Targeted ablation of the WW domain-containing oxidoreductase
tumor suppressor leads to impaired steroidogenesis. Endocrinology.
150:1530–1535. 2009. View Article : Google Scholar :
|
|
24
|
Aqeilan RI and Croce CM: WWOX in
biological control and tumorigenesis. J Cell Physiol. 212:307–310.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Qin HR, Iliopoulos D, Semba S, Fabbri M,
Druck T, Volinia S, Croce CM, Morrison CD, Klein RD and Huebner K:
A role for the WWOX gene in prostate cancer. Cancer Res.
66:6477–6481. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nunez MI, Rosen DG, Ludes-Meyers JH, Abba
MC, Kil H, Page R, Klein-Szanto AJ, Godwin AK, Liu J, Mills GB, et
al: WWOX protein expression varies among ovarian carcinoma
histotypes and correlates with less favorable outcome. BMC Cancer.
5:642005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kosla K, Pluciennik E, Kurzyk A,
Jesionek-Kupnicka D, Kordek R, Potemski P and Bednarek AK:
Molecular analysis of WWOX expression correlation with
proliferation and apoptosis in glioblastoma multiforme. J
Neurooncol. 101:207–213. 2011. View Article : Google Scholar :
|
|
28
|
Kuroki T, Yendamuri S, Trapasso F,
Matsuyama A, Aqeilan RI, Alder H, Rattan S, Cesari R, Nolli ML,
Williams NN, et al: The tumor suppressor gene WWOX at FRA16D is
involved in pancreatic carcinogenesis. Clin Cancer Res.
10:2459–2465. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Płuciennik E, Nowakowska M, Wujcicka WI,
Sitkiewicz A, Kazanowska B, Zielińska E and Bednarek AK: Genetic
alterations of WWOX in Wilms’ tumor are involved in its
carcinogenesis. Oncol Rep. 28:1417–1422. 2012.
|
|
30
|
Żelazowski MJ, Płuciennik E, Pasz-Walczak
G, Potemski P, Kordek R and Bednarek AK: WWOX expression in
colorectal cancer - a real-time quantitative RT-PCR study. Tumor
Biol. 32:551–560. 2011. View Article : Google Scholar
|
|
31
|
Li YP, Wu CC, Chen WT, Huang YC and Chai
CY: The expression and significance of WWOX and β-catenin in
hepatocellular carcinoma. APMIS. 121:120–126. 2013. View Article : Google Scholar
|
|
32
|
Xiong Z, Hu S and Wang Z: Cloning of WWOX
gene and its growth-inhibiting effects on ovarian cancer cells. J
Huazhong Univ Sci Technolog Med Sci. 30:365–369. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Qu J, Lu W, Li B, Lu C and Wan X: WWOX
induces apoptosis and inhibits proliferation in cervical cancer and
cell lines. Int J Mol Med. 31:1139–1147. 2013.PubMed/NCBI
|
|
34
|
Bednarek AK, Keck-Waggoner CL, Daniel RL,
Laflin KJ, Bergsagel PL, Kiguchi K, Brenner AJ and Aldaz CM: WWOX,
the FRA16D gene, behaves as a suppressor of tumor growth. Cancer
Res. 61:8068–8073. 2001.PubMed/NCBI
|
|
35
|
Gourley C, Paige AJ, Taylor KJ, Ward C,
Kuske B, Zhang J, Sun M, Janczar S, Harrison DJ, Muir M, et al:
WWOX gene expression abolishes ovarian cancer tumorigenicity in
vivo and decreases attachment to fibronectin via integrin alpha3.
Cancer Res. 69:4835–4842. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nowakowska M, Pospiech K, Lewandowska U,
Piastowska-Ciesielska AW and Bednarek AK: Diverse effect of WWOX
overexpression in HT29 and SW480 colon cancer cell lines. Tumor
Biol. 35:9291–9301. 2014. View Article : Google Scholar
|
|
37
|
Lewandowska U, Zelazowski M, Seta K,
Byczewska M, Pluciennik E and Bednarek AK: WWOX, the tumor
suppressor gene affected in multiple cancers. J Physiol Pharmacol.
60(Suppl 1): 47–56. 2009.
|
|
38
|
Płuciennik E, Kośla K, Wójcik-Krowiranda
K, Bieńkiewicz A and Bednarek AK: The WWOX tumor suppressor gene in
endometrial adenocarcinoma. Int J Mol Med. 32:1458–1464. 2013.
|
|
39
|
Pecorelli S: Revised FIGO staging for
carcinoma of the vulva, cervix, and endometrium. Int J Gynaecol
Obstet. 105:103–104. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mo B, Vendrov AE, Palomino WA, DuPont BR,
Apparao KB and Lessey BA: ECC-1 cells: A well-differentiated
steroid-responsive endometrial cell line with characteristics of
luminal epithelium. Biol Reprod. 75:387–394. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pfaffl MW, Horgan GW and Dempfle L:
Relative expression software tool (REST) for group-wise comparison
and statistical analysis of relative expression results in
real-time PCR. Nucleic Acids Res. 30:e362002. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sakuragi N, Nishiya M, Ikeda K, Ohkouch T,
Furth EE, Hareyama H, Satoh C and Fujimoto S: Decreased E-cadherin
expression in endometrial carcinoma is associated with tumor
dedifferentiation and deep myometrial invasion. Gynecol Oncol.
53:183–189. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Płuciennik E, Kusińska R, Potemski P,
Kubiak R, Kordek R and Bednarek AK: WWOX - the FRA16D cancer gene:
Expression correlation with breast cancer progression and
prognosis. Eur J Surg Oncol. 32:153–157. 2006. View Article : Google Scholar
|
|
44
|
Wang X, Chao L, Ma G, Chen L, Zang Y and
Sun J: The prognostic significance of WWOX expression in patients
with breast cancer and its association with the basal-like
phenotype. J Cancer Res Clin Oncol. 137:271–278. 2011. View Article : Google Scholar
|
|
45
|
Spoelstra NS, Manning NG, Higashi Y,
Darling D, Singh M, Shroyer KR, Broaddus RR, Horwitz KB and Richer
JK: The transcription factor ZEB1 is aberrantly expressed in
aggressive uterine cancers. Cancer Res. 66:3893–3902. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Singh M, Spoelstra NS, Jean A, Howe E,
Torkko KC, Clark HR, Darling DS, Shroyer KR, Horwitz KB, Broaddus
RR, et al: ZEB1 expression in type I vs type II endometrial
cancers: A marker of aggressive disease. Mod Pathol. 21:912–923.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yan H and Sun Y: Evaluation of the
mechanism of epithelial-mesenchymal transition in human ovarian
cancer stem cells transfected with a WW domain-containing
oxidoreductase gene. Oncol Lett. 8:426–430. 2014.PubMed/NCBI
|
|
48
|
Palumbo JS, Kombrinck KW, Drew AF, Grimes
TS, Kiser JH, Degen JL and Bugge TH: Fibrinogen is an important
determinant of the metastatic potential of circulating tumor cells.
Blood. 96:3302–3309. 2000.PubMed/NCBI
|
|
49
|
Sahni A, Arévalo MT, Sahni SK and
Simpson-Haidaris PJ: The VE-cadherin binding domain of fibrinogen
induces endothelial barrier permeability and enhances
transendothelial migration of malignant breast epithelial cells.
Int J Cancer. 125:577–584. 2009. View Article : Google Scholar : PubMed/NCBI
|