Introduction
Despite advances in breast cancer care and regimens
it still imposes a large burden on health care systems around the
world, especially when metastatic disease is detected. Breast
cancer is associated with latent disease and high relapse rate
which can often present clinically as bone metastases (1). The majority of breast cancer related
bone metastases present as the osteolytic phenotype, which is
identified by loss of bone density accompanied by an increase in
osteoclast numbers (2).
The variability in metastatic cancer patterns is
undoubtedly influenced by the molecular and cellular
characteristics of both the tumour cells and the tissue in which
they invade (3). It was Stephen
Paget, through autopsy data, who first established a link between
breast cancer metastases and the bone, giving rise to his ‘seed and
soil’ hypothesis (4). As this
theory has evolved the metastatic cascade has been shown to be a
highly inefficient multistep process which involves a wide variety
of factors including integrins, matrix metalloproteinases and
tumour secreted factors (5,6).
Invasion into the bone results in the release of a variety of
factors, in addition to those produced by the tumour cells, which
generate a feedback loop to the tumour cells enhancing tumour cell
dormancy, survival and growth in the bone marrow and the
microenvironment (7). However,
much still remains unknown of how these factors interact with each
other and the disseminating tumour cells to culminate in bone
metastases and how best these can be targeted in therapies.
Members of the tumour necrosis factor receptor
superfamily (TNFRSF) osteoprotegrin (OPG), receptor activator of
nuclear κB (RANK) and RANK ligand (RANKL) have been shown to be
integral molecular regulators in the bone remodelling cycle. The
RANKL:OPG ratio is a major determinant of bone mass, both
physiologically and patho-physiologically (8). Osteoblasts have been shown to
incorporate both pro- and antibone resorptive signals and thus
control the bone remodelling response by altering the expression of
RANKL and secretion of its inhibitor OPG (9,10).
RANK, expressed on the surface of osteoclasts, through binding to
RANKL, expressed on the surface of osteoblasts, promotes osteoclast
differentiation and maturation, thus promoting bone resorption.
OPG, a soluble decoy receptor for RANKL, secreted by osteoblasts,
inhibits RANK interaction thus promoting osteoblast survival and
hence bone formation. However, OPG, RANK and RANKL have also been
linked to tumourigenesis in a variety of cancers which have a
predisposition to form bone metastases. Both circulating RANKL and
OPG have previously been identified as novel biomarker candidates
for predicting bone metastases in breast cancer patients (11,12).
Quantitative PCR and immunohistochemistry have shown negative
correlations between estrogen receptor status and levels of OPG,
RANK and RANKL (13).
There has also been some in vitro evidence to
suggest that endogenously produced OPG, from breast cancer cells or
bone marrow stromal cells, can also promote breast cancer cell
survival through inhibition of TNF related apoptosis inducing
ligand (TRAIL) (14,15). This inhibition occurs as OPG acts
as a decoy receptor for the TRAIL receptor, though with less
affinity than that seen with RANKL, therefore blocking the
apoptotic pathway. This prevention of apoptosis through TRAIL
inhibition has also been shown, in the MDA-MB-231 breast cancer
cell line, to result in the up regulation of RANKL thus
contributing to the ‘vicious’ bone cycle between tumour cells and
bone cells by further enhancing osteolysis and the release of
growth factors which can further enhance tumour growth (16).
The bone microenvironment is a complex combination
of cells, growth factors and cytokines. Trying to isolate the
factors which are crucial components in facilitating the
establishment of bone metastases is a substantial challenge. One of
the factors, which have been shown to influence tumourigenesis
traits and cancer progression is hepatocyte growth factor (HGF),
also known as scatter factor (17–19).
Despite its discovery 30 years ago its wide and complex influences
on cancer cells, the metastatic cascade and tumour
microenvironments remain under intense investigation for potential
new targeted therapies (20,21).
In the present study the targeting of OPG and RANK
in bone metastasis derived breast cancer cells (MDA-MB-231 cells)
was explored. These manipulated cells were then exposed to the
influences of HGF and a bone protein-like environment to explore
the potential implications on HGF signalling thus potentially
altering disease progression.
Materials and methods
Ethics statement
All research involving human tissue was carried out
under the Panel B Bro Taf Research Ethics Committee for the Bro Taf
Health Board, Cardiff, UK. All data were analysed anonymously and
informed written consent was given (Bro Taf Health Board,
2007).
Cell lines and treatments
Human breast cancer MDA-MB-231 cells were purchased
from the American Type Culture Collection (ATCC, Rockville, MD,
USA). MDA-MB-231 cells were maintained in Dubecco's modified
Eagle's medium (DMEM) (PAA Laboratories Ltd., Somerset, UK)
supplemented with penicillin, streptomycin and 10% foetal calf
serum (PAA Laboratories Ltd.) and incubated at 37°C, 5%
CO2 and 95% humidity. Hepatocyte growth factor was a
kind gift from Dr T. Nakamura (Osaka University Medical School,
Osaka, Japan). Bone proteins were extracted from fresh human bone
tissues collected immediately after hip replacement under the local
health board ethics committee guidelines. Bones were crushed at ice
cold temperatures and subsequently processed in a Bioraptor
sonicator (Wolf Laboratories, York, UK) to extract matrix proteins
(22). Throughout this study HGF
was used at a final concentration of 40 ng/ml, whilst the BME
extract from the femoral heads was used at a final concentration of
50 μg/ml.
Generation of MDA-MB-231 breast cancer
cells with suppressed OPG or RANK expression
OPG and RANK expression were targeted in human
MDA-MB-231 breast cancer cells using ribozyme transgenes
specifically generated to target and cleave each transcript. This
methodology has been previously reported (23,24).
Briefly, ribozyme transgene sequences were designed based on Zukers
predicted secondary mRNA structure using Zukers RNA Mfold program
(25) and were synthesised by
Sigma-Aldrich (Poole, Dorset, UK) (Table I). Ribozymes were subsequently
cloned into a pEF6/V5-His-TOPO plasmid vector (Invitrogen, Paisley,
UK). Both control pEF6 plasmids, containing no insert, and plasmids
containing the relevant ribozyme transgene were transfected
separately into MDA-MB-231 breast cancer cells using
electroporation. Following transfection, these cells underwent a
selection period and subsequent verification of OPG or RANK
knockdown. Cells containing the ribozyme transgenes were termed
MDA-MB-231OPGKD or MDA-MB-231RANKKD and were
compared throughout the study to control MDA-MB-231 cells
containing the closed control plasmid, termed
MDA-MB-231pEF6.
 | Table IPrimers designed for ribozyme
synthesis. |
Table I
Primers designed for ribozyme
synthesis.
| Target | Ribozyme | Primer name | Primer sequence
(5′-3′) |
|---|
| | T7F |
TAATACGACTCACTATAGGG |
| | RBBMR |
TTCGTCCTCACGGACTCATCAG |
| | RBTPF |
CTGATGAGTCCGTGAGGACGAA |
| OPG | OPG ribozyme 1 | OPGRIB1F |
CTGCAGCTCCTTGCACACGGGGCTGCAGTATACTGATGAGTCCGTGAGGA |
| | OPGRIB1R |
ACTAGTACACAGACAGCTGGCACACCAGTGACGAGTGTTTCGTCCTCACGGACT |
| OPG ribozyme 2 | OPGRIB2F |
CTGCAGACACTGCAATTTGTGTGTTTTCTACTGGGTGCTTTACTGATGAGTCCGTGAGGA |
| | OPGRIB2R |
ACTAGTTCTTCTCAAATGAGACGTCATTTCGTCCTCACGGACT |
| OPG ribozyme 3 | OPGRIB3F |
CTGCAGGGTAACATCTATTCCACATTTTGAGTTCTGATGAGTCCGTGAGGA |
| | OPGRIB3R |
ACTAGTTCCGGAAACAGTGAATTTCGTCCTCACGGACT |
| RANK | RANK ribozyme
1 | RANKRIB1F |
CTGCAGCGCGCGGGGCCATGGCGCGGCTGATGAGTCCGTGAGGA |
| | RANKRIB1R |
ACTAGTGCCGCGGCGCCGCCAGCCTGTTTCGTCCTCACGGACT |
| RANK ribozyme
2 | RANKRIB2F |
CTGCAGCTCATAATGCTTCTCACTGGCTGATGAGTCCGTGAGGA |
| | RANKRIB2R |
ACAGTCTTTGCAGATCGCTCCTCCATGTTTCGTCCTCACGGACT |
| RANK ribozyme
3 | RANKRIB3F |
CTGCAGGTACTTTCCTGGTTCACATTTGTCTGATGAGTCCGTGAGGA |
| | RANKRIB3R |
ACTAGTAGCATTATGAGCATCTGGGACGGTGCTGTTTCGTCCTCACGGACT |
| RANK ribozyme
4 | RANKRIB4F |
CTGCAGTGCTGACCAAAGTTTGCCGTGTGTGCTGATGAGTCCGTGAGGA |
| | RANKRIB4R |
ACTAGTGGAGTCCTCAGGTGACAGTTGTGTCAGTTTCGTCCTCACGGAC |
| RANK ribozyme
5 | RANKRIB5F |
CTGCAGCTGGCATCTTCGCCTTGTGCGTAGGCTGATGAGTCCGTGAGGA |
| | RANKRIB5R |
ACTAGTGTCAGGGCACATGTGTAGGAGGTGGTTTCGTCCTCACGGACT |
RNA extraction and reverse
transcription-polymerase chain reaction (RT-PCR)
Cells were grown to confluence in a
25-cm2 flask before RNA was extracted using total RNA
isolation (TRI) reagent (Sigma) in accordance with the supplied
protocol. RNA was subsequently quantified using a spectrophotometer
(Implen Nanophotometer, Muchen, Germany) configured to detect
single strand RNA (μg/μl). RNA was standardised to 500 ng and used
as a template to generate cDNA using high capacity cDNA reverse
transcription kit (Applied Biosystems, Manchester, UK). Following
cDNA synthesis, sample quality and uniformity was normalised
against GAPDH expression (primer details in Table II). The amplifluor system
(Intergen Inc., New York, NY, USA) was utilised with qPCR Master
Mix (ABgene, Surrey, UK). Conditions for qPCR were; 15 min initial
95°C period followed by 60 cycles of 95°C for 15 sec, 55°C for 60
sec and 72°C for 20°C sec.
 | Table IIPrimers for conventional RT-PCR and
real-time qPCR. |
Table II
Primers for conventional RT-PCR and
real-time qPCR.
| Gene | Primer name | Primer sequence
(5′-3′) | Optimal annealing
temperature (°C) | Product size
(bp) |
|---|
| OPG | OPGF8 |
GAACCCCAGAGCGAAATACA | 55 | 509 |
| OPGR8 |
CGGTAAGCTTTCCATCAAGC | | |
| OPGF1 |
GTTCTGCTTGAAACATAGGAG | 55 | 115 |
| OPGZR1 |
ACTGAACCTGACCGTACACGTCTCATTTGAGAAGAACC | | |
| RANK | RANKF9 |
CAGAGCACAGTGGGTTCAGA | 55 | 462 |
| RANKR9 |
GATGATGTCGCCCTTGAAGT | | |
| RANKF2 |
TCTGATGCCTTTTCCTCCAC | 55 | 119 |
| RANKZR2 |
ACTGAACCTGACCGTACATGGCAGAGAAGAACTGCAAA | | |
| RANKL | RANKLF9 |
GACTCCATGAAAATGCAGAT | 55 | 500 |
| RANKLR9 |
TCCTTTCATCAGGGTATGAG | | |
| RANKLF1 |
AAGGAGCTGTGCAAAAGGAA | 55 | 74 |
| RANKLZR1 |
ACTGAACCTGACCGTACAATCCACCATCGCTTTCTCTG | | |
| GAPDH | GAPDHF10 |
AGCTTGTCATCAATGGAAAT | 55 | 593 |
| GAPDHR10 |
CTTCACCACCTTCTTGATGT | | |
| GAPDHF |
CTGAGTACGTCGTGGAGTC | 55 | 93 |
| GAPDHZR |
ACTGAACCTGACCGTACACAGAGATGATGATGACCCTTTTG | | |
| PDPL | PDPLF |
GAATCATCGTTGTGGTTATG | 55 | |
| PDPLZR |
ACTGAACCTGACCGTACACTTTCATTTGCCTATCACAT | | |
SDS-PAGE and western blotting
Protein was extracted from a confluent
75-cm2 tissue culture flask of MDA-MB-231 cells. Cells
were detached and lysed in a buffer comprising 50 mM Tris-base, 5
mM EGTA, 150 mM NaCl, 1% Triton X-100, 100 μg/ml PMSF, 10 μg/ml
aprotinin, 10 μg/ml leupeptin, 5 mM sodium vanadate and 50 mM
sodium fluoride on a rotor wheel for 1 h before removal of
insolubles through centrifugation at 13,000 g. The Bio-Rad DC
protein assay kit (Bio-Rad Laboratories, CA, USA) was used to
quantify protein levels in each sample and samples were
subsequently standardised to 2 mg/ml and diluted in 2X concentrate
Laemmli sample buffer (Sigma) before being boiled for 5 min.
Samples were loaded onto a 10% acrylamide gel and separated
electrophoretically. Following separation the proteins were blotted
onto a PVDF membrane (Merck-Millipore, Feltham, UK). Proteins were
detected using the Merck-Millipore SNAP i.d. protein detection
system. OPG expression was detected using anti-OPG antibody
[R&D Systems, Abingdon, UK (BAF805)], RANK expression was
detected using anti-RANK antibody [Santa Cruz Biotechnology, Inc.,
CA, USA (sc-9072)]. To assess uniformity of the samples GAPDH
expression was also detected using anti-GAPDH antibody [Santa Cruz
Biotechnology, Inc. (sc-32233)]. Following binding of the primary
antibody, the membranes were probed with peroxidase conjugated
anti-goat (OPG), anti-rabbit (RANK) or anti-mouse (GADPH) secondary
antibodies (Sigma). Expression was visualised using the Luminata
chemiluminescence detection kit (Merck-Millipore) and detected
using a UVIProChem camera system (UVItec Ltd., Cambridge, UK).
In vitro cell proliferation assay
An in vitro cell proliferation assay was used
to examine the impact of OPG or RANK suppression on cell
proliferation. Cells were seeded into two 96-well plates at a
seeding density of 3×103 cells/well with or without
treatment and incubated for 1 and 5 days. Following incubation,
cells were fixed in 4% formaldehyde (v/v) and stained with 0.5%
crystal violet (w/v). Subsequently, 10% acetic acid (v/v) was used
to extract the crystal violet stain and cell density determined
through spectrophotometric analysis using a Bio-Tek Elx800
multi-plate reader (Bio-Tek Instruments Inc., VT, USA).
In vitro Matrigel matrix adhesion
assay
Cell-matrix adhesion was assessed using a modified
in vitro Matrigel adhesion assay (26). In brief, wells in a 96-well plate
were pre-coated with 5 μg of Matrigel (BD Matrigel matrix, Matrigel
basement membrane matrix, Biosciences). Cells were seeded at
4.5×104 cells/well with or without treatment and left to
adhere to the Matrigel for 40 min at 37°C with 5% CO2.
Adherent cells were fixed in 4% formaldehyde (v/v) and stained with
0.5% crystal violet (w/v). Subsequently, adherent cells were
visualised under the microscope and representative images captured
for analysis.
In vitro cell motility assay
Cell motility was assessed using a cytodex-2 bead
motility assay as previously described (27,28).
In brief, 1×106 cells were incubated in 10 ml of
complete medium supplemented with 20 mg of cytodex-2 beads. The
following day, beads were washed twice with complete medium before
being resuspended, added to the 96-well plate with or without
treatment and incubated for 4 h at 37°C with 5% CO2.
Migrated cells were fixed in 4% formaldehyde (v/v) and stained with
0.5% crystal violet (w/v). Subsequently, adherent cells were
visualised under the microscope and representative images captured
for analysis.
In vitro Matrigel cell invasion
assay
Cell invasiveness was assessed using an in
vitro Matrigel invasion assay modified from refs. 29,30.
In brief, transwell inserts containing 8-μm pores (Falcon, 24-well
format, Greiner Bio-One, Germany) were placed in a 24-well plate
(Nunc, Greiner Bio-One) and coated with 50 μg of Matrigel (BD
Matrigel matrix, Matrigel basement membrane matrix, Biosciences).
Subsequently 2×104 cells/insert were added to the insert
and 1 ml of medium was added to the bottom of the 24-well plate to
sustain any invaded cells. The plate was incubated for 3 days at
37°C with 5% CO2 after which inserts were cleaned to
remove any non-invaded cells, before invaded cells were fixed in 4%
formaldehyde (v/v) and stained with 0.5% crystal violet (w/v).
Subsequently, invaded cells were visualised under the microscope
and representative images captured for analysis.
Statistical analysis
The Sigma plot 11.0 statistical software package was
used to assess statistical differences between the OPG or RANK
suppressed MDA-MB-231 cells compared to the pEF6 vector control
MDA-MB-231 cells using the Student's two tailed t-test or
non-parametric Mann-Whitney U test. Experimental procedures were
repeated a minimum of 3 independent times. Data represent mean
values ± SEM, p-values of ≤0.05 were regarded as statistically
significant.
Results
Expression of OPG and RANK has previously been
established in three breast cancer cell lines (31). There are also potential links
between the expression profiles of OPG, RANK, the pro-tumourigenic
stimuli HGF and the bone microenvironment.
Suppression of molecules of interest
using ribozyme transgenes
OPG or RANK expression was successfully targeted in
MDA-MB-231 breast cancer cells following transfection with anti-OPG
or anti-RANK ribozyme transgenes contained within a pEF6 plasmid.
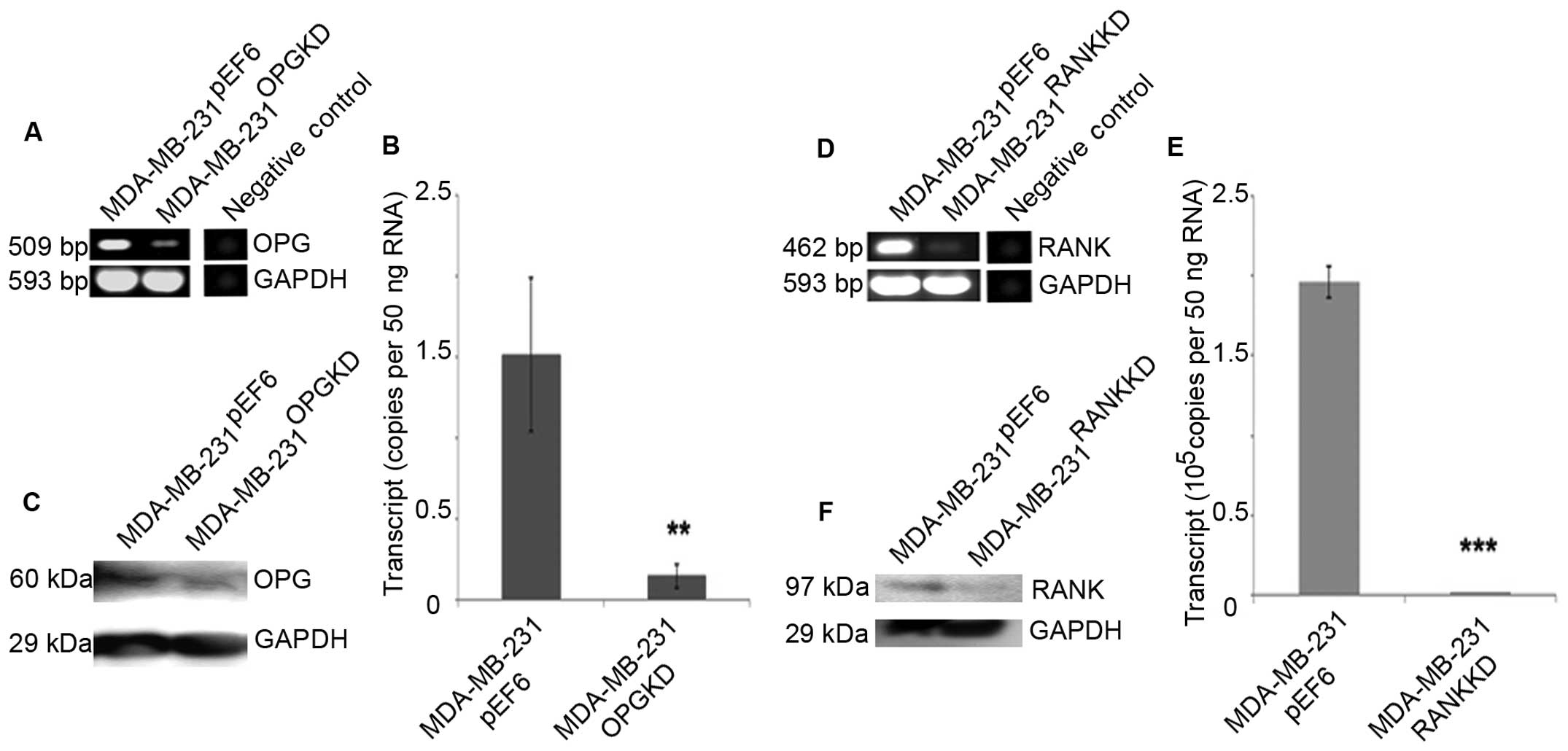
Reduced OPG transcript expression was seen in
MDA-MB-231OPGKD cells compared to the
MDA-MB-231pEF6 cells using both RT-PCR and qPCR
(Fig. 1A and B). The result was
subsequently confirmed at a protein level using western blotting
(Fig. 1C).
Reduced RANK transcript expression was seen in
MDA-MB-231RANKKD cells compared to the
MDA-MB-231pEF6 cells using both RT-PCR and qPCR
(Fig. 1D and E). This was
subsequently confirmed at a protein level using western blotting
(Fig. 1F).
Impact on MDA-MB-231 breast cancer cell
proliferation
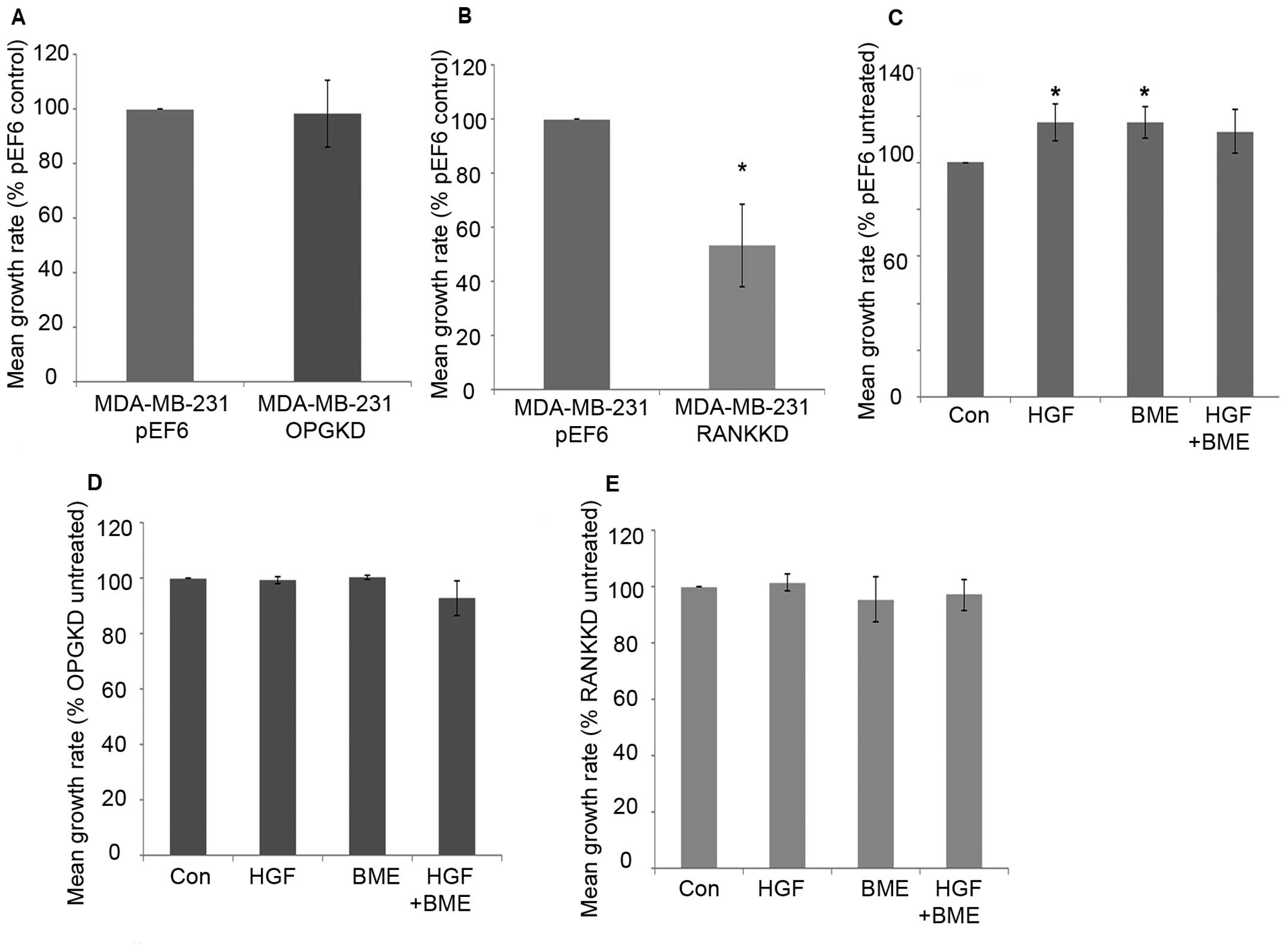
MDA-MB-231 breast cancer cell proliferation over 5
days was not significantly altered after suppression of OPG
compared to the control cells (Fig.
2A). Suppression of RANK in MDA-MB-231 breast cancer cells
resulted in a statistically significant decrease in cell
proliferation after 5-day incubation compared to the control cells
(Fig. 2B, p=0.029). Individual 40
ng/ml HGF and 50 μg/ml BME treatments significantly increased
MDA-MB-231pEF6 cell proliferation after 5-day incubation
compared to the untreated control (Fig. 2C, p=0.029). A similar pattern was
seen after incubation with a combined 40 ng/ml HGF and 50 μg/ml BME
treatment; however, this did not reach statistical significance.
MDA-MB-231OPGKD (Fig.
2D) and MDA-MB-231RANKKD (Fig. 2E) cell proliferation was less
responsive to HGF and BME treatments compared to those observed in
the MDA-MB-231pEF6 cells. No statistically significant
changes were observed in either of the suppressed cell lines
compared to their respective untreated controls.
Impact on MDA-MB-231 breast cancer
cell-matrix adhesion
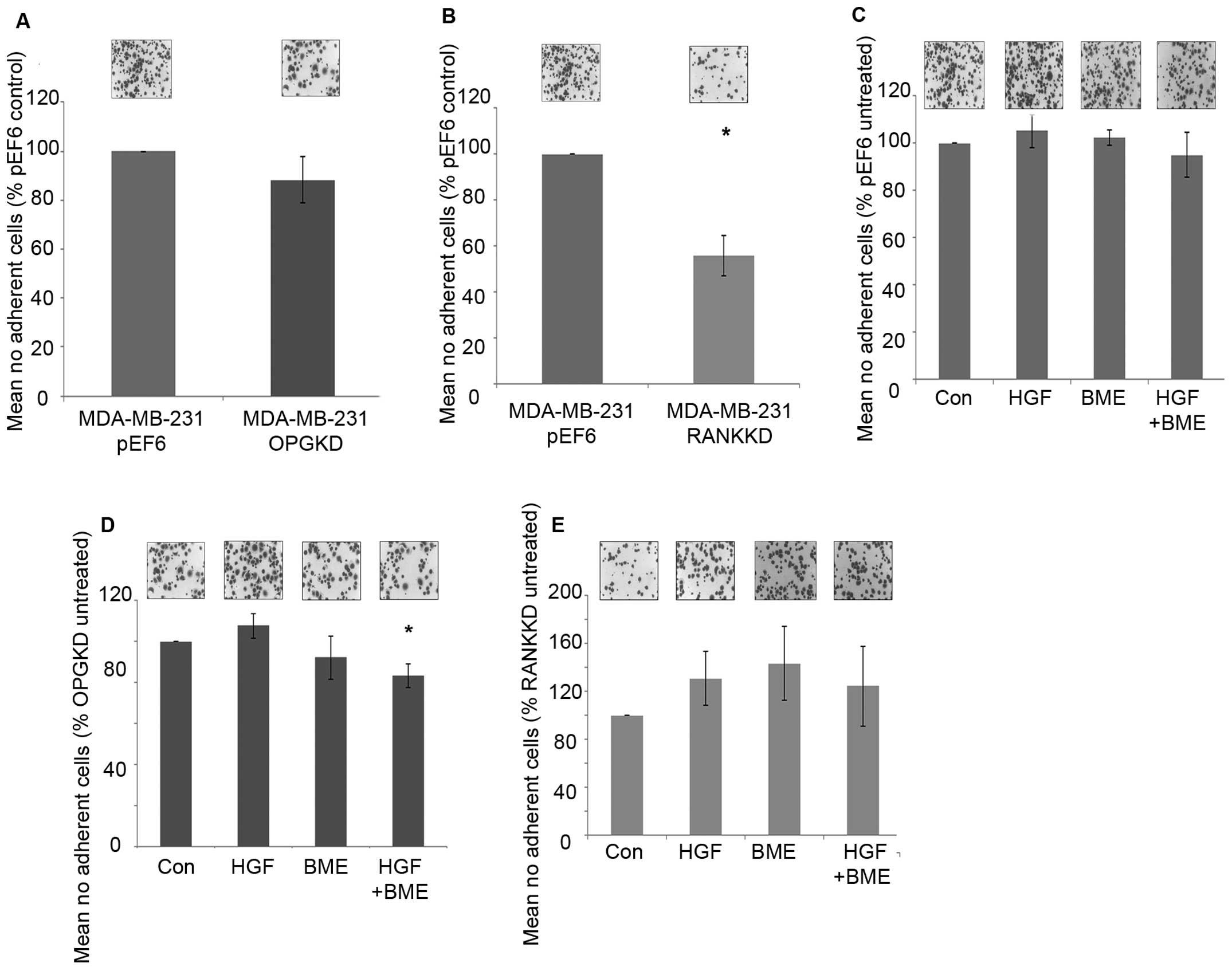
Suppression of OPG in MDA-MB-231 breast cancer cells
did not appear to affect cell-matrix adhesion (Fig. 3A). In contrast, suppression of RANK
in MDA-MB-231 breast cancer cells resulted in a statically
significant decrease in cell-matrix adhesion in vitro
(Fig. 3B, p=0.029).
MDA-MB-231pEF6 cell-matrix adhesion appeared unaffected
under the influence of treatment with 40 ng/ml HGF and/or 50 μg/ml
BME (Fig. 3C). A similar trend was
seen in the MDA-MB-231OPGKD cells treated individually
with 40 ng/ml HGF or 50 μg/ml BME (Fig. 3D), however, when these treatments
were combined cell-matrix adhesion was significantly reduced
compared to the untreated cells (p=0.024). In the
MDA-MB-231RANKKD cells treatment with 40 ng/ml HGF
and/or 50 μg/ml BME did not appear to significantly affect
cell-matrix adhesion (Fig.
3E).
Impact on MDA-MB-231 breast cancer cell
motility
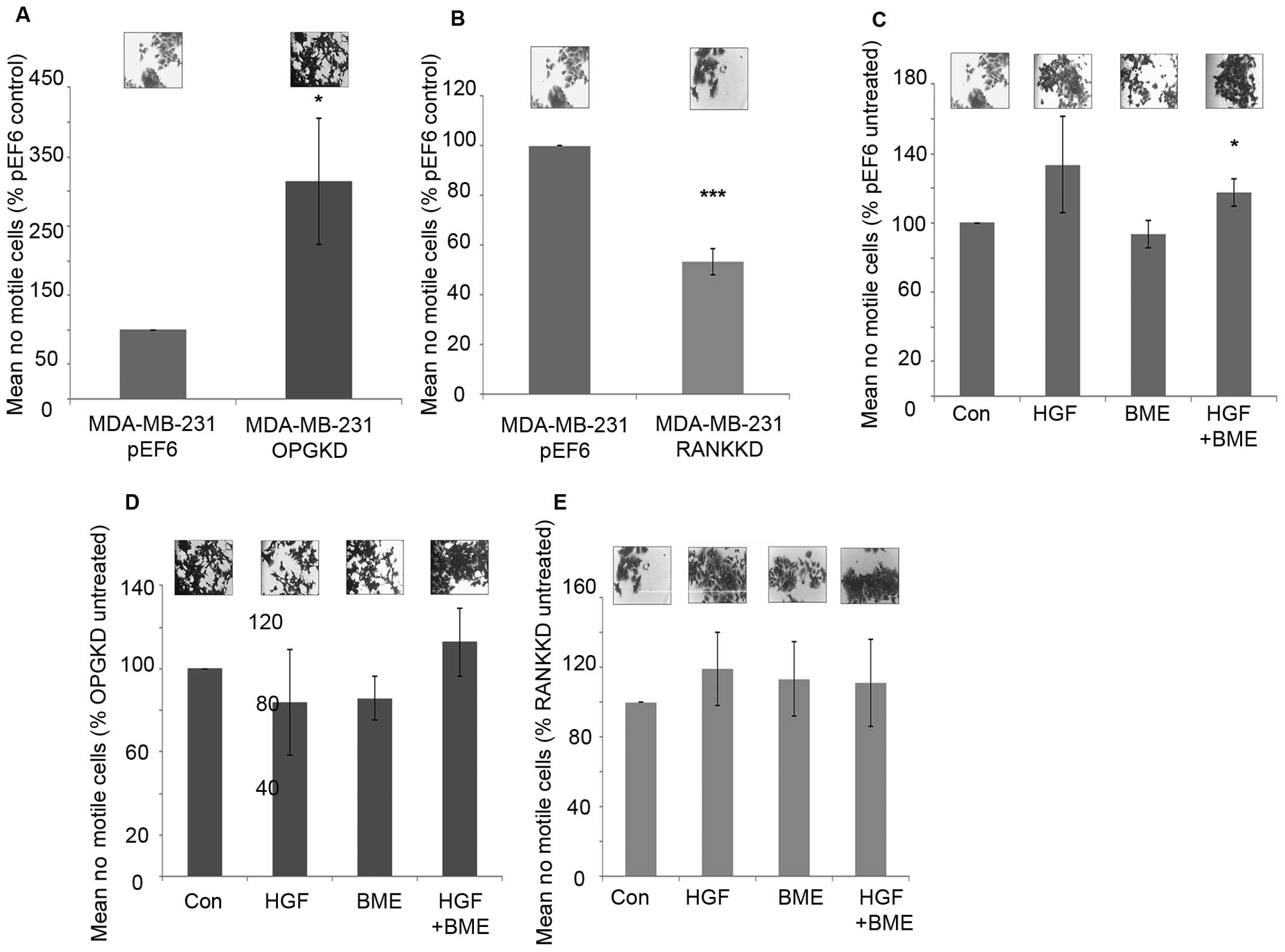
Suppression of OPG expression in MDA-MB-231 cells
resulted in significantly increased cell motility in vitro
compared to the control cells (Fig.
4A, p=0.029). In contrast, suppression of RANK in MDA-MB-231
breast cancer cells resulted in significantly decreased cell
motility in vitro (Fig. 4B,
p≤0.001). When MDA-MB-231pEF6 cells were treated with 40
ng/ml HGF cell motility was increased, however, this did not pass
the statistical threshold (Fig.
4C). However, no noticeable effect was seen on
MDA-MB-231pEF6 cell motility when cells were treated
with 50 μg/ml BME. When both treatments were combined
MDA-MB-231pEF6 cell motility was significantly increased
compared to the untreated control cells (Fig. 4C, p=0.029). In contrast, treatment
of MDA-MB-231OPGKD cells with 40 ng/ml HGF or 50 μg/ml
BME did not appear to impact MDA-MB-231 cell motility compared to
the untreated control (Fig. 4D).
When these treatments were combined increased cell motility was
observed compared to the untreated control cells, however, this
trend also failed to reach the statistically significant threshold.
Similar responses to the exogenous HGF and BME treatments were also
seen in the MDA-MB-231RANK KD cells, in which cell
motility was only marginally affected by these factors (Fig. 4E).
Impact on MDA-MB-231 breast cancer cell
invasion
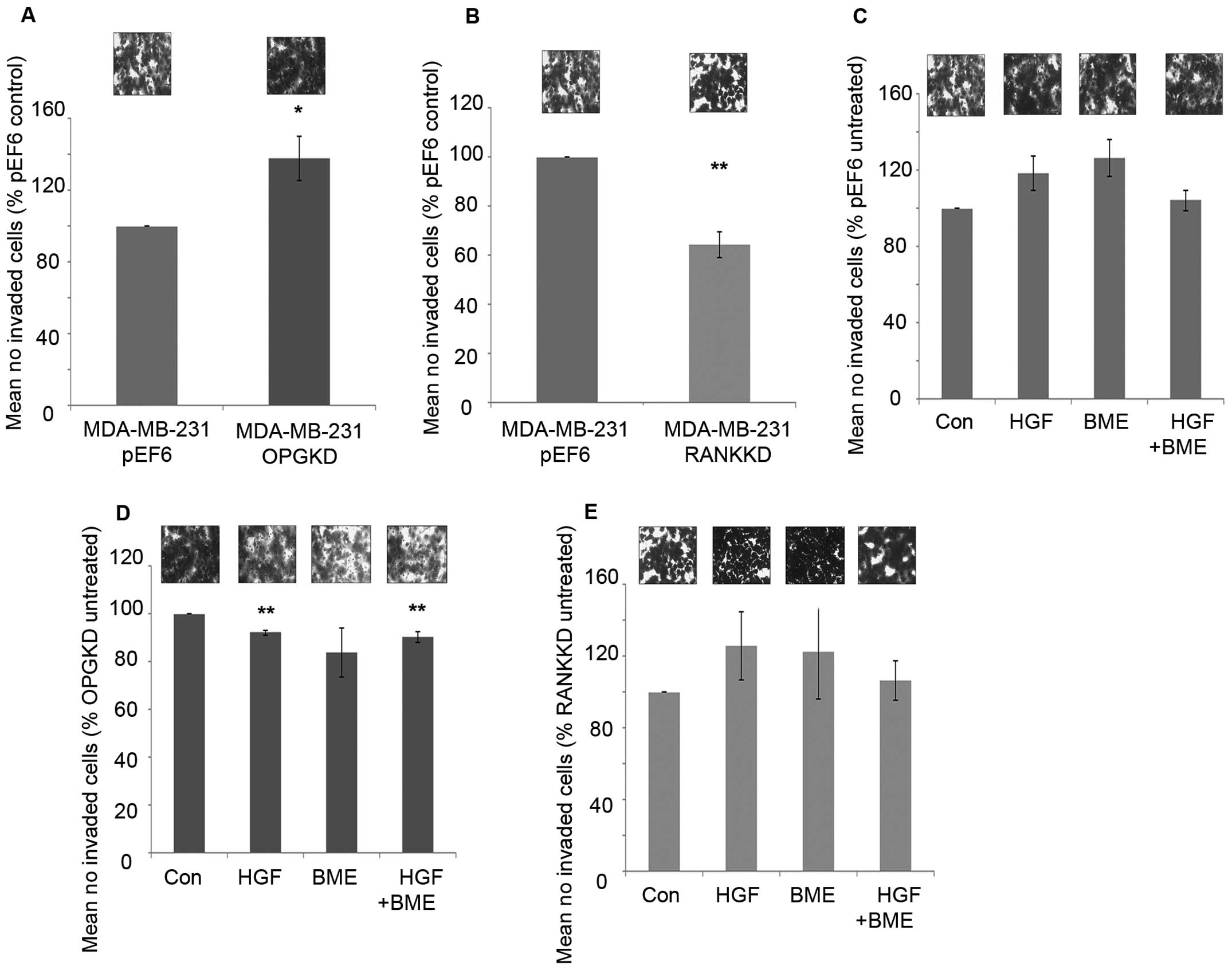
Suppression of OPG resulted in significantly
increased cell invasiveness compared to the control cells (Fig. 5A, p=0.037). However, suppression of
RANK in MDA-MB-231 cells resulted in a significant decrease in
breast cancer cell invasion in vitro compared to the control
cells (Fig. 5B, p=0.002).
Treatment of MDA-MB-231pEF6 cells with 40 ng/ml HGF or
50 μg/ml BME increased cell invasion in vitro compared to
untreated cells, however, these trends did not reach the
statistically significant threshold (Fig. 5C). When these treatments were added
in combination no noticeable effect on MDA-MB-231 cell invasion was
observed. In contrast, when MDA-MB-231OPGKD cells were
treated with 40 ng/ml HGF or a combination of 40 ng/ml HGF and 50
μg/ml BME significant reductions in cell invasion were observed
compared to the untreated cells (Fig.
5D, p=0.002 and 0.013, respectively). The largest reduction in
cell invasion was observed under the individual 50 μg/ml BME
treatment; however, this trend did not reach statistical
significance. MDA-MB-231RANKKD cell invasion under both
individual exogenous treatments increased in vitro compared
to the untreated control cells in a similar trend to that observed
in the MDA-MB-231pEF6 control cells, though none of
these changes reached a significant level (Fig. 5E). However, interestingly a
combined treatment appeared to have little impact on breast cancer
cell invasion.
Discussion
With the combined efforts of surgeons, oncologists
and research, treatment options for primary breast cancer have
improved. However, one aspect of the disease which still remains
poorly understood and controlled is its metastatic spread,
particularly to the bone. Through the recent licensing of
Denosumab, the neutralising RANKL antibody, some progress has been
achieved; however, there still remains no preventative measures or
screening tools which can identify those most at risk.
Whilst many previous studies have considered the
role OPG plays in the inhibition of TRAIL and thus apoptosis
(15), few reports consider the
molecular implications in breast cancer progression to the bone. Of
interest from this study was the lack of proliferation response to
the exogenous HGF and BME treatments in the OPG suppressed cells
which had been seen in the control cells. Of similar interest was
that this lack of response to the exogenous stimuli (HGF and BME)
was also seen in the cell-matrix adhesion assays. This study has
also highlighted the potential roles suppression of OPG may have in
increasing breast cancer cell motility and invasion in addition to
its role in preventing apoptosis. Suppression of OPG resulted in
significantly more motile MDA-MB-231 breast cancer cells compared
to the untreated control. However, also of interest was that the
exogenous treatments did not appear to have any further effect on
this cell function. Though suppression of OPG resulted in
significantly increased cell invasion, of note was that all the
exogenously added treatments resulted in decreased cell invasion.
This is of particular interest because, though not reaching
statistical significance in the control cell line, treatment with
HGF or BME resulted in increases in cell invasion. This highlights
that OPG may play an integral role in breast cancer cells homing to
the bone environment. This present data therefore suggest that
expression of OPG may result in the suppression of these aggressive
cancer cell traits and may also contribute to the regulation of the
MDA-MB-231 breast cancer cell response to various environmental
stimuli, including HGF, once bone metastases have already been
established. This study implies in vitro the targeting of
OPG also results in a response suppression to the oncogenic factor
HGF. This is an interesting observation given a number of reports
which demonstrate the potential prognosis effect of its receptor,
c-MET expression and its phosphorylated version could have on
breast cancer survival (32).
However, an in vivo study by Zinonos et al (33) suggested that pharmacological
inhibition of OPG though beneficial for bone health (reduction in
osteolysis) also resulted in an increase in formation of soft
tissue metastases. This was supported by Weichhaus et al
(34) demonstrating that
suppression of OPG in a chick embryo model reduced metastasis.
These data and that from elsewhere in the scientific literature
therefore suggest that the targeting of OPG in breast cancer may be
a double edged sword (35,36).
In contrast, the suppression of RANK in MDA-MB-231
breast cancer cells resulted in decreased responses in all the
traits studied. Cell proliferation was significantly decreased
after 5-day incubation compared to the control. Similar lack of
response to the exogenous treatments was seen in the RANK
suppressed cells. Interestingly, individual HGF or BME treatments
increased cell-matrix adhesion and cell invasion in the RANK
suppressed cells, though these did not pass the threshold for
statistical significance. Most previous studies have overexpressed
RANK in breast cancer models and reported increases in aggressive
cell behaviour, including increased cell migration and invasion as
well as greater metastatic bone colonisation (37). Casimiro et al (38) in their study found a link between
bone-seeking RANK positive subclones of MDA-MB-231 cells and
increased cell migration and invasion through the RANKL JNK and ERK
1/2 signalling pathway. This demonstrates that the three molecules,
OPG, RANK and RANKL, originally linked to regulation of bone
turnover have other roles, potentially even pro-metastatic ones in
breast cancer. The data reported here suggest that the targeting of
RANK affects breast cancer cell behaviour associated with a
metastatic phenotype (i.e., migration and invasion) in its own
right, changes which subsequently remained unaltered when exposed
to a bone-like environment. This therefore opens the possibility to
explore the combination of dual therapies which combine targeting
of breast cancer cell expressed RANKL (Denosumab) and RANK.
The human body is an intricate combination of a
variety of cells and factors which could never be replicated in a
2-D model, possibly accounting for the disparity between the in
vitro results and our previously published clinical data.
Isolating OPG and RANK in this model system has demonstrated,
particularly with OPG that they may play roles in bone metastases
associated with breast cancer. Further scientific study is now
necessary to fully understand the downstream molecules of OPG which
influence this tumourigenic behaviour beyond the inhibition of
TRAIL-induced apoptosis.
Acknowledgements
The authors would like to thank Cancer Research
Wales for supporting this study.
References
|
1
|
Coleman RE: Clinical features of
metastatic bone disease and risk of skeletal morbidity. Clin Cancer
Res. 12:S6243–S6249. 2006. View Article : Google Scholar
|
|
2
|
Roodman GD: Mechanisms of bone metastasis.
N Engl J Med. 350:1655–1664. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Coleman RE: Metastatic bone disease:
Clinical features, pathophysiology and treatment strategies. Cancer
Treat Rev. 27:165–176. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Paget S: The distribution of secondary
growths in cancer of the breast. 1889. Cancer Metastasis Rev.
8:98–101. 1989.PubMed/NCBI
|
|
5
|
Siclari VA, Guise TA and Chirgwin JM:
Molecular interactions between breast cancer cells and the bone
microenvironment drive skeletal metastases. Cancer Metastasis Rev.
25:621–633. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fidler IJ and Poste G: The ‘seed and soil’
hypothesis revisited. Lancet Oncol. 9:8082008. View Article : Google Scholar
|
|
7
|
Weilbaecher KN, Guise TA and McCauley LK:
Cancer to bone: A fatal attraction. Nat Rev Cancer. 11:411–425.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Boyce BF and Xing L: Functions of
RANKL/RANK/OPG in bone modeling and remodeling. Arch Biochem
Biophys. 473:139–146. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hakeda Y, Kobayashi Y, Yamaguchi K, Yasuda
H, Tsuda E, Higashio K, Miyata T and Kumegawa M: Osteoclastogenesis
inhibitory factor (OCIF) directly inhibits bone-resorbing activity
of isolated mature osteoclasts. Biochem Biophys Res Commun.
251:796–801. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yasuda H, Shima N, Nakagawa N, Mochizuki
SI, Yano K, Fujise N, Sato Y, Goto M, Yamaguchi K, Kuriyama M, et
al: Identity of osteoclastogenesis inhibitory factor (OCIF) and
osteoprotegerin (OPG): A mechanism by which OPG/OCIF inhibits
osteoclastogenesis in vitro. Endocrinology. 139:1329–1337.
1998.PubMed/NCBI
|
|
11
|
Ibrahim T, Sacanna E, Gaudio M, Mercatali
L, Scarpi E, Zoli W, Serra P, Ricci R, Serra L, Kang Y, et al: Role
of RANK, RANKL, OPG, and CXCR4 tissue markers in predicting bone
metastases in breast cancer patients. Clin Breast Cancer.
11:369–375. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mercatali L, Ibrahim T, Sacanna E, Flamini
E, Scarpi E, Calistri D, Ricci M, Serra P, Ricci R, Zoli W, et al:
Bone metastases detection by circulating biomarkers: OPG and
RANK-L. Int J Oncol. 39:255–261. 2011.PubMed/NCBI
|
|
13
|
Van Poznak C, Cross SS, Saggese M, Hudis
C, Panageas KS, Norton L, Coleman RE and Holen I: Expression of
osteoprotegerin (OPG), TNF related apoptosis inducing ligand
(TRAIL), and receptor activator of nuclear factor kappaB ligand
(RANKL) in human breast tumours. J Clin Pathol. 59:56–63. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Neville-Webbe HL, Cross NA, Eaton CL,
Nyambo R, Evans CA, Coleman RE and Holen I: Osteoprotegerin (OPG)
produced by bone marrow stromal cells protects breast cancer cells
from TRAIL-induced apoptosis. Breast Cancer Res Treat. 86:269–279.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Holen I, Cross SS, Neville-Webbe HL, Cross
NA, Balasubramanian SP, Croucher PI, Evans CA, Lippitt JM, Coleman
RE and Eaton CL: Osteoprotegerin (OPG) expression by breast cancer
cells in vitro and breast tumours in vivo - a role in tumour cell
survival? Breast Cancer Res Treat. 92:207–215. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nicolin V and Narducci P: Soluble TRAIL
could enhance bone destruction acting on Rank-ligand in
estrogen-independent human breast cancer cell line MDA-MB-231. Acta
Histochem. 112:189–192. 2010. View Article : Google Scholar
|
|
17
|
Michalopoulos G, Houck KA, Dolan ML and
Leutteke NC: Control of hepatocyte replication by two serum
factors. Cancer Res. 44:4414–4419. 1984.PubMed/NCBI
|
|
18
|
Nakamura T, Nawa K and Ichihara A: Partial
purification and characterization of hepatocyte growth factor from
serum of hepatectomized rats. Biochem Biophys Res Commun.
122:1450–1459. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Russell WE, McGowan JA and Bucher NL:
Biological properties of a hepatocyte growth factor from rat
platelets. J Cell Physiol. 119:193–197. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jiang WG, Martin TA, Parr C, Davies G,
Matsumoto K and Nakamura T: Hepatocyte growth factor, its receptor,
and their potential value in cancer therapies. Crit Rev Oncol
Hematol. 53:35–69. 2005. View Article : Google Scholar
|
|
21
|
Parikh RA, Wang P, Beumer JH, Chu E and
Appleman LJ: The potential roles of hepatocyte growth factor
(HGF)-MET pathway inhibitors in cancer treatment. Onco Targets
Ther. 7:969–983. 2014.PubMed/NCBI
|
|
22
|
Davies S and Jiang WG: ALCAM, activated
leukocyte cell adhesion molecule, influences the aggressive nature
of breast cancer cells, a potential connection to bone metastasis.
Anticancer Res. 30:1163–1168. 2010.PubMed/NCBI
|
|
23
|
Sanders AJ, Parr C, Mason MD and Jiang WG:
Suppression of hepatocyte growth factor activator inhibitor-1 leads
to a more aggressive phenotype of prostate cancer cells in vitro.
Int J Mol Med. 20:613–619. 2007.PubMed/NCBI
|
|
24
|
Yuan Z, Sanders AJ, Ye L, Wang Y and Jiang
WG: Knockdown of human antigen R reduces the growth and invasion of
breast cancer cells in vitro and affects expression of cyclin D1
and MMP-9. Oncol Rep. 26:237–245. 2011.PubMed/NCBI
|
|
25
|
Zuker M: Mfold web server for nucleic acid
folding and hybridization prediction. Nucleic Acids Res.
31:3406–3415. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jiang WG, Hiscox S, Hallett MB, Horrobin
DF, Mansel RE and Puntis MC: Regulation of the expression of
E-cadherin on human cancer cells by gamma-linolenic acid (GLA).
Cancer Res. 55:5043–5048. 1995.PubMed/NCBI
|
|
27
|
Jiang WG, Hiscox S, Singhrao SK, Nakamura
T, Puntis MC and Hallett MB: Inhibition of HGF/SF-induced membrane
ruffling and cell motility by transient elevation of cytosolic free
Ca2+. Exp Cell Res. 220:424–433. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rosen EM, Carley W and Goldberg ID:
Scatter factor regulates vascular endothelial cell motility. Cancer
Invest. 8:647–650. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Albini A, Iwamoto Y, Kleinman HK, Martin
GR, Aaronson SA, Kozlowski JM and McEwan RN: A rapid in vitro assay
for quantitating the invasive potential of tumor cells. Cancer Res.
47:3239–3245. 1987.PubMed/NCBI
|
|
30
|
Parish CR, Jakobsen KB and Coombe DR: A
basement-membrane permeability assay which correlates with the
metastatic potential of tumour cells. Int J Cancer. 52:378–383.
1992. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Owen S, Ye L, Sanders AJ, Mason MD and
Jiang WG: Expression profile of receptor activator of nuclear-κB
(RANK), RANK ligand (RANKL) and osteoprotegerin (OPG) in breast
cancer. Anticancer Res. 33:199–206. 2013.
|
|
32
|
Raghav KP, Wang W, Liu S, Chavez-MacGregor
M, Meng X, Hortobagyi GN, Mills GB, Meric-Bernstam F, Blumenschein
GR Jr and Gonzalez-Angulo AM: cMET and phospho-cMET protein levels
in breast cancers and survival outcomes. Clin Cancer Res.
18:2269–2277. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zinonos I, Luo KW, Labrinidis A, Liapis V,
Hay S, Panagopoulos V, Denichilo M, Ko CH, Yue GG, Lau CB, et al:
Pharmacologic inhibition of bone resorption prevents cancer-induced
osteolysis but enhances soft tissue metastasis in a mouse model of
osteolytic breast cancer. Int J Oncol. 45:532–540. 2014.PubMed/NCBI
|
|
34
|
Weichhaus M, Segaran P, Renaud A, Geerts D
and Connelly L: Osteoprotegerin expression in triple-negative
breast cancer cells promotes metastasis. Cancer Med. 3:1112–1125.
2014. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Body JJ, Greipp P, Coleman RE, Facon T,
Geurs F, Fermand JP, Harousseau JL, Lipton A, Mariette X, Williams
CD, et al: A phase I study of AMGN-0007, a recombinant
osteoprotegerin construct, in patients with multiple myeloma or
breast carcinoma related bone metastases. Cancer. 97(Suppl):
887–892. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chanda D, Isayeva T, Kumar S, Siegal GP,
Szafran AA, Zinn KR, Reddy VV and Ponnazhagan S: Systemic
osteoprotegerin gene therapy restores tumor-induced bone loss in a
therapeutic model of breast cancer bone metastasis. Mol Ther.
16:871–878. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Blake ML, Tometsko M, Miller R, Jones JC
and Dougall WC: RANK expression on breast cancer cells promotes
skeletal metastasis. Clin Exp Metastasis. 31:233–245. 2014.
View Article : Google Scholar
|
|
38
|
Casimiro S, Mohammad KS, Pires R,
Tato-Costa J, Alho I, Teixeira R, Carvalho A, Ribeiro S, Lipton A,
Guise TA, et al: RANKL/RANK/MMP-1 molecular triad contributes to
the metastatic phenotype of breast and prostate cancer cells in
vitro. PLoS One. 8:e631532013. View Article : Google Scholar : PubMed/NCBI
|