Introduction
Exosomes are small RNA and protein containing
extracellular vesicles that are able to mediate hetero- and
homotypic intercellular communication (1). These natural nanovectors are formed
through inward budding of endosomal membranes, giving rise to
intracellular multivesicular bodies that integrate into the plasma
membrane, and are eventually released (2). Different cell types have been shown
to produce exosomes of biologic significance into the extracellular
space and the biologic fluids, including B cells, dendritric cells,
T cells, platelets, stem cells, and cancer cells (3–8). We
have recently demonstrated a role of exosomes as mediators of
platelet lysate activity in bone regeneration through the ability
to influence osteogenic differentiation and promote cell
proliferation and the migration of mesenchymal stromal cell
(9). In cancer, exosomes can
facilitate tumour progression by supplying the tumour niche with
molecules that favour the progression of oncogenic processes, such
as proliferation, invasion and metastasis, modulation of immune
response, and drug resistance (10–12).
Recently, Corcoran et al (13) have demonstrated the ability of
prostate cancer cells to transfer multidrug resistance (MDR)
phenotype via microvesicles/exosomes, suggesting that, in the
context of tumour microenvironment, the development of the MDR
phenotype can be mediated, at least in part, by the transfer of
P-glycoprotein (P-gp) and microRNAs by exosomes (13,14).
Similar results have been obtained in breast cancer, where the
transfer of P-gp and miRNAs by exosomes modulates cell cycle
distribution and drug-induced apoptosis (15,16).
Osteosarcoma (OS) is the most common primary bone
cancer in children and adolescents. Despite aggressive treatment
regimens, the outcome is unsatisfactory, particularly in patients
with metastatic and/or recurrent disease (17). Treatment failure is commonly due to
the development of chemoresistance (18), which appears to be mediated by a
variety of mechanisms (19).
In this study we investigated the potential role of
exosomes to transfer a drug resistance phenotype in human OS cells.
For this purpose we purified and characterized exosomes from an MDR
OS cell line (20) and we showed,
for the first time, that exosomes transfer functional MDR-1 mRNA
and its product P-glycoprotein to drug-sensitive cells in
vitro. This intercellular transfer provides an additional
pathway for the cellular acquisition and dissemination of
drug-resistant traits indicating exosomes as important mediators in
the spread of MDR in human osteosarcoma.
Materials and methods
Drug
A stock solution of doxorubicin (DXR)
(Sigma-Aldrich, Milan, Italy) was prepared in dimethylsulfoxide (5
mg/ml) and stored at −20°C. Appropriate concentrations of DXR
solution were freshly diluted before each experiment.
Cell culture
The human OS cell line MG-63 was obtained from the
American Type Culture Collection and validated in May 2014 by
short-tandem repeat profiling of extracted genomic DNA generated by
ATCC-LGC standards. MDR cell line MG-63DXR30 was established from
the parental MG-63 (20). Cells
were grown in Iscove's modified Dulbecco's medium (IMDM) (Gibco,
Thermo Fisher Scientific, Waltham, MA, USA), supplemented with 10%
heat inactivated fetal bovine serum (FBS) (Sigma-Aldrich), 100 U/ml
penicillin, and 0.1 mg/ml streptomycin (Lonza, Milan, Italy).
Drug-resistant variant MG-63DXR30, was continuously cultured in the
presence of the selective drug concentration (30 ng/ml
doxorubicin). All cell lines were maintained at 37°C in a
humidified 5% CO2 atmosphere.
Co-culture assays
The effect of MG-63DXR30 secretome on MG-63
viability was evaluated by the Boyden chamber assay with 0.4-μm
pore membrane filters (Costar, Corning Inc., NY, USA). MG-63 or
MG-63DXR30 cells were seeded (5×103 cells/well) into the
upper chamber, and MG-63 cells were applied at equal proportion to
the lower chamber. Twenty-four hours after seeding, 10 ng/ml of
doxorubicin was added to the lower chamber. After 72 h, cell
viability was assessed using the Alamar Blue assay (Invitrogen,
Thermo Fisher Scientific). The fluorescence was read at 535–590 nm
using a microplate-reader (Tecan Infinite F200pro, Tecan, Milan,
Italy). The results were expressed as relative fluorescence units
(RFU).
Exosome isolation and purification
MG-63 and MG-63DXR30 cells were cultured until 70%
confluence. Cells were washed with phosphate-buffered saline (PBS)
and incubated for two consecutive periods with IMDM supplemented
with 10% FBS depleted of exosomes (FDE) obtained via
ultracentrifugation (9). Following
collection of the supernatant, the exosomes were concentrated by
differential centrifugation: 500 × g for 10 min (two times), 2,000
× g for 15 min (two times), and 10,000 × g for 30 min (two times)
at 4°C to remove floating cells and cellular debris. The
supernatant was then ultracentrifuged at 110,000 × g for 1 h at
4°C. The exosome pellet was resuspended in PBS and centrifuged at
110,000 × g for 1 h at 4°C (Beckman Coulter, Milan, Italy). The
exosome pellet was resuspended in PBS and stored at −80°C until
use. Exosome quantity was determined by the Bradford method
(Bio-Rad, Milan, Italy). The exosomes extracted from the
supernatant of MG-63 and from the medium of MG-63DXR30 were named
Exo/S and Exo/DXR, respectively.
Electron microscopy
Exosomes were resuspended in 2% paraformaldehyde
(PFA) and loaded onto formvar/carboncoated grids. Next, exosomes
were fixed in 1% glutaraldehyde, washed, and counterstained with a
solution of uranyloxalate, pH 7.0, embedded in a mixture of 4%
uranylacetate and 2% methylcellulose before observation with a
Zeiss-EM 109 electron microscope (Zeiss, Jena, Germany).
Western blot analysis
Exosomes and cell pellets were treated with RIPA
lysis buffer (1% Triton X-100, 10% Na-deoxycholate, 5 M NaCl, 1 M
Tris-HCl pH 7.4, 0.5 M EGTA pH 8.0, 1 M NaF) and protease inhibitor
cocktail (Roche, Milan, Italy) for 20 min at 4°C. Nuclei and cell
debris were removed by centrifugation. The protein concentration
was determined using the Bradford assay (Bio-Rad). The total
cellular proteins and exosomal proteins were resolved by 10%
SDS-polyacrylamide gel and transferred to a nitrocellulose membrane
(Thermo Fisher Scientific). The membrane was blocked with 5% bovine
serum albumin (BSA) (Sigma-Aldrich) in T-TBS (0.1 M Tris-HCl pH
8.0, 1.5 M NaCl and 1% Tween-20) for 1 h at room temperature.
Subsequently, the membranes were incubated with rabbit polyclonal
CD63 (sc-15363) (1:500), mouse polyclonal calnexin (sc-23954)
(1:500) and mouse polyclonal MDR-1/P-gp (sc-55510) (1:200) (Santa
Cruz Biotechnology, Santa Cruz, CA, USA) antibodies overnight at
4°C. After vigorous washing in T-TBS, the membranes were incubated
with the secondary antibody for 1 h at room temperature.
Anti-rabbit antibody (NA934VS) (1:1,000) for CD63, anti-mouse
antibody (NA931VS) (1:2,000) for calnexin and MDR-1/P-gp (GE
Healthcare, Milan, Italy), all conjugated to horseradish peroxidase
were diluted in T-TBS containing 5% BSA and used as secondary
antibodies. Immunocomplexes were detected with the ECL western blot
analysis system (Amersham Pharmacia Biotech, Piscataway, NJ, USA).
Reversible Ponceau S (Sigma-Aldrich) staining was used to assess
equal gel loading.
Exosome labelling and uptake
Exosomes were labelled using the PKH26 Red
Fluorescent Cell Linker kit (Sigma-Aldrich) according to the
manufacturer's instructions with minor modifications (9). Two microgram (2 μg) of the PKH26
labelled exosomes, or the same volume of the PKH26-PBS control,
were resuspended in IMDM supplemented with 10% FDE and added to
9×103 MG-63 cells maintained at 37°C in a humidified
atmosphere with 5% CO2. All samples were
ultracentrifuged at 110,000 × g for 1 h at 4°C before being added
to the cells. After 4 and 24 h of incubation, uptake was stopped by
washing and fixation in 3.7% PFA for 10 min. Cells were then
stained with a fluorescein isothiocyanate (FITC)-conjugated
phalloidin (Sigma-Aldrich) and visualised with a Nikon Eclipse
E800M fluorescence microscope (Nikon, Tokyo, Japan). All cells per
high power-field were counted, and the percentage of PKH26
positively stained cells was determined. Five representative high
power-fields per sample were evaluated.
Cell viability
Cell viability was evaluated by the acid phosphatase
assay (Sigma-Aldrich) as previously described (21). MG-63 cells were seeded in 96-well,
flat-bottomed tissue culture plates at a density of
2×103 cells/well. After 24 h, cells were treated with
1.5 μg of Exo/S or Exo/DXR or PBS (control), in the presence of
IMDM + 10% FDE. Four hours after exosome addition, cells were
treated with increasing concentrations of doxorubicin (1–60 ng/ml)
for 72 h. Briefly, the cells were washed and incubated at 37°C with
100 μl of buffer containing 0.1 M sodium acetate pH 5.0, 0.1%
Triton X-100, and 5 mM p-nitrophenil phosphate. After 2 h, the
reaction was stopped with the addition of 10 μl of 1 N NaOH, and
colour development was assayed at 405 nm using a microplate reader
(Tecan Infinite F200pro). Data are reported as cell survival in
respect to untreated cells (set=100%). All experiments were
performed three times in triplicate.
Wound-healing assay
Confluent MG-63 and MG-63DXR30 cell monolayers were
scratched with a sterile 100-μl pipette tip and incubated with or
without Exo/S or Exo/DXR. Cell migration was monitored for 24 h
under a Nikon Eclipse-TE 2000-S microscope (Nikon). The widths of
the ‘wound’ (scratched areas) were measured by the NIS-Elements
Image Software BR 4.00.00 (Nikon) and the proportion of wound
healing was calculated by the following formula: 100% − (width
after 24 h/width at the beginning) × 100%.
Quantitative real-time polymerase chain
reaction (qRT-PCR)
Cells were seeded in 12-well plates
(1.5×104 cells/well). The following day, cells were
treated with 7.5 μg of Exo/S or Exo/DXR or PBS (control) for 72 h.
Total RNA from cells and exosomes (n=3) was extracted using
TRIzol® reagent (Invitrogen, Life Technologies, Monza,
Italy) (9). To confirm that the
RNA was confined to exosomes, Exo/S and Exo/DXR were treated with
0.4 μg/μl RNase A (Sigma-Aldrich) for 1 h at 37°C. Total mRNA was
reverse transcribed using the MULV Reverse Transcriptase kit
(Applied Biosystems, Thermo Fisher Scientific). The expression of
MDR-1 (AF016535.1) was evaluated by quantitative real-time
polymerase chain reaction (qRT-PCR) using the LightCycler
instrumentation and the Universal Probe Library system (Roche
Applied Science, Monza, Italy). Probes and primers were selected by
a web-based assay design software (Probe Finder https://www.roche-applied-science.com):
MDR-1-f 5′-GCCATCAGTCCTGTTCTTGG-3′; MDR-1-r 5′-GCTTTTGCATACGCTA
AGAG TTC-3′. The results were expressed as the ratio between gene
of interest and reference gene (GAPDH: NM_002046.3; GAPDH-f
5′-AGCCACATCGCTCAGACAC-3′; GAPDH-r 5′-GCCCAATACGACCAAATCC-3′)
according to the 2-ΔΔCT method (22).
Immunofluorescence assay
MG-63 cells (5×103 cells/cm2)
were seeded in IMDM + 10% FDE. After 24 h, cells were treated with
5 μg of Exo/S or Exo/DXR or PBS (negative control) for 72 h, and
processed as previously described (23). At termination, cells were fixed in
3.7% PFA for 10 min, permeabilized in 0.1% Triton X-100/PBS for 4
min, and blocked using 1% BSA/PBS for 30 min. Cells were stained
with an anti-MDR-1 antibody (sc-55510) (Santa Cruz Biotechnology)
at 1:50 diluted in PBS containing 0.1% BSA at 22°C for 10–12 h,
followed by incubation with Alexa Fluor 568-conjugated goat
anti-mouse secondary antibody (A11004) (Invitrogen) at 1:1,000
dilution in PBS containing 1% BSA at 22°C for 30 min. MG-63DXR30
cells were used as a positive control. Images were acquired with a
Nikon Eclipse E800M fluorescence microscope (Nikon). Ten
non-overlapping fields/image were taken (three images/sample were
collected) and analysed using NIS-Elements Image software BR4.00.00
(Nikon). Changes of fluorescence intensity were calculated dividing
the red fluorescent intensity by the number of cells on each
field.
Statistical analysis
Statistical analysis was performed by StatView™
5.0.1 software (SAS Institute, Cary, NC, USA). Quantitative results
were expressed as mean ± the standard deviation. The Wilcoxon
signed rank test was used to evaluate the effects of exosomes
treatment on MDR-1/P-gp expression by real-time PCR and
immunofluorescence, and to assess MG-63DXR30 effects on MG-63
viability by co-culture assays. The Mann-Whitney U test was applied
to evaluate the quantity of exosome release and to analyze the
effects of exosomes on cell viability and migration. Experiments
were performed in triplicate. p<0.05 was considered
statistically significant.
Results
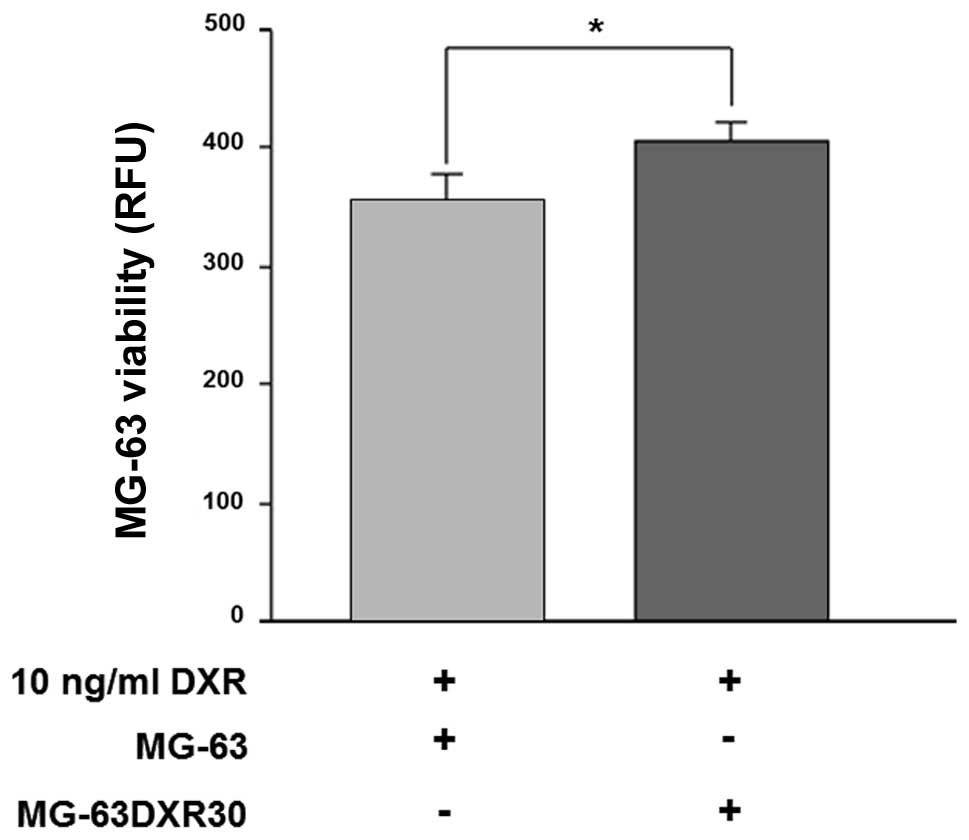
MG-63DXR30 cells transmit doxorubicin
resistance to MG-63 cells
MG-63 were co-cultured with MG-63DXR30 in a 1:1
ratio, for 72 h, in the presence of doxorubicin. As shown in
Fig. 1, after drug exposure,
survival of MG-63 cells was significantly increased (p=0.0039).
These results suggested that MG-63DXR30 transmit chemoresistance to
recipient cells, and such effect could be ascribed to exosomes.
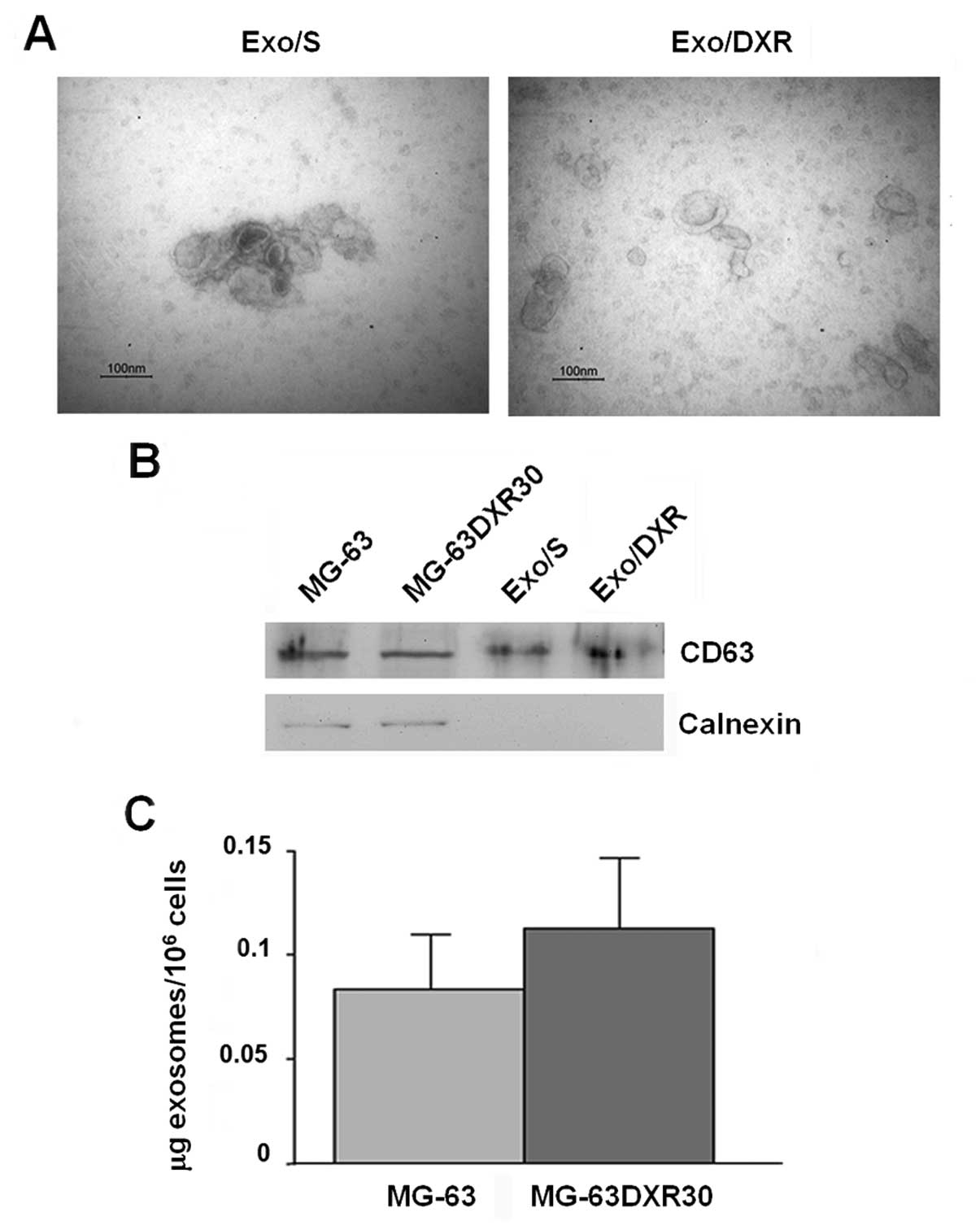
Identification and characterization of
exosomes
To investigate exosome correlation with resistance
transmission, we collected exosomes from supernatant of MG-63 and
MG-63DXR30 OS cell lines through a series of centrifugation and
ultracentrifugation steps. Transmission electron microscopy
analysis showed that the nanovesicles isolated from OS cells were
morphologically homogeneous, ranging from 30 to 100 nm in size,
with a typical round or cup shape appearance (Fig. 2A). Exosome purity was assessed by
western blot analysis. As shown in Fig. 2B, they expressed exosome-related
protein CD63 and, as expected, were negative for the endoplasmic
reticulum protein calnexin (15).
Ponceau S staining served as loading control (data not shown).
Similar amounts of exosomes were secreted by MG-63 and MG-63DXR30
cells (Fig. 2C).
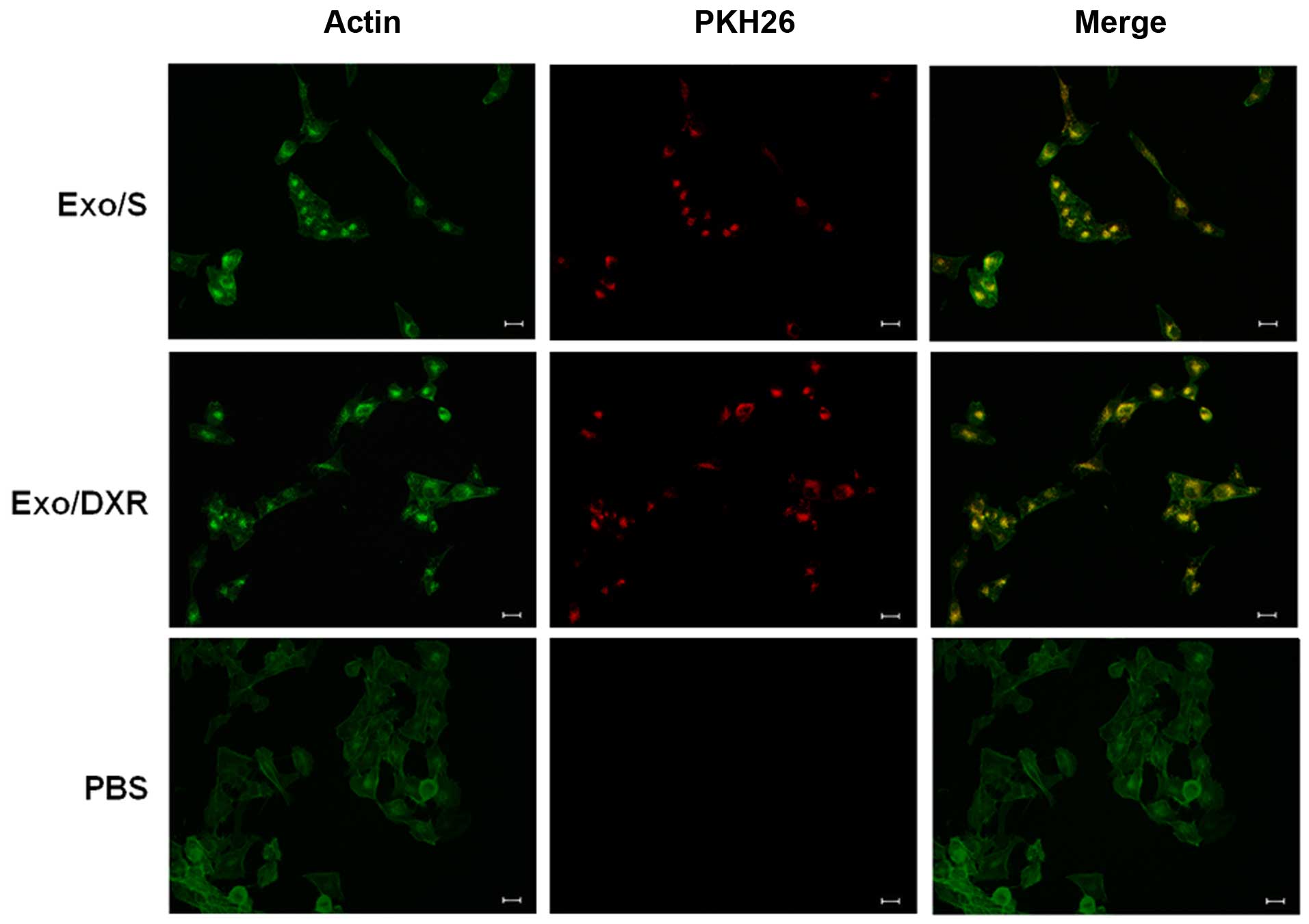
Uptake of exosomes
To examine whether exosomes from MG-63DXR30
(Exo/DXR) and MG-63 (Exo/S) could be taken up in MG-63 cells, PKH26
labeled exosomes were incubated with MG-63 cells at two different
time points and examined using fluorescence microscopy. After 4 h,
a few exosomes were already taken up by MG-63 (data not shown). As
shown in Fig. 3, after incubation,
MG-63 cells were able to take up similar amount of Exo/DXR
(97.7±1.1%) and Exo/S (97±1.4%). In particular, PKH26 signal was
detected in the perinuclear region, suggesting the adsorption and
internalization of exosomes. No fluorescent signal was detected in
the control (PBS).
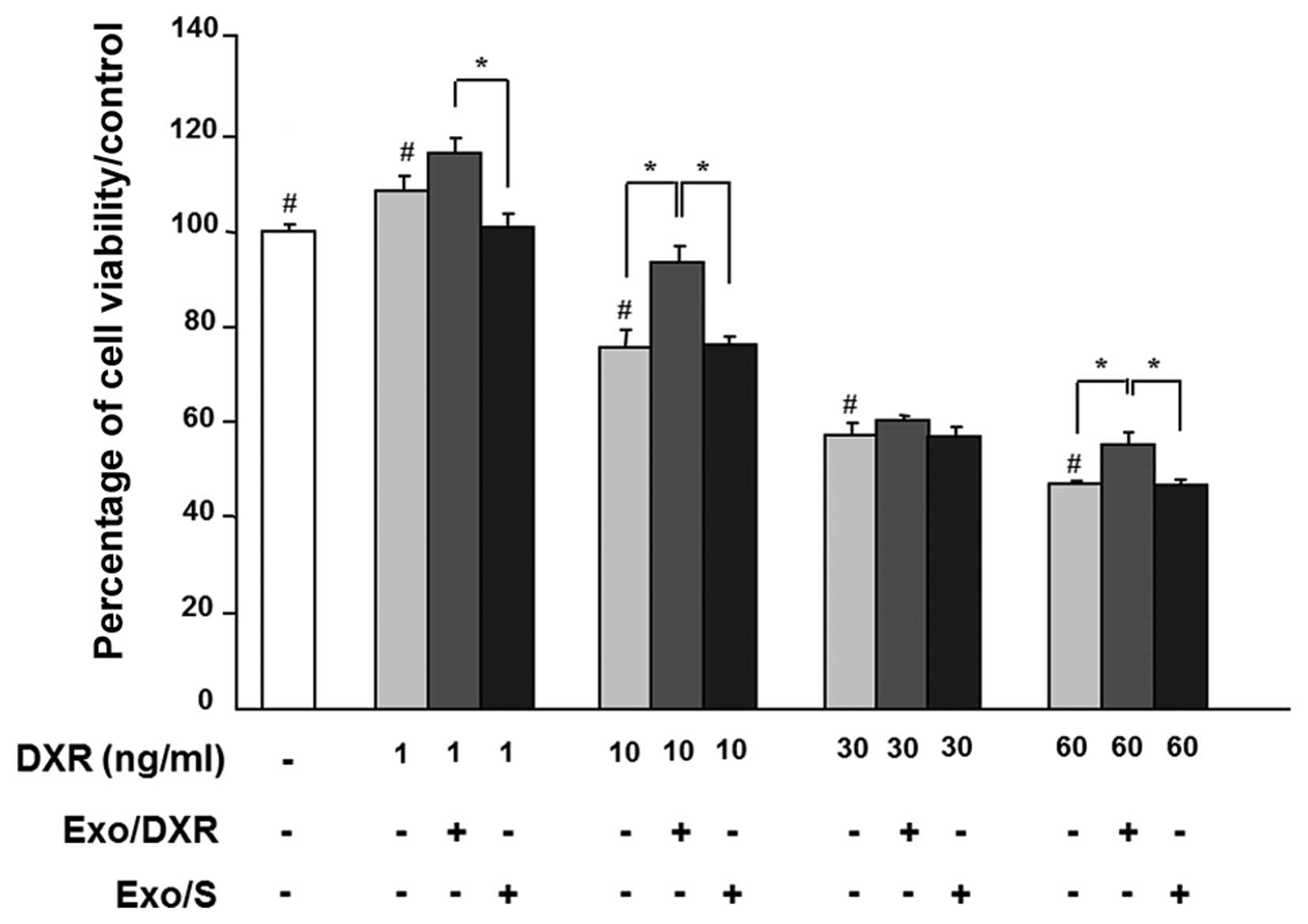
Exo/DXR decrease the sensitivity of MG-63
cells to doxorubicin
Cell viability was examined in exosome-treated MG-63
in the presence of increasing concentrations of doxorubicin (1–60
ng/ml) for 72 h. As expected, doxorubicin affected MG-63 viability
in a dose-dependent way. Incubation of MG-63 cells with 1.5 μg of
Exo/DXR decreased the sensitivity of cells to doxorubicin in all
concentration tested. In particular, cells cultured at 10 ng/ml of
doxorubicin and exposed to Exo/DXR showed a significant increase
(p=0.0095) in viability compared to cells incubated with
doxorubicin alone (Fig. 4). On the
contrary, MG-63 viability was not modified when cells where treated
with Exo/S (Fig. 4).
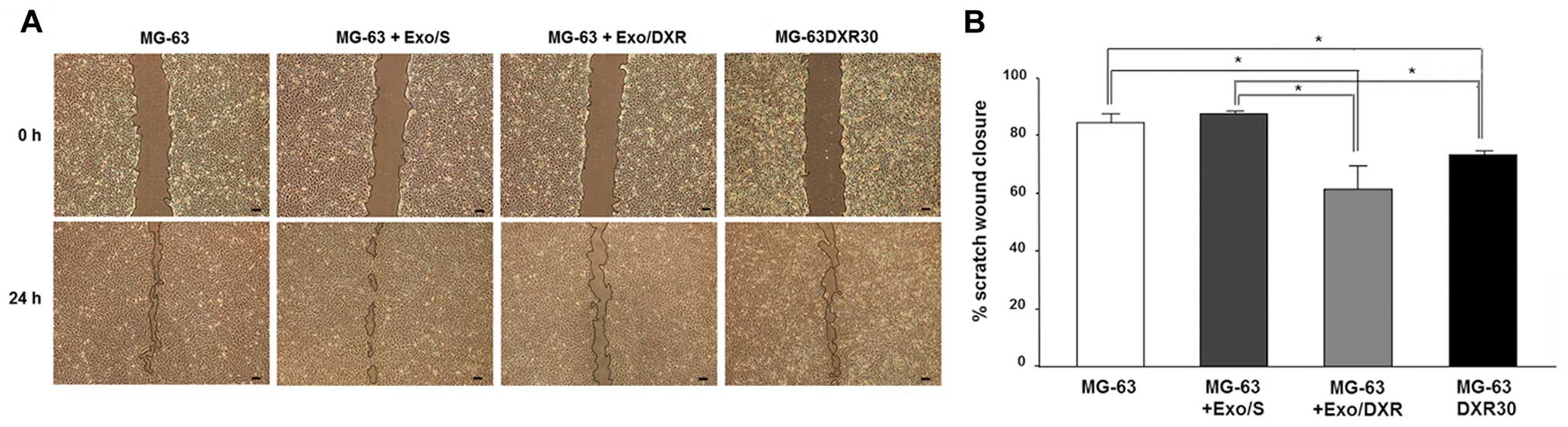
Exo/DXR decrease MG-63 cell motility
The effects of Exo/DXR on MG-63 motility were
evaluated by the wound-healing assay. After 24 h the migration of
MG-63DXR30 was significantly lower than that of parental MG-63
cells (p=0.02). MG-63 treated with Exo/DXR exhibited a significant
decrease of wound closure compared to MG-63 and MG-63 incubated
with Exo/S (p=0.02), whereas no difference was detected between
Exo/DXR treated cells and MG-63DXR30 (Fig. 5A and B).
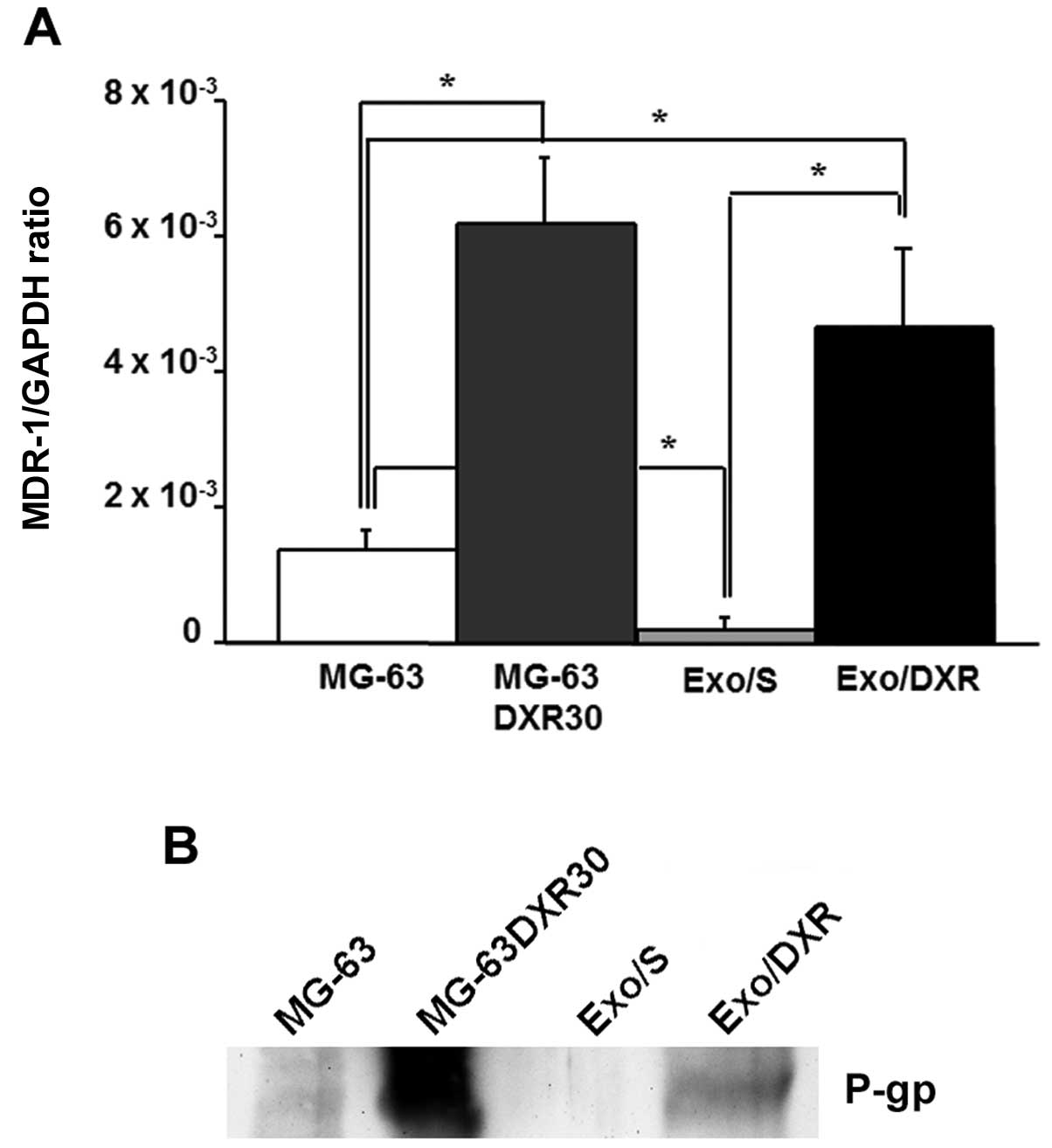
Exo/DXR expressed MDR-1 mRNA and
P-gp
To verify the presence of MDR-1 in exosome samples,
we extracted total RNA from Exo/DXR, Exo/S and their cells of
origin. The expression of MDR-1 was evaluated by qRT-PCR. Results
demonstrated that Exo/DXR expressed higher levels of MDR-1 mRNA
compared to Exo/S (p=0.03) (Fig.
6A). The western blot analysis was consistent with the results
from qRT-PCR. Exo/DXR and their donor cells, but not MG-63 and
Exo/S, showed high levels of P-gp expression by western blot
analysis (Fig. 6B). Ponceau S
staining served as loading control (data not shown).
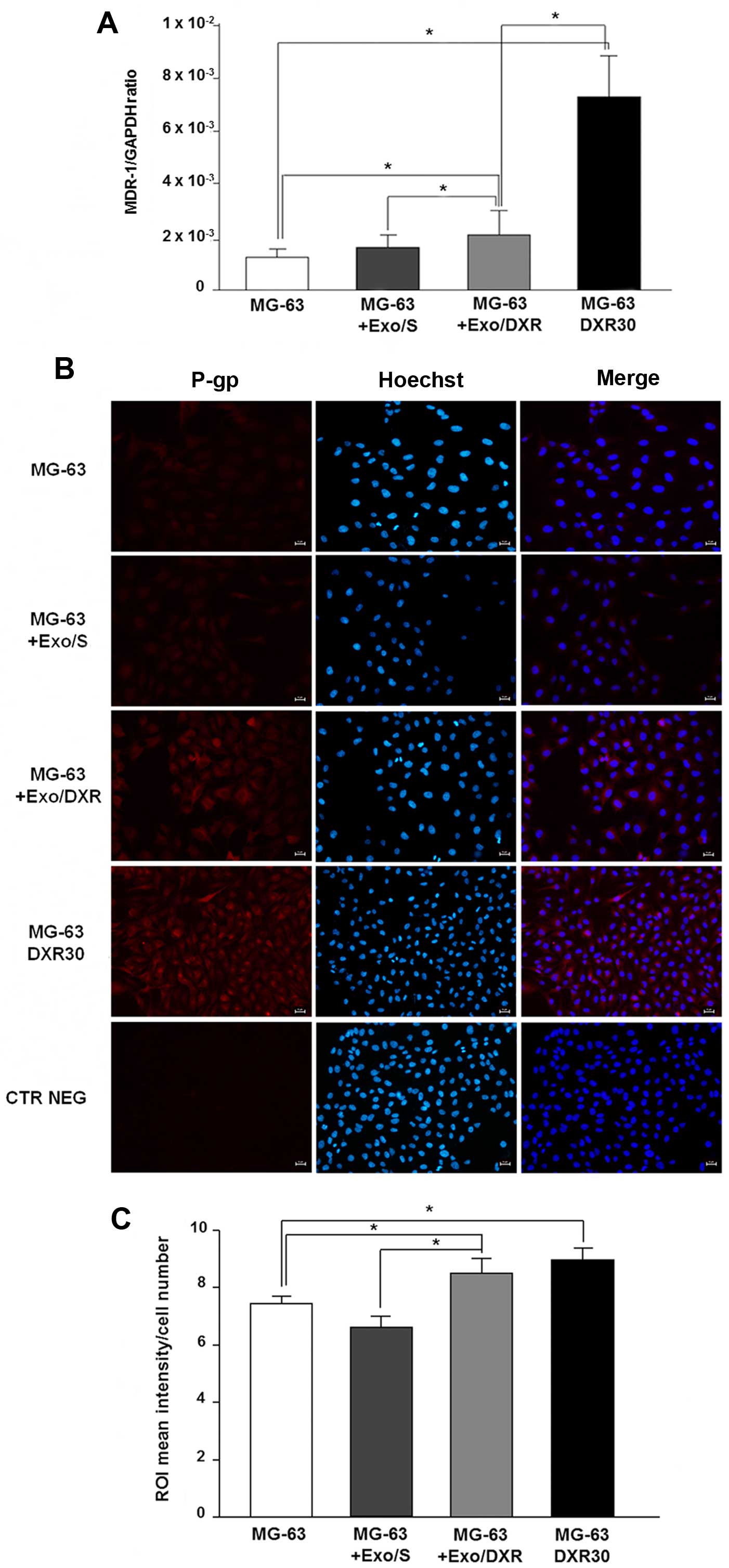
Exo/DXR transfer MDR-1 mRNA and P-gp to
recipient cells
The ability of Exo/DXR to transfer MDR-1 mRNA and
P-gp was assessed by qRT-PCR and immunofluorescence analysis.
Incubation of MG-63 cells with Exo/DXR induced a significant
increase of MDR-1 mRNA levels compared to untreated cells (p=0.02).
On the contrary, no substantial change in MDR-1 expression was
observed when MG-63 cells were treated with Exo/S (Fig. 7A). As expected, MG-63DXR30
expressed significantly higher levels of MDR-1 mRNA compared to
MG-63 cells (p=0.0006) (Fig. 7A).
Immunofluorescence analysis confirmed a significant increase of
P-gp expression in MG-63 treated with Exo/DXR (p=0.009) (Fig. 7B and C). Interestingly, MG-63
treated with Exo/DXR and MG-63DXR30 cells expressed similar levels
of P-gp. The treatment with Exo/S did not affect P-gp protein
levels (Fig. 7B and C).
Discussion
OS is the most common primary malignant bone tumour
in children and adolescents (17).
Although neoadjuvant chemotherapy and improved surgical techniques
have increased the survival rate of OS ≤65–75%, this is still
unsuccessful in 30–40% of patients with localised tumours and in
80–85% of patients with metastatic disease at presentation
(24,25). MDR, both intrinsic and acquired, is
still a major concern regarding the clinical management of OS
patients and a key issue in the failure of current treatment
(19). The MDR phenotype can be
mediated by several mechanisms, including increased
energy-dependent efflux of chemotherapeutic drugs (26). The principal transmembrane
transporter responsible for this mechanism is P-glycoprotein
(P-gp), a member of the ATP-binding cassette protein superfamily,
encoded by the MDR-1 gene, which lowers intracellular drug
concentrations to sub-lethal levels (27,28).
Although P-gp is involved in resistance or poor response to
chemotherapy (29), other
undefined cellular factors also seem to participate in modulating
drug cytotoxicity. A growing body of evidence has demonstrated that
several features of host microenvironment play also a role,
including extracellular pH, temperature, oxygen supply, and
extracellular matrix (30–32). Moreover, the intercellular transfer
of MDR represents an additional mechanism for the cellular
acquisition and spreading of drug-resistant traits (33). Acquired MDR was recently found to
be mediated by exosomes released by drug resistant cells (34). Such phenomenon was observed in
several tumour models, including ovarian cancer (35), prostate cancer (14), breast cancer (15), and melanoma (36). In this study, we reported that MDR
phenotype can also be induced in human OS via exosomes derived from
MDR cells. MG-63DXR30 derived-exosomes are able to decrease MG-63
sensitivity to doxorubicin as well as to transfer phenotypic
characteristics representative of their cell line of origin to
recipient cells, including cell motility.
In agreement with previous studies (13,15),
we demonstrated that P-gp is contained in MG-63DXR30
derived-exosomes and that MDR can be transferred to sensitive cells
by the delivery of P-gp in recipient cells through exosomes.
Moreover, exosomes are known to affect target cells
by transferring mRNAs and microRNAs (37). The presence of mRNA and microRNA,
termed ‘exosomal shuttle RNA’, in exosomes suggests that genetic
material exchange could be an additional level of exosome-mediated
intercellular communication (38).
In particular, recent studies have described how the transfer of
specific miRNA via exosomes potentially contributes to drug
resistance in prostate cancer and breast cancer (14,16).
In this study, we demonstrated, for the first time,
that MDR-1 mRNA is highly expressed within Exo/DXR, and is
transferred to and accumulated in OS sensitive cells after exosome
treatment. The presence of selective MDR-1 mRNA in Exo/DXR suggests
the intriguing possibility that this mRNA could be an additional
factor that participate in drug resistance acquisition of sensitive
cells.
In conclusion, this study corroborates the evidence
that exosomes from MDR cells are capable of transferring
chemo-resistance by horizontal transfer of RNAs, including the
specific mRNA of P-gp.
Acknowledgements
This study was supported by the Italian Ministry of
Health, ‘5 per mille’ 2012 grant awarded to Professor Nicola
Baldini.
References
|
1
|
Théry C, Zitvogel L and Amigorena S:
Exosomes: Composition, biogenesis and function. Nat Rev Immunol.
2:569–579. 2002.PubMed/NCBI
|
|
2
|
Simons M and Raposo G: Exosomes -
vesicular carriers for inter-cellular communication. Curr Opin Cell
Biol. 21:575–581. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Raposo G, Nijman HW, Stoorvogel W,
Liejendekker R, Harding CV, Melief CJ and Geuze HJ: B lymphocytes
secrete antigen-presenting vesicles. J Exp Med. 183:1161–1172.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zitvogel L, Regnault A, Lozier A, Wolfers
J, Flament C, Tenza D, Ricciardi-Castagnoli P, Raposo G and
Amigorena S: Eradication of established murine tumors using a novel
cell-free vaccine: Dendritic cell-derived exosomes. Nat Med.
4:594–600. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Blanchard N, Lankar D, Faure F, Regnault
A, Dumont C, Raposo G and Hivroz C: TCR activation of human T cells
induces the production of exosomes bearing the TCR/CD3/zeta
complex. J Immunol. 168:3235–3241. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Heijnen HF, Schiel AE, Fijnheer R, Geuze
HJ and Sixma JJ: Activated platelets release two types of membrane
vesicles: Microvesicles by surface shedding and exosomes derived
from exocytosis of multivesicular bodies and alpha-granules. Blood.
94:3791–3799. 1999.PubMed/NCBI
|
|
7
|
Fontana S, Saieva L, Taverna S and
Alessandro R: Contribution of proteomics to understanding the role
of tumor-derived exosomes in cancer progression: State of the art
and new perspectives. Proteomics. 13:1581–1594. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
De Jong OG, Van Balkom BW, Schiffelers RM,
Bouten CV and Verhaar MC: Extracellular vesicles: potential roles
in regenerative medicine. Front Immunol. 5:6082014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Torreggiani E, Perut F, Roncuzzi L, Zini
N, Baglìo SR and Baldini N: Exosomes: Novel effectors of human
platelet lysate activity. Eur Cell Mater. 28:137–151.
2014.PubMed/NCBI
|
|
10
|
Yang C and Robbins PD: The roles of
tumor-derived exosomes in cancer pathogenesis. Clin Dev Immunol.
2011:8428492011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Filipazzi P, Bürdek M, Villa A, Rivoltini
L and Huber V: Recent advances on the role of tumor exosomes in
immunosuppression and disease progression. Semin Cancer Biol.
22:342–349. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gong J, Jaiswal R, Mathys JM, Combes V,
Grau GE and Bebawy M: Microparticles and their emerging role in
cancer multidrug resistance. Cancer Treat Rev. 38:226–234. 2012.
View Article : Google Scholar
|
|
13
|
Corcoran C, Rani S, O'Brien K, O'Neill A,
Prencipe M, Sheikh R, Webb G, McDermott R, Watson W, Crown J, et
al: Docetaxel-resistance in prostate cancer: Evaluating associated
phenotypic changes and potential for resistance transfer via
exosomes. PLoS One. 7:e509992012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Corcoran C, Rani S and O'Driscoll L:
miR-34a is an intracellular and exosomal predictive biomarker for
response to docetaxel with clinical relevance to prostate cancer
progression. Prostate. 74:1320–1334. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lv MM, Zhu XY, Chen WX, Zhong SL, Hu Q, Ma
TF, Zhang J, Chen L, Tang JH and Zhao JH: Exosomes mediate drug
resistance transfer in MCF-7 breast cancer cells and a probable
mechanism is delivery of P-glycoprotein. Tumour Biol.
35:10773–10779. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen WX, Liu XM, Lv MM, Chen L, Zhao JH,
Zhong SL, Ji MH, Hu Q, Luo Z, Wu JZ, et al: Exosomes from
drug-resistant breast cancer cells transmit chemoresistance by a
horizontal transfer of microRNAs. PLoS One. 9:e952402014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Heare T, Hensley MA and Dell'Orfano S:
Bone tumors: Osteosarcoma and Ewing's sarcoma. Curr Opin Pediatr.
21:365–372. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Baldini N, Scotlandi K, Barbanti-Bròdano
G, Manara MC, Maurici D, Bacci G, Bertoni F, Picci P, Sottili S,
Campanacci M, et al: Expression of P-glycoprotein in high-grade
osteosarcomas in relation to clinical outcome. N Engl J Med.
333:1380–1385. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li S, Sun W, Wang H, Zuo D, Hua Y and Cai
Z: Research progress on the multidrug resistance mechanisms of
osteosarcoma chemotherapy and reversal. Tumour Biol. 36:1329–1338.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Roncuzzi L, Pancotti F and Baldini N:
Involvement of HIF-1α activation in the doxorubicin resistance of
human osteosarcoma cells. Oncol Rep. 32:389–394. 2014.PubMed/NCBI
|
|
21
|
Salerno M, Avnet S, Bonuccelli G, Hosogi
S, Granchi D and Baldini N: Impairment of lysosomal activity as a
therapeutic modality targeting cancer stem cells of embryonal
rhabdomyosarcoma cell line RD. PLoS One. 9:e1103402014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
23
|
Su L, Mruk DD, Lui WY, Lee WM and Cheng
CY: P-glycoprotein regulates blood-testis barrier dynamics via its
effects on the occludin/zonula occludens 1 (ZO-1) protein complex
mediated by focal adhesion kinase (FAK). Proc Natl Acad Sci USA.
108:19623–19628. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mankin HJ, Hornicek FJ, Rosenberg AE,
Harmon DC and Gebhardt MC: Survival data for 648 patients with
osteosarcoma treated at one institution. Clin Orthop Relat Res.
429:286–291. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bacci G, Briccoli A, Rocca M, Ferrari S,
Donati D, Longhi A, Bertoni F, Bacchini P, Giacomini S, Forni C, et
al: Neoadjuvant chemotherapy for osteosarcoma of the extremities
with metastases at presentation: Recent experience at the Rizzoli
Institute in 57 patients treated with cisplatin, doxorubicin, and a
high dose of methotrexate and ifosfamide. Ann Oncol. 14:1126–1134.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gottesman MM, Fojo T and Bates SE:
Multidrug resistance in cancer: Role of ATP-dependent transporters.
Nat Rev Cancer. 2:48–58. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Borst P and Elferink RO: Mammalian ABC
transporters in health and disease. Annu Rev Biochem. 71:537–592.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ambudkar SV, Kimchi-Sarfaty C, Sauna ZE
and Gottesman MM: P-glycoprotein: From genomics to mechanism.
Oncogene. 22:7468–7485. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Baldini N, Scotlandi K, Serra M, Picci P,
Bacci G, Sottili S and Campanacci M: P-glycoprotein expression in
osteosarcoma: A basis for risk-adapted adjuvant chemotherapy. J
Orthop Res. 17:629–632. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Harguindey S, Orive G, Luis Pedraz J,
Paradiso A and Reshkin SJ: The role of pH dynamics and the
Na+/H+ antiporter in the etiopathogenesis and
treatment of cancer. Two faces of the same coin - one single
nature. Biochim Biophys Acta. 1756:1–24. 2005.PubMed/NCBI
|
|
31
|
Shicang Y, Guijun H, Guisheng Q, Yuying L,
Guoming W and Ruiling G: Efficacy of chemotherapeutic agents under
hypoxic conditions in pulmonary adenocarcinoma multidrug resistant
cell line. J Chemother. 19:203–211. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen KG and Sikic BI: Molecular pathways:
Regulation and therapeutic implications of multidrug resistance.
Clin Cancer Res. 18:1863–1869. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Levchenko A, Mehta BM, Niu X, Kang G,
Villafania L, Way D, Polycarpe D, Sadelain M and Larson SM:
Intercellular transfer of P-glycoprotein mediates acquired
multidrug resistance in tumor cells. Proc Natl Acad Sci USA.
102:1933–1938. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao L, Liu W, Xiao J and Cao B: The role
of exosomes and ‘exosomal shuttle microRNA’ in tumorigenesis and
drug resistance. Cancer Lett. 356B:339–346. 2015. View Article : Google Scholar
|
|
35
|
Safaei R, Larson BJ, Cheng TC, Gibson MA,
Otani S, Naerdemann W and Howell SB: Abnormal lysosomal trafficking
and enhanced exosomal export of cisplatin in drug-resistant human
ovarian carcinoma cells. Mol Cancer Ther. 4:1595–1604. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Federici C, Petrucci F, Caimi S, Cesolini
A, Logozzi M, Borghi M, D'Ilio S, Lugini L, Violante N, Azzarito T,
et al: Exosome release and low pH belong to a framework of
resistance of human melanoma cells to cisplatin. PLoS One.
9:e881932014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ramachandran S and Palanisamy V:
Horizontal transfer of RNAs: Exosomes as mediators of intercellular
communication. Wiley Interdiscip Rev RNA. 3:286–293. 2012.
View Article : Google Scholar :
|