Introduction
Lung cancer is one of the leading causes of
cancer-related mortality worldwide, among which non-small cell lung
cancer (NSCLC) accounts for ~85% of lung cancer (1). The population with NSCLC has grown
fast over the past decades in China (2). Despite the considerable advances in
medical and surgical treatment of NSCLC patients, the prognosis of
NSCLC remains unsatisfactory and the 5-year survival rate of
patients with NSCLC is <16% (3). Tumor metastasis is frequent, and a
great challenge in the clinical treatment of NSCLC, and mostly
responsible for the low 5-year survival rate (4). Therefore, there is an urgent need to
find potential molecular mechanisms involved in NSCLC metastasis,
which may contribute to establish novel diagnostic markers and
novel therapeutic targets for NSCLC.
MicroRNAs (miRNAs) are a class of small
non-protein-coding RNAs of ~22 nucleotides in size that negatively
regulate mRNA stability and/or repress mRNA translation by binding
to the 3′-untranslated region (3′-UTR) (5,6).
miRNAs has been reported to play pivotal roles in a wide range of
cellular processes including proliferation, cycle, differentiation,
apoptosis and metastasis (7).
miRNAS are dysregulated in many cancers and involve in the
initiation and progression of various cancer types, and function
either as oncomiRs or tumor suppressor miRNAs, based on the
regulated tumor forms and their targeted genes (8,9). For
NSCLC, numerous miRNAs have been identified to be involved in NSCLC
procession and metastasis, and can act as potent therapeutic
targets or diagnosis marker for NSCLC (10–12).
MicroRNA-107 (miR-107), located on chromosome 10,
has been shown to be downregulated, and function as a tumor
suppressor in several types of cancer, such as glioma (13), breast cancer (14), gastric cancer (15), cervical cancer (16) and renal clear cell carcinoma
(17). Previous studies showed
that the expression of miR-107 was reduced in NSCLC tissues, and
overexpression of miR-107 was able to induce cell cycle arrest in
human NSCLC cell lines (18), and
increase cisplatin chemosensitivity of A549 non-small cell lung
cancer cell line (19). However,
the biological roles, especially with regard to migration and
invasion, have not yet been thoroughly validated in NSCLC.
Therefore, the aim of this study was to investigate the role of
miR-107 on the procession and metastasis of NSCLC. In this study,
we verified that miR-107 plays an inhibitory role in tumor growth
and metastasis in NSCLC cells by targeting BDNF and indirectly
regulating the P13K/AKT signaling pathway.
Materials and methods
Cell lines and tissue samples
All of the NSCLC cell lines used in this study were
obtained from the Cell Culture Center of the Shanghai Institute for
Biological Sciences of Chinese Academy of Science (Shanghai,
China), including four NSCLC cell lines (A549, H1299, SPCA1 and
H358) and the normal lung cell line (BEAS-2B). The cells were grown
in monolayer in Dulbecco's modified Eagle's medium (DMEM, Gibco,
Grand Island, NY, USA) supplemented with 10% fetal bovine serum
(FBS, Gibco) and maintained at 37°C in humidified air with 5%
CO2.
A total of 30 tumor tissue specimens and
corresponding adjacent normal lung tissues were obtained from
patients who underwent curative resection for NSCLC at the First
Hospital, Jilin University (Changchun, China) between August 2014
and September 2015. Relevant clinical data of NSCLC patients were
collected and are listed in Table
I. None of the patients received chemotherapy or radiotherapy
before surgery. This study was approved by the Ethics Committee of
the First Hospital, Jilin University. All patients signed a written
consent for the use of their specimens and disease information.
 | Table ICorrelation between
clinicopathological features and miR-107 expression in 30 NSCLC
tissues. |
Table I
Correlation between
clinicopathological features and miR-107 expression in 30 NSCLC
tissues.
| | miR-107
expression | |
|---|
| |
| |
|---|
| No. of
Variables | cases | Low (n %) | High (n %) | P-value |
|---|
| Age (years) | | | | 0.845 |
| <55 | 16 | 9 (56.3) | 7 (43.7) | |
| ≥55 | 14 | 7 (50.0) | 7 (50.0) | |
| Gender | | | | 0.488 |
| Male | 17 | 10 (58.9) | 7 (41.1) | |
| Female | 13 | 6 (46.2) | 7 (53.8) | |
| TNM stage | | | | <0.01 |
| I–II | 20 | 7 (35.0) | 13 (65.0) | |
| III–IV | 10 | 9 (90.0) | 1 (10.0) | |
| Tumor size | | | | 0.372 |
| <5 cm | 18 | 10 (55.6) | 8 (44.4) | |
| ≥5 cm | 12 | 6 (50.0) | 6 (50.0) | |
| Lymph node
metastasis | | | | <0.01 |
| No | 19 | 6 (31.6) | 13 (68.4) | |
| Yes | 11 | 10 (90.9) | 1 (9.1) | |
miRNA, siRNA, plasmid construction, and
transfection
miR-107 mimic (miR-107), and corresponding miRNA
negative control (miR-NC) were synthesized by GenePharma (Shanghai,
China). siRNAs against BDNF (si-BDNF) and corresponding scramble
negative control (si-NC) were designed and synthesized by RiboBio
(Guangzhou, China). The coding domain sequence of human BDNF mRNA
was amplified by PCR, and inserted into pcDNA 3.0 vector
(Invitrogen, Grand Island, NY, USA), named as pBDNF. Transfection
was performed using lipofectamine 2000 (Invitrogen) according to
the manufacturer's instructions.
RNA extraction and real-time PCR
Total RNA of the cultured cells and the tissues was
extracted using TRIzol (Invitrogen) according to the manufacturer's
instructions. The purity and concentration of total RNA were
determined by a dual-beam ultraviolet spectrophotometer (Eppendorf,
Hamburg, Germany). Then, a total of 3 μg of mRNA was reverse
transcribed to single-stranded cDNAs using a
PrimeScript® RT reagent kit (Takara Biotechnology Co.,
Ltd., Dalin, China). qRT-PCR for miR-107 and BNDF were performed
using SYBR premix real-time PCR Reagent (Takara) under an ABI7900
real-time PCR system (Applied Biosystems, Foster City, CA, USA).
The primers for miR-107 and U6 were brought from Applied
Biosystems. The primers for BDNF and β-actin used in this study
were as described previously (20). U6 RNA was used to normalize the
miR-107 RNA levels, and β-actin was used to normalize the level of
BDNF mRNAs. The comparative 2−ΔΔCt method was employed
for relative quantification.
Cell proliferation, migration and
invasion assays
Cell proliferation was determined by Cell Counting
Kit-8 assay (CCK-8, Dojindo, Kumamoto, Japan) according to the
manufacturer's instructions. Briefly, transfected cells were seeded
into 24-well plates at a density of 5×103 cells/well and
cultured for 24–72 h. At indicated time (24, 48 and 72 h), 10 μl
CCK8 solution were added to each well until visual color conversion
occurred. The absorbance at 450 nm was read on a microplate reader
(Bio-Tek Instruments Inc., Winooski, VT, USA).
To examine the migration ability of cells in
vitro, a wound-healing assay was performed after transfection.
Briefly, transfected cells (2×104 cells/well) were
seeded into 24-well tissue culture plates. When cells were grown to
a density of 70–80%, the linear wound of cellular monolayer were
created by 200 μl pipette tips. The wound closure was observed and
photographed at 0 and 24 h using an IX51 inverted light microscope
(Olympus, Tokyo, Japan).
For Transwell migration assays, 5×104
cells were suspended in serum-free medium and seeded into upper
Transwell chambers coated with Matrigel (BD Biosciences, Bedford,
MA, USA), then 600 μl medium containing 10% FBS was added to the
lower chamber. After incubation for 48 h in a humidified atmosphere
of 5% CO2 at 37°C, cells that migrated onto the lower
surface of the membrane were fixed with 100% methanol and stained
with 0.1% crystal violet, while the non-invading cells on the upper
membrane surface were removed with cotton swabs. Cells on the lower
surface were photographed and counted at five randomly selected
fields with a magnification of ×200 by microscopy (Olympus).
MicroRNA target prediction and
luciferase-reporter activity assay
miRNA targets were predicted using the algorithms
TargetSan (https://www.targetscan.org) miRanda
(http://www.microrna.org/) and PicTar (http://pictar.mdc-berlin.de/). The human BDNF 3′UTR
oligonucleotides containing the wild-type (Wt) or mutant (Mut)
binding site of miR-107 were cloned into the pGL3-control vector
(Ambion, Austin, TX, USA) at the NheI and XhoI sites.
For luciferase assays, cells were co-transfected with miR-107 mimic
or miR-NC and Wt or Mut plasmid using Lipofectamine 2000 reagent.
After 48-h transfection, luciferase activity was measured using the
dual-luciferase assay system (Promega, Madison, WI, USA).
Renilla-luciferase was used for normalization.
Western blotting
Total cellular and tissue proteins were extracted
using RIPA lysis buffer containing proteinase inhibitor (Sigma,
USA). Concentrations of total cellular protein were determined
using a BCA assay kit (Pierce, Rockford, IL, USA). Equal amounts of
proteins (25 μg/sample) were separated with 10% SDS-PAGE and
transferred onto a nitrocellulose membrane (Bio-Rad, Munich,
Germany), followed by probing overnight at 4°C with antibodies
against BDNF (1:1,000; Santa Cruz Biotechnology, CA, USA), AKT
(1:1,000; Santa Cruz Biotechnology), p-AKT (1:1,000; Santa Cruz
Biotechnology), PI3K (1:1000; Santa Cruz Biotechnology) and p-PI3K
(1:1,000; Cell Signaling Technology, CA, USA). The membranes were
then incubated for 2 h at room temperature with the secondary
HRP-conjugated antibodies (1:5,000; Cell Signaling Technology).
Monoclonal mouse β-actin antibody (1:1000; Cell Signaling
Technology) was used as an internal control. Proteins in the
membrane were detected by the enhanced chemiluminescence system
(ECL kit, Millipore, USA) and its band images were analyzed with
the Bio-Rad ChemiDoc XRS system (Bio-Rad).
Xenograft tumor model
Young male athymic nude mice (6-week-old) were
purchased from the Model Animal Research Center of Jilin University
(Changchun, China). Nude mice were manipulated and cared for
according to NIH Animal Care and Use Committee guidelines in the
Experiment Animal Center of the Jilin University (Changchun,
China).
Approximately 2×106 A549 cells stably
carrying miR-107 or miR-NC was injected subcutaneously into the
lower flanks of 8 nude mice. Tumor volumes were measured every 5
days from the sixth day post-injection onward for 30 days before
the animals were sacrificed. Tumor volume was measured every week
by measuring the length (L), width (W), and height (H) with
calipers and using the formula: Volume (V) = [π/6 × L × W × H].
Thirty days after inoculation, mice were sacrificed. Tumor tissues
were dissected, and the volume and weight were measured.
Statistical analysis
Statistical analysis was performed using the SPSS
software package (SPSS Standard version 19.0, SPSS Inc., USA). Data
are shown as mean ± standard deviation (SD) of at least three
separate experiments. Statistical significance was analyzed using
Student's t-test or one-way ANOVA. The relationship between miR-107
level and clinical and pathological variables was analysed using
Pearson's χ2 test. The correlations between miR-107
expression and BDNF mRNA expression were analyzed using Pearson
analysis. P-value <0.05 was considered as statistically
significant.
Results
miR-107 is downregulated in both NSCLC
cells and clinical specimens
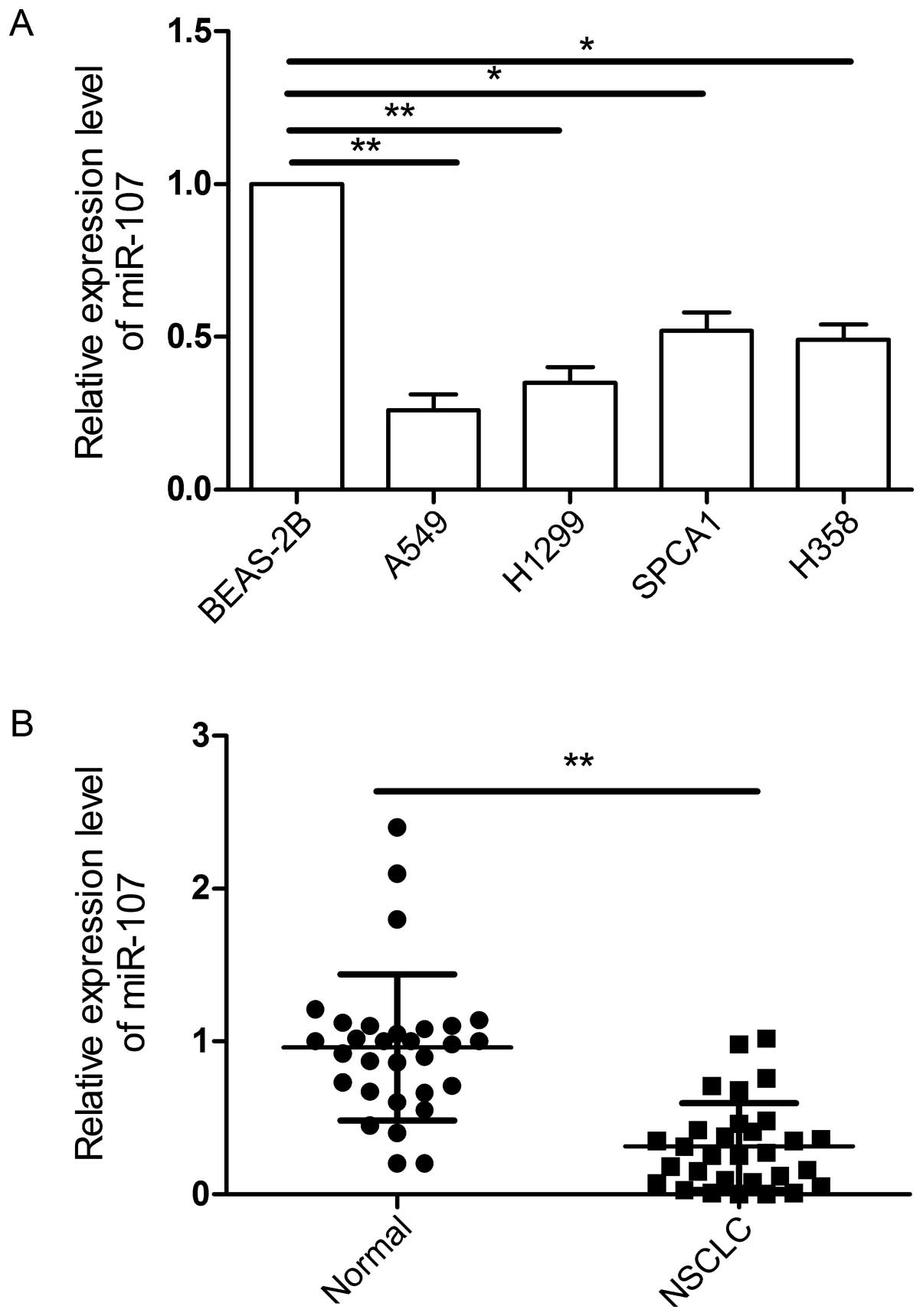
To examine miR-107 expression levels, we first
applied qRT-PCR technology and examined miR-107 expression in four
NSCLC cell lines (A549, H1299, SPCA1 and H358) and a normal lung
cell line (BEAS-2B). The result showed that miR-107 was aberrantly
downregulated in all NSCLC cell lines, as compared to normal lung
cell line (Fig. 1A,
*P<0.05). To validate whether aberrant downregulation
of miR-107 was also the case in clinical specimens, we then
examined miR-107 expression in 30 NSCLC tissues and matched
adjacent normal tissues. It was found that the expression of
miR-107 in NSCLC tissues was significantly lower than those of
their matched adjacent normal tissues (P<0.01) (Fig. 1B). To further investigate the
clinicopathological significance of miR-107 level in NSCLC
patients, 30 NSCLC patients were divided into 2 subgroups base on
the mean (0.31) of all NSCLC samples: low miR-107 group (<0.31,
16 cases) and a high miR-107 group (>0.31, 14 cases). Then
correlations between miR-107 expression and clinicopathologic
parameters were analyzed by Pearson's χ2 test. It was
found that the reduction of miR-107 expression was significantly
associated with lymph node metastasis and the TNM stage, but not
with age, gender or tumor size.
miR-107 inhibits NSCLC cell growth in
vitro and in vivo
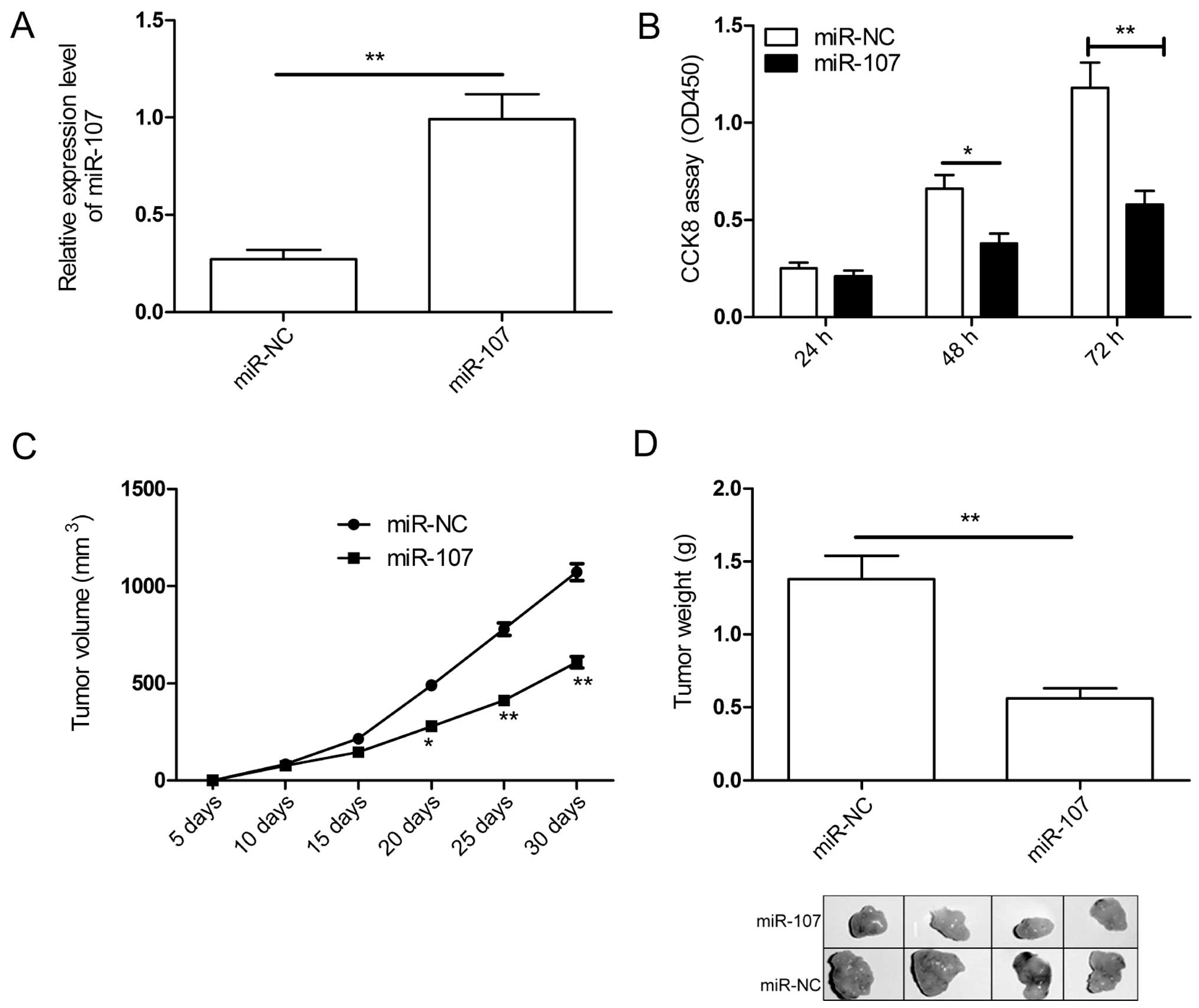
To determine if miR-107 has the propensity to
suppress tumorigenesis, we introduced the miR-107 mimic into A549
cells, to alter the level of total miR-107 in A549 cells, which
express the lowest level of miR-107 among four NSCLC cell lines
(Fig. 1A). qRT-PCR confirmed the
elevated level of miR-107 in the transfected A549 cells (Fig. 2A). Then cell proliferation was
determined in A549 cells transfected with miR-107 mimic or miR-NC.
The CCK-8 assay showed cell proliferation was obviously suppressed
in A59 cells after manipulation of miR-107 mimic at 48- and 72-h
time-points, while no significant difference was found at 24 h
time-point (Fig. 2B). Besides, we
explored the effect of miR-107 overexpression on in vivo
growth of NSCLC tumors. The human A549 cells stably expressing
miR-107 or miR-NC was implanted subcutaneously into nude mice to
allow tumor formation. Tumors grew slower in the A549/miR-107 group
than in the A549/miR-NC group (Fig.
2C). At day 30 post-injection, the mice were sacrificed, and
tumor tissues were dissected, and weighed. A significant decrease
in weight (Fig. 2D) was observed
in mice injected with A549/miR-107 compared to the group injected
with A549/NC.
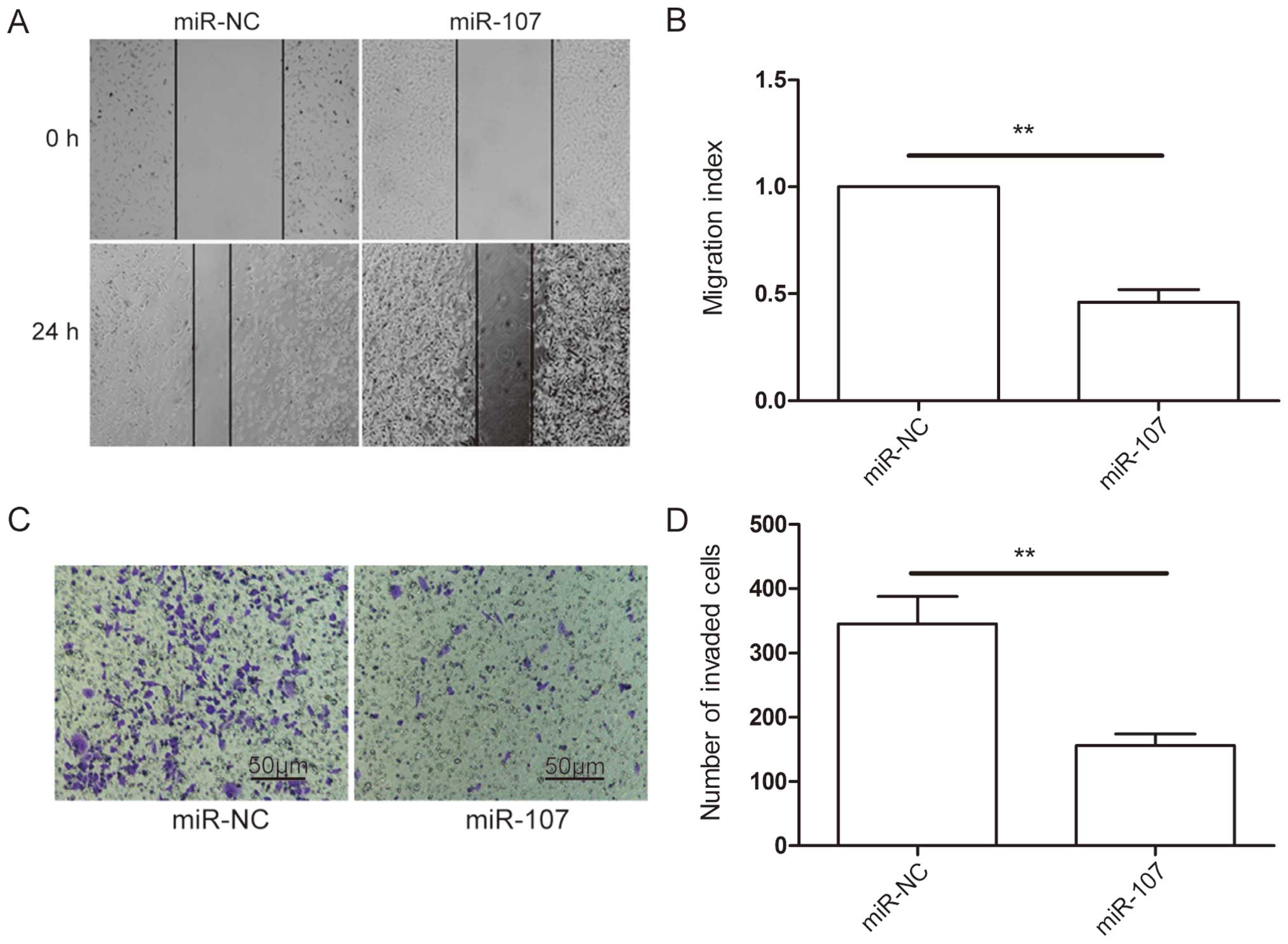
miR-107 inhibits NSCLC cell
metastasis
The above results showed that reduced expression of
miR-107 was associated with lymph node metastasis (Table I), suggesting that it may regulate
the metastasis process. To determine if this indeed is the case, we
carried out wound-healing and Transwell invasion assays in A549
cells transfected with miR-107 mimic or miR-NC. The results showed
that ectopic overexpression of miR-206 caused a suppression of cell
migration (Fig. 3A and B) and
invasion (Fig. 3C and D)
capability in A549 cells. These results suggested that miR-107
inhibits NSCLC metastasis.
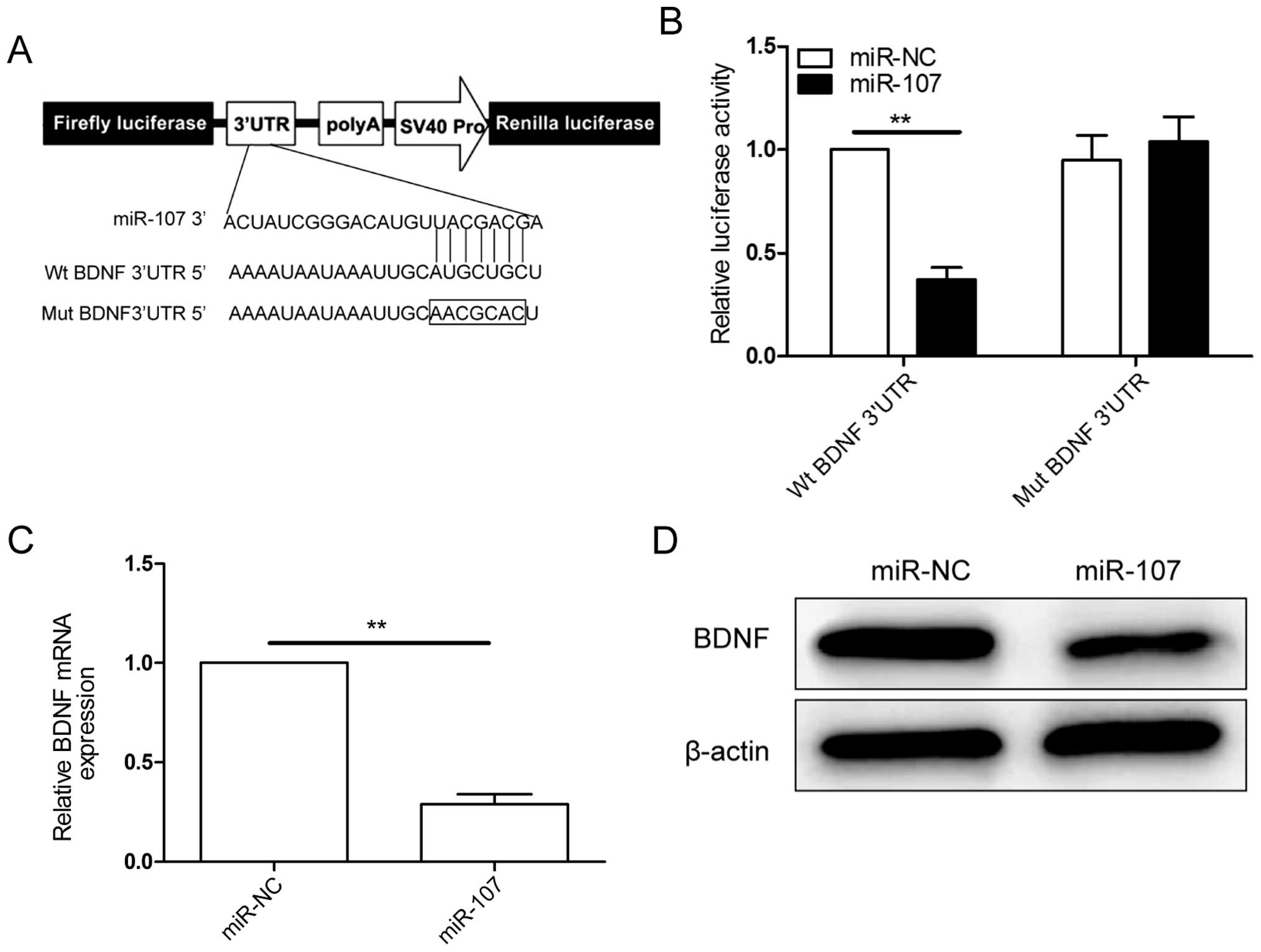
BDNF is a direct target of miR-107
Potential targets of miR-107 were predicted using
three bioinformatic databases (TargetScan, miRanda and PicTar),
BDNF was chosen as a target gene of miR-107, based on a putative
target sequences at 299–305 bp of BDNF (Fig. 4A). To verify whether BDNF is a
direct target of miR-107 in NSCLC cells, a human BDNF 3′UTR
fragment containing the binding sites of miR-107 (Fig. 4A) or the mutant sites were cloned
into the pGL3 vector, and miR-107 mimic or miR-NC were
co-transfected into A549 cells and cultured for 48 h, then
luciferase activities were measured. It was found that
overexpression of miR-107 obviously suppressed the luciferase
activity of wild-type BDNF 3′UTR, but the activity of the
mutant-type BDNF 3′UTR was not changed (Fig. 4B), suggesting that BDNF is a direct
target of miR-107 in NSCLC cells. Then, qRT-PCR and western blot
analysis confirmed that overexpression of miR-107 markedly
inhibited BDNF expression on mRNA level (Fig. 4C) and protein level (Fig. 4D) in A549 cells. These results
indicated that miR-107 can bind directly to BDNF and inhibits its
expression.
Inverse correlation between BDNF and
miR-107 expression in NSCLC patients
We next examined the BDNF mRNA expression in tumor
tissues and the corresponding adjacent normal lung tissues in a
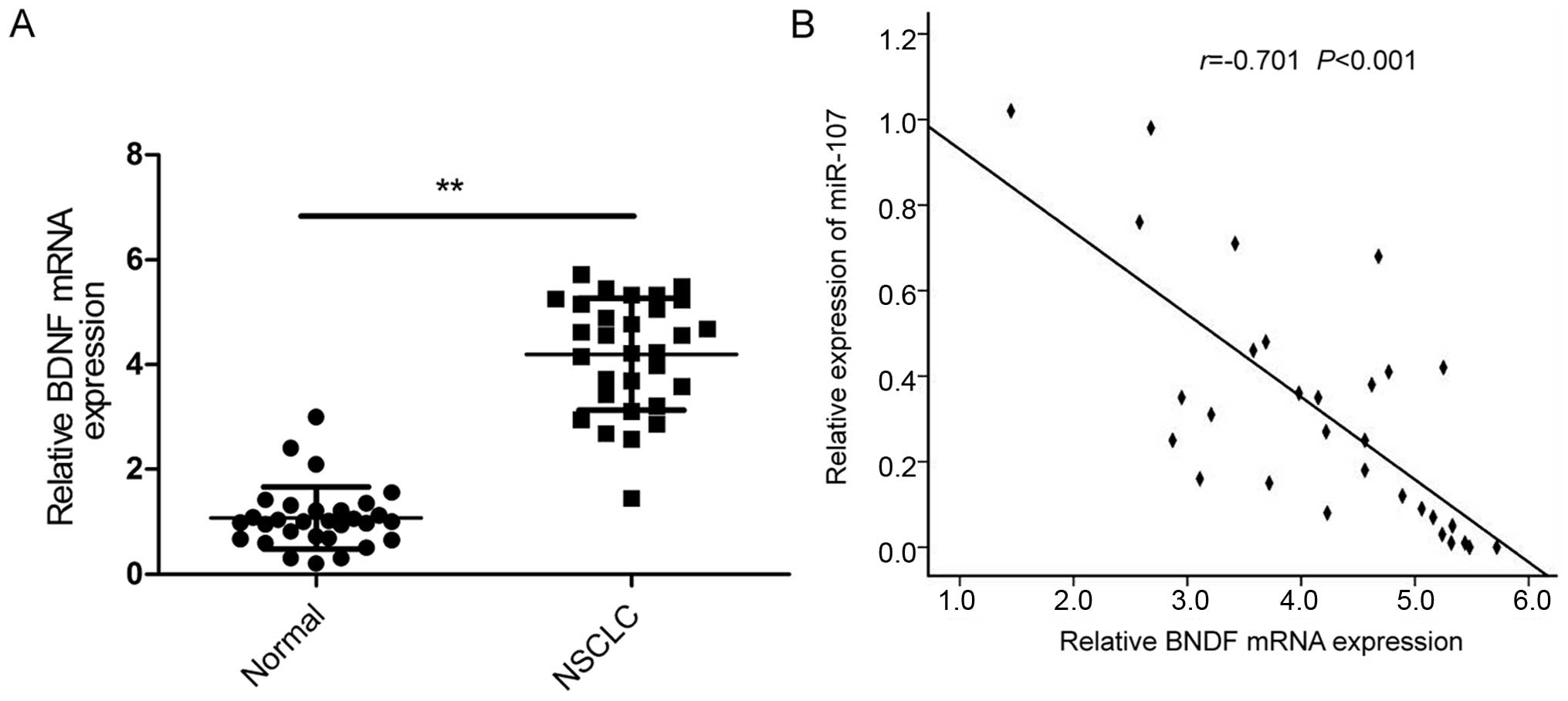
total of 30 patients with NSCLC by qRT-PCR (Fig. 5A). The data showed that BDNF
expression was significantly increased in NSCLC tissues compared to
the adjacent normal tissues. Pearson correlation analysis revealed
that the expression of miR-107 was inversely correlated with BDNF
in the 30 patients with NSCLC (r=−0.701; P<0.001; Fig. 5B).
Overexpression of BDNF reverses the tumor
suppressive effect of miR-107 in NSCLC
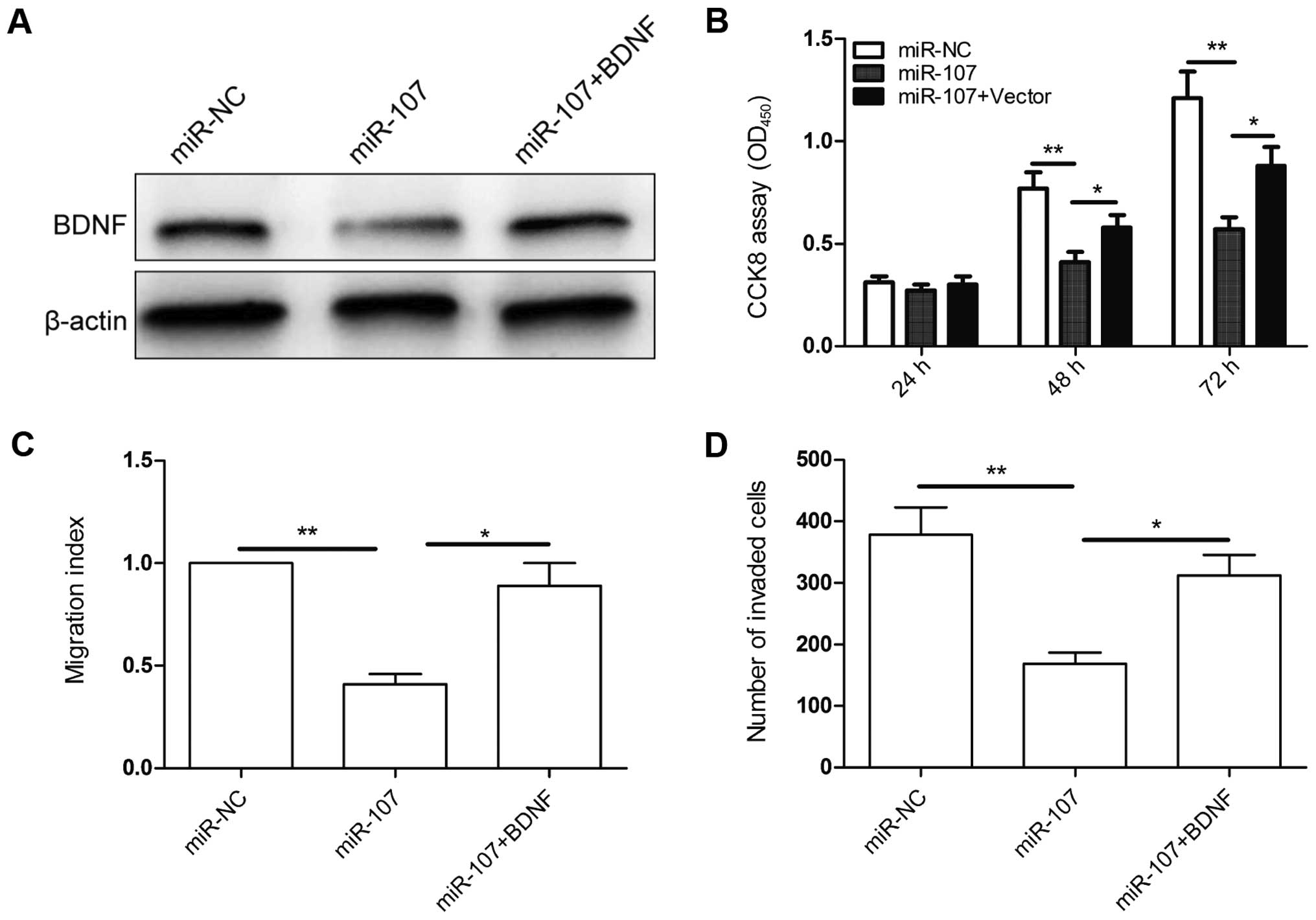
To evaluate if BDNF is responsible for the
functional effects of miR-107 in NSCLC cells, we generated a BDNF
overexpressing vector the pBDNF, and transfected it into miR-107 or
miR-NC overexpressed A549 cells. The transfection efficiency was
verified by western blot assay (Fig.
6A). Then, we carried out CCK8, would healing, and Transwell
invasion assays to evaluate the effect of BDNF overexpression on
cell proliferation, migration and invasion in the above cells. BDNF
overexpression reversed the inhibition effect on cell
proliferation, migration and invasion in A549 cells induced by
miR-107 overexpression (Fig.
6B–D). Therefore, our data clearly demonstrated that miR-107
inhibits NSCLC cell proliferation, migration, and invasion by
targeting BDNF.
miR-107 inhibits the PI3K/AKT
signaling
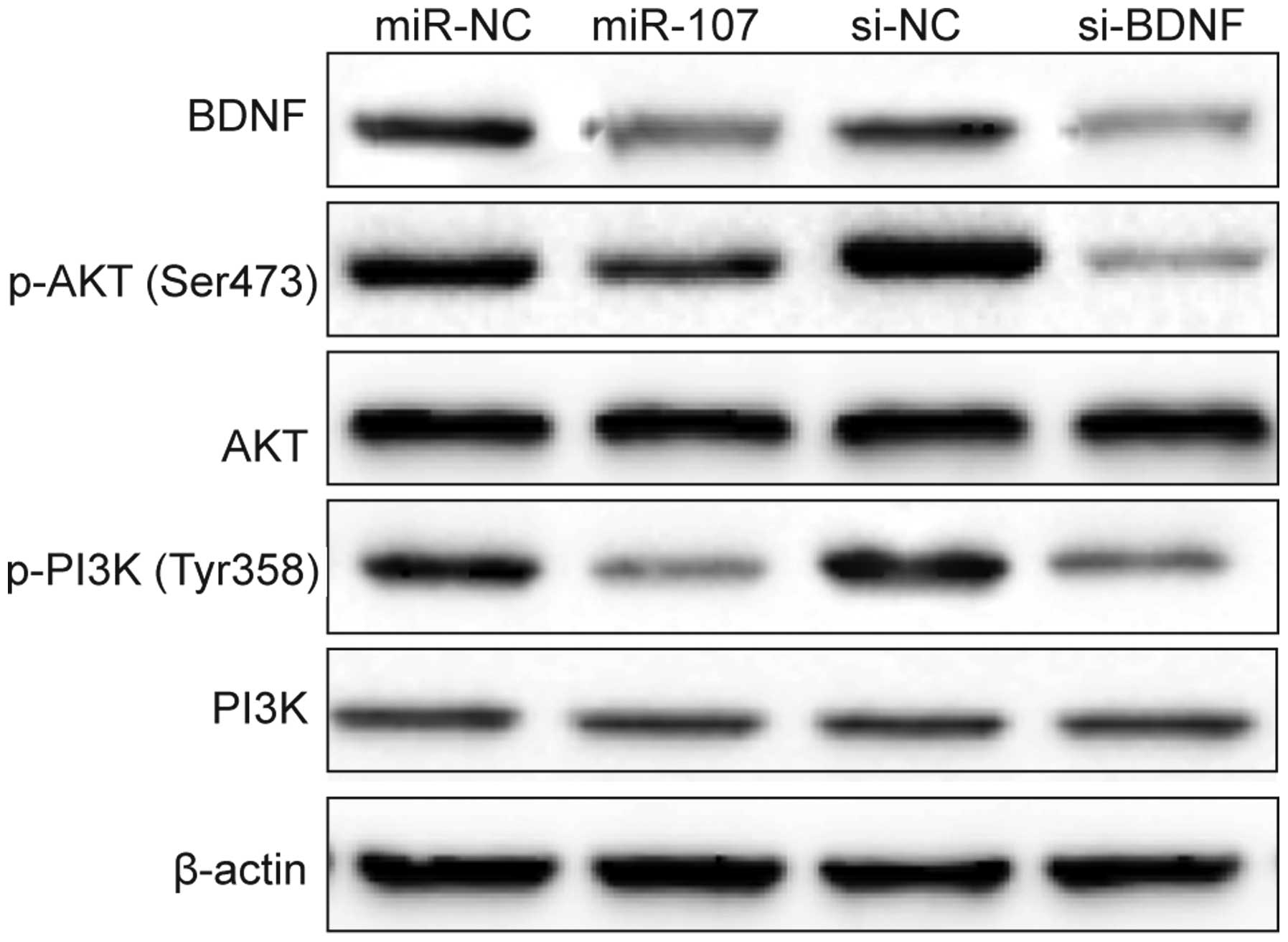
It has been shown that BDNF activation can trigger
PI3K/AKT pathways (21), which
regulates cell proliferation, apoptosis invasion, and inflammation
in various cancers (22).
Therefore, we investigated the possibility that miR-107 regulates
this pathway by targeting BDNF. PI3K, p-PI3K, AKT and p-AKT protein
expression was detected in A549 cells transfected with miR-107
mimic/miR-NC or si-BDNF/si-NC by western blotting. It was found
that miR-107 mimics decreased BDNF expression and the
phosphorylation levels of PI3K (p-PI3K) and AKT (p-AKT) expression,
without change of total PI3K and AKT expression (Fig. 7). Consistent with this result, we
observed that downregulation of BDNF by si-BDNF also decreased BDNF
expression and p-PI3K and p-AKT expression, but had no effect on
total PI3K and AKT expression (Fig.
7). These results might suggest that miR-107 exerts it
suppressive role in NSCLC cells by repressing BDNF and indirectly
regulating PI3K/AKT signaling pathway.
Discussion
A large number of studies have indicated that miRNAs
may play an important role in NSCLC initiation and development
(10–12). We demonstrated that miR-107 is
frequently downregulated in human NSCLC tissues and cancer cell
lines, which is consistent with previous results (18). In addition, we found that
downregulation of miR-107 was particularly significant in tumors
with lymphatic metastasis, and advanced TNM stage. These results
supported the opinion of a previous study that low expression of
miR-107 was significantly correlated with TNM stage, regional lymph
node involvement, and tumor differentiation (23). We also showed that miR-107
significantly inhibited NSCLC proliferation, migration and
invasion. These results suggested that miR-107 has a crucial role
in NSCLC growth and metastasis.
miR-107, located on chromosome 10, has been shown to
be involved in various biological processes, including
adipogenesis, hypoxia, angiogenesis and proliferation and cell
cycle (24). miR-107 has been
showed to function as a tumor suppressor in multiple cancer, such
as glioma (13), breast cancer
(14), gastric cancer (15), cervical cancer (16) and renal clear cell carcinoma
(17). On the contrary, miR-107
was significantly upregulated in cancer tissues and cell lines, and
miR-107 overexpression was able to promote cell proliferation in
HepG2 cells, suggesting miR-107 as oncogene in liver cancer
(25). Previously studies showed
that the expression of miR-107 was reduced in NSCLC tissues
(23), and overexpression of
miR-107 was able to induce cell cycle arrest in human NSCLC cell
lines (18), and increase
cisplatin chemosensitivity of A549 (19). However, the potential roles and
mechanism involved in NSCLC metastasis remains largely unknown. In
this study, we showed that miR-107 inhibited NSCLC growth in
vitro and in vivo, as well as NSCLC metastasis. Our
results together with previous studies indicated that miR-107
function as a tumor suppressor miRNA in NSCLC.
Brain-derived neurotrophic factor (BDNF), a member
of the neurotrophin family, has been showed to play an important
role in the development and regeneration of the neurons (26). Binding of BDNF to its major
receptor, tropomyosin-related receptor kinase B (TrkB) with high
affinity and specificity (27),
caused the activation of multiple downstream signaling pathway,
such as PI3K/AKT, RAS/ERK, PLC/PKC, AMPK/ACC and JAK/STAT pathways
(28). Recently studies have
demonstrated that BDNF was able to promote tumorigenesis and
progression in several human malignancies, such as clear cell renal
cell carcinoma (29), breast
(30), colon cancer (31), colorectal cancer (32), and neuroblastoma (33), suggesting that BDNF was closely
associated with tumor progression. For NSCLC, It was reported that
the expression of BDNF was upregulated in NSCLC tissues, and was
associated with poor prognosis in non-small cell lung cancer
(34), and that BDNF facilitates
tumorigenesis of NSCLC (35), and
NSCLC metastasis (36). These
studies suggested that BDNF functions as an oncogene in NSCLC. We
confirmed that BDNF is a target of miR-107 in regulating NSCLC by
luciferase activity assay, qRT-PCR and western blotting. In
addition, we also found that BDNF expression was upregulated, and
inversely correlated with miR-107 in NSCLC tissues. Enforced
overexpression of BDNF effectively reversed the tumor suppressive
functions of miR-107 on NSCLC proliferation, migration and
invasion. Of note, we found that miR-107 overexpression or
downregulation of BDNF was able to inhibit activation of PI3K/AKT
signaling pathway. These results might suggest that miR-107 exerted
it suppressive role in NSCLC by targeting BDNF and indirectly
regulating PI3K/AKT signaling pathway.
In conclusion, this study showed that miR-107 was
down-regulated in NSCLC cell lines and tissues, and was associated
with lymph node metastasis and TNM stage. Overexpression of miR-107
significantly decreased the proliferation, migration and invasion
of NSCLC cells in vitro and suppressed tumor growth in
vivo. We also identified a likely novel mechanism of miR-107 to
suppress tumor growth and metastasis by inhibiting BDNF and
indirectly regulating PI3K/AKT signaling pathway. Thus, miR-107
functions as a tumor suppressor in NSCLC by repressing BDNF. The
identification of miR-107 and its target gene in non-small lung
cell cancer may contribute to understanding the potential molecular
mechanisms of NSCLC, and may have diagnostic as well as therapeutic
value for non-small cell lung cancer.
Acknowledgements
This study was supported by The National Youth
Science Foundation (81401883).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang L, Parkin DM, Ferlay J, Li L and Chen
Y: Estimates of cancer incidence in China for 2000 and projections
for 2005. Cancer Epidemiol Biomarkers Prev. 14:243–250.
2005.PubMed/NCBI
|
|
3
|
Schabath MB, Nguyen A, Wilson P, Sommerer
KR, Thompson ZJ and Chiappori AA: Temporal trends from 1986 to 2008
in overall survival of small cell lung cancer patients. Lung
Cancer. 86:14–21. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li C and Hong W: Research status and
funding trends of lung cancer biomarkers. J Thorac Dis. 5:698–705.
2013.PubMed/NCBI
|
|
5
|
Fabian MR, Sonenberg N and Filipowicz W:
Regulation of mRNA translation and stability by microRNAs. Annu Rev
Biochem. 79:351–379. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Guo H, Ingolia NT, Weissman JS and Bartel
DP: Mammalian microRNAs predominantly act to decrease target mRNA
levels. Nature. 466:835–840. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
McManus MT: MicroRNAs and cancer. Semin
Cancer Biol. 13:253–258. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Farazi TA, Spitzer JI, Morozov P and
Tuschl T: miRNAs in human cancer. J Pathol. 223:102–115. 2011.
View Article : Google Scholar :
|
|
10
|
Boeri M, Sestini S, Fortunato O, Verri C,
Suatoni P, Pastorino U and Sozzi G: Recent advances of
microRNA-based molecular diagnostics to reduce false-positive lung
cancer imaging. Expert Rev Mol Diagn. 15:801–813. 2015.PubMed/NCBI
|
|
11
|
Skrzypski M, Dziadziuszko R and Jassem J:
MicroRNA in lung cancer diagnostics and treatment. Mutat Res.
717:25–31. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guan P, Yin Z, Li X, Wu W and Zhou B:
Meta-analysis of human lung cancer microRNA expression profiling
studies comparing cancer tissues with normal tissues. J Exp Clin
Cancer Res. 31:542012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen L, Li ZY, Xu SY, Zhang XJ, Zhang Y,
Luo K and Li WP: Upregulation of miR-107 inhibits glioma
angiogenesis and VEGF expression. Cell Mol Neurobiol. 36:113–120.
2016. View Article : Google Scholar
|
|
14
|
Zhang L, Ma P, Sun LM, Han YC, Li BL, Mi
XY, Wang EH and Song M: MiR-107 down-regulates SIAH1 expression in
human breast cancer cells and silencing of miR-107 inhibits tumor
growth in a nude mouse model of triple-negative breast cancer. Mol
Carcinog. 55:768–777. 2016. View
Article : Google Scholar
|
|
15
|
Zhang M, Wang X, Li W and Cui Y: miR-107
and miR-25 simultaneously target LATS2 and regulate proliferation
and invasion of gastric adenocarcinoma (GAC) cells. Biochem Biophys
Res Commun. 460:806–812. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou C, Li G, Zhou J, Han N, Liu Z and Yin
J: miR-107 activates ATR/Chk1 pathway and suppress cervical cancer
invasion by targeting MCL1. PLoS One. 9:e1118602014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Song N, Ma X, Li H, Zhang Y, Wang X, Zhou
P and Zhang X: microRNA-107 functions as a candidate tumor
suppressor gene in renal clear cell carcinoma involving multiple
genes. Urol Oncol. 33:205.e1–11. 2015. View Article : Google Scholar
|
|
18
|
Takahashi Y, Forrest AR, Maeno E,
Hashimoto T, Daub CO and Yasuda J: MiR-107 and miR-185 can induce
cell cycle arrest in human non small cell lung cancer cell lines.
PLoS One. 4:e66772009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang Z, Zhang L, Yin ZY, Fan XL, Hu B,
Wang LQ and Zhang D: miR-107 regulates cisplatin chemosensitivity
of A549 non small cell lung cancer cell line by targeting cyclin
dependent kinase 8. Int J Clin Exp Pathol. 7:7236–7241.
2014.PubMed/NCBI
|
|
20
|
Yan H, Wu W, Ge H, Li P and Wang Z:
Up-regulation of miR-204 enhances anoikis sensitivity in epithelial
ovarian cancer cell line via brain-derived neurotrophic factor
pathway in vitro. Int J Gynecol Cancer. 25:944–952. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mao XY, Zhou HH, Li X and Liu ZQ:
Huperzine A alleviates oxidative glutamate toxicity in hippocampal
HT22 cells via activating BDNF/TrkB-dependent PI3K/Akt/mTOR
signaling pathway. Cell Mol Neurobiol. 36:915–925. 2016. View Article : Google Scholar
|
|
22
|
Huang H, Zhong R, Xia Z, Song J and Feng
L: Neuroprotective effects of rhynchophylline against ischemic
brain injury via regulation of the Akt/mTOR and TLRs signaling
pathways. Molecules. 19:11196–11210. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhong KZ, Chen WW, Hu XY, Jiang AL and
Zhao J: Clinicopathological and prognostic significance of
microRNA-107 in human non-small cell lung cancer. Int J Clin Exp
Pathol. 7:4545–4551. 2014.
|
|
24
|
Ristori E, Lopez-Ramirez MA, Narayanan A,
Hill-Teran G, Moro A, Calvo CF, Thomas JL and Nicoli S: A
Dicer-miR-107 interaction regulates biogenesis of specific miRNAs
crucial for neurogenesis. Dev Cell. 32:546–560. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang JJ, Wang CY, Hua L, Yao KH, Chen JT
and Hu JH: miR-107 promotes hepatocellular carcinoma cell
proliferation by targeting Axin2. Int J Clin Exp Pathol.
8:5168–5174. 2015.PubMed/NCBI
|
|
26
|
McAllister AK: Neurotrophins and neuronal
differentiation in the central nervous system. Cell Mol Life Sci.
58:1054–1060. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Arévalo JC and Wu SH: Neurotrophin
signaling: Many exciting surprises! Cell Mol Life Sci.
63:1523–1537. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sandhya VK, Raju R, Verma R, Advani J,
Sharma R, Radhakrishnan A, Nanjappa V, Narayana J, Somani BL,
Mukherjee KK, et al: A network map of BDNF/TRKB and BDNF/p75NTR
signaling system. J Cell Commun Signal. 7:301–307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
De la Cruz-Morcillo MA, Berger J,
Sánchez-Prieto R, Saada S, Naves T, Guillaudeau A, Perraud A,
Sindou P, Lacroix A, Descazeaud A, et al: p75 neurotrophin receptor
and pro-BDNF promote cell survival and migration in clear cell
renal cell carcinoma. Oncotarget. Apr 22–2016.(Epub ahead of
print). PubMed/NCBI
|
|
30
|
Kang HJ, Kim JM, Kim SY, Kim SW, Shin IS,
Kim HR, Park MH, Shin MG, Yoon JH and Yoon JS: A Longitudinal study
of BDNF promoter methylation and depression in breast cancer.
Psychiatry Investig. 12:523–531. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang SM, Lin C, Lin HY, Chiu CM, Fang CW,
Liao KF, Chen DR and Yeh WL: Brain-derived neurotrophic factor
regulates cell motility in human colon cancer. Endocr Relat Cancer.
22:455–464. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tanaka K, Okugawa Y, Toiyama Y, Inoue Y,
Saigusa S, Kawamura M, Araki T, Uchida K, Mohri Y and Kusunoki M:
Brain-derived neurotrophic factor (BDNF)-induced
tropomyosin-related kinase B (Trk B) signaling is a potential
therapeutic target for peritoneal carcinomatosis arising from
colorectal cancer. PLoS One. 9:e964102014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kaplan DR, Matsumoto K, Lucarelli E and
Thiele CJ; Eukaryotic Signal Transduction Group. Induction of TrkB
by retinoic acid mediates biologic responsiveness to BDNF and
differentiation of human neuroblastoma cells. Neuron. 11:321–331.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Okamura K, Harada T, Wang S, Ijichi K,
Furuyama K, Koga T, Okamoto T, Takayama K, Yano T and Nakanishi Y:
Expression of TrkB and BDNF is associated with poor prognosis in
non-small cell lung cancer. Lung Cancer. 78:100–106. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang SY, Hui LP, Li CY, Gao J, Cui ZS and
Qiu XS: More expression of BDNF associates with lung squamous cell
carcinoma and is critical to the proliferation and invasion of lung
cancer cells. BMC Cancer. 16:1712016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang S, Guo D, Luo W, Zhang Q, Zhang Y,
Li C, Lu Y, Cui Z and Qiu X: TrkB is highly expressed in NSCLC and
mediates BDNF-induced the activation of Pyk2 signaling and the
invasion of A549 cells. BMC Cancer. 10:432010. View Article : Google Scholar : PubMed/NCBI
|