Introduction
Gliomas account for approximately 70% of all human
primary brain tumors in adults, with astrocytomas representing the
largest group (1). According to
increasing malignancy, astrocytomas are classified into pilocytic
astrocytomas (WHO grade I) and malignant astrocytomas grade II, III
or IV (2,3). Due to its largely heterogeneous
phenotype, the most aggressive grade IV tumor, with a patient's
median survival time of only 12–15 months, was historically termed
glioblastoma multiforme (GBM) (4,5),
accompanied by the idea that gliomas may arise from immature
precursor cells of the nervous system (‘glioblasts’).
Following the mayor concepts of the cancer stem cell
theory (reviewed for example in ref. 6), the expression, regulation and
functional role of stem cell markers has been widely investigated.
For gliomas, the expression of stem/progenitor cell markers like
CD133, Musashi-1, Nestin, Nanog and PDGFR has been described by us
and others (7,8) and reviewed (9). Additionally, glioma cells with stem
cell properties have been isolated from tumor samples or
established glioma cell lines, based on the expression of stem cell
markers (e.g. CD133) or their ability to survive under defined stem
cell conditions, and have been investigated in vitro and
in vivo concerning their tumor-initiating potential,
migratory and proliferative capacity and resistance to chemo- and
radiotherapy (reviewed in ref. 10).
In general, the concepts of how cancer stem cells
gain their ability to self-renew and proliferate are hardly
understood. One reason could be the re-acquisition of embryonic
properties enabling pluripotency, which might be comparable to the
reprogramming of differentiated somatic cells to induced
pluripotent stem cells (iPSCs) by introduction of embryonic stem
cell transcription factors. This experimental procedure was firstly
described by Takahashi and Yamanaka (11), who introduced 24 candidate factors
by retroviral transfection into murine fibroblasts. Based on these
candidates, they were able to show, that the combination of four
factors, OCT4 (octamer binding transcription factor; synonyms Oct3,
POUF5), SOX2 (sex determining region Y-box 2), MYC and KLF4
(Krüppel-like factor 4), efficiently induced a reprogramming of the
somatic fibroblasts yielding pluripotent iPSCs (11,12).
OCT4, MYC, KLF4 and SOX2 as well as the readout for efficient
reprogramming, Nanog (Tir nan Og), are transcription factors that
regulate the expression of a multitude of genes downstream. They
are all involved in the maintenance and regulation of stem cells at
same and different time-points of development (13–20).
Their expression has also been reported in cancer, being
predominantly expressed by the cancer stem cell population
(21–28).
Nevertheless, whether gliomas originate from a
malignantly transformed stem/progenitor cell or if stem cell
properties (and marker expression) are regained within the process
of malignant transformation of e.g. differentiated glial cells is
still unclear.
Thus, to evaluate whether one of the mentioned stem
cell markers or transcription factors involved in maintenance and
regulation of stem cells could probably be identified as a ‘master
marker’ during glioma progression and recurrence, we investigated
their particular transcription in human astrocytomas of different
malignancy grades and matched primary and recurrent glioblastoma
samples. We also analyzed the correlation between gene expression
patterns, showed their intratumoral localization and proliferative
state [related to GFAP (glial fibrillary acidic protein)
distribution, an astroglial marker, and MIB-1/Ki-67, a
proliferation marker], compared their co-expression amongst each
other and with the neural stem cell marker Musashi-1 [Musashi
homolog-1 (Drosophila)], and analyzed their regulation upon
extrinsic chemotherapeutic stress and intrinsic overexpression of
SOX2, KLF4 and OCT4 in vitro.
Materials and methods
Tissue samples and cell cultures
Human astrocytomas of WHO grade I-IV [6 WHO I, 5 WHO
II, 5 WHO III, 21 WHO IV (not matched, 14 primary and 7 recurrent)]
and matched primary and recurrent glioblastomas (14 different
pairs, WHO IV) were surgically dissected tissues from the
Department of Neurosurgery (Kiel, Germany). All samples were
obtained in accordance with the approved ethical standards of the
ethics committee of the University of Kiel and the Helsinki
Declaration of 1975 (D 536/15) and after informed consent of the
donors. The diagnosis was established by a neuropathologist.
Commercial human glioma cell lines were obtained from Deutsches
Krebsforschungszentrum (Heidelberg, Germany; U118 and U343) or from
the American Type Culture Collection (ATCC; Manassas, VA, USA;
A172, T98G and U373). Long-term human primary cultures (A764 and
A767) we obtained by dissociation of glioblastoma tissue and
repetitive subcultivations, whereas short-term human primary
cultures (14/07, 25/07, 76/12 and 96/14) were only used for up to
three passages after preparation from tumor tissues. When an
adequate amount of tumor tissue was available, matched samples of
solid/cultured primary human glioblastomas were used for
experiments. All cell lines were cultured in Dulbecco's modified
Eagle's medium (DMEM; Invitrogen, Carlsbad, CA, USA) supplemented
with 10% fetal calf serum (FCS). Cells were routinely checked for
mycoplasma contamination by PCR.
Quantitative RT-PCR
RNA was isolated using TRIzol reagent (Invitrogen)
following the manufacturer's instructions for tissue or cell
cultures. DNA was digested by RNase-Free DNase (Fermentas, St.
Leon-Rot, Germany; Promega, Madison, WI, USA) and cDNA was
synthesized by reverse transcription (Fermentas). Real-time PCR was
performed using TaqMan Master Mix and Primer Probes (Applied
Biosystems, Foster City, CA, USA): hGAPDH (Hs99999905_m1),
hGFAP (Hs00157674_m1), hOCT4 (Hs00999632_g1),
hNanog (Hs02387400_g1), hMusashi-1 (Hs00159291_m1),
hSOX2 (Hs00602736_s1), hKLF4 (HS00358836_m1) and
hMYC (Hs00153408_m1). Cycle of threshold values
(CT) were measured by an ABI 7500 Fast cycler (Applied
Biosystems) or the MyiQ™ Single Color Real-Time PCR detection
system (Bio-Rad Laboratories, Munich, Germany). All genes of
interest were normalized to the house-keeping gene
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) yielding
ΔCT values: CT (gene of interest) -
CT (GAPDH). As ΔCT values mirror the
exponential course of the PCR, a ΔCT of 3.33 corresponds
to a 10-fold lower expression compared to GAPDH, a ΔCT
value of 6.67 corresponds to a 100-fold lower expression and so on.
For statistical analysis, the relative gene expression compared to
GAPDH (2−ΔCT) was employed. To include undetectable
samples in mean expressions, these sample values were defined as 21
(= ΔCT that would be obtained after the maximum cycle
number). The expression differences between primary and recurrent
glioblastomas were calculated as n-fold ΔΔCT values =
2−ΔCT primary - DCT recurrent and displayed in a grey
shade encoded manner. A relative expression value of 1 (equal
expression in primary and recurrent tumor) was assigned to 30%
grey. Expression values >1 (higher expression in recurrent than
primary tumor) were assigned with increasing grey shading, reaching
black at 3-fold (or higher) induction. Expression values <1
(indicating lower expression in recurrent than primary
glioblastomas) were assigned with lighter grey shadings, increasing
to white shading for values reaching 0. The induction of gene
expression upon stimulation is displayed as relative gene
expression, n-fold expression changes were calculated as ΔΔCT
values = 2−ΔCT control - ΔCT stimulus.
Immunohistochemistry
For immunohistochemistry, 10-μm cryo-sections were
fixed with an ice-cold acetone-methanol mixture (1:1) for 10 min,
blocked for autofluorescence with sudan black (1% in 70% ethanol)
and for unspecific antibody-binding with 0.5% glycine/0.5% bovine
serum albumin. Primary antibodies were applied overnight at 4°C,
secondary antibodies were incubated at 37°C for 1 h, and nuclei
were counterstained with DAPI (Sigma-Aldrich, Hamburg, Germany).
Slides were embedded with Thermo Scientific™ Shandon™ Immu-Mount™
(Thermo Shandon, Inc., Pittsburgh, PA, USA). Between each step,
slides were washed 3× for 5 min with Tris-buffered saline with 0.1%
Tween (TBS-T). Primary antibodies were anti-GFAP (mouse, 1:100;
Dako, Glostrup, Denmark), anti-MIB-1 (Ki-67 clone, mouse, 1:100;
Dako), anti-OCT4 (rabbit, 1:200; Cell Signaling Technology,
Danvers, MA, USA), anti-SOX2 (rabbit, 1:100; Santa Cruz
Biotechnology, Santa Cruz, CA, USA), anti-Musashi-1 (1:100, mouse;
R&D Systems, Minneapolis, MN, USA), anti-Nanog (1:500, rabbit;
Thermo Fisher Scientific, Rockford, IL, USA) and anti-KLF4 (1:250,
mouse; Pierce/Thermo Fisher Scientific). When antibodies were
derived from different species, they were applied simultaneously
and for the antibody controls, both primary antibodies were
omitted. If the primary antibodies were derived from the same
species, first one primary antibody and its secondary antibody were
stained, then unspecific binding was blocked by F(ab) fragments
derived from the very same species [donkey anti-mouse and
anti-rabbit F(ab) fragments, 1:100, from Dianova, Hamburg, Germany]
and the second primary and secondary antibodies were applied. In
this case, antibody controls were performed by omitting both
primary antibodies or, in order to control F(ab) blocking
efficiency, one of them, respectively. Secondary antibodies were
donkey anti-mouse or anti-rabbit IgGs labelled with Alexa Fluor 488
or Alexa Fluor 555 (1:800-1:1000; Invitrogen).
Stimulation and transfection
experiments
For stimulation and transfection experiments, T98G
and A172 glioblastoma cells were seeded subconfluently on 6- or
12-well plates or 75-mm2 culture flasks and grown
overnight. Media were changed to DMEM + 10% FCS supplemented with
temozolomide or camptothecin (both Sigma-Aldrich) or corresponding
concentrations of DMSO as solvent control, cells were incubated for
the indicated times and harvested for RNA isolation. For
transfection, media were changed to Opti-MEM (Invitrogen), and
transfection was performed using Lipofectamine (Invitrogen). The
vectors pcDNA3.3eGFP (mock, Addgene plasmid 26822), pcDNA3.3_KLF4
(Addgene plasmid 26815), pcDNA3.3_OCT4 (Addgene plasmid 26816) and
pcDNA3.3_SOX2 (Addgene plasmid 26817) were generated by Derrick
Rossi (Harvard University Department of Stem Cell and Regenerative
Biology, Boston, MA, USA) (29),
and kindly provided by deposition at Addgene, Inc., (Cambridge, MA,
USA). Media were changed to DMEM + 10% FCS 6 h after transfection,
and cells were harvested for RNA isolation after the indicated
times.
Statistical analysis
To test significance of expression increase or
decrease the Student's t-test was performed. For matched samples
(primary-recurrence from the same patient; samples from the same
experiment) a paired test was used. Astrocytoma samples of
different grades from different patients were analyzed by an
unpaired test. For the calculation of mean expression values of
tumor samples, undetectable samples were set ΔCT = 21,
as this would commonly represent the detection limit if GAPDH is
used as house-keeping gene and 40 amplification cycles are run. For
Pearson correlation analysis, expression levels of all investigated
stem cell markers of the different astrocytoma malignancy grades, a
cohort of mixed primary and recurrent glioblastomas (not matched)
and the respective groups of primary and recurrent glioblastomas of
matched patient samples were correlated using GraphPad Prism,
P-values indicate as follows: *p<0.05,
**p<0.01, ***p<0.001 and
****p<0.0001.
Results
Expression of stem cell factors in
different malignancy grades of astrocytomas and primary and
recurrent GBM
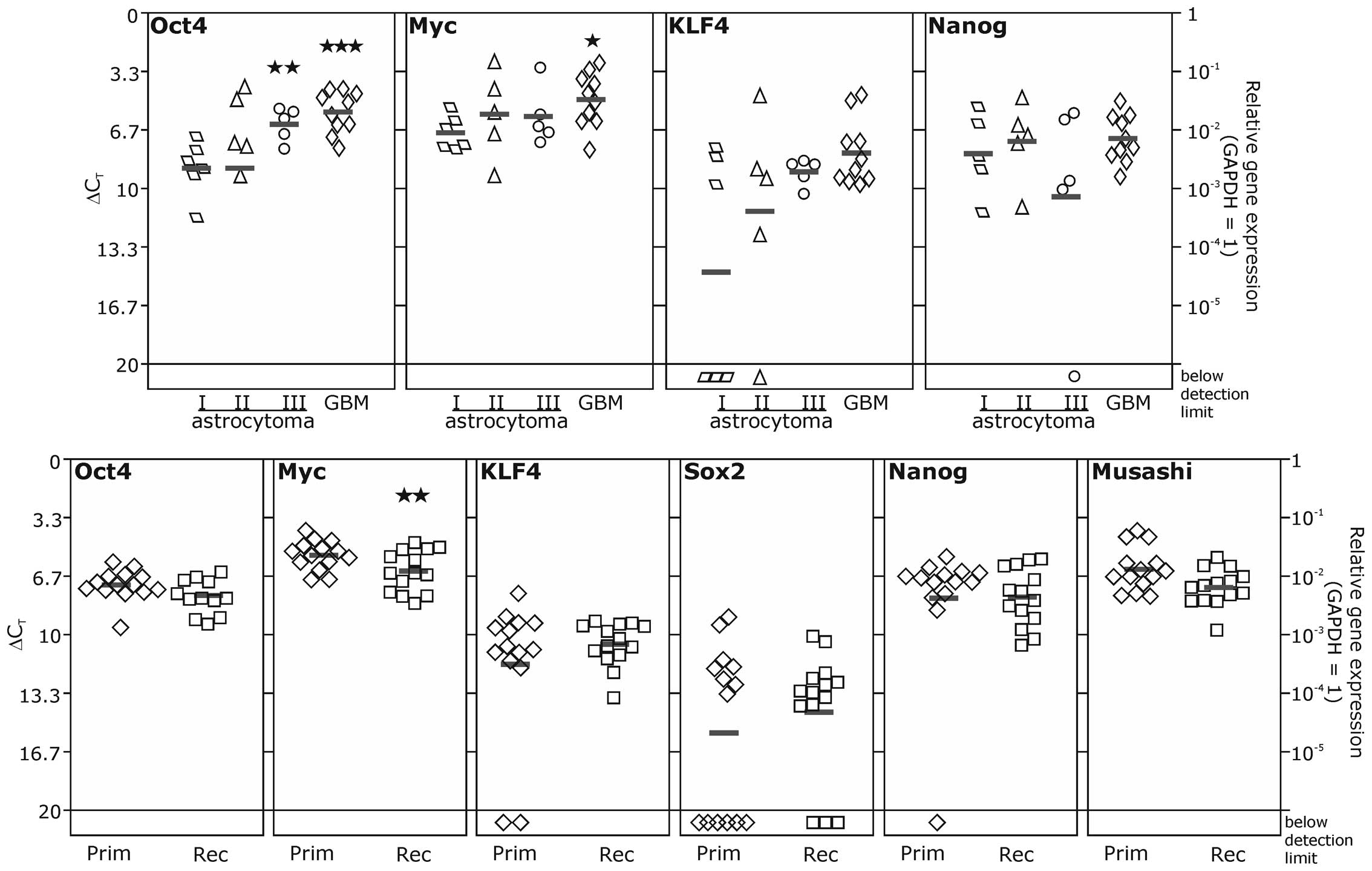
We first investigated the expression of the stem
cell factors in astrocytomas of WHO grade I–IV, with IV
representing the glioblastoma multiforme (GBM), and matched primary
and recurrent glioblastomas (Fig.
1). However, as increasing expression levels of SOX2 in
astrocytomas with higher malignancy have already been described
earlier by us and others (7,30),
we restricted our analysis in astrocytomas to OCT4, Nanog, KLF4 and
MYC. These were detectable on moderate to high levels in
astrocytomas of all malignancy grades. However, for Nanog and KLF4,
we found also a lack of expression in distinct samples. Comparable
results were obtained for the transcription of Nanog, KLF4 and MYC
in matched primary and recurrent glioblastoma samples. In these
samples, we also investigated the transcription of SOX2, which was
considerably lower, and could not be detected in 9/28
primary/recurrent glioblastoma samples. For OCT4 and MYC we
observed a significant increase in mRNA expression in highly
malignant astrocytomas (grade III and IV for OCT4 and grade IV for
MYC) in comparison to the benign grade I astrocytomas. A trend to
higher transcription levels in high grade astrocytomas was also
visible for KLF4. However, due to high variances within the groups,
these results were not statistically robust. In contrast, Nanog
mRNA expression was hardly altered in astrocytomas of different
grades in our sample cohort. In matched samples of primary and
recurrent glioblastomas mean expression levels were not altered
during progression from primary to recurrent glioblastomas except
for MYC, which was found slightly decreased in recurrent samples.
However, regarding the regulation profile of each patient's
primary-recurrent pair, we found distinct patterns of upregulation
and downregulation of stem cell markers (Table I, n-fold expression changes between
individual primary-recurrent pairs, which are numbered
consecutively). Based on these observations, we distinguished one
group of primary/recurrent pairs with one or more markers being
clearly upregulated (>2-fold; Table
I, upper part, pairs 1–6) from a second group showing no clear
upregulation, or even downregulation of one or more stem cell
markers in the recurrent tumor (Table
I, lower part, pairs 7–14).
 | Table IIndividual expression differences of
matched primary and recurrent glioblastomas. |
Table I
Individual expression differences of
matched primary and recurrent glioblastomas.
Interestingly, when analyzing a possible correlation
of mRNA expression of individual investigated stem cell markers in
both astrocytomas of WHO grade I–IV and matched primary and
recurrent glioblastomas (Table
II), significant positive correlations were restricted to
individual marker combinations for most grades and the primary
glioblastomas of the primary/recurrent pairs. In detail,
astrocytomas grade I showed significant correlations between
MYC and Nanog as well as MYC and KLF4 expression. In
astrocytomas grade II we also observed a significant positive
correlation between MYC and Nanog, whereas in astrocytomas grade
III there was no correlation between the markers. In astrocytomas
grade IV (glioblastomas) a significant positive correlation between
KLF4 and MYC expression was detectable (Table IIA). Thus, we could not observe a
specific correlation pattern consistent to all astrocytoma
malignancy grades. However, one has to keep in mind that the sample
groups of astrocytomas grade I–III are quite small due to the
limited availability of tumor tissue. Our further analysis of the
cohort of matched primary/recurrent glioblastoma pairs revealed in
the primary samples only a significant correlation between OCT4 and
KLF4 expression, but remarkably, in recurrences several stem cell
markers showed statistically significant correlations between their
mRNA expression levels (e.g. OCT4-MYC, Nanog-MYC and Nanog-OCT4;
Table IIB).
 | Table IICorrelation analysis between mRNA
expression of stem cell markers and recurrent glioblastomas. |
Table II
Correlation analysis between mRNA
expression of stem cell markers and recurrent glioblastomas.
Taken together, embryonal and neural stem cell
markers are expressed on mRNA level in astrocytomas of different
WHO grades, and primary and recurrent glioblastomas. While OCT4 and
MYC expression was elevated with increasing malignancy of
astrocytomas, MYC expression was slightly reduced in recurrent
tumors compared to the corresponding primary tumor. In addition, a
correlation analysis of individual stem cell marker mRNA expression
revealed sporadic significant positive correlations between
Nanog-MYC or KLF4-MYC expression in glioma progression. Remarkably,
significant positive correlations between Nanog or OCT4 with other
embryonic stem cell factors were frequently found in glioma
recurrence, potentially identifying these markers as highly
important in glioma progression and recurrence. Apart from this,
the regulation pattern of the matched patient samples revealed that
upregulation or downregulation of stem cell markers in the progress
of primary to recurrent glioblastoma occurs in an individual,
probably patient-specific manner.
In situ expression and localization of
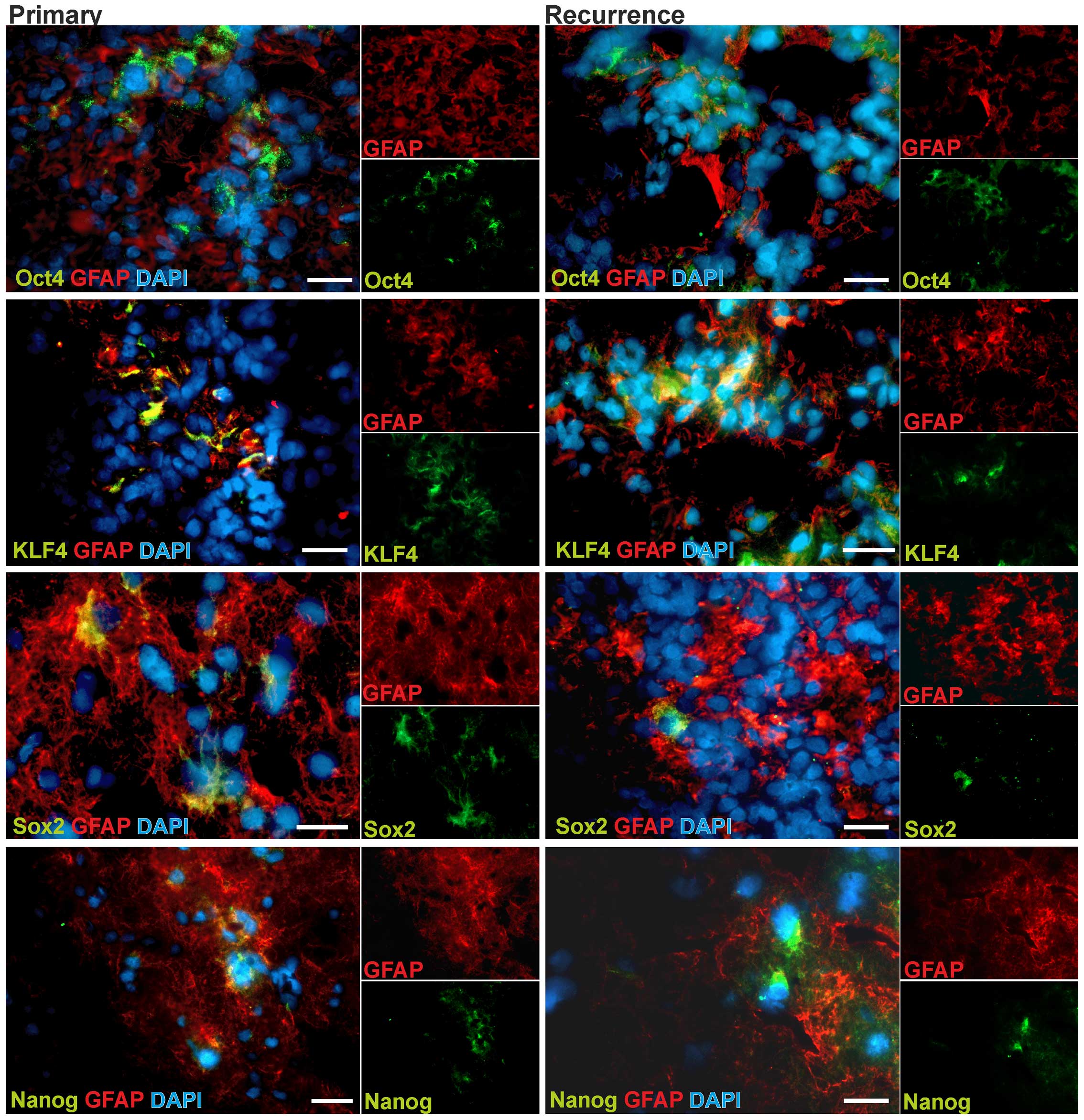
stem cell factors
To investigate the localization and regional
distribution and their potential alteration in glioblastoma
progression, we prepared fluorescence stainings of OCT4, SOX2, KLF4
and Nanog using 3 pairs of matched primary and recurrent
glioblastoma samples. The protein expression of MYC has carefully
been described elsewhere to be evenly throughout the tumor
(31). To identify tumor cells in
more differentiated tumor regions, we co-stained for GFAP (glial
fibrillary acidic protein). As exemplarily shown in Fig. 2, all investigated markers could be
detected on protein levels in glioblastoma cells. Remarkably, only
a small subpopulation of tumor cells showed positive staining for
the respective stem cell markers, and these cells were commonly
found in small groups in GFAP positive tumor regions. To
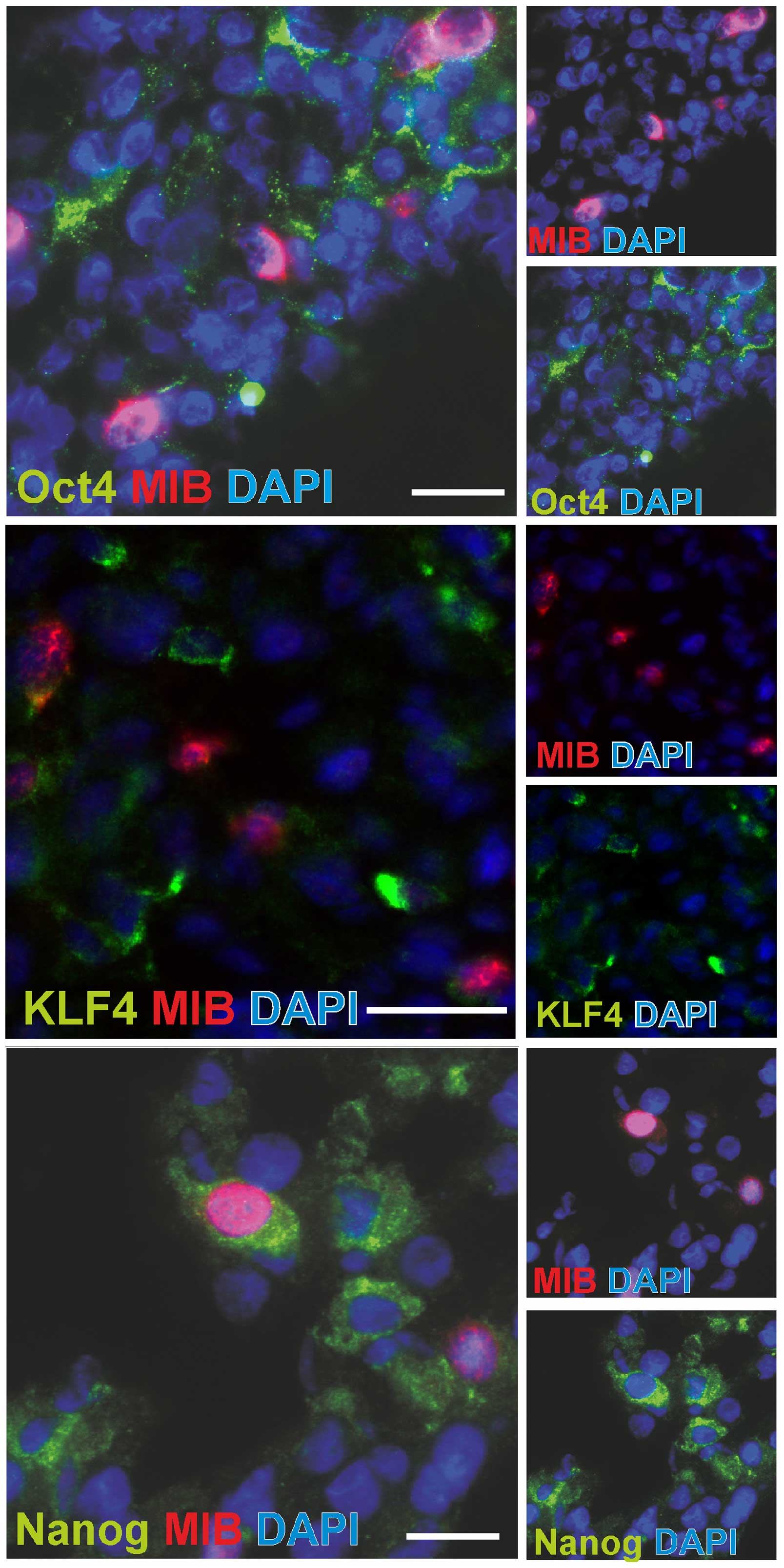
characterize the proliferative status of glioma cells expressing
embryonic stem cell markers, we performed fluorescence
double-stainings of OCT4, KLF4 and Nanog with MIB-1/Ki-67, which is
a nuclear marker for all active cell cycle phases. Cells with
positive staining for OCT4, Nanog or KLF4 did hardly show any
MIB-1-signal in the nucleus. Very rare examples of double positive
cells are shown in Fig. 3.
However, the vast majority of stem cell marker positive cells did
not show staining of the proliferation marker MIB-1 (compare also
Table III).
 | Table IIIEvaluation of embryonic stem cell
factor co-expression. |
Table III
Evaluation of embryonic stem cell
factor co-expression.
| GFAP | MIB | Sox2 | Musashi | KLF4 | Nanog |
|---|
|
|
|
|
|
|
|
|---|
| pos | neg | pos | neg | pos | neg | pos | neg | pos | neg | pos | neg |
|---|
| Oct4 |
| pos | ● | + | ● | ++ | ++ | ● | + | + | + | ● | ● | + |
| neg | +++ | | ++ | | + | | ● | | ● | | + | |
| Nanog |
| pos | ● | + | ● | ++ | ++ | ● | + | ++ | + | + | | |
| neg | +++ | | ++ | | + | | ● | | + | | | |
| KLF4 |
| pos | + | + | ● | ++ | + | ● | ● | ● | | | | |
| neg | +++ | | ++ | | ++ | | ● | | | | | |
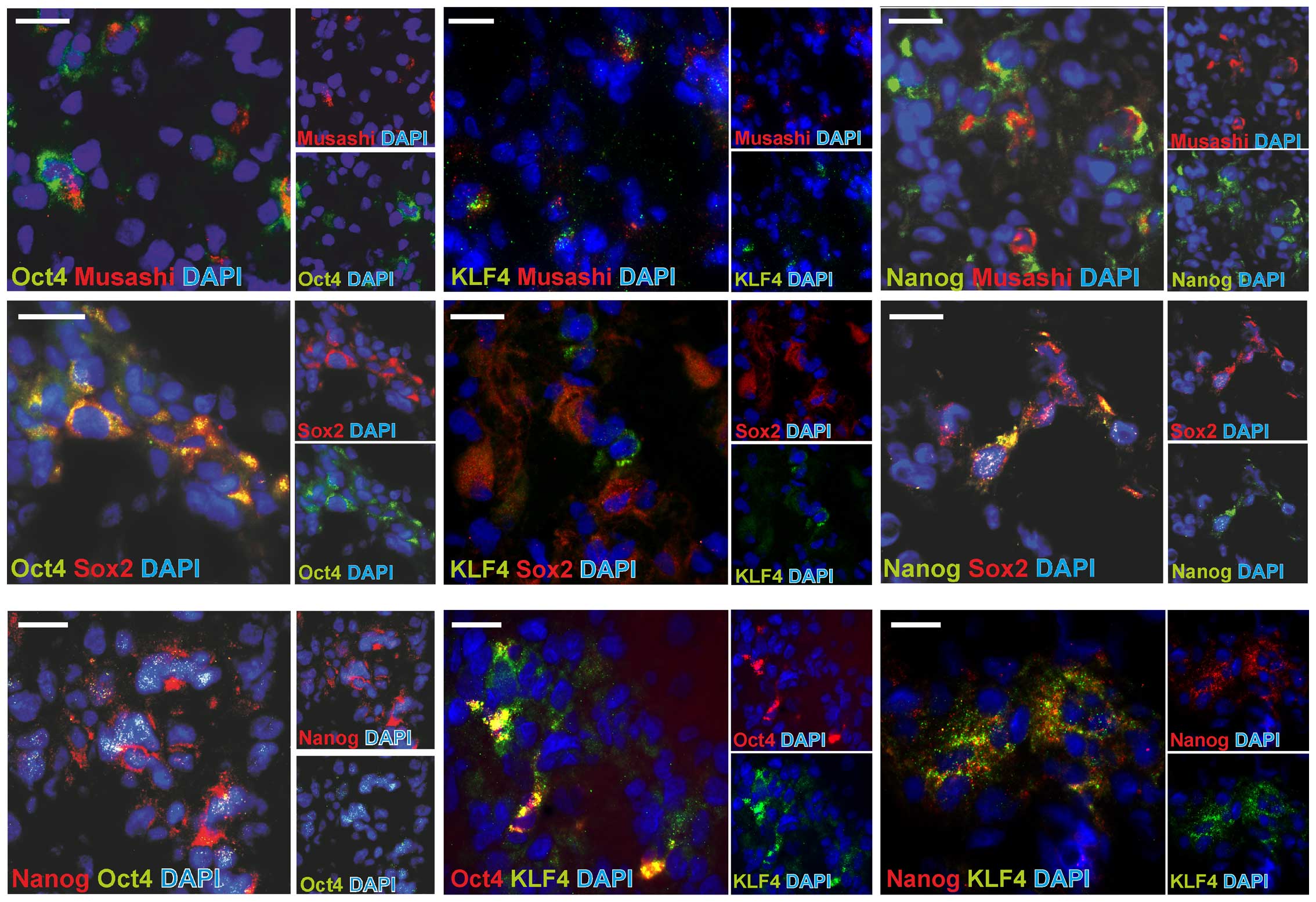
Co-expression of stem cell markers
To comprehensively analyze the co-expression
patterns of embryonic stem cell markers and also compare it to
known neural/glioma stem cell markers, we performed fluorescence
double-stainings of combinations of the markers OCT4, KLF4 and
Nanog with SOX2 and Musashi-1, which are well documented
neural/glioma stem cell markers. Representative examples are shown
in Fig. 4, and results are
summarized in Table III, which
also includes the co-staining results with GFAP and MIB-1 in a
semi-quantitative evaluation of 4 different glioblastoma samples,
respecting the intra- and inter-individual variances. In fact,
OCT4, Nanog and KLF4 were partly co-expressed with the other stem
cell markers (OCT4, Nanog, KLF4, SOX2 and Musashi-1), but were also
found to be expressed in single positive cells. Remarkably, most
stem cell marker-expressing cells were also SOX2-positive, but
KLF4-negative. When comparing these data to mRNA correlation
analysis of investigated stem cell markers (Table IIB), it became apparent that Nanog
and OCT4, which showed a positive mRNA correlation to KLF4,
respectively, were also frequently co-expressed by the same cell
types. Additionally, the missing correlation between KLF4 and SOX2
mRNA expression was underlined by an infrequent KLF4-SOX2
co-staining. Contrasting these findings, Nanog and OCT4, which
positively correlated in mRNA expression, were rarely co-expressed.
Additionally, a frequently observed co-staining of OCT4 and Nanog
with SOX2 was not reflected in a positive correlation of the mRNA
expression. Summarized, a heterogeneous picture results which leads
to the statement that despite the evidence of a specific role of
individual stem cell markers in glioma progression and recurrence,
a ‘master marker’ which is common to all glioblastoma cells with
stem cell characteristics, could not be identified.
Expression of stem cell factors in
glioblastoma short- and long-term cultures and cell lines
For further in vitro investigations, we
analyzed the mRNA expression of the stem cell markers OCT4, SOX2,
KLF4, MYC, Nanog and Musashi-1 in five commercially available
glioma cell lines (A172, U118, U343, U373 and T98G) and two
long-term cultures (A764 and A767) as well as four short-term human
glioblastoma cultures (14/07, 25/07, 76/12 and 96/14). We found
these markers to be expressed on moderate levels in nearly all
investigated cell lines as well as in long- and short-term
cultures, apart from T98G and 14/07 that rarely expressed Nanog
(T98G and 14/07) and SOX2 (T98G), and also comparably low levels of
Musashi-1 (T98G; Fig. 5A and B).
Interestingly, mRNA expression levels of different stem cell
markers showed a surprising range of variation within commercial
cell lines and short- and long-term cultures, but within this range
cell lines were comparable to long- and short-term cultures. Thus,
we chose established glioma cell lines, which are available
unlimitedly and therefore solve the problem of limited access to
tumor tissue, for further experiments. Experiments mainly focused
on the ability to regulate stem cell factors upon extrinsic
chemotherapeutic stress and upon intrinsic overexpression of
respective stem cell transcription factors.
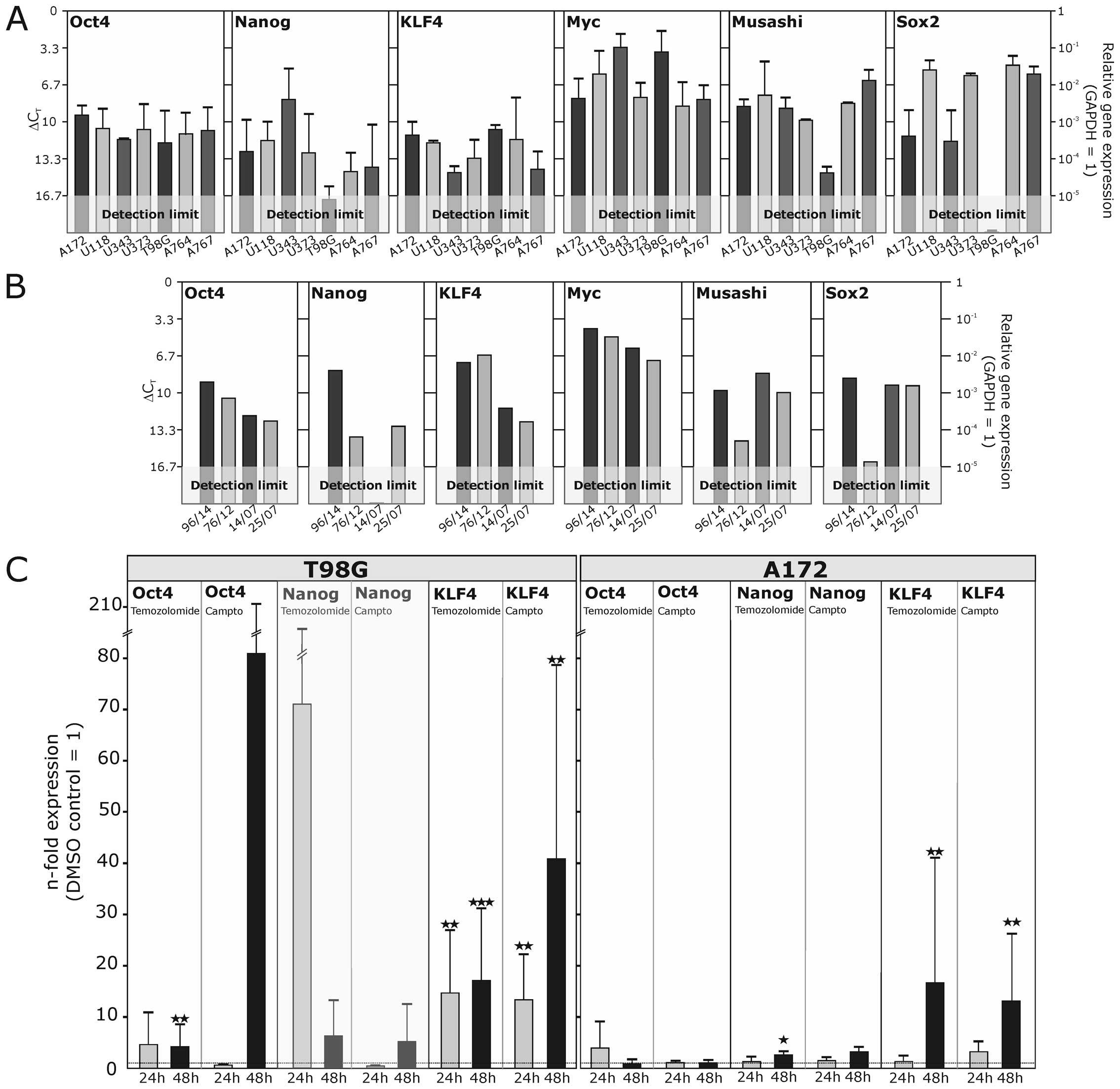 | Figure 5Transcription of stem cell markers in
glioblastoma cell lines and short- and long-term primary cultures
as well as regulation upon stimulation. (A) The mRNA expression of
different commercially available or long-term self-generated
glioblastoma cell lines was analyzed by quantitative RT-PCR. While
some genes are quite evenly expressed on high, moderate or low
levels, like MYC, OCT4 and KLF4, other genes like Nanog, Musashi-1
and SOX2, are more differentially expressed in different cell
lines. (B) In short-term glioblastoma primary cultures mRNA
expression levels of stem cell markers were comparable to those in
commercial cell lines and long-term cultures concerning expression
intensity and variability. (C) The glioblastoma cell lines T98G and
A172 were stimulated with the cytotoxic drugs temozolomide (400
μg/ml) or camptothecin (50 μg/ml) for 24 and 48 h, respectively,
and the regulation of stem cell marker transcription was analyzed
by quantitative RT-PCR. Altered gene expression was partly observed
for OCT4, Nanog and KLF4, which are displayed as n-fold expression
in comparison to the solvent control treated with equal volumes of
DMSO. Most robust expression changes were observed for KLF4, which
was significantly induced in both cell lines after stimulation with
temozolomide or camptothecin. Stimulations were performed in n=3
independent experiments, and a paired Student's t-test was used for
statistical analysis. |
Regulation of stem cell factors by
chemotherapeutics
To investigate the influence of chemotherapeutics on
the expression levels of stem cell markers, we chose A172 and T98G
as exemplary glioma cell lines because A172 showed moderate to high
expression levels of stem cell markers, and T98G showed low
expression levels for Nanog and SOX2 (comparable to low expression
levels of the short-term cultures 14/07 and 76/12). These cell
lines were stimulated for 24 or 48 h with 50 μg/ml camptothecin, or
400 μg/ml temozolomide, the most commonly used chemotherapeutic in
glioblastoma treatment; corresponding DMSO concentrations were used
for stimulation of solvent controls and used to determine
expression changes. For MYC, SOX2 and Musashi-1 we did not observe
any significant regulation of the mRNA expression level (data not
shown). In contrast, in T98G cells, the expression level of OCT4
was significantly elevated 4.2-fold (±4.3 SD) after 48 h (Fig. 5C). This effect was even more
pronounced upon stimulation with camptothecin, but due to high
variations not significant. In A172 cells, OCT4 mRNA expression was
regulated neither by temozolomide nor by camptothecin. Nanog, which
was hardly detectable in unstimulated T98G cells, was found to be
increased. However, the proximity to the detection limit has to be
kept in mind, thus the data are transparently shaded. In
temozolomide-treated A172 cells (48 h), Nanog was significantly but
only slightly increased (2.6-fold±0.7 SD), whereas camptothecin
yielded no significant effect. Contrasting these slight or little
robust expressional changes of OCT4 and Nanog, KLF4 was
significantly induced by both temozolomide and camptothecin in T98G
and A172 cells. Showing a significant induction after 24 h already,
temozolomide yielded a 17.1-fold (±14.0 SD), and camptothecin a
40.9-fold (±37.8 SD) upregulation after 48 h. In A172 cells, an
upregulation to 17.7-fold (±24.4 SD) by temozolomide, and to 13.1
(±13.2 SD) by camptothecin was induced after 48 h.
Summarized, we observed an induction of the
embryonic stem cell markers Nanog and KLF4 (and partly OCT4) in
glioma cell lines upon chemotherapeutic stress. Surprisingly, this
effect is more pronounced in T98G cells, where baseline expression
of stem cell markers is comparably low. Among the investigated stem
cell markers, KLF4 is most robustly regulated in both glioma cell
lines upon extrinsic cytotoxic challenge with camptothecin or
temozolomide.
Regulation of stem cell markers by
overexpression of transcription factors SOX2, KLF4 or OCT4
Having observed this limited but specific plasticity
regarding stem cell marker regulation upon extrinsic challenge, we
investigated, if stem cell marker expression might be regulated by
intrinsic alteration of stem cell transcription factors SOX2, KLF4
and OCT4. Thus, we performed overexpression experiments with stem
cell marker low expressing T98G cells using expression vectors for
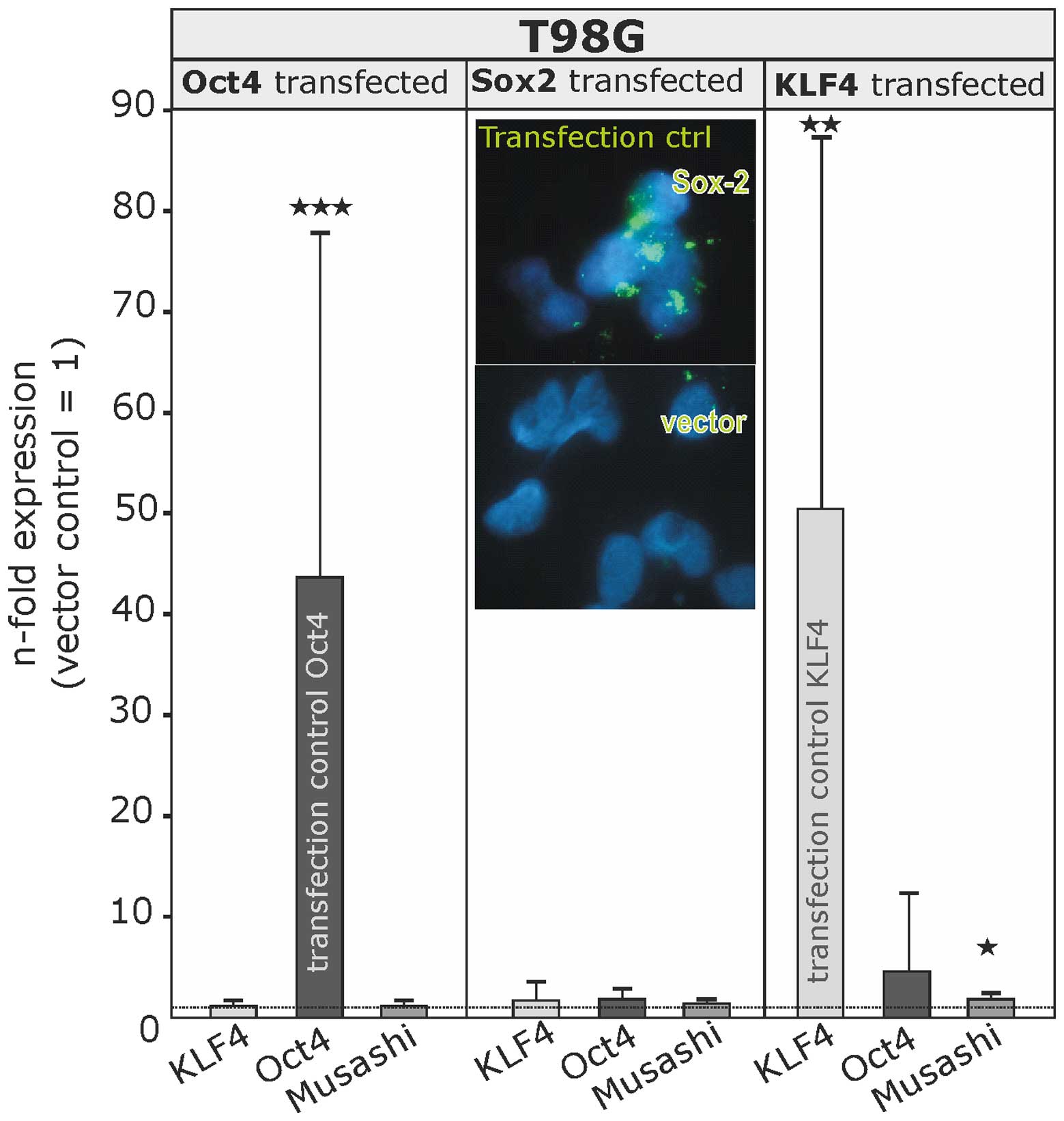
the respective transcription factors (Fig. 6). Successful transfection was
proven either by quantitative RT-PCR (OCT4, 43.6-fold induction
±34.2 SD; KLF4, 50.4-fold induction ±36.8 SD) or in case of SOX2 by
immunocytochemistry (insert in Fig.
6).
The overexpression of these transcription factors
did not yield a significant regulation (induction or reduction) of
other stem cell markers investigated (SOX2, KLF4, OCT4, MYC, Nanog
and Musashi-1), apart from Musashi-1, which was only very slightly
induced (1.8-fold±0.7 SD) upon KLF4 overexpression.
Taken together, the embryonic stem cell
transcription factors SOX2, MYC, OCT4, KLF4 and Nanog are expressed
in astrocytomas of different malignancy grades as well as in
primary and recurrent glioblastomas in a very distinct, probably
patient specific expression pattern. Positive correlations between
mRNA expression levels of stem cell markers were more pronounced in
recurrent glioblastomas, while in situ co-expression occurs
in a complex manner, which did not necessarily match the mRNA
correlation results. In vitro, stem cell markers were
expressed in established cell lines as well as in glioblastoma
short- and long-term cultures. The factors were partly regulated by
extrinsic chemotherapeutic challenge but not by intrinsic
overexpression of the respective transcription factors.
Discussion
One major concept of tumor development, progression
and recurrence is the cancer stem cell hypothesis that postulates
the existence of a subpopulation of cancer cells with stem cell
properties (like unlimited self-renewal and asymmetric division)
within a heterogeneous tumor. Targeting these stem-like cancer
cells would be highly desirable, as they are supposed to give rise
to recurrences and metastases, e.g. after resection and adjuvant
treatment. However, although the existence of tumor cells with stem
cell character and the initiation of tumors, recurrences and
metastases by tumor cells with stem cell properties have been
proven for a multitude of tumors (32–35),
to date strategies for clear identification in situ and
subsequent systematic targeting of these putatively
recurrence-initiating cells in vivo are still missing. In
glioma, CD133 was the first marker being identified for glioma
stem-like and initiating cells. CD133 is a pentaspan glycoprotein,
which is physiologically expressed by neural, hematopoietic and
endothelial stem cells (36,37).
However, later investigation showed that CD133-negative glioma
cells may also have cancer stem cell characteristics such as highly
tumorigenic potential (reviewed in ref. 38), and can switch to CD133 expression
when they meet the appropriate environment and conditions (39,40).
Thus, glioma stem-like cells might be better identified and
targeted by their ability to divide unlimitedly and give rise to
multifaceted progeny, which can be compared to the pluripotency of
embryonic stem cells.
Regarding the fact that as few as four transcription
factors can induce pluripotency in differentiated fibroblasts
(11), we analyzed mRNA expression
of the transcription factors KLF4, OCT4, MYC and SOX2, as well as
Nanog, the read out of successful iPSC generation, in astrocytomas
of different malignancy grades as well as in matched primary and
recurrent glioblastomas by quantitative RT-PCR. We showed, that for
most genes transcription was detectable on moderate to high levels
in all investigated tumor grades and stages. However, in some
individual cases, KLF4, SOX2 and/or Nanog were not detectable. We
observed a significant increase in mean mRNA expression for OCT4 in
highly malignant astrocytomas (grade III and IV) in comparison to
the benign grade I astrocytoma and a slight but significant
increase of MYC in grade IV astrocytomas (GBM). For KLF4, an
increased transcription was also observed, but due to individual
variance statistical robustness could not be proven. An increased
expression of SOX2 with increased malignancy of astrocytomas has
been reported by us in an earlier study (7). These findings are in line with
analysis of Guo et al (41), showing increased protein levels of
OCT4 and SOX2 in malignant astrocytomas. They also show increasing
expression of Nanog, however, we could not confirm these findings
on mRNA level in our sample cohort. Other studies even pointed to
higher Nanog levels in astrocytomas and oligodendrogliomas of grade
III compared to glioblastomas (42).
Regarding the mean mRNA level in primary and
recurrent glioblastomas, we did not observe any significant changes
except for MYC, and this mean reduction was comparably mild and
with high interindividual variance (ranging from 19 to 217%).
Interestingly, regarding the individual differences between the
matched primary and recurrent samples from the same patients
(compare Table I), the cohort
could be divided into one group with clear upregulation
(>2-fold) of at least one stem cell marker and one group showing
predominantly decreased marker transcription. The frequent decrease
of stem cell factor expression in recurrences is surprising, as
glioma aggressiveness and ability to relapse is often addicted to
stem cell properties. Especially the expression of MYC, which was
decreased in 11/14 pairs of our matched primary-recurrent cohort,
is known to be significantly increased in CD133-positive glioma
stem cells. Depletion of CD133 inhibits growth of glioma stem
cells, induces apoptosis, reduces ability to glioma sphere
formation and inhibits the tumor formation in a murine xenograft
model (43). Additionally, high
expression levels especially of Nanog, but also of MYC and KLF4
were associated with shorter survival times in a cohort of low and
high grade gliomas of different histopathological subtypes,
indicating that these embryonic stem cell factors significantly
attribute to the clinical outcome of gliomas (8). However, as we for the first time
address matched primary and recurrent gliomas, our observations of
frequently decreased stem cell factor expression emphasizes a
broader view on individualized stem cell factor expression and
regulation during progression.
A correlation analysis between stem cell marker
expression levels within the investigated malignancy grades and the
primary and recurrent glioblastoma cohort revealed distinct
positive correlations between individual stem cell marker pairs.
Interestingly, in recurrences we observed most frequently
significant positive correlations between stem cell marker
expressions apart from SOX2.
These correlative findings could, at least in parts,
be underlined by co-stainings of the stem cell factors with SOX2,
Musashi-1 and in comparison to each other. Here, we observed on the
protein level double stainings for each possible combination but
also frequent single expression patterns. Remarkably, especially
the significantly positive correlated embryonic stem cell marker
pairs like KLF4-OCT4 and KLF4-Nanog were also frequently
co-expressed. In contrast to this, less frequent co-expression was
observed for the pair Nanog-OCT4, although their expression also
positively correlated. Frequent co-expression was observed for SOX2
with the other embryonic stem cell factors, although there was no
correlation observed on mRNA level. In earlier studies, a
co-expression of Nanog and CD133, and its positive correlation to
malignancy grade was reported (44). However, based on our observations,
a ‘master marker’, which uniquely identifies all glioma cells with
stem cell characteristics, cannot be defined.
Concerning the intratumoral distribution of
embryonic stem cell factors, we observed immunoreactivity for KLF4,
OCT4, Nanog and SOX2 in small groups or clusters of cells even
within GFAP-positive tumor glioblastoma regions, irrespective of
primary or recurrent glioblastoma samples. However, in general
embryonic stem cell-like expression signature is more likely
observed in poorly differentiated tumors, and the SOX2 expression
is known to decrease upon differentiation in vitro (45,46).
Concerning co-stainings with the proliferation marker MIB-1/Ki-67,
glioma cells expressing the embryonic transcription factor SOX2,
like those expressing neural stem cell markers CD133, Nestin and
Musashi-1, are rarely actively proliferating (7). Accordingly, we hardly observed any
glioma cells expressing OCT4, KLF4 or Nanog with nuclear staining
for MIB-1/Ki-67. The majority of stem cell factor positive cells
was negative for MIB-1/Ki-67 and also negative for GFAP (though in
GFAP-positive regions). These findings hint to an undifferentiated,
non-proliferative/resting status of embryonic stem cell factor
expressing cells, which is supported by the findings of Holmberg
et al (47) concerning
SOX2.
In gliomas and other tumors, the expression and
regulation of embryonic stem cell markers seems to be critical for
the maintenance of tumor stem-like/initiating cells (48–51).
The induction of the stem cell factors MYC, KLF4, OCT4, Nanog and
SOX2 by hypoxia has been reported for various established cancer
cell lines (52). This report also
shows an upregulation of OCT4, Nanog and MYC in primary human
glioma cells in hypoxic conditions, and an enhanced formation of
neurospheres (52). Further
investigations demonstrated that signaling of the receptor tyrosine
kinase MET/hepatocyte growth factor receptor HGFR could induce the
expression of OCT4, KLF4 and Nanog (53). We could additionally show that
OCT4, KLF4 and Nanog are at least partly induced upon extrinsic
chemotherapeutic treatment. These effects were more prominent in
the glioma cell line T98G, which initially showed low expression
levels of Nanog, Musashi-1 and SOX2. In this cell line we performed
also overexpression experiments with KLF4, OCT4 and SOX2. However,
although silencing of OCT4 also provoked downregulation of SOX2 and
reduced the tumorigenicity in vitro and in vivo
(48), we could not observe any
robust regulatory influence of intrinsic overexpression on the
expression on any other embryonic or neural stem cell marker.
However, recent studies have shown that glioma stem-like cells
could be generated by expression of SOX2, OCT4 and Nanog in patient
derived glioma cell lines showing neurosphere formation and
chemotherapy resistance (54).
Taken together, these results implicate that the expression of stem
cell factors is altered due to the environmental conditions, so
that these conditions, e.g. hypoxia, therapeutic challenge, may
influence on the glioma progression.
In conclusion, the embryonic stem cell factors KLF4,
OCT4, SOX2, MYC and Nanog are differentially expressed in human
astrocytomas and glioblastomas and individually regulated in
glioblastoma progression. Their mRNA expression levels partly
correlate with each other, especially in recurrent glioblastomas.
Staining of stem cell factors in glioblastoma sections occurs solo
or in combinations and is restricted to a heterogeneous
subpopulation of glioblastoma cells with limited proliferation
activity. In correlation analysis and co-stainings, a
‘master-marker’ defining the complete glioma stem cell subset could
not be defined. However, the clear inductions of some stem cell
markers and positive correlations especially in recurrent
glioblastomas underline the importance of embryonic stem cell
factors in glioma progression and recurrence. Extrinsic application
of chemotherapeutics but not intrinsic inductions of stem cell
transcription factors regulate subsets of stem cell markers. As
stem cell markers and transcription factors control self-renewal
and tumorigenic potential and therefore glioma progression and
recurrence, they may serve as a promising future target for
diagnostics and novel therapeutic approaches.
Acknowledgements
We thank Judith Becker, Martina Burmester, Fereshteh
Ebrahim, Jörg Krause, Miriam Lemmers and Brigitte Rehmke for expert
technical assistance. Derrick Rossi kindly provided the expression
vectors. The present study was supported by grants of the Deutsche
Forschungsgemeinschaft (ME758/10-1 and HE3400/5-1), the State of
Schleswig-Holstein (Molecular Imaging in the North MOIN), the
popgen 2.0 network (P2N, a grant from the German Ministry for
Education and Research 01EY1103), and the Family Mehdorn
foundation.
Abbreviations:
|
GAPDH
|
glycerinaldehyde-3-phosphate-dehydrogenase
|
|
GBM
|
glioblastoma multiforme
|
|
GFAP
|
glial fibrillary acidic protein
|
|
iPSCs
|
induced pluripotent stem cells
|
|
KLF4
|
Krüppel-like factor 4
|
|
Musashi-1
|
Musashi (Drosophila)
homolog-1
|
|
Nanog
|
‘Tir nan Og’
|
|
OCT4
|
octamer binding transcription
factor
|
|
qRT-PCR
|
quantitative reverse
transcription-polymerase chain reaction
|
|
SOX2
|
sex determining region Y-box 2
|
|
WHO
|
World Health Organization
|
References
|
1
|
Ricard D, Idbaih A, Ducray F, Lahutte M,
Hoang-Xuan K and Delattre JY: Primary brain tumours in adults.
Lancet. 379:1984–1996. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Walker C, Baborie A, Crooks D, Wilkins S
and Jenkinson MD: Biology, genetics and imaging of glial cell
tumors. Br J Radio. 84:S90–S106. 2011. View Article : Google Scholar
|
|
4
|
Bailey P and Cushing H: A classification
of the tumours of the glioma group on a histogenetic basis, with a
correlated study of prognosis. Lippincott-Raven; Philadelphia:
1926
|
|
5
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al; European Organisation for Research and Treatment of
Cancer Brain Tumor and Radiotherapy Groups; National Cancer
Institute of Canada Clinical Trials Group. Radiotherapy plus
concomitant and adjuvant temozolomide for glioblastoma. N Engl J
Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sell S: On the stem cell origin of cancer.
Am J Pathol. 176:2584–494. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ma YH, Mentlein R, Knerlich F, Kruse ML,
Mehdorn HM and Held-Feindt J: Expression of stem cell markers in
human astrocytomas of different WHO grades. J Neurooncol. 86:31–45.
2008. View Article : Google Scholar
|
|
8
|
Elsir T, Edqvist PH, Carlson J, Ribom D,
Bergqvist M, Ekman S, Popova SN, Alafuzoff I, Ponten F, Nistér M
and Smits A: A study of embryonic stem cell-related proteins in
human astrocytomas: Identification of Nanog as a predictor of
survival. Int J Cancer. 134:1123–1131. 2014. View Article : Google Scholar
|
|
9
|
Dell'Albani P: Stem cell markers in
gliomas. Neurochem Res. 33:2407–2415. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hamerlink P: Cancer stem cells and
glioblastoma. Glioma Cell Biology. Mentlein R and Sedo A:
Springer-Verlag; Wien, New York: pp. 3–23. 2014
|
|
11
|
Takahashi K and Yamanaka S: Induction of
pluripotent stem cells from mouse embryonic and adult fibroblast
cultures by defined factors. Cell. 126:663–676. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Okita K, Ichisaka T and Yamanaka S:
Generation of germline-competent induced pluripotent stem cells.
Nature. 448:313–317. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Okamoto K, Okazawa H, Okuda A, Sakai M,
Muramatsu M and Hamada H: A novel octamer binding transcription
factor is differentially expressed in mouse embryonic cells. Cell.
60:461–472. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Warren L, Manos PD, Ahfeldt T, Loh YH, Li
H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, et al: Highly
efficient reprogramming to pluripotency and directed
differentiation of human cells with synthetic modified mRNA. Cell
Stem Cell. 7:618–630. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gubbay J, Collignon J, Koopman P, Capel B,
Economou A, Münsterberg A, Vivian N, Goodfellow P and Lovell-Badge
R: A gene mapping to the sex-determining region of the mouse Y
chromosome is a member of a novel family of embryonically expressed
genes. Nature. 346:245–250. 1990. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zappone MV, Galli R, Catena R, Meani N, De
Biasi S, Mattei E, Tiveron C, Vescovi AL, Lovell-Badge R,
Ottolenghi S, et al: Sox2 regulatory sequences direct expression of
a (beta)-geo transgene to telencephalic neural stem cells and
precursors of the mouse embryo, revealing regionalization of gene
expression in CNS stem cells. Development. 127:2367–2382.
2000.PubMed/NCBI
|
|
17
|
Graham V, Khudyakov J, Ellis P and Pevny
L: SOX2 functions to maintain neural progenitor identity. Neuron.
39:749–765. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Segre JA, Bauer C and Fuchs E: Klf4 is a
transcription factor required for establishing the barrier function
of the skin. Nat Genet. 22:356–360. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chambers I, Colby D, Robertson M, Nichols
J, Lee S, Tweedie S and Smith A: Functional expression cloning of
Nanog, a pluripotency sustaining factor in embryonic stem cells.
Cell. 113:643–655. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mitsui K, Tokuzawa Y, Itoh H, Segawa K,
Murakami M, Takahashi K, Maruyama M, Maeda M and Yamanaka S: The
homeoprotein Nanog is required for maintenance of pluripotency in
mouse epiblast and ES cells. Cell. 113:631–642. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gidekel S, Pizov G, Bergman Y and Pikarsky
E: Oct-3/4 is a dose-dependent oncogenic fate determinant. Cancer
Cell. 4:361–370. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Peng S, Maihle NJ and Huang Y:
Pluripotency factors Lin28 and Oct4 identify a sub-population of
stem cell-like cells in ovarian cancer. Oncogene. 29:2153–2159.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chiou SH, Wang ML, Chou YT, Chen CJ, Hong
CF, Hsieh WJ, Chang HT, Chen YS, Lin TW, Hsu HS, et al:
Coexpression of Oct4 and Nanog enhances malignancy in lung
adenocarcinoma by inducing cancer stem cell-like properties and
epithelial-mesenchymal transdifferentiation. Cancer Res.
70:10433–10444. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Taub R, Kirsch I, Morton C, Lenoir G, Swan
D, Tronick S, Aaronson S and Leder P: Translocation of the c-myc
gene into the immunoglobulin heavy chain locus in human Burkitt
lymphoma and murine plasmacytoma cells. Proc Natl Acad Sci USA.
79:7837–7841. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hemmati HD, Nakano I, Lazareff JA,
Masterman-Smith M, Geschwind DH, Bronner-Fraser M and Kornblum HI:
Cancerous stem cells can arise from pediatric brain tumors. Proc
Natl Acad Sci USA. 100:15178–15183. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang J, Sakariassen PØ, Tsinkalovsky O,
Immervoll H, Bøe SO, Svendsen A, Prestegarden L, Røsland G, Thorsen
F, Stuhr L, et al: CD133 negative glioma cells form tumors in nude
rats and give rise to CD133 positive cells. Int J Cancer.
122:761–768. 2008. View Article : Google Scholar
|
|
27
|
Zhao W, Hisamuddin IM, Nandan MO, Babbin
BA, Lamb NE and Yang VW: Identification of Krüppel-like factor 4 as
a potential tumor suppressor gene in colorectal cancer. Oncogene.
23:395–402. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jeter CR, Badeaux M, Choy G, Chandra D,
Patrawala L, Liu C, Calhoun-Davis T, Zaehres H, Daley GQ and Tang
DG: Functional evidence that the self-renewal gene NANOG regulates
human tumor development. Stem Cells. 27:993–1005. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wan F, Herold-Mende C, Campos B, Centner
FS, Dictus C, Becker N, Devens F, Mogler C, Felsberg J, Grabe N, et
al: Association of stem cell-related markers and survival in
astrocytic gliomas. Biomarkers. 16:136–143. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang J, Wang H, Li Z, Wu Q, Lathia JD,
McLendon RE, Hjelmeland AB and Rich JN: c-Myc is required for
maintenance of glioma cancer stem cells. PLoS One. 3:e37692008.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Faria MH, Khayat AS, Burbano RR and
Rabenhorst SH: c-MYC amplification and expression in astrocytic
tumors. Acta Neuropathol. 116:87–95. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Barker N, Ridgway RA, van Es JH, van de
Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR,
Sansom OJ and Clevers H: Crypt stem cells as the cells-of-origin of
intestinal cancer. Nature. 457:608–611. 2009. View Article : Google Scholar
|
|
34
|
Ignatova TN, Kukekov VG, Laywell ED,
Suslov ON, Vrionis FD and Steindler DA: Human cortical glial tumors
contain neural stem-like cells expressing astroglial and neuronal
markers in vitro. Glia. 39:193–206. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
O'Connor ML, Xiang D, Shigdar S, Macdonald
J, Li Y, Wang T, Pu C, Wang Z, Qiao L and Duan W: Cancer stem
cells: A contentious hypothesis now moving forward. Cancer Lett.
344:180–187. 2014. View Article : Google Scholar
|
|
36
|
Galli R, Binda E, Orfanelli U, Cipelletti
B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F and Vescovi
A: Isolation and characterization of tumorigenic, stem-like neural
precursors from human glioblastoma. Cancer Res. 64:7011–7021. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Singh SK, Clarke ID, Terasaki M, Bonn VE,
Hawkins C, Squire J and Dirks PB: Identification of a cancer stem
cell in human brain tumors. Cancer Res. 63:5821–5828.
2003.PubMed/NCBI
|
|
38
|
Beier CP and Beier D: CD133 negative
cancer stem cells in glioblastoma. Front Biosci (Elite Ed).
3:701–710. 2011. View
Article : Google Scholar
|
|
39
|
Nishide K, Nakatani Y, Kiyonari H and
Kondo T: Glioblastoma formation from cell population depleted of
Prominin1-expressing cells. PLoS One. 4:e68692009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Varlakhanova NV, Cotterman RF, deVries WN,
Morgan J, Donahue LR, Murray S, Knowles BB and Knoepfler PS: myc
maintains embryonic stem cell pluripotency and self-renewal.
Differentiation. 80:9–19. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Guo Y, Liu S, Wang P, Zhao S, Wang F, Bing
L, Zhang Y, Ling EA, Gao J and Hao A: Expression profile of
embryonic stem cell-associated genes Oct4, Sox2 and Nanog in human
gliomas. Histopathology. 59:763–775. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Clement V, Sanchez P, de Tribolet N,
Radovanovic I, Ruiz i Altaba A and Altaba R: HEDGEHOG-GLI1
signaling regulates human glioma growth, cancer stem cell
self-renewal, and tumorigenicity. Curr Biol. 17:165–172. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Weina K and Utikal J: SOX2 and cancer:
Current research and its implications in the clinic. Clin Transl
Med. 3:192014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Niu CS, Li DX, Liu YH, Fu XM, Tang SF and
Li J: Expression of NANOG in human gliomas and its relationship
with undifferentiated glioma cells. Oncol Rep. 26:593–601.
2011.PubMed/NCBI
|
|
45
|
Ben-Porath I, Thomson MW, Carey VJ, Ge R,
Bell GW, Regev A and Weinberg RA: An embryonic stem cell-like gene
expression signature in poorly differentiated aggressive human
tumors. Nat Genet. 40:499–507. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hattermann K, Held-Feindt J, Lucius R,
Müerköster SS, Penfold ME, Schall TJ and Mentlein R: The chemokine
receptor CXCR7 is highly expressed in human glioma cells and
mediates antiapoptotic effects. Cancer Res. 70:3299–3308. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Holmberg J, He X, Peredo I, Orrego A,
Hesselager G, Ericsson C, Hovatta O, Oba-Shinjo SM, Marie SKN,
Nistér M, et al: Activation of neural and pluripotent stem cell
signatures correlates with increased malignancy in human glioma.
PLoS One. 6:e184542011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ikushima H, Todo T, Ino Y, Takahashi M,
Saito N, Miyazawa K and Miyazono K: Glioma-initiating cells retain
their tumorigenicity through integration of the Sox axis and Oct4
protein. J Biol Chem. 286:41434–41441. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kumar SM, Liu S, Lu H, Zhang H, Zhang PJ,
Gimotty PA, Guerra M, Guo W and Xu X: Acquired cancer stem cell
phenotypes through Oct4-mediated dedifferentiation. Oncogene.
31:4898–4911. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zbinden M, Duquet A, Lorente-Trigos A,
Ngwabyt SN, Borges I, Ruiz i Altaba A and Altaba A: NANOG regulates
glioma stem cells and is essential in vivo acting in a
cross-functional network with GLI1 and p53. EMBO J. 29:2659–2674.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhu XY, Wang L, Luan SH, Zhang HS, Huang
WT and Wang NH: The PGI-KLF4 pathway regulates self-renewal of
glioma stem cells residing in the mesenchymal niches in human
gliomas. Neoplasma. 61:401–410. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Mathieu J, Zhang Z, Zhou W, Wang AJ,
Heddleston JM, Pinna CMA, Hubaud A, Stadler B, Choi M, Bar M, et
al: HIF induces human embryonic stem cell markers in cancer cells.
Cancer Res. 71:4640–4652. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Jun HJ, Bronson RT and Charest A:
Inhibition of EGFR induces a c-MET-driven stem cell population in
glioblastoma. Stem Cells. 32:338–348. 2014. View Article : Google Scholar
|
|
54
|
Olmez I, Shen W, McDonald H and Ozpolat B:
Dedifferentiation of patient-derived glioblastoma multiforme cell
lines results in a cancer stem cell-like state with
mitogen-independent growth. J Cell Mol Med. 19:1262–1272. 2015.
View Article : Google Scholar : PubMed/NCBI
|