Introduction
Osteosarcoma (OS) is the most common primary
malignant bone tumor, and affects all ages, but especially two
age-groups, namely adolescents and, to a lesser extent, the elderly
(7th and 8th decades) (1,2). OS can arise in any bone but
preferentially affects the metaphyses of long bones, such as the
distal femur, proximal tibia, and proximal humerus (3). OS is highly aggressive with rapid
growth, local invasion and early metastasis. In the past, OS was
rarely curable, but it now has a 5-year overall survival rate of
70–80%, due to modern multimodal therapeutic strategies (4,5);
however, in the last two decades, there has been no substantial
improvement in the prognosis of OS patients with metastases or
relapse, and pulmonary metastasis remains the main cause of death
in OS patients (6,7). Therefore, it is essential to find new
valuable pathogenic factors associated with OS, which can deepen
the understanding of this neoplasm, help early diagnosis and
improve prognosis.
Provirus integrating site Moloney murine leukemia
virus (PIM) proteins are a family of short-lived, highly conserved
serine/threonine kinases with three members: PIM1, PIM2 and PIM3.
These kinases are highly homologous at the amino acid level, and
many of their functions overlap with each other. PIM kinases have
unique structural properties and are mainly regulated by
transcription and translation (8,9). The
Janus-activated kinase/signal transducers and activators of
transcription (JAK/STAT) pathway, which is activated by various
cytokines and hormones, plays an important role in regulating the
expression of PIM proteins (8).
Using transgenic mouse models, PIM kinases have been shown to have
carcinogenic potential, especially when they collaborate with other
oncogenes such as c-Myc (avian myelocytomatosis viral oncogene
homolog), N-Myc, Bcl2 (B-cell CLL/lymphoma 2), GFI1 (growth factor
independent 1 transcriptional repressor), and Frat1 (frequently
rearranged in advanced T-cell lymphomas-1) in tumorigenesis
(10). Moreover, expression of the
three PIM kinases is found to be increased in various tumors,
mainly in hematopoietic malignancies (11), and have recently been shown to be
over-expressed in numerous solid tumors (e.g., prostate cancers,
gastric carcinoma, pancreatic cancers, bladder cancer, Ewing's
sarcoma and liposarcoma) (12–18),
their expression patterns correlate with the diagnosis and
prognosis of the various pathologies (15,19).
PIM kinases mediate their oncogenic activity in
tumor cells by modifying a variety of cellular substrates, and the
responses are either isoform-specific or common to the three
kinases (8). PIM kinases regulate
cell cycle progression by directly phosphorylating p21
(cyclin-dependent kinase inhibitor 1A), p27 (cyclin-dependent
kinase inhibitor 1B), Cdc25A (Cell division cycle 25A), and Cdc25C
(Cell division cycle 25C) (20–23).
They can block cell apoptosis through the regulation of BAD
(Bcl-2-associated agonist of cell death) (24,25),
and can also affect cell motility by modifying NFATc1 (nuclear
factor of activated T-cells, cytoplasmic 1), CXCR4 (C-X-C chemokine
receptor type 4), GSK3B (glycogen synthase kinase 3β) and FOXP3
(forkhead box P3) (26–28). Additionally, PIM kinases may be
involved in early tumorigenesis, primarily by interaction of PIM1
with nuclear mitotic apparatus protein (NUMA), which has been shown
to result in genomic instability (29).
Inhibition of PIM kinases has become a promising
approach for cancer therapy (30,31).
This is because they have unique structural features that enable
the design of highly selective inhibitors (32), which do not cause serious side
effects, supported by the fact that mice deficient for all three
PIM family members are viable and fertile (33). Furthermore, inhibitors of PIM
kinases can sensitize cancer cells to chemotherapy and may
synergize with other antitumor agents (34).
However, the roles of PIM kinases in OS remain
largely unknown. In this study, we examined all three PIM kinases
in 43 paraffin-embedded OS samples and analyzed their expression
patterns associated with clinicopathological features to elucidate
their clinical significance. Furthermore, we performed in
vitro experiments to evaluate their biological effects on OS
cells.
Materials and methods
Clinical specimens
This study was approved by the Research Ethics
Committee of Shengjing Hospital (IRB number, 2015PS203K). A total
of 45 cases of paraffin-embedded OS tumor specimens were obtained
from the department of pathology, Shengjing Hospital of China
Medical University, between 2007 and 2015. Among them, one case of
extra-skeletal OS and one case of sclerotic OS were excluded. Thus,
43 patients were included in this study. All 43 patients had
primary OS, and all specimens were stained with hematoxylin and
eosin (H&E) to verify the targeted tissues. Clinical stages of
these OS patients were classified according to the guidelines of
the American Joint Committee on Cancer. Clinical information was
obtained by reviewing medical records. A total of 21 patients
received adjuvant chemotherapy, four patients received
postoperative radiation therapy, two patients received both
adjuvant chemotherapy and radiation therapy, seven patients
received no adjuvant treatment, and the adjuvant treatment for nine
patients was unknown. Alkaline phosphatase results of five patients
were not available. Follow-up was terminated April 25, 2016. The
median time of follow-up was 37 months (6–86 months). Overall
survival was defined as the interval between surgery and death or
the last observation taken.
Immunohistochemistry and evaluation of
staining
The immunohistochemical analysis steps were as
follows: after heat-induced antigen retrieval, tissue sections were
deparaffinized in xylene and rehydrated in graded alcohols.
Antigens were retrieved by boiling the tissue sections in citrate
buffer (pH 6.0) for 3 min, followed by successive rinses in
phosphate-buffered saline (PBS). Endogenous peroxidase was
inhibited by incubating the sections in 3% hydrogen peroxide for 30
min. Non-specific staining was blocked by incubation with 5–10%
bovine serum albumin at room temperature for 20 min. Slides were
incubated overnight with anti-PIM1 goat polyclonal antibody (1:250;
Santa Cruz, Dallas, TX, USA), anti-PIM2 (1:400; GeneTex, Irvine,
CA, USA) and anti-PIM3 (1:400; Abcam, Cambridge, MA, USA) rabbit
polyclonal antibodies at 4°C. After washing with PBS, slides were
incubated with the corresponding secondary antibody for 30 min at
room temperature. Slides were then incubated with avidin
horseradish peroxidase and the DAB (Diaminobenzidine, JSGB-BIO,
Beijing, China) substrate. Finally, sections were counterstained
with hematoxylin. PBS was used to replace anti-PIM antibodies in
negative controls.
Immunostaining results were independently evaluated
by two observers (Shuai Mou and Ding Ding) who were blinded to the
clinicopathological features. The extent of immunoreactivity for
PIM kinases was considered by both staining intensity and staining
extent, as previously described (35); the staining intensity was scored as
0 (negative), 1 (mild), 2 (moderate), or 3 (strong). The percentage
of positive cells in the whole tumor area was scored as 0 (0%), 1
(1–25%), 2 (26–50%), 3 (51–75%), or 4 (76–100%). The sum of the
staining intensity and extent scores was used as the final staining
scores (0–7), and we considered samples having a final score of ≥3
as positive.
Cell lines and cell culture
Three OS cell lines, including MG-63, U2OS and
MNNG/HOS were obtained from the cell bank of Shanghai Biology
Institute, Chinese Academy of Science. MG-63 and MNNG cells were
cultured in DMEM (Hyclone, UT, USA) with 10% fetal bovine serum
(Biological Industries) and 1% penicillin and streptomycin
(Hyclone). U2OS cells were grown in RPMI 1640 medium (Hyclone) with
10% fetal bovine serum (Biological Industries, Haemek, Israel) and
1% penicillin and streptomycin (Hyclone).
Cell proliferation assay
Human OS cells (1×104/well) were plated
in 96-well plates in DMEM containing 10% FBS at a density of
1×105 cells/ml and incubated for 24, 36, 48 or 60 h. At
the end of the incubations, cellular proliferation was measured by
the 3-(4,5)-(dimethylthiazol-zyl)-3,5-diphenyltetrazolium
bromide (MTT) assay. MTT crystals were dissolved in 150 μl of DMSO.
After a 4-h incubation at 37°C, the optical densities at 490 nm
were measured using a Microplate Reader (Bio-Rad, CA, USA).
Cell transfection
To knock down the expression of PIM kinases, three
siRNA, which specifically targeted PIM1, PIM2 and PIM3 (PIM1:
5′-GGUGUGUGGAGAUAUUCCUTT-3′, PIM2: 5′-CUGCUUGACUGGUUUGAGATT-3′ and
PIM3: 5′-UCGUGCACCGCGACAUUAATT-3′), and negative control siRNA
(siCont) were designed and synthesized by Genepharma (Shanghai,
China). For overexpression, three pcDNA3.1/V5-HisC plasmids for
expressing human PIM kinase genes were kindly provided by Professor
Päivi M. Ojala (University of Helsinki, Helsinki, Finland), as
previously described (36). Cells
were transfected with siRNA or plasmids using Lipofectamine 2000
(Invitrogen, CA, USA), according to the manufacturer's protocol.
For RNA extraction and western blot assays, cells were used at 24
or 48 h after transfection.
Cell migration and invasion assay
Assays were performed using modified Boyden chambers
with polycarbonate nucleopore membranes. Pre-coated filters (6.5 mm
in diameter, 8-μm pore size, Matrigel 100 μg/cm2) were
rehydrated with 100 μl medium. Then, 1×105 cells in 100
μl serum-free DMEM supplemented with 0.1% bovine serum albumin were
placed in the upper part of each chamber, whereas the lower
compartments were filled with 600 μl DMEM containing 10% serum.
After incubation for 24 h at 37°C, non-invaded cells were removed
from the upper surface of the filter using a cotton swab, and the
invaded cells on the lower surface of the filter were fixed,
stained, photographed and counted under high-power
magnification.
Real-time PCR analysis
Total RNA was extracted from the cells using TRIzol
reagent (Invitrogen). The concentration and purity of the total-RNA
were calculated by absorbance at 260 and 280 nm. Single-strand cDNA
synthesis was performed using the PrimeScript RT Reagent kit
(Takara Bio, Shiga, Japan). Real-time PCR was performed using SYBR
Premix Ex Taq (Takara Bio) on an Mx 3000P real-time PCR system
(Applied Biosystems, MA, USA) using the following conditions: 50
cycles of 95°C for 10 sec and 60°C for 30 sec. All the reactions
were repeated at least three times. PCR primers were designed using
Primer 5.0 software and the sequences are listed in Table I.
 | Table IPrimer sequences for detection of
mRNA expression. |
Table I
Primer sequences for detection of
mRNA expression.
| Genes | Primers | Primer sequence
(5′-3′) |
|---|
| PIM1 | Forward |
GCTCGGTCTACTCAGGCATC |
| Reverse |
GCTCCCCTTTCCGTGATGAA |
| PIM2 | Forward |
CGTGGAGTTGTCCATCGTG |
| Reverse |
AAGGGAATGTCCCCACACAC |
| PIM3 | Forward |
GTACAGTCTGCTTGTGGGCT |
| Reverse |
GAAAGAACCCCCATCTGCGA |
| Cyclin D1 | Forward |
CCGAGGAGCTGCTGCAAATGGAGCT |
| Reverse |
TGAAATCGTGCGGGGTCATTGCGGC |
| MMP2 | Forward |
CGCATCTGGGGCTTTAAACAT |
| Reverse |
TCAGCACAAACAGGTTGCAG |
| GAPDH | Forward |
CTCTGCTCCTCCTGTTCGAC |
| Reverse |
GCGCCCAATACGACCAAATC |
Western blot analysis
Total protein was extracted in RIPA lysis buffer
(Beyotime, Shanghai, China), quantified by the BCA protein assay
(Beyotime), and equal amounts of protein were separated by SDS-PAGE
(sodium dodecyl sulfate-polyacrylamide gel electrophoresis) and
transferred to PVDF (polyvinylidene fluoride) membranes (Millipore,
MA, USA). Membranes were blocked with 5% nonfat milk in PBS for 1 h
at room temperature and then incubated with PIM1, PIM2, PIM3,
Cyclin D1 (Santa Cruz) and MMP2 (Santa Cruz) antibodies overnight
at 4°C. After extensive washing, membranes were re-probed with
horseradish peroxidase-conjugated secondary antibodies (Boster
Wuhan, China). Membranes were developed using ECL or ECL plus
(Thermo Fisher Scientific, MA, USA), according to the
manufacturer's instructions.
Statistical analysis
All statistical analyses were performed using
SPSS19.0. The Chi-square test was applied to analyze the
correlation between the expression of PIM kinases and the
clinicopathological features. Immunohistochemistry (IHC) scores of
PIM kinases was calculated by multiplying the percentage and
intensity scores, and the cut-off point was considered by a final
score of ≥3 as positive. The Kaplan-Meier method was used for
survival analysis, and differences in survival were estimated using
the log-rank test. Both univariate and multivariate analyses were
carried out to identify independent prognostic factors for survival
using the Cox proportional hazards model. The two-tailed, unpaired
Student's t-test was used to compare differences between
experimental and control groups in the in vitro assays.
Results are shown as mean values with 95% confidence intervals.
Error bars represent standard deviations. A value of P<0.05 was
considered statistically significant.
Results
All three PIM kinases are expressed in
the majority of OS tumor samples
To explore whether PIM kinases were expressed in OS,
we performed immunohistochemistry to detect the protein expression
of the three kinase proteins in 43 paraffin-embedded OS samples. In
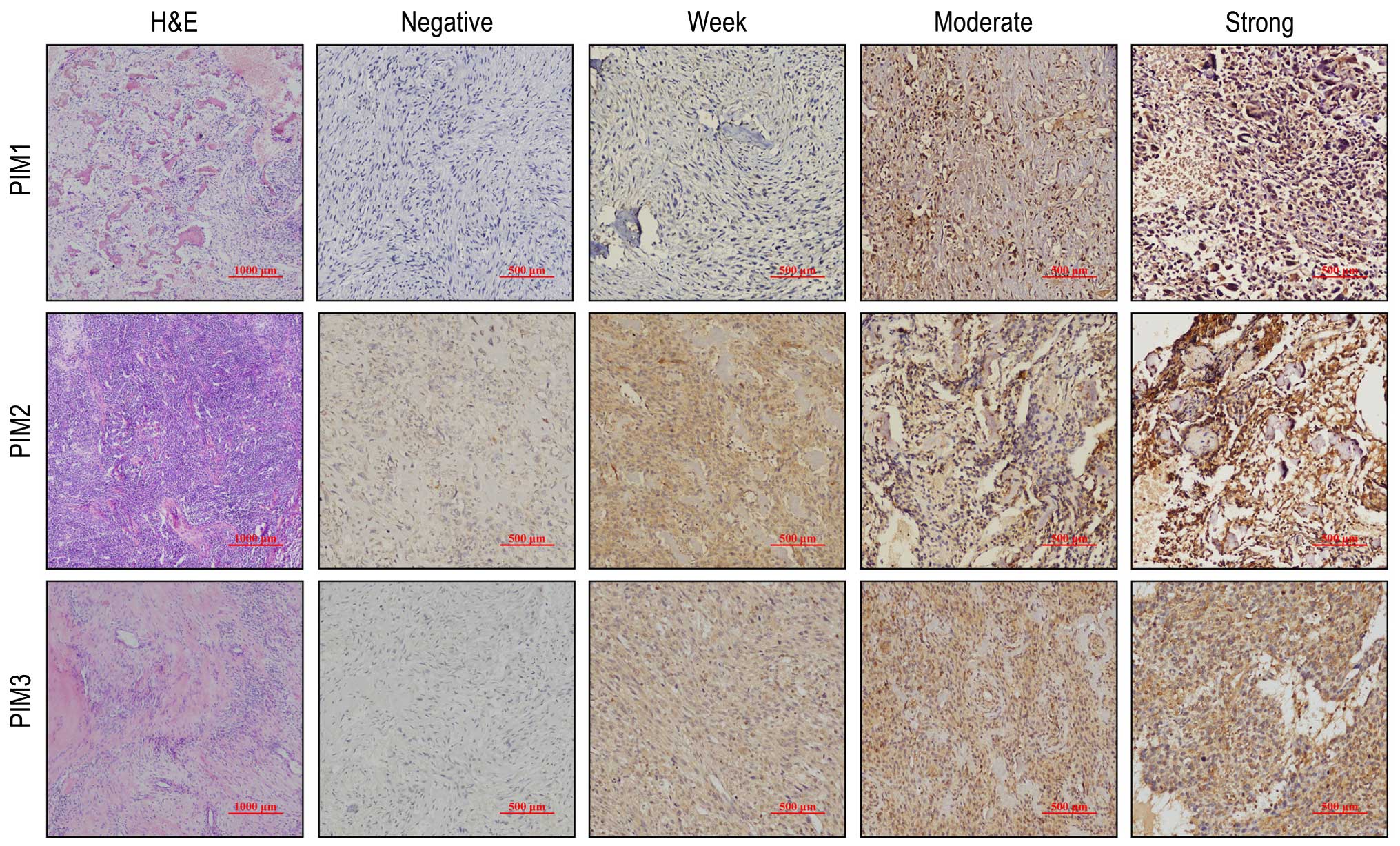
the OS tissues, expression of PIM1, PIM2 and PIM3 was observed as a
brown-yellow color, localized both in the nuclei and cytoplasm. The
expression intensities of PIM kinases varied from negative-
to-weak, medium and strong staining (Fig. 1). According to our IHC score (see
Materials and methods), 76.7% (33/43), 74.4% (32/43) and 69.7%
(30/43) of cases were PIM1-positive, PIM2-positive and
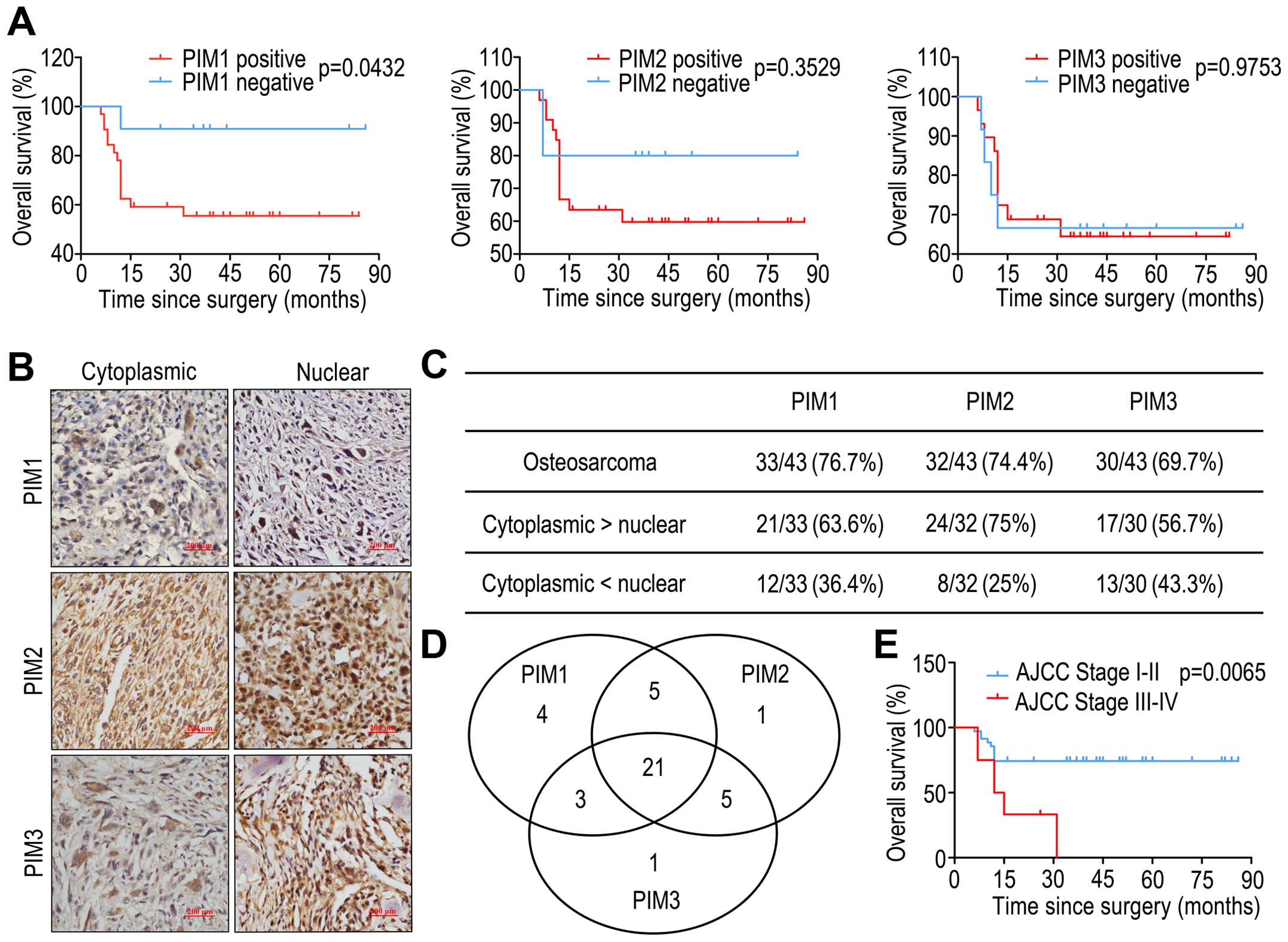
PIM3-positive, respectively (Fig.
2C). Of the 43 cases, 21 cases expressed all three PIM kinases,
and 3 cases did not express the PIM kinases. Of the remaining
cases, 11 cases were positive for either PIM1 alone or co-expressed
with PIM2 or PIM3, 11 cases were positive for PIM2 alone or in
combination with PIM1 or PIM3, and 9 cases were positive for PIM3
alone or together with PIM1 or PIM2 (Fig. 2D).
Investigation of the expression patterns
of PIM kinases and the clinical significance in OS
In order to elucidate the clinical significance of
PIM kinase expression, we evaluated the association between the
positive expression of the three PIM kinases and the
clinicopathological characteristics, including sex, age, location,
pathologic subtype, tumor size, tumor stage and alkaline
phosphatase (ALP). However, there was no significant correlation
with these clinicopathological parameters and positive expression
of the three PIM kinases (Table
II). We also observed the association of the expression of the
three PIM kinases and the clinical outcome of OS patients.
Kaplan-Meier analysis showed that only positive expression of PIM1
showed a poorer prognosis than patients with negative expression of
PIM1 (log-rank test, P=0.0432 in overall survival), but there were
no significant differences between positive and negative expression
of PIM2 and PIM3 (Fig. 2A).
Kaplan-Meier analysis by AJCC tumor stage is shown in Fig. 2E. We then performed univariate and
multivariate Cox regression survival analysis, with the variables
including sex, age, pathologic subtype, tumor size, tumor stage,
and positive expression of ALP, PIM1, PIM2 and PIM3. In the
univariate analysis, only the tumor stage of OS was a significant
predictor of poor prognosis, while the other indicators, including
PIM1, PIM2 and PIM3 expression were not good predictors for overall
survival of OS patients, according to both univariate and
multivariate analysis (data not shown).
 | Table IICorrelation between the Pim kinase
expression and different clinicopathological features in 43
osteosarcoma patients.a |
Table II
Correlation between the Pim kinase
expression and different clinicopathological features in 43
osteosarcoma patients.a
| | PIM1 | PIM2 | PIM3 |
|---|
| |
|
|
|
|---|
|
Characteristics | No. of
patients | Positive | P-value | Positive | P-value | Positive | P-value |
|---|
| Gender |
| Male | 16 | 13 | 0.590 | 11 | 0.512 | 10 | 0.424 |
| Female | 27 | 20 | | 21 | | 20 | |
| Age (years) |
| <30 | 29 | 21 | 0.333 | 22 | 0.755 | 22 | 0.210 |
| ≥30 | 14 | 12 | | 10 | | 8 | |
| Site of primary
disease |
| Femur | 17 | 13 | 0.957 | 13 | 0.931 | 9 | 0.097 |
| Tibia | 10 | 8 | | 7 | | 7 | |
| Other | 16 | 12 | | 12 | | 14 | |
| AJCC Stage |
| I–II | 35 | 25 | 0.165 | 27 | 0.392 | 24 | 0.721 |
| III–IV | 8 | 8 | | 5 | | 6 | |
| Metastasis |
| Absence | 35 | 25 | 0.165 | 27 | 0.392 | 24 | 0.721 |
| Presence | 8 | 8 | | 5 | | 6 | |
| Tumor size, cm |
| ≤8 | 29 | 23 | 0.566 | 21 | 0.665 | 20 | 0.869 |
| >8 | 14 | 10 | | 11 | | 10 | |
| Histologic
type |
| Conventional | 37 | 29 | 0.529 | 26 | 0.312 | 24 | 0.155 |
| Special | 6 | 4 | | 6 | | 6 | |
| ALP |
| Normal range | 24 | 18 | 0.809 | 17 | 0.601 | 18 | 0.482 |
| Beyond normal
range | 14 | 10 | | 11 | | 9 | |
Subcellular distribution of PIM kinases was found to
be related to their biological function and disease stage (37,38),
we therefore investigated the nuclear and cytoplasmic expression
patterns of the three PIM kinases in our OS samples (Fig. 2B). Nuclear staining of positive
expression of PIM1, PIM2 and PIM3 was 36.4% (12/33), 25% (8/32) and
43.3% (13/30), respectively (Fig.
2C). However, we observed that neither nuclear nor cytoplasmic
expression of the three PIM kinases was significantly correlated
with OS tumor stage or prognosis (data not shown).
PIM kinases promote proliferation of OS
cells through cyclin D1 regulation
We then examined the biological effects of PIM
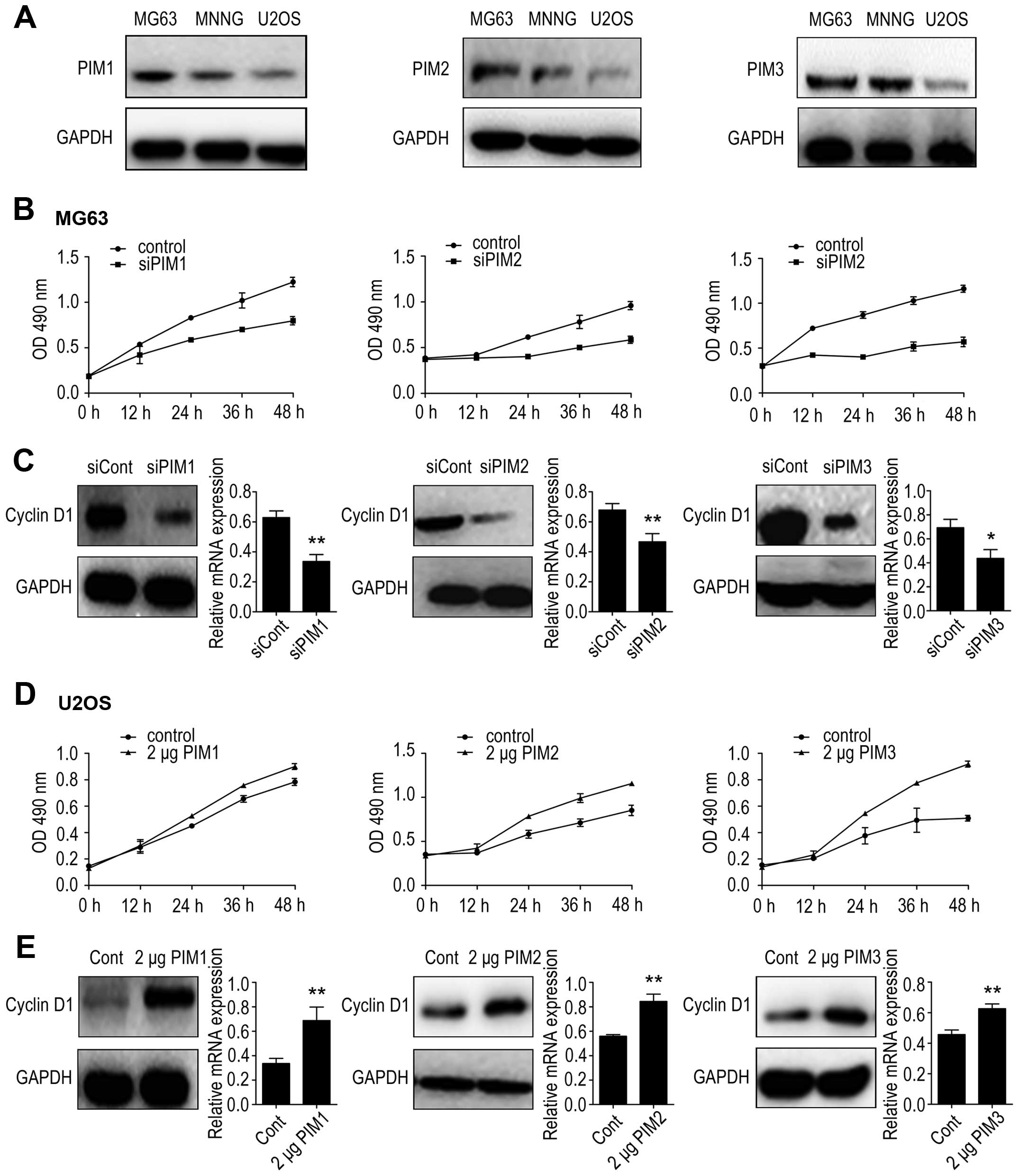
kinases on OS cells. Firstly, we measured the basal protein levels
of the three PIM kinases in the three OS cell lines by western
blotting. We found that the protein levels of PIM1, PIM2 and PIM3
were relatively higher in MG-63 cells than those in MNNG/HOS cells
or U2OS cells (Fig. 3A). Thus, we
chose the MG-63 cell line for use in RNA interference studies and
the U2OS cell line for use in plasmid overexpression studies.
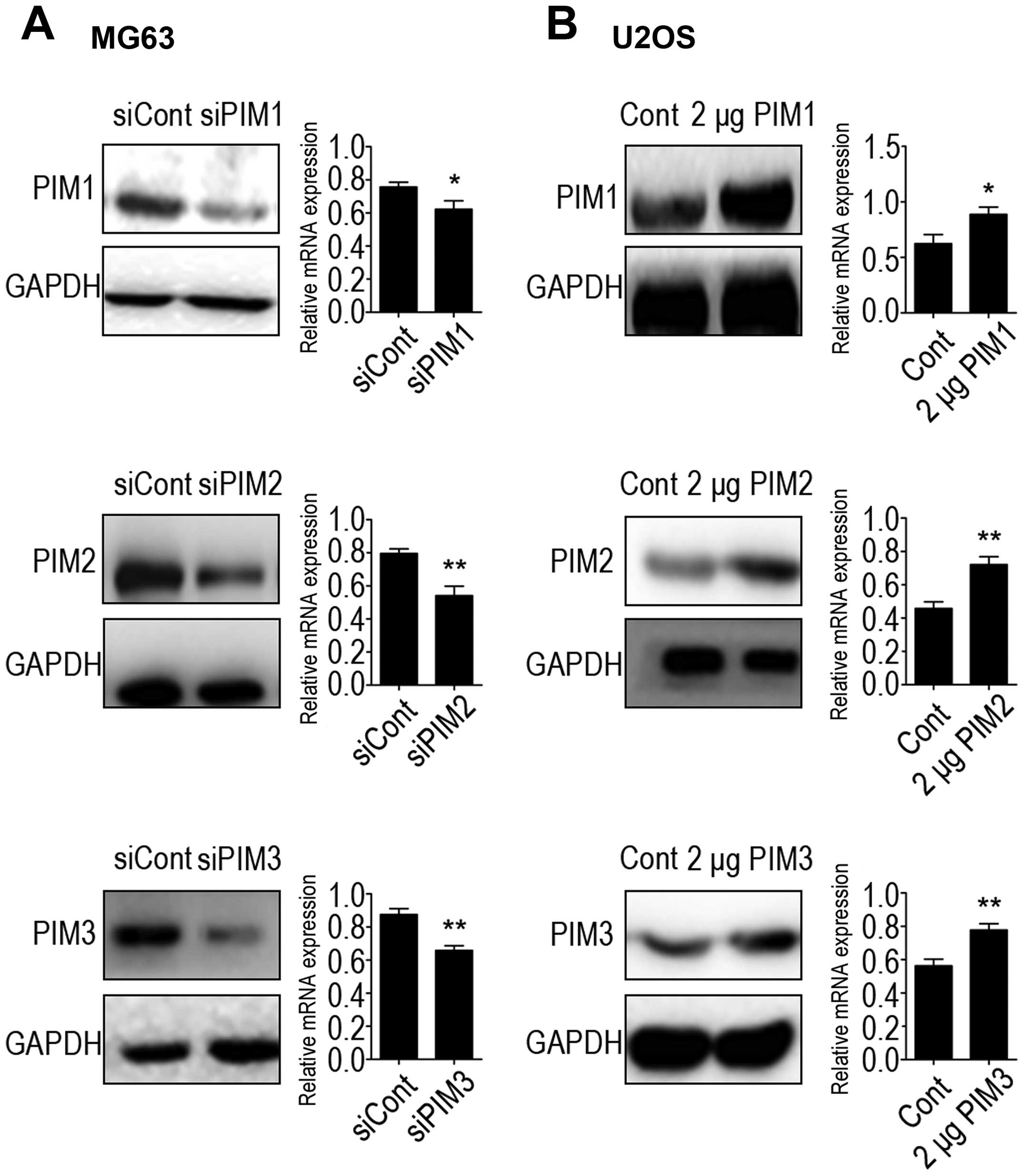
Downregulation and increased expression of PIM1, PIM2 and PIM3 was
verified at both the mRNA and protein expression levels (Fig. 4). Cell proliferation rates were
measured using the MTT assay and results were determined at 0, 12,
24, 36 and 48 h post-transfection. Results showed that PIM1
knockdown inhibited MG-63 cell growth to 69 and 65% of the controls
at 36 and 48 h, respectively; PIM2 knockdown inhibited MG-63 cell
growth to 64 and 61% of the controls at 36 and 48 h, respectively,
and PIM3 knockdown inhibited MG-63 cell growth to 55 and 54% of the
controls at 36 and 48 h, respectively (Fig. 3B). Conversely, PIM1, PIM2 and PIM3
overexpression in U2OS cells exhibited a significantly higher rate
of proliferation at 48 h post-transfection (Fig. 3D).
It has been recently reported that the association
between the three PIM kinases can mediate tumor cell proliferation
and cyclin D1 expression (39–41).
To further investigate these underlying mechanisms, we examined the
protein and mRNA levels of cyclin D1 after PIM knockdown or
overexpression. PIM1 depletion decreased cyclin D1 expression in
MG-63 cells (Fig. 3C); accordingly
PIM1 overexpression increased cyclin D1 expression in U2OS cells.
Similar results were also observed in the case of PIM2 and PIM3
(Fig. 3E). Taken together, these
results demonstrated that all three PIM kinases affected the
proliferation of OS cells, possibly via cyclin D1 regulation.
PIM kinases promote the migratory and
invasive potential of OS cells by modulating MMP2
We also investigated the effects of depletion of PIM
kinases on OS cell motility; cell migration and invasion were
examined by Transwell assays. Results showed that, compared with
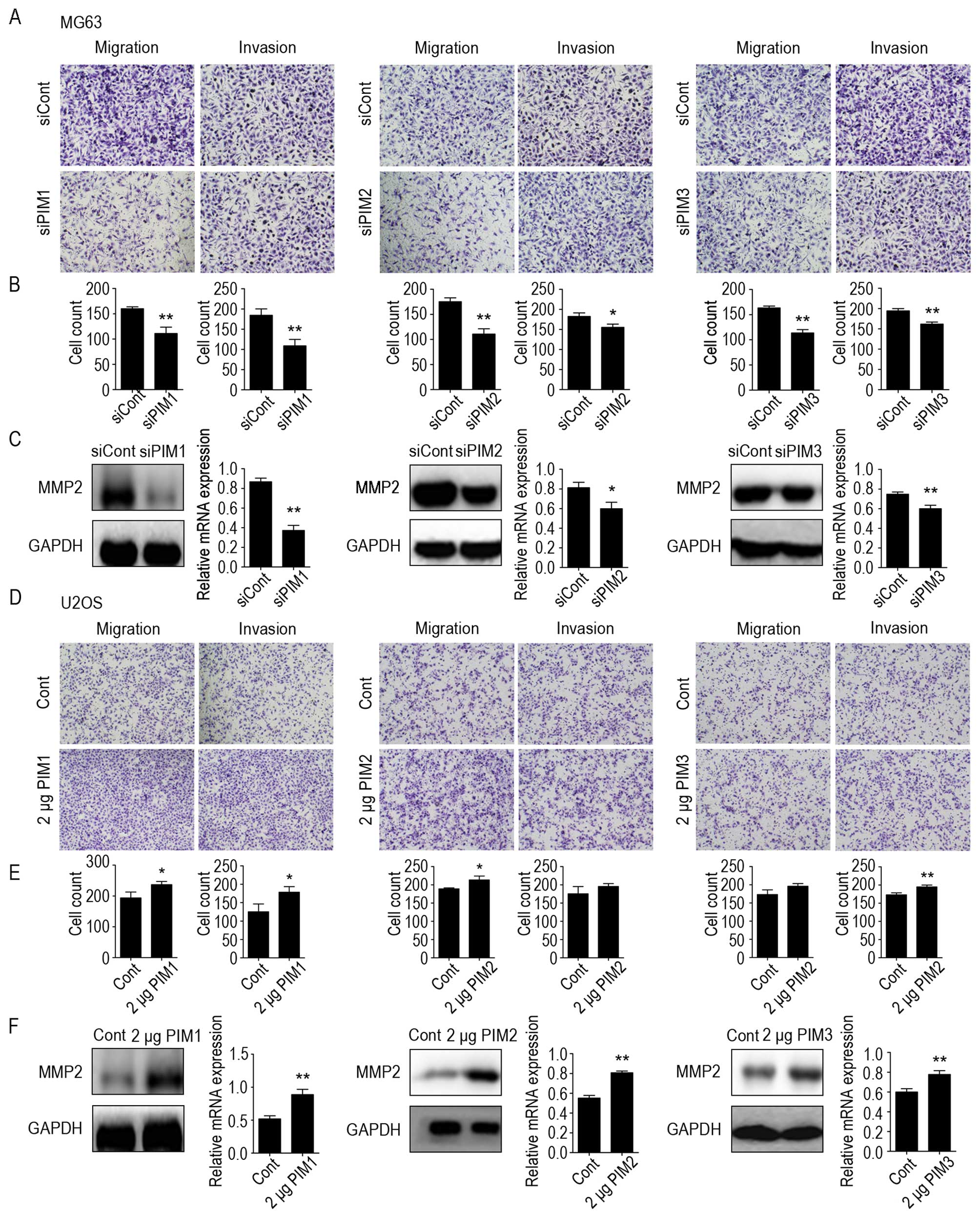
the control cells, PIM1, PIM2 and PIM3 knockdown significantly
inhibited MG-63 cell migration by 69, 63 and 70%, respectively
(Fig. 5A and B). Moreover,
deficiency of the three kinases markedly inhibited the invasiveness
of MG-63 cells by 59, 85 and 83%, respectively (Fig. 5A and B). Accordingly, upregulation
of PIM1, PIM2 and PIM3 expression promoted the migration and
invasion of U2OS cells (Fig. 5D and
E).
It is well known that matrix metalloproteinases 2
(MMP2) can facilitate enhanced tumor cell invasion and metastasis
potential (42). Therefore, we
investigated the expression levels of MMP2 in PIM kinase-knockdown
MG63 cells and in PIM kinase-overexpressing U2OS cells by western
blotting and real-time PCR analysis. Results revealed that MMP-2
protein and mRNA levels decreased in PIM kinase-knockdown cells
compared with the negative controls, whereas the cells
over-expressing PIM kinase showed an increase in MMP2 levels
(Fig. 5C and F). These results
suggest that all three PIM kinases can enhance the motility of OS
cells, at least in part, by modulating MMP-2.
Discussion
PIM kinases have been identified as proto-oncogenes
implicated in tumorigenesis, and their expression and functions
have been demonstrated in various tumors (8,10).
However, their role in OS has not been clearly defined. In this
study, we found that all three PIM kinases were frequently
expressed in OS tissues. We also showed that positive expression of
PIM1 was associated with poorer prognosis for overall survival of
OS patients, while the positive expression of PIM2 and PIM3 was not
found to predict prognosis. The subcellular distributions of the
three PIM kinases were not correlated with the disease stage or
clinical outcome. In addition, our in vitro study
demonstrated that knockdown of PIM kinases effectively inhibited OS
cell proliferation, migration and invasion, whereas overexpression
of PIM kinases resulted in increased OS cell growth and
motility.
The literature on expression of PIM kinases in OS is
extremely limited, Lu et al reported that the PIM1 gene
locus (6p12–21) is a common amplification event in OS (43). More recently, during the course of
this work, Liao et al reported five out of seven OS tissue
samples had high expression of PIM1 protein, and 77.2% (88/144) OS
specimens from 70 patients exhibited PIM1 immunostaining on the OS
cell nucleus (44). Both of these
studies indicated that PIM1 is expressed in OS and may therefore
play an important role. Nevertheless, in this present study, we
demonstrated that all three PIM kinases were expressed in a
majority of OS samples, but only positive expression of PIM1 was
associated with poorer prognosis for overall survival of OS
patients. The prognostic value of PIM1 in OS patients was also
reported in the previous study (44). However, our univariate and
multivariate Cox regression analyses indicated the three PIM
kinases are not good predictors for overall survival of OS
patients. This conflict may be due to our small sample size, as OS
is relative rare (1,45,46).
Therefore, an additional study with a larger number of cases is
needed to confirm the clinical significance of the expression of
the three PIM kinases, particularly PIM2 and PIM3, in OS
patients.
Some studies have reported that nuclear accumulation
of PIM1 is necessary for its biologic effects and may correlate
with tumor progression and disease stage (37,38).
In the previous study, Liao et al reported that the cellular
34 kDa PIM1 isoform staining in OS tissues was predominantly
nuclear (44). However, in our
study, nuclear PIM1 localization was not dominant in our OS
samples, and PIM1 nuclear staining accounted for 36.4% in the PIM1
positive group. Furthermore, we observed all three PIM kinases are
distributed to both the nucleus and cytoplasm, and their expression
patterns are uncorrelated with OS tumor stage or prognosis (data
not shown). Notably, the shuttling of subcellular distribution of
PIM kinases between the nucleus and the cytoplasm is complicated
and not well understood (38).
Cytoplasmic sequestration of PIM1 may also be relevant and can
promote cancer cell survival (47). Thus, further investigations are
needed to clarify the functions of different subcellular
distributions of PIM kinases and the underlying mechanisms,
particularly in OS.
PIM kinases are associated with cell proliferation
as they prevent apoptosis and promote cell survival (8). Consistently, our in vitro
study revealed that all three PIM kinases can promote OS cell
proliferation, while silencing PIM kinases can decrease cell
proliferation rates. The underlying mechanism may be related to the
association of PIM kinase with cyclin D1. Previous studies have
found that when PIM kinases were inhibited, the expression of
cyclin D1 was reduced and the change in cyclin D1 expression could
influence cell cycle progression and cancer cell proliferation
(39–41). Furthermore, we found that
knock-down of any of the three PIM kinases resulted in decreased
cyclin D1 expression in OS cells, whereas overexpression of PIM
kinases in OS cells increased cyclin D1 expression. Together, these
results indicate that all three PIM kinases may have a role in the
growth of OS cells.
We also investigated the association of the three
PIM kinases with the motility of OS cells. Our results showed that
OS cells in which the three PIM kinases had been knocked down had
lower migration and invasion. In contrast, overexpression of any
PIM kinase family member had the opposite effect, displaying
increased migration and invasion of OS cells. These results suggest
that PIM kinases may affect OS metastasis. In agreement with this,
previous studies have suggested a role for PIM kinases in cell
motility and metastasis, for example, PIM1 was able to promote
migration in leukemia and prostate cancer cells (48), PIM2 was shown to induce human liver
cell malignant transformation and increase the migration rate
(49), and PIM3 can promote
ovarian cancer cell migration (50). In addition, absence of PIM kinases
blocks the process of bone invasion induced by the
3-methylcholanthrene-induced sarcoma in vivo (51). However, the regulatory mechanisms
involved in controlling cell motility by the three PIM kinases are
not well known; in this study, we found that PIM kinase depletion
downregulated MMP2 expression, whereas cells overexpressing PIM
kinase showed an increase in MMP2 levels, suggesting that MMP2 may
play a role in regulating OS cell motility by PIM kinases.
Recently, multiple substrates of PIM kinases that
promoted cancer cell metastasis were identified, including CXCR4,
NFATc1, GSK3B and FOXP3 (26). Of
these, CXCR4 was the first to be confirmed and the most studied.
PIM1, but not PIM2 and PIM3, was shown to regulate homing and
migration of leukemia cells via modulation of stromal-derived
factor-1alpha (SDF-1/CXCL12)-CXCR4 signaling (27,52).
Subsequent research has demonstrated that PIM1 and PIM3, but not
PIM2, can phosphorylate CXCR4 and promote metastatic properties of
prostate cancer. Importantly, overexpressing PIM1 or PIM3 may take
advantage of the CXCL12/CXCR4 chemokine pathway and stimulate the
formation of lung metastases from orthotopically-induced prostate
tumors in mice (53). It is known
that CXCR4 expression plays an important role in the pulmonary
metastatic process in OS (54,55).
Therefore, the association between PIM kinases and CXCR4 suggests
that PIM kinases may also be involved in the lung metastasis of OS,
but this remains to be elucidated. Unfortunately, in this study, we
were unable to verify the expression of PIM kinases in OS
metastasis, especially lung metastasis, which are the main cause of
death in OS patients. Future studies are needed to substantiate
this intriguing possibility.
In conclusion, in this study, we show for the first
time, that all three PIM kinases are frequently expressed in OS.
Although the expression patterns and the functions of PIM kinases
in OS are not completely understood, we found that positive
expression of PIM1 was associated with poorer prognosis, which
supports the use of PIM1 as a potential prognostic biomarker of OS.
Moreover, in our in vitro study, we found that all three PIM
kinases can significantly affect OS cell proliferation, migration
and invasion, which suggests that PIM kinases have multiple
biological functions on OS cells, and could therefore serve as
potential therapeutic targets in OS. However, more studies are
needed to elucidate the oncogenic roles of PIM kinases and their
clinical significance in OS.
Acknowledgements
This study was supported by Outstanding Scientific
Fund of Shengjing Hospital and two grants from the National Natural
Science Foundation of China (81370981 and 81300714). We acknowledge
Professor Päivi M. Ojala (University of Helsinki, Helsinki,
Finland) for providing us with the PIM pcDNA3.1/V5-HisC
plasmids.
References
|
1
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004: Data
from the Surveillance, Epidemiology, and End Results Program.
Cancer. 115:1531–1543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jawad MU, Cheung MC, Clarke J, Koniaris LG
and Scully SP: Osteosarcoma: Improvement in survival limited to
high-grade patients only. J Cancer Res Clin Oncol. 137:597–607.
2011. View Article : Google Scholar
|
|
3
|
Bielack SS, Kempf-Bielack B, Delling G,
Exner GU, Flege S, Helmke K, Kotz R, Salzer-Kuntschik M, Werner M,
Winkelmann W, et al: Prognostic factors in high-grade osteosarcoma
of the extremities or trunk: An analysis of 1,702 patients treated
on neoadjuvant cooperative osteosarcoma study group protocols. J
Clin Oncol. 20:776–790. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hegyi M, Semsei AF, Jakab Z, Antal I, Kiss
J, Szendroi M, Csoka M and Kovacs G: Good prognosis of localized
osteosarcoma in young patients treated with limb-salvage surgery
and chemotherapy. Pediatr Blood Cancer. 57:415–422. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Anderson ME: Update on survival in
osteosarcoma. Orthop Clin North Am. 47:283–292. 2016. View Article : Google Scholar
|
|
6
|
Zwaga T, Bovee JV and Kroon HM:
Osteosarcoma of the femur with skip, lymph node, and lung
metastases. Radiographics: A review publication of the Radiological
Society of North America Inc. 28:277–283. 2008. View Article : Google Scholar
|
|
7
|
Miller BJ, Cram P, Lynch CF and Buckwalter
JA: Risk factors for metastatic disease at presentation with
osteosarcoma: An analysis of the SEER database. J Bone Joint Surg
Am. 95:e892013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Narlik-Grassow M, Blanco-Aparicio C and
Carnero A: The PIM family of serine/threonine kinases in cancer.
Med Res Rev. 34:136–159. 2014. View Article : Google Scholar
|
|
9
|
Nawijn MC, Alendar A and Berns A: For
better or for worse: The role of Pim oncogenes in tumorigenesis.
Nat Rev Cancer. 11:23–34. 2011. View
Article : Google Scholar
|
|
10
|
Aguirre E, Renner O, Narlik-Grassow M and
Blanco-Aparicio C: Genetic modeling of PIM proteins in cancer:
Proviral tagging and cooperation with oncogenes, tumor suppressor
genes, and carcinogens. Front Oncol. 4:1092014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Alvarado Y, Giles FJ and Swords RT: The
PIM kinases in hematological cancers. Expert Rev Hematol. 5:81–96.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guo S, Mao X, Chen J, Huang B, Jin C, Xu Z
and Qiu S: Overexpression of Pim-1 in bladder cancer. J Exp Clin
Cancer Res. 29:1612010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Reiser-Erkan C, Erkan M, Pan Z, Bekasi S,
Giese NA, Streit S, Michalski CW, Friess H and Kleeff J:
Hypoxia-inducible proto-oncogene Pim-1 is a prognostic marker in
pancreatic ductal adenocarcinoma. Cancer Biol Ther. 7:1352–1359.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Brault L, Gasser C, Bracher F, Huber K,
Knapp S and Schwaller J: PIM serine/threonine kinases in the
pathogenesis and therapy of hematologic malignancies and solid
cancers. Haematologica. 95:1004–1015. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nga ME, Swe NN, Chen KT, Shen L, Lilly MB,
Chan SP, Salto-Tellez M and Das K: PIM-1 kinase expression in
adipocytic neoplasms: Diagnostic and biological implications. Int J
Exp Pathol. 91:34–43. 2010. View Article : Google Scholar
|
|
16
|
Holder SL and Abdulkadir SA: PIM1 kinase
as a target in prostate cancer: Roles in tumorigenesis, castration
resistance, and docetaxel resistance. Curr Cancer Drug Targets.
14:105–114. 2014. View Article : Google Scholar
|
|
17
|
Warnecke-Eberz U, Bollschweiler E, Drebber
U, Metzger R, Baldus SE, Hölscher AH and Mönig S: Prognostic impact
of protein overexpression of the proto-oncogene PIM-1 in gastric
cancer. Anticancer Res. 29:4451–4455. 2009.PubMed/NCBI
|
|
18
|
Albertson DJ, Schmidt RL, Bearss JJ, Tripp
SR, Bearss DJ and Liu T: Patterns and significance of PIM kinases
in urothelial carcinoma. Appl Immunohistochem Mol Morphol.
23:717–723. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Peng YH, Li JJ, Xie FW, Chen JF, Yu YH,
Ouyang XN and Liang HJ: Expression of pim-1 in tumors, tumor stroma
and tumor-adjacent mucosa co-determines the prognosis of colon
cancer patients. PLoS One. 8:e766932013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mochizuki T, Kitanaka C, Noguchi K,
Muramatsu T, Asai A and Kuchino Y: Physical and functional
interactions between Pim-1 kinase and Cdc25A phosphatase.
Implications for the Pim-1-mediated activation of the c-Myc
signaling pathway. J Biol Chem. 274:18659–18666. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Morishita D, Katayama R, Sekimizu K,
Tsuruo T and Fujita N: Pim kinases promote cell cycle progression
by phosphorylating and down-regulating p27Kip1 at the
transcriptional and posttranscriptional levels. Cancer Res.
68:5076–5085. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang Y, Wang Z and Magnuson NS: Pim-1
kinase-dependent phosphorylation of p21Cip1/WAF1
regulates its stability and cellular localization in H1299 cells.
Mol Cancer Res. 5:909–922. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bachmann M, Kosan C, Xing PX, Montenarh M,
Hoffmann I and Möröy T: The oncogenic serine/threonine kinase Pim-1
directly phosphorylates and activates the G2/M specific phosphatase
Cdc25C. Int J Biochem Cell Biol. 38:430–443. 2006. View Article : Google Scholar
|
|
24
|
Yan B, Zemskova M, Holder S, Chin V, Kraft
A, Koskinen PJ and Lilly M: The PIM-2 kinase phosphorylates BAD on
serine 112 and reverses BAD-induced cell death. J Biol Chem.
278:45358–45367. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Aho TL, Sandholm J, Peltola KJ, Mankonen
HP, Lilly M and Koskinen PJ: Pim-1 kinase promotes inactivation of
the pro-apoptotic Bad protein by phosphorylating it on the Ser112
gatekeeper site. FEBS Lett. 571:43–49. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Santio NM, Salmela M, Arola H, Eerola SK,
Heino J, Rainio EM and Koskinen PJ: The PIM1 kinase promotes
prostate cancer cell migration and adhesion via multiple signalling
pathways. Exp Cell Res. 342:113–124. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Grundler R, Brault L, Gasser C, Bullock
AN, Dechow T, Woetzel S, Pogacic V, Villa A, Ehret S, Berridge G,
et al: Dissection of PIM serine/threonine kinases in
FLT3-ITD-induced leukemogenesis reveals PIM1 as regulator of
CXCL12-CXCR4-mediated homing and migration. J Exp Med.
206:1957–1970. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rainio EM, Sandholm J and Koskinen PJ:
Cutting edge: Transcriptional activity of NFATc1 is enhanced by the
Pim-1 kinase. J Immunol. 168:1524–1527. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bhattacharya N, Wang Z, Davitt C, McKenzie
IF, Xing PX and Magnuson NS: Pim-1 associates with protein
complexes necessary for mitosis. Chromosoma. 111:80–95. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Magnuson NS, Wang Z, Ding G and Reeves R:
Why target PIM1 for cancer diagnosis and treatment? Future Oncol.
6:1461–1478. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Arunesh GM, Shanthi E, Krishna MH, Sooriya
Kumar J and Viswanadhan VN: Small molecule inhibitors of PIM1
kinase: July 2009 to February 2013 patent update. Expert Opin Ther
Pat. 24:5–17. 2014. View Article : Google Scholar
|
|
32
|
Qian KC, Wang L, Hickey ER, Studts J,
Barringer K, Peng C, Kronkaitis A, Li J, White A, Mische S, et al:
Structural basis of constitutive activity and a unique nucleotide
binding mode of human Pim-1 kinase. J Biol Chem. 280:6130–6137.
2005. View Article : Google Scholar
|
|
33
|
Mikkers H, Nawijn M, Allen J, Brouwers C,
Verhoeven E, Jonkers J and Berns A: Mice deficient for all PIM
kinases display reduced body size and impaired responses to
hematopoietic growth factors. Mol Cell Biol. 24:6104–6115. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xie Y, Xu K, Linn DE, Yang X, Guo Z,
Shimelis H, Nakanishi T, Ross DD, Chen H, Fazli L, et al: The
44-kDa Pim-1 kinase phosphorylates BCRP/ABCG2 and thereby promotes
its multimerization and drug-resistant activity in human prostate
cancer cells. J Biol Chem. 283:3349–3356. 2008. View Article : Google Scholar
|
|
35
|
Zemskova MY, Song JH, Cen B, Cerda-Infante
J, Montecinos VP and Kraft AS: Regulation of prostate stromal
fibroblasts by the PIM1 protein kinase. Cell Signal. 27:135–146.
2015. View Article : Google Scholar :
|
|
36
|
Cheng F, Weidner-Glunde M, Varjosalo M,
Rainio EM, Lehtonen A, Schulz TF, Koskinen PJ, Taipale J and Ojala
PM: KSHV reactivation from latency requires Pim-1 and Pim-3 kinases
to inactivate the latency-associated nuclear antigen LANA. PLoS
Pathog. 5:e10003242009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ionov Y, Le X, Tunquist BJ, Sweetenham J,
Sachs T, Ryder J, Johnson T, Lilly MB and Kraft AS: Pim-1 protein
kinase is nuclear in Burkitt's lymphoma: Nuclear localization is
necessary for its biologic effects. Anticancer Res. 23A:167–178.
2003.
|
|
38
|
Brault L, Menter T, Obermann EC, Knapp S,
Thommen S, Schwaller J and Tzankov A: PIM kinases are progression
markers and emerging therapeutic targets in diffuse large B-cell
lymphoma. Br J Cancer. 107:491–500. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhu X, Xu JJ, Hu SS, Feng JG, Jiang LH,
Hou XX, Cao J, Han J, Ling ZQ and Ge MH: Pim-1 acts as an oncogene
in human salivary gland adenoid cystic carcinoma. J Exp Clin Cancer
Res. 33:1142014. View Article : Google Scholar
|
|
40
|
Yang Q, Chen LS, Neelapu SS, Miranda RN,
Medeiros LJ and Gandhi V: Transcription and translation are primary
targets of Pim kinase inhibitor SGI-1776 in mantle cell lymphoma.
Blood. 120:3491–3500. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Quan J, Zhou L and Qu J: Knockdown of
Pim-3 suppresses the tumorigenicity of glioblastoma by regulating
cell cycle and apoptosis. Cell Mol Biol. 61:42–50. 2015.PubMed/NCBI
|
|
42
|
Wang Z, Cao CJ, Huang LL, Ke ZF, Luo CJ,
Lin ZW, Wang F, Zhang YQ and Wang LT: EFEMP1 promotes the migration
and invasion of osteosarcoma via MMP-2 with induction by AEG-1 via
NF-κB signaling pathway. Oncotarget. 6:14191–14208. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lu XY, Lu Y, Zhao YJ, Jaeweon K, Kang J,
Xiao-Nan L, Ge G, Meyer R, Perlaky L, Hicks J, et al: Cell cycle
regulator gene CDC5L, a potential target for 6p12-p21 amplicon in
osteosarcoma. Mol Cancer Res. 6:937–946. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liao Y, Feng Y, Shen J, Gao Y, Cote G,
Choy E, Harmon D, Mankin H1, Hornicek F1 and Duan Z1: Clinical and
biological significance of PIM1 kinase in osteosarcoma. J Orthop
Res. 34:1185–1194. 2016. View Article : Google Scholar
|
|
45
|
Gurney J, Swensen A and Bulterys M:
Malignant bone tumors. Cancer Incidence and Survival Among Children
and Adolescents: United States SEER Program 1975–1995. National
Cancer Institute, SEER Program; Ries L, Smith M, Gurney J, Linet M,
Tamra T, Young J and Bunin G: Bethesda, MD: NIH Pub. No. 99-4649;
pp. 99–110. 1999
|
|
46
|
Wagle S, Park SH, Kim KM, Moon YJ, Bae JS,
Kwon KS, Park HS, Lee H, Moon WS, Kim JR, et al: DBC1/CCAR2 is
involved in the stabilization of androgen receptor and the
progression of osteosarcoma. Sci Rep. 5:131442015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kim JH, Kim WS, Yun Y and Park C:
Epstein-Barr virus latent membrane protein 1 increases
chemo-resistance of cancer cells via cytoplasmic sequestration of
Pim-1. Cell Signal. 22:1858–1863. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Santio NM, Vahakoski RL, Rainio EM,
Sandholm JA, Virtanen SS, Prudhomme M, Anizon F, Moreau P and
Koskinen PJ: Pim-selective inhibitor DHPCC-9 reveals Pim kinases as
potent stimulators of cancer cell migration and invasion. Mol
Cancer. 9:2792010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ren K, Duan W, Shi Y, Li B, Liu Z and Gong
J: Ectopic over-expression of oncogene Pim-2 induce malignant
transformation of nontumorous human liver cell line L02. J Korean
Med Sci. 25:1017–1023. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhuang H, Zhao MY, Hei KW, Yang BC, Sun L,
Du X and Li YM: Aberrant expression of PIM-3 promotes proliferation
and migration of ovarian cancer cells. Asian Pac J Cancer Prev.
16:3325–3331. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Narlik-Grassow M, Blanco-Aparicio C,
Cecilia Y, Peregrina S, Garcia-Serelde B, Muñoz-Galvan S, Cañamero
M and Carnero A: The essential role of PIM kinases in sarcoma
growth and bone invasion. Carcinogenesis. 33:1479–1486. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Decker S, Finter J, Forde AJ, Kissel S,
Schwaller J, Mack TS, Kuhn A, Gray N, Follo M, Jumaa H, et al: PIM
kinases are essential for chronic lymphocytic leukemia cell
survival (PIM2/3) and CXCR4-mediated microenvironmental
interactions (PIM1). Mol Cancer Ther. 13:1231–1245. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Santio NM, Eerola SK, Paatero I,
Yli-Kauhaluoma J, Anizon F, Moreau P, Tuomela J, Härkönen P and
Koskinen PJ: Pim kinases promote migration and metastatic growth of
prostate cancer xenografts. PLoS One. 10:e01303402015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Oda Y, Yamamoto H, Tamiya S, Matsuda S,
Tanaka K, Yokoyama R, Iwamoto Y and Tsuneyoshi M: CXCR4 and VEGF
expression in the primary site and the metastatic site of human
osteosarcoma: analysis within a group of patients, all of whom
developed lung metastasis. Mod Pathol. 19:738–745. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Laverdiere C, Hoang BH, Yang R, Sowers R,
Qin J, Meyers PA, Huvos AG, Healey JH and Gorlick R: Messenger RNA
expression levels of CXCR4 correlate with metastatic behavior and
outcome in patients with osteosarcoma. Clin Cancer Res.
11:2561–2567. 2005. View Article : Google Scholar : PubMed/NCBI
|