Introduction
Hepatocellular carcinoma (HCC) represents the fifth
most commonly diagnosed malignancy and the third leading cause of
cancer-related death worldwide (1). Due to the extensive research efforts,
understanding of the molecular mechanisms involved in the
tumorigenic transformation of HCC has been clarified gradually.
However, the molecular targets for HCC therapy remain insufficient
(2). Recent studies suggest that
human cancer can be considered as a stem cell disorder. According
to these studies, tumors may be viewed as abnormal growth which is
driven by a small population of cancer cells called cancer stem
cells (CSCs) that are endowed with the ability of self-renewal
(3,4).
Previous studies have identified the
CD133+ phenotype as the molecular marker of liver cancer
stem cells (LCSCs) (5). The LCSCs
were found to show a greater colony-forming efficiency and greater
ability to form a tumor in vivo. In addition, the LCSCs are
believed to be responsible for the high resistance to
chemotherapeutic drugs (6).
Therefore, to treat the HCC more effectively, it is imperative that
LCSCs are specifically targeted.
Tumor necrosis factor (TNF)-related apoptosis
inducing ligand (TRAIL) is a cytokine, which belongs to the TNF
superfamily (7). As the TRAIL
triggers apoptosis selectively in tumor cells without damaging the
normal tissue cells, it has been tested in clinical trials for
cancer therapy (8). However,
unfortunately, some cancer cells (especially CSCs) are resistant to
TRAIL and the mechanism of this resistance is not fully understood
(9). Therefore, it is urgent to
identify the mechanisms and take strategies to decrease the
resistance of CSCs to TRAIL.
MicroRNAs (miRNAs), small functional RNAs of 19–25
nt, are a class of short non-coding RNA molecules and play
important regulatory roles by binding to the target mRNAs at the
3′-untranslated region (3′-UTR) (10,11).
Increasing evidence shows that miRNAs have significant roles in a
number of biological processes such as proliferation,
differentiation, metabolism and apoptosis. Aberrant expression of
miRNAs is associated with multiple human diseases including cancer
(12,13). In the present study, we found the
miR-25 was dysregulated in LCSCs. We therefore investigated the
relationship between the miR-25 and the treatment of TRAIL in
LCSCs, which may serve as a potential target for HCC therapy.
Materials and methods
Animals
Nude mice (BALB/c, nu/nu; 4–5 week old; 18–23 g in
weight) were obtained from the Shanghai Laboratory Animal Center
(Shanghai, China). The animals were kept in ventilated,
pathogen-free racks under a 12 h light-dark photoperiodicity with
controlled humidity (50–60%) and temperature (22–24°C). The animals
were kept with free access to food and water. Animal care and the
experimental procedures in this study were approved by the Animal
Care Committee of the First Affiliated Hospital, School of
Medicine, Zhejiang University and complied with the recommendations
of the Chinese guidelines for the care and use of laboratory
animals.
Cell lines
The HCC cells lines of HepG2, Huh7, PLC and the
normal liver cell line of L-O2 were obtained from the Institute of
Biochemistry and Cell Biology, Chinese Academy of Sciences
(Shanghai, China). Cells were cultured in DMEM basic medium
containing 10% fetal bovine serum (FBS) (both from Gibco, Grand
Island, NY, USA) at 37°C in a humidified 5% CO2
incubator. To evaluate the role of anti-miR-25 in HCC in
vivo, the stable HepG2 cell line with miR-25 knockdown was
generated. Briefly, we purchased the recombinant lentivirus
contained miR-25 antisense nucleotide sequence from the Shanghai
Genechem Co., Ltd. (Shanghai, China). The routine HepG2 cells were
then transfected with 5×105 transducing units of
lentivirus, and the cells were selected with 1 μg/ml puromycin for
2 weeks.
Measurement of miR-25 expression
level
The expression level of miR-25 was measured by
reverse transcription and quantitative real-time polymerase chain
reaction (qRT-PCR). Briefly, total RNA was extracted from the cells
using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to
the manufacturer’s instructions. The reverse transcription reaction
for miR-25 was performed by using the One Step PrimeScript miRNA
cDNA synthesis kit (Takara, Dalian, China). Subsequently, the
qRT-PCR reaction was performed by SYBR® Premix Ex Taq™
II (Takara). The expression of miR-25 was determined using the
2−ΔΔCT analysis method (14) taking the U6 snRNA as the internal
control.
Identification and sorting of LCSCs and
non-CSCs
HCC cells were collected and washed twice. Cell
samples were then stained with anti-CD133-FITC (Miltenyi Biotec,
Gladbach, Germany) for 20 min at room temperature. The percentages
of CD133+ cells were analyzed and sorted as the LCSCs
using the fluorescent-activated cell sorting equipment (Beckman
Coulter, Brea, CA, USA).
Transfection
Cell-based experiments were carried out by
transfection of 50 pmol/ml RNA oligonucleotides including miR-25
mimics, miR-25 antisense-oligonucleotides (anti-miR-25), control
RNA oligonucleotides (miR-NC) and small interfering RNA of
phosphatase and tensin homologue (PTEN) (PTEN siRNA). All of these
RNAs were purchased from RiboBio Co., Ltd. (Guangzhou, China) and
transfected into the cells using Lipofectamine™ 2000 reagent
(Invitrogen) according to the manufacturer’s instructions.
Cell viability assay
The sorted LCSCs and non-CSCs were seeded in 96-well
plates at 5×103 cells/well and transfected with 50
pmol/ml RNA oligonucleotides. Twenty-four hours post-transfection,
cells were incubated in different concentrations of TRAIL for 48 h.
Subsequently, 20 μl
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
(5 mg/ml; Sigma-Aldrich, St. Louis, MO, USA) was added to the
medium and incubated at 37°C for 4 h followed by lysing in dimethyl
sulfoxide at room temperature for 30 min. The absorbance in each
well was measured at 570 nm using a microplate reader (Sunrise
Microplate Reader; Tecan, Männedorf, Switzerland).
Luciferase reporter assay
Fragment of PTEN 3′-UTR containing the seed region
of miR-25 was cloned into the pGL3 Luciferase Reporter Vectors
(Promega, Madison, WI, USA) named pGL3-PTEN. The mutant PTEN
reporter was created by mutating the seed regions of the miR-25
binding sites (GUGCAAU) by using the site-directed mutagenesis kit
(Takara, Shiga, Japan) named pGL3-mutant PTEN. For luciferase
reporter assay, LCSCs were seeded in 48-well plates followed by
co-transfection with 50 pmol/ml miR-25 mimics or inhibitors
together with 2 μg/ml Firefly luciferase reporters and 100 ng/ml
Renilla luciferase pRL-TK vector (Promega). Luciferase
activity was measured 48 h after transfection by using the
Dual-Luciferase Reporter assay system (Promega) according to the
manufacturer’s instructions.
Mitochondria isolation
For detecting the release of cytochrome c,
second mitochondria derived activator of caspase/direct IAP binding
protein with low pI (Smac/DIABLO) and apoptosis-inducing factor
(AIF) in cytoplasm, mitochondria of LCSCs were isolated using the
Mitochondria Isolation kit for Mammalian Cells (Thermo Fisher
Scientific, Rockford, IL, USA) according to the manufacturer’s
guidance.
Immunoprecipitation
Cells were lysed in RIPA buffer and centrifuged at
12,000 × g for 10 min. The resulting supernatants were collected
and incubated with primary antibody of Bad (Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA) overnight at 4°C.
Subsequently, the protein A agarose beads were added and incubated
for 2 h. After washing the beads with cold RIPA buffer, the
co-precipitated proteins were removed from the beads by boiling in
sodium dodecyl sulfate (SDS) sample buffer.
Western blot analysis
Fifty micrograms of the extracted and the
co-precipitated proteins were separated by 10% SDS-polyacrylamide
gel electrophoresis (PAGE) and transferred to a PVDF membrane
(Millipore, Billerica, MA, USA). The membrane was then blocked with
5% non-fat milk and incubated with various antibodies followed by
incubation of horseradish peroxidase-conjugated antibody (Santa
Cruz Biotechnology, Inc.). Signals were detected using enhanced
chemiluminescence (ECL) reagents (Thermo Fisher Scientific)
according to the manufacturer’s guidance.
Detection of apoptosis and reactive
oxygen species (ROS)
LCSCs were treated with RNAs and TRAIL. Then, the
cells were stained with PI and Annexin V (Sigma-Aldrich) for
apoptosis detection and dihydroethidium (DHE; Molecular Probes,
Camarillo, CA, USA) for measurement of ROS. Both apoptosis and ROS
were detected on the flow cytometry according to the manufacturer’s
guidance, respectively.
Tumor growth in nude mice
An equal number (5×106) of HepG2 cells
transfected with lentivirus anti-miR-25 or lentivirus control were
harvested and washed. The experimental animals were divided into
four groups (8 mice/group). For xenografts, two groups of mice were
subcutaneously injected with lentivirus control HepG2 cells (for
lenti control group and TRAIL group), and two groups of mice were
subcutaneously injected with lentivirus anti-miR-25 HepG2 cells
(for anti-miR-25 group and TRAIL plus anti-miR-25 group). The tumor
volume (V) was calculated based on the following equation: 1/2 ×
length × width2. The mice in TRAIL group and TRAIL plus
anti-miR-25 group received intraperitoneal (i.p.) injections of 40
μg/kg TRAIL per two days when the tumors reached a mean volume of
100 mm3 (15). The
tumor formation was monitored every three days using calipers. Nude
mice were euthanized at the experimental end-point (31 days
post-injection).
To measure the population of CSCs in tumor tissues,
we purified the cells from the tumor. Briefly, the tumor xenografts
were cut and digested with 1 mg/ml of collagenase type III (5 ml/g
tissue; Worthington Biochemical, Lakewood, NJ, USA) in 10% FBS
containing DMEM medium at 37°C for 2 h. The resulting cells were
then stained with anti-CD133-FITC for 20 min before analyzing on
the FACS vantage.
Statistical analysis
The data are presented as the mean ± standard
deviation and carried out by at least three independent
experiments. Statistical analysis was performed by Student’s t-test
using SPSS 15.0 software. Values of P<0.05 were considered
statistically significant.
Results
miR-25 is overexpressed in LCSCs
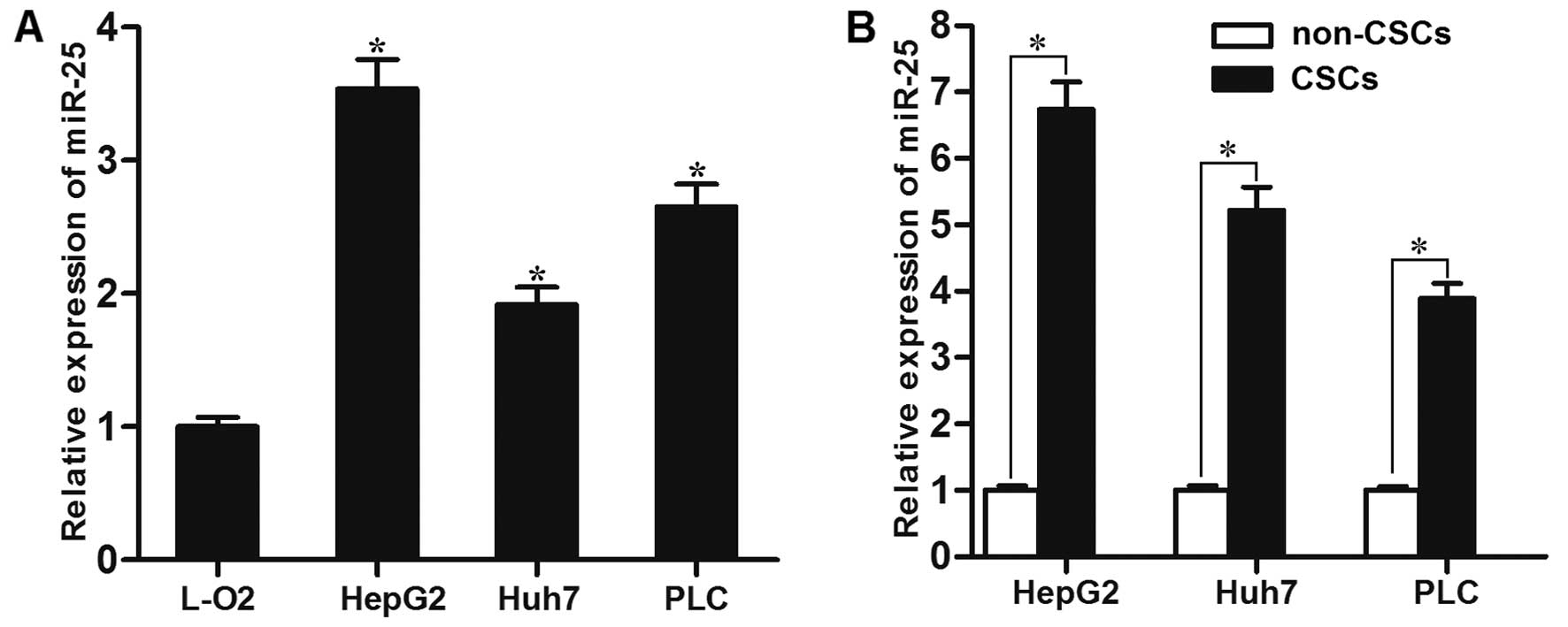
To investigate the role of miR-25 in HCC, its
expression levels between normal liver cell line and HCC cell lines
were compared by qRT-PCR. The expression of miR-25 was
significantly higher in HCC cell lines than that in the L-O2 cells
(Fig. 1A). We further evaluated
the expression of miR-25 in the CSCs of HepG2, Huh7 and PLC, as
well as in the non-CSCs of these cell lines. As shown in Fig. 1B, miR-25 was upregulated overall in
all tested HCC cell lines. Taken together, these results suggested
that the upregulation of miR-25 may be involved in LCSCs.
miR-25 is associated with the sensitivity
of LCSCs to TRAIL in vitro
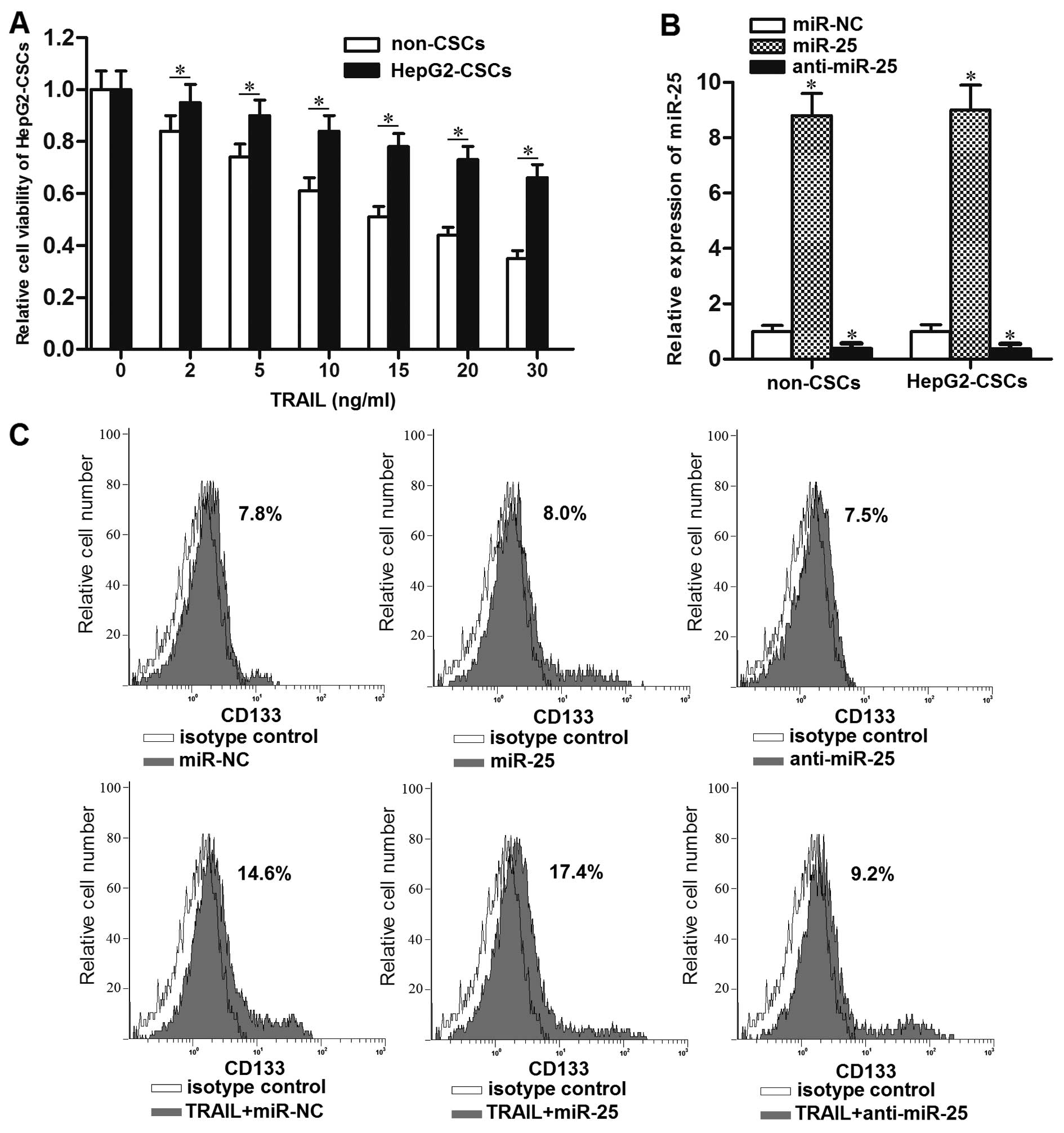
In order to evaluate the sensitivity of LCSCs and
non-CSCs to TRAIL, we sorted them in HepG2 cell line using the
anti-CD133 antibody. Subsequently, the HepG2-CSCs and non-CSCs were
treated with different concentrations of TRAIL. The sensitivity to
TRAIL was significantly lower in HepG2-CSCs than that in the
non-CSCs (Fig. 2A). The results
suggest the high resistance of LCSCs to TRAIL. To evaluate the role
of miR-25 in LCSCs, we transfected the HepG2-CSCs and non-CSCs with
the miR-25 mimic or inhibitors. Transfection of miR-25 mimics
significantly upregulated the expression level of miR-25 in both
the HepG2-CSCs and non-CSCs, whereas the expression of miR-25 was
significantly downregulated due to the transfection of anti-miR-25
(Fig. 2B). Interestingly, the flow
cytometric analysis showed that the percentage of CD133+
HepG2-CSCs population was increased after the single treatment of
TRAIL. On the other hand, although changing the expression of
miR-25 did not influence the percentage of HepG2-CSCs population
significantly, the anti-miR-25 significantly inhibited the effect
of TRAIL on enriching the HepG2-CSCs population, whereas the miR-25
mimics even increased the enrichment of HepG2-CSCs population
induced by TRAIL (Fig. 2C and D).
We consider that the treatment of TRAIL alone probably killed the
TRAIL-sensitive non-CSCs, whereas the TRAIL-resistant HepG2-CSCs
survived under the TRAIL treatment. To demonstrate the effect of
miR-25 on the sensitivity of LCSCs to TRAIL, we next performed a
cell viability assay. The HCC-CSCs (HepG2-CSCs, Huh7-CSCs and
PLC-CSCs) transfected with anti-miR-25 showed significantly higher
sensitivity to TRAIL than those in the miR-NC group (Fig. 2E). However, the cells transfected
with miR-25 mimics showed the opposite effect on the sensitivity of
TRAIL compared with those in the miR-NC groups. Taken together,
these results indicated that knockdown of miR-25 could increase the
sensitivity of LCSCs to TRAIL-induced cell death in
vitro.
Knockdown of miR-25 increases the
antitumor effect of TRAIL on HCC in vivo
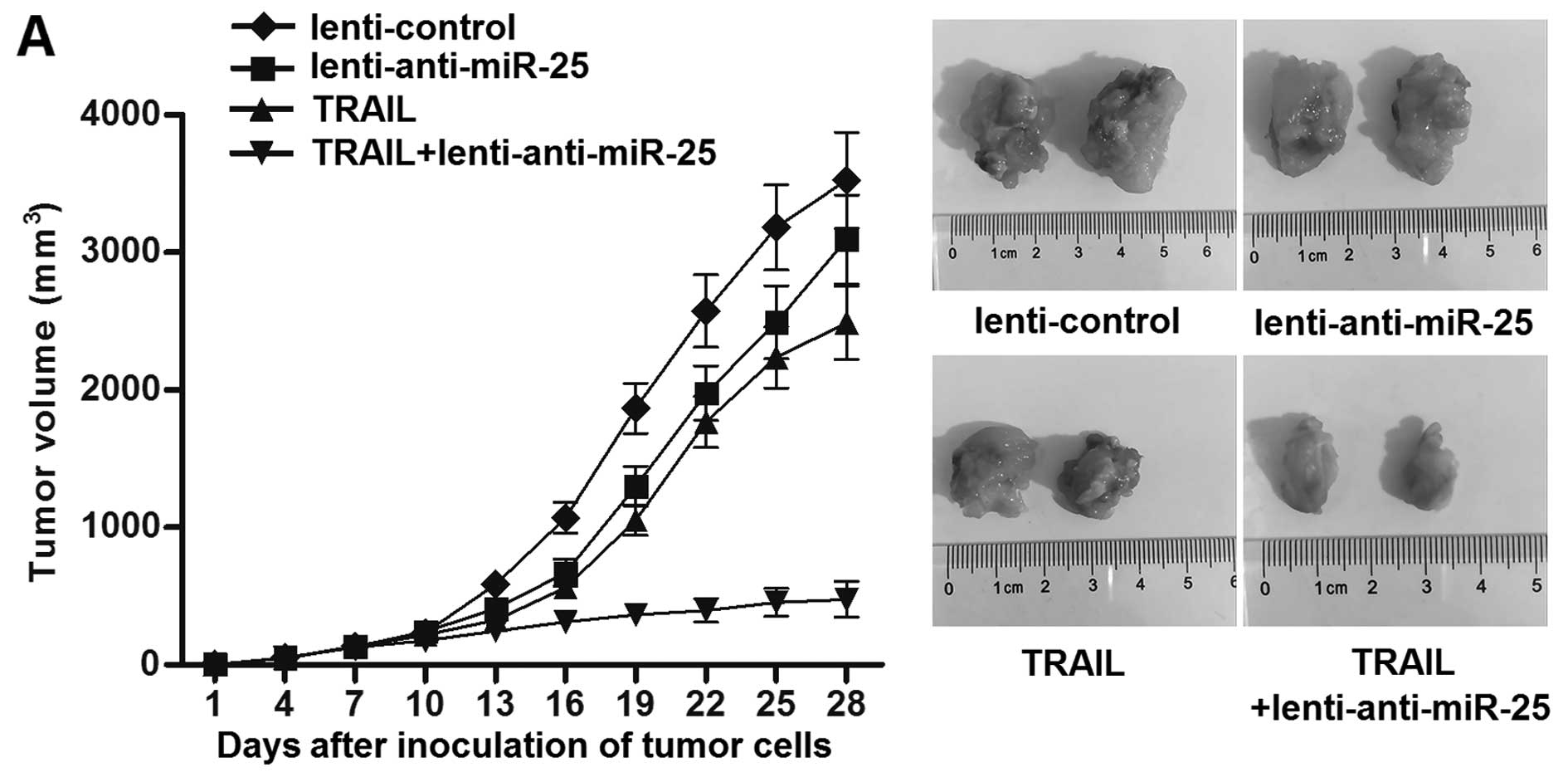
To investigate the effect of miR-25 inhibitors on
TRAIL treatment in vivo, a total of 5×106 of
HepG2 cells transfected with lentivirus anti-miR-25 or lentivirus
control were inoculated to the nude mice. Then, the tumor volumes
were calculated every three days. The results showed that the
growth of HepG2 cells transfected with lentivirus anti-miR-25 was
significantly slower than the cells transfected with lentivirus
control when they were treated with equal dose of TRAIL in
vivo (Fig. 3A). Furthermore,
the results of flow cytometry indicated that the percentage of CSC
population in lentivirus control tumor tissues was significantly
higher than the lentivirus anti-miR-25 tumor tissues when the mice
were treated with equal dose of TRAIL in vivo (Fig. 3B and C). Taken together, these
results indicated that knockdown of miR-25 could enhance the
curative effect of TRIAL and increase the sensitivity of LCSCs to
TRAIL in vivo.
PTEN is the target of miR-25 in
HepG2-CSCs
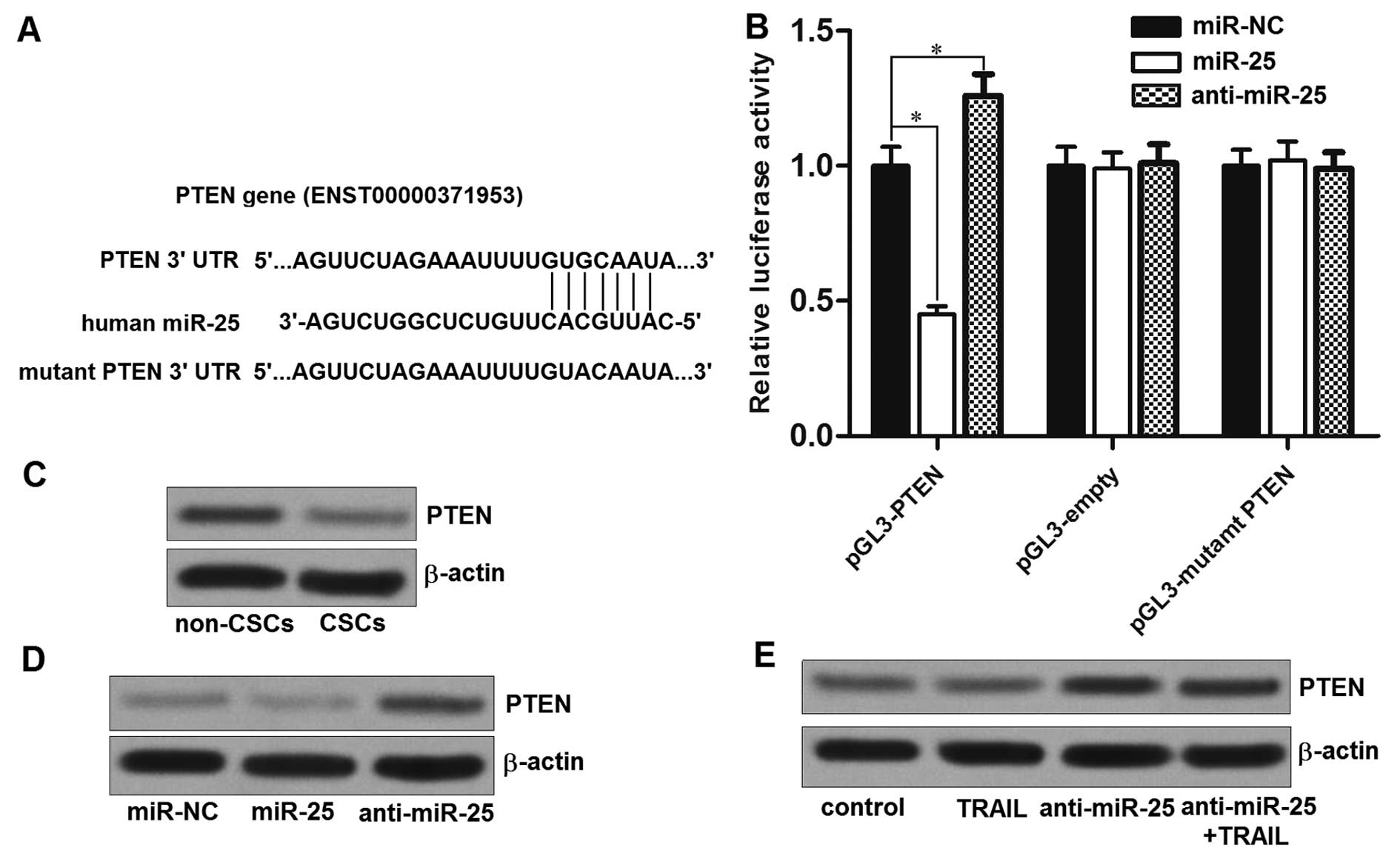
To understand how miR-25 regulated the sensitivity
of TRAIL to LCSCs, three different public databases (TargetScan,
miRanda and PicTar) were used. All of these databases showed a
prediction that a highly conserved sequence in the 3′-UTR of the
PTEN mRNA was targeted by miR-25 (Fig.
4A). To investigate whether miR-25 directly targets the PTEN
mRNA, we cloned the PTEN 3′-UTR sequence into the pGL3 luciferase
reporter vectors. The results of dual luciferase reporter assays
showed that co-transfection of miR-25 mimics and luciferase
reporter with wild-type of PTEN 3′-UTR significantly decreased the
luciferase activities, whereas the anti-miR-25 increased the
luciferase activities. Moreover, the mutant or empty plasmids
exhibited no response to miR-25 or anti-miR-25 (Fig. 4B). These results suggest that
miR-25 directly regulates the PTEN in HepG2-CSCs. Consistent with
this, we found the expression level of PTEN was significantly lower
in HepG2-CSCs than that in the corresponding non-CSCs (Fig. 4C), because the expression of miR-25
is overexpressed in HepG2-CSCs. To confirm that miR-25 regulates
the expression of PTEN, the PTEN protein was measured after the
miR-25 or anti-miR-25 was transfected into HepG2-CSCs. Transfection
of miR-25 decreased the expression of PTEN (Fig. 4D). Whereas, the transfection of
anti-miR-25 increased the expression of PTEN. In addition, we
detected the expression of PTEN in the lenti-control tumor tissues
(mice in control group and TRAIL group) and lenti anti-miR-25 tumor
tissues (mice in anti-miR-25 group and anti-miR-25 plus TRAIL
group) in vivo. We found that the protein levels of PTEN in
miR-25 knockdown tumor cells were significantly increased compared
with the control tumor cells (Fig.
4E). Taken together, we demonstrated that the PTEN gene is a
functional target of miR-25 in HCC.
Knockdown of miR-25 increases the
sensitivity of HepG2-CSCs to TRAIL through PTEN pathway
To investigate the role of PTEN in the anti-miR-25
promoted cell death in HepG2-CSCs, we knocked down the expression
of PTEN by its specific siRNA, and the transfection efficiency of
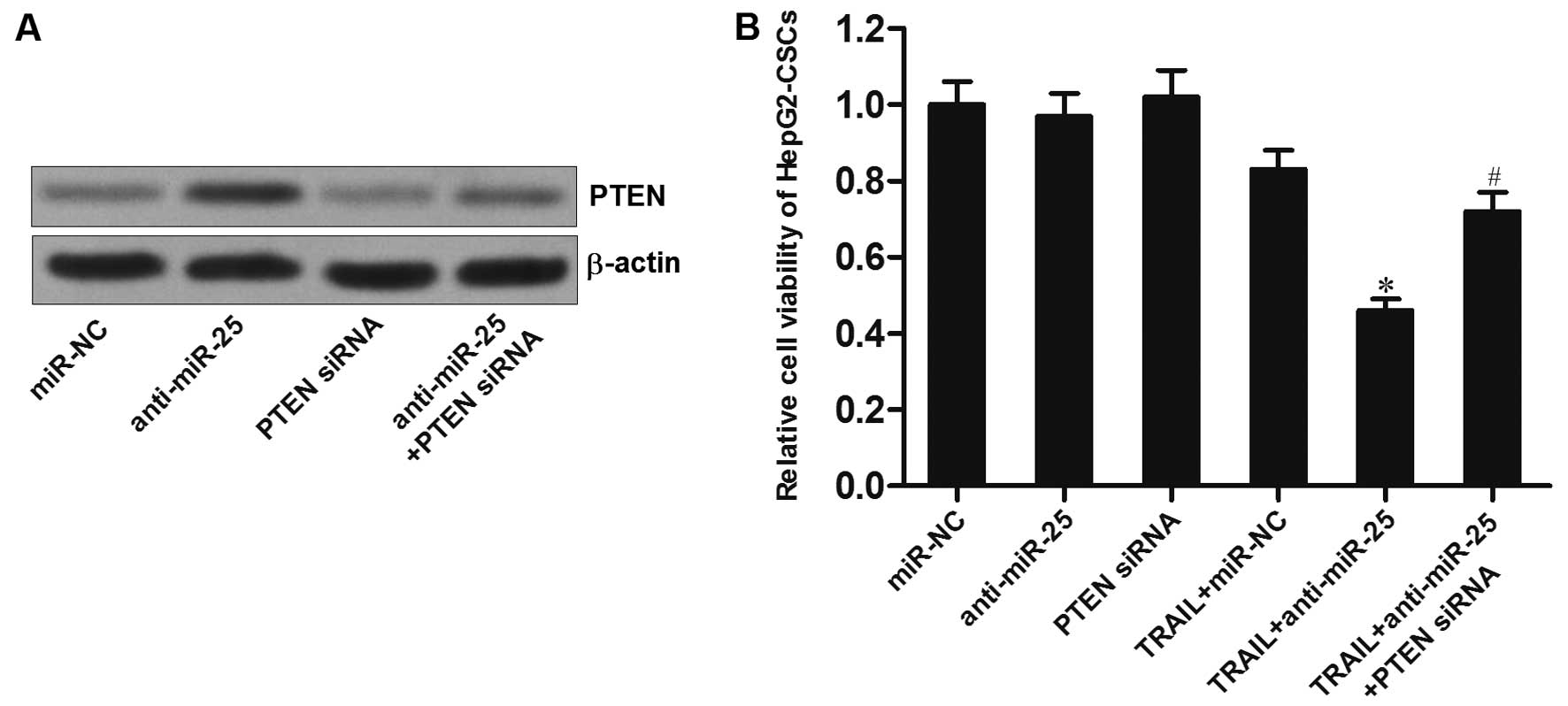
PTEN siRNA is shown in Fig. 5A. We
observed that although the anti-miR-25 significantly enhanced the
cytotoxicity of TRAIL to HepG2-CSCs, the promotion of anti-miR-25
was significantly inhibited when the PTEN siRNA was introduced
(Fig. 5B). Furthermore, as the
anti-miR-25 inhibited the enrichment of HepG2-CSC population
induced by TRAIL, transfection of PTEN siRNA abolished the effect
of anti-miR-25 in HepG2 cells (Fig. 5C
and D). Taken together, these results indicate that the
anti-miR-25 increases the sensitivity of HepG2-CSCs to TRAIL by
upregulating the expression of PTEN.
Knockdown of miR-25 increases the
sensitivity of HepG2-CSCs to TRAIL through apoptosis pathway
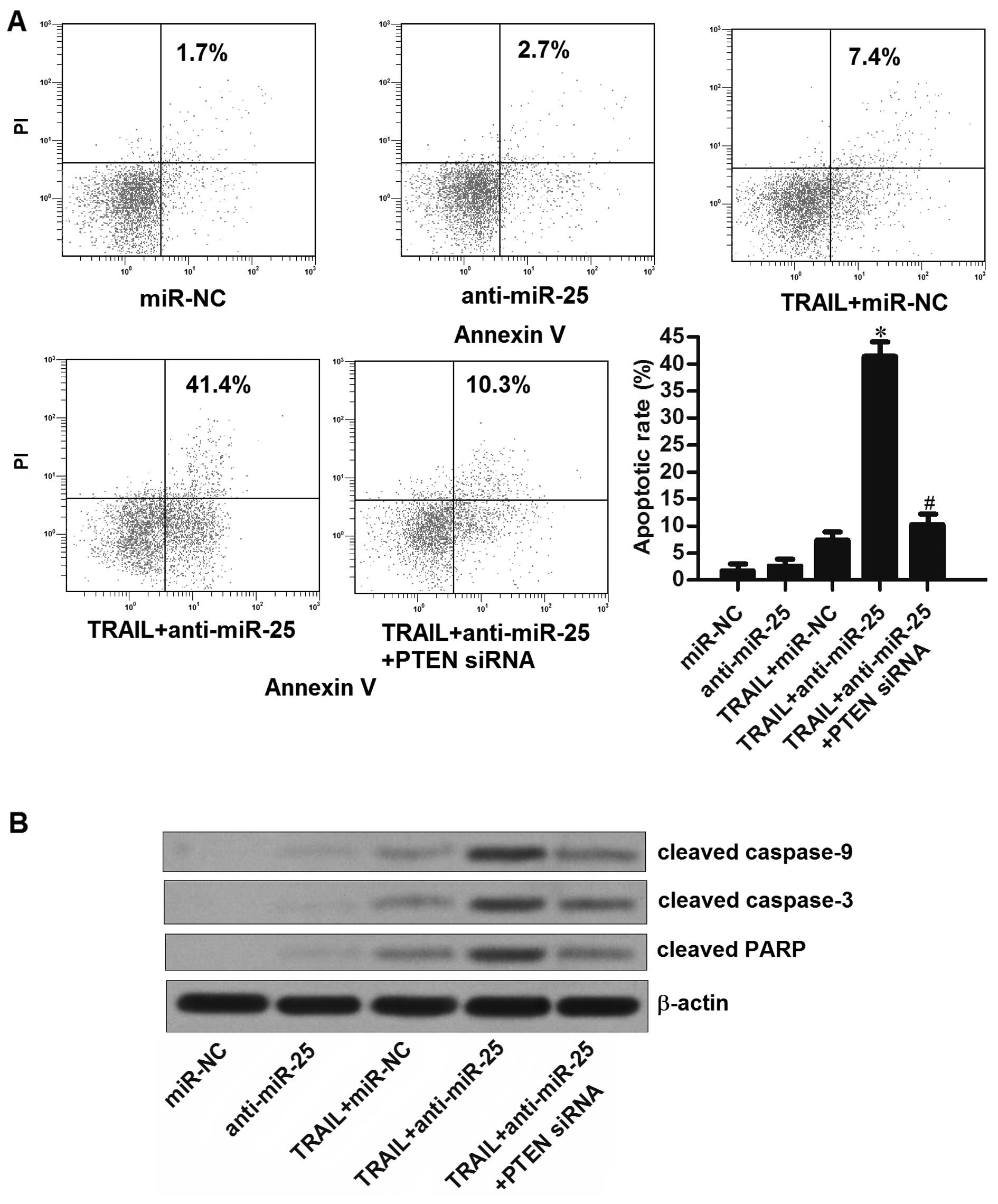
We observed that although the anti-miR-25 did not
induce apoptosis of HepG2-CSCs directly, it significantly enhanced
the effect of TRAIL on inducing the cell death through the
apoptosis pathway. In addition, the transfection of PTEN siRNA
rescued the HepG2-CSCs from apoptosis induced by the combination
with TRAIL and anti-miR-25 (Fig.
6A). Furthermore, we found knockdown of miR-25 promoted the
activation of caspase-9 and caspase-3 induced by the TRAIL
treatment in the HepG2-CSCs. Moreover, the transfection of PTEN
siRNA inhibited the cleavage of these caspases induced by the
combination with TRAIL and anti-miR-25 (Fig. 6B). These results indicate that
knockdown of miR-25 in HepG2-CSCs enhanced the TRAIL-induced
apoptosis through the PTEN pathway.
Knockdown of miR-25 enhances the
TRAIL-induced apoptosis via PTEN/PI3K/Akt/Bad signaling
pathway
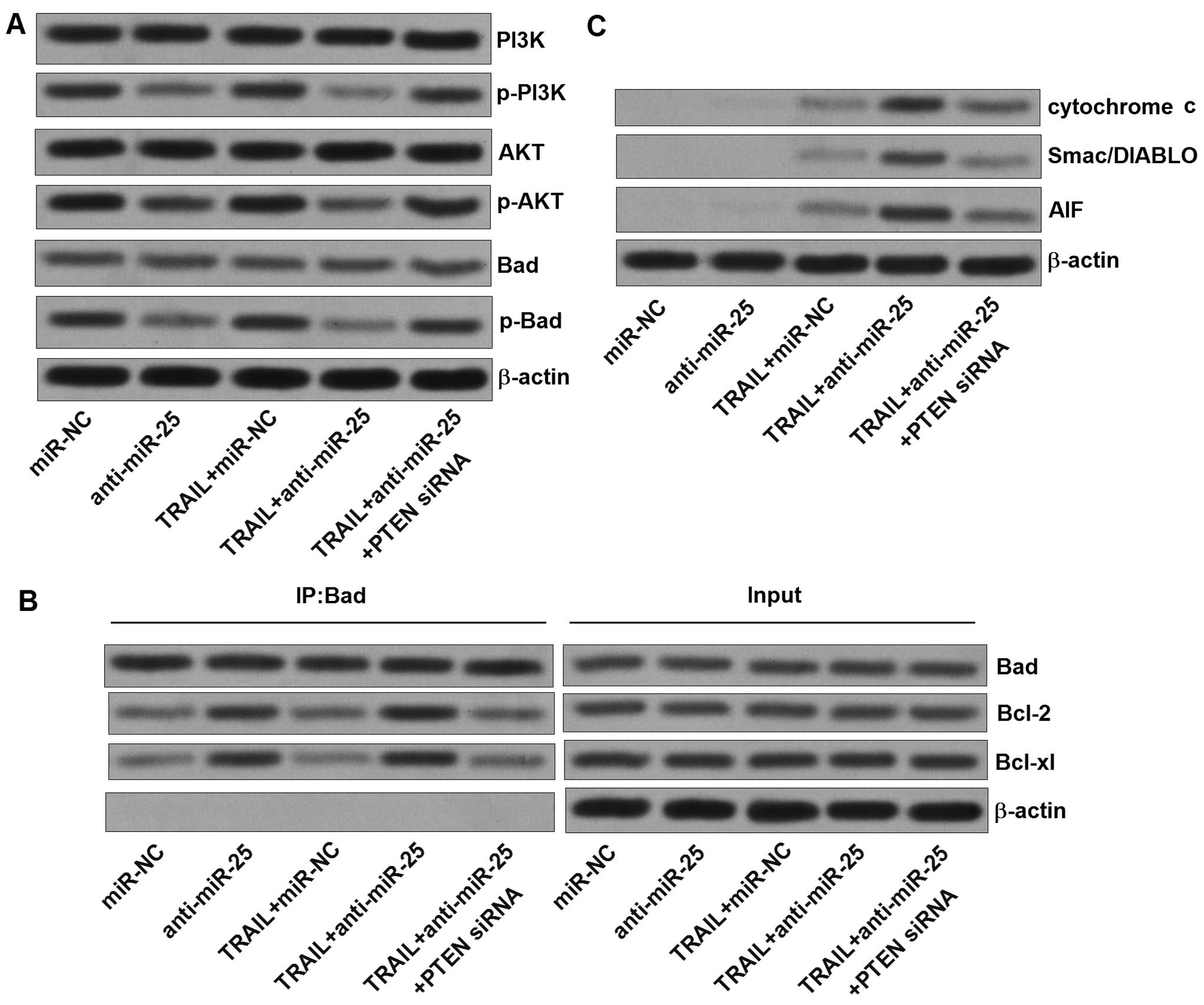
Since the PTEN is a natural inhibitor of PI3K and
negatively regulates the Akt (16), we next investigated the
relationship between the anti-miR-25 and the PTEN/PI3K/Akt
signaling. Transfection of anti-miR-25 inhibited the
phosphorylation of PI3K and Akt (Fig.
7A). In contrast, the single TRAIL treatment did not alter the
activity of PI3K or Akt. Previous studies have demonstrated that
Bad is one of the substrates for Akt (17). As we observed, the anti-miR-25
induced significant inhibition of Bad phosphorylation, which may be
the molecular mechanism by which the anti-miR-25 promotes the
apoptosis in HepG2-CSCs (18). The
dephosphorylation of Bad induced by the anti-miR-25, knockdown of
miR-25 significantly increased the Bad-Bcl-2 heterodimer and the
Bad-Bcl-xL heterodimer in HepG2-CSCs (Fig. 7B), subsequently inactivating the
anti-apoptotic proteins of Bcl-2 and Bcl-xL. Studies have
demonstrated that the inactivation of Bcl-2 and Bcl-xL promotes the
release of mitochondria-derived apoptogenic compounds (19). We therefore detected the cytochrome
c, Smac/DIABLO, and AIF in the cytoplasm of HepG2-CSCs. The
levels of all of these apoptogenic compounds were significantly
increased in the cytoplasm after the HepG2-CSCs were co-treated
with TRAIL and anti-miR-25 (Fig.
7C). In addition, ROS which is an important apoptosis inducer
(20) was also generated after the
combination treatment with TRAIL and anti-miR-25 (Fig. 7D). Taken together, our data
strongly suggests that the anti-miR-25 promotes the TRAIL-induced
apoptosis which is regulated by the PTEN/PI3K/Akt/Bad signaling
pathway.
Discussion
Previous studies have demonstrated that miRNAs are
associated with the sensitivity of cancer cells to the antitumor
treatment. For instance, overexpression of miR-122 in HCC could
reverse the resistance to doxorubicin by inhibiting the glucose
metabolism of cancer cells (21).
Knockdown of miR-221 could enhance the TRAIL-induced apoptosis by
upregulating the expression of BIM gene in breast cancer (22). The tumor suppressor of miR-101
could sensitize the HCC cells to the induction of apoptosis via
targeting the gene of Mcl-1 (23).
It is clear that the dysregulation of miRNAs should be considered
responsible for the low-sensitivity of cancer cells to the
chemotherapy.
miR-25 has been reported to act as an onco-miRNA in
various cancers, such as ovarian cancer, breast cancer and gastric
cancer. High levels of miR-25 in plasma predicted poor prognosis in
patients with gastric cancer (24–26).
However, the role of miR-25 in the cancer stem cells of HCC remains
unclear. In the present study, we demonstrate that the miR-25 is
dysregulated in the LCSCs. Moreover, we proved that the knockdown
of miR-25 could significantly increase the sensitivity of
HepG2-CSCs to the treatment of TRAIL in vitro and in
vivo. Interestingly, knockdown of miR-25 could inhibit the
effect of TRAIL on enriching the HepG2-CSCs population, because the
absence of miR-25 promoted the killing of HepG2-CSCs induced by
TRAIL. These findings may provide a potential use of miR-25
antagonist as a therapeutic strategy for reducing the
drug-resistance of LCSCs.
PTEN is a well-known tumor suppressor in multiple
cancers, which inhibits the tumorigenicity by negatively regulating
the highly oncogenic PI3K/Akt-signaling pathway (27). Previous studies have demonstrated
that the gene of PTEN is dysregulated in various cancers, and the
change of PTEN is one of the critical factors for tumorigenesis
(28,29). Recently, it was reported that PTEN
is regulated by miRNAs in cancers, and the miRNAs/PTEN axis has
been confirmed to be associated with the tumor growth, migration,
invasion and chemosensitivity (30–32).
In this study we also indicated that the gene of PTEN is
downregulated in the CSCs of HepG2. Moreover, the miR-25/PTEN axis
determined the sensitivity of HepG2-CSCs to TRAIL.
BCL2 associated agonist of cell death (Bad) is a
pro-apoptosis protein belonging to the Bcl-2 family, which is also
a substrate of Akt. The non-phosphorylated Bad promotes apoptosis
by conjugating to the anti-apoptotic proteins of Bcl-2 and Bcl-xL,
subsequently inactivating them. However, when Bad is phosphorylated
by Akt, it will separate from the heterodimer of Bcl-2/Bcl-xL, and
bind to 14-3-3 scaffold proteins in the cytoplasm with an inactive
form. Thus, the free Bcl-2 and Bcl-xL could inhibit the apoptosis
pathway (18,33). Our present study demonstrates that
the knockdown of miR-25 increases the expression of PTEN, thus
inhibiting the PI3K/Akt signaling pathway followed by keeping the
Bad in a non-phosphorylated form. Benefiting from this, TRAIL is
able to induce the mitochondrial apoptosis (34) in HepG2-CSCs, leading to the release
of mitochondria-derived apoptogenic compounds such as cytochrome
c, Smac/DIABLO, AIF and the ROS. Finally, the combination
with anti-miR-25 and TRAIL induces the cleavage of caspases and the
occurrence of apoptosis.
In conclusion, we have provided several pieces of
evidence to prove that the miR-25 is associated with the
sensitivity of liver cancer stem cells to TRAIL-induced apoptosis.
Mechanistically, we demonstrate that the knockdown of miR-25
promotes TRAIL-induced apoptosis by inhibiting the PI3K/Akt/Bad
signaling pathway through the miR-25/PTEN axis. The combination
with anti-miR-25 and TRAIL may represent a novel strategy for the
treatment of LCSCs.
Acknowledgements
This study is supported by the Medical and Health
Technology Plan of Zhejiang Province (grant no. 2013KYB117).
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Villanueva A, Newell P, Chiang DY,
Friedman SL and Llovet JM: Genomics and signaling pathways in
hepatocellular carcinoma. Semin Liver Dis. 27:55–76. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Reya T, Morrison SJ, Clarke MF and
Weissman IL: Stem cells, cancer, and cancer stem cells. Nature.
414:105–111. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Clarke MF and Fuller M: Stem cells and
cancer: Two faces of eve. Cell. 124:1111–1115. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Suetsugu A, Nagaki M, Aoki H, Motohashi T,
Kunisada T and Moriwaki H: Characterization of CD133+
hepatocellular carcinoma cells as cancer stem/progenitor cells.
Biochem Biophys Res Commun. 351:820–824. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ma S, Lee TK, Zheng BJ, Chan KW and Guan
XY: CD133+ HCC cancer stem cells confer chemoresistance
by preferential expression of the Akt/PKB survival pathway.
Oncogene. 27:1749–1758. 2008. View Article : Google Scholar
|
|
7
|
Joshi P, Jeon YJ, Laganà A, Middleton J,
Secchiero P, Garofalo M and Croce CM: MicroRNA-148a reduces
tumorigenesis and increases TRAIL-induced apoptosis in NSCLC. Proc
Natl Acad Sci USA. 112:8650–8655. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Walczak H, Miller RE, Ariail K, Gliniak B,
Griffith TS, Kubin M, Chin W, Jones J, Woodward A, Le T, et al:
Tumoricidal activity of tumor necrosis factor-related
apoptosis-inducing ligand in vivo. Nat Med. 5:157–163. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Piggott L, Omidvar N, Martí Pérez S,
French R, Eberl M and Clarkson RW: Suppression of apoptosis
inhibitor c-FLIP selectively eliminates breast cancer stem cell
activity in response to the anti-cancer agent, TRAIL. Breast Cancer
Res. 13:R882011. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gargalionis AN and Basdra EK: Insights in
microRNAs biology. Curr Top Med Chem. 13:1493–1502. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ou Y, Zhai D, Wu N and Li X:
Downregulation of miR-363 increases drug resistance in
cisplatin-treated HepG2 by dysregulating Mcl-1. Gene. 572:116–122.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
15
|
Ma H, Yao Q, Zhang AM, Lin S, Wang XX, Wu
L, Sun JG and Chen ZT: The effects of artesunate on the expression
of EGFR and ABCG2 in A549 human lung cancer cells and a xenograft
model. Molecules. 16:10556–10569. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Stambolic V, Suzuki A, de la Pompa JL,
Brothers GM, Mirtsos C, Sasaki T, Ruland J, Penninger JM,
Siderovski DP and Mak TW: Negative regulation of PKB/Akt-dependent
cell survival by the tumor suppressor PTEN. Cell. 95:29–39. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Brazil DP and Hemmings BA: Ten years of
protein kinase B signalling: A hard Akt to follow. Trends Biochem
Sci. 26:657–664. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Datta SR, Dudek H, Tao X, Masters S, Fu H,
Gotoh Y and Greenberg ME: Akt phosphorylation of BAD couples
survival signals to the cell-intrinsic death machinery. Cell.
91:231–241. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ye L, Yuan G, Xu F, Sun Y, Chen Z, Chen M,
Li T, Sun P, Li S and Sun J: The small-molecule compound BM-1197
inhibits the antiapoptotic regulators Bcl-2/Bcl-xL and triggers
apoptotic cell death in human colorectal cancer cells. Tumour Biol.
36:3447–3455. 2015. View Article : Google Scholar
|
|
20
|
Tian X, Zhao L, Song X, Yan Y, Liu N, Li
T, Yan B and Liu B: HSP27 inhibits homocysteine-induced endothelial
apoptosis by modulation of ROS production and mitochondrial
caspase-dependent apoptotic pathway. BioMed Res Int.
2016:48478742016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pan C, Wang X, Shi K, Zheng Y, Li J, Chen
Y, Jin L and Pan Z: MiR-122 reverses the doxorubicin-resistance in
hepatocellular carcinoma cells through regulating the tumor
metabolism. PLoS One. 11:e01520902016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ye Z, Hao R, Cai Y, Wang X and Huang G:
Knockdown of miR-221 promotes the cisplatin-inducing apoptosis by
targeting the BIM-Bax/Bak axis in breast cancer. Tumour Biol.
37:4509–4515. 2016. View Article : Google Scholar
|
|
23
|
He H, Tian W, Chen H and Deng Y:
MicroRNA-101 sensitizes hepatocellular carcinoma cells to
doxorubicin-induced apoptosis via targeting Mcl-1. Mol Med Rep.
13:1923–1929. 2016.PubMed/NCBI
|
|
24
|
Feng S, Pan W, Jin Y and Zheng J: MiR-25
promotes ovarian cancer proliferation and motility by targeting
LATS2. Tumour Biol. 35:12339–12344. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Z, Wang N, Liu P, Chen Q, Situ H, Xie
T, Zhang J, Peng C, Lin Y and Chen J: MicroRNA-25 regulates
chemoresistance-associated autophagy in breast cancer cells, a
process modulated by the natural autophagy inducer
isoliquiritigenin. Oncotarget. 5:7013–7026. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li BS, Zhao YL, Guo G, Li W, Zhu ED, Luo
X, Mao XH, Zou QM, Yu PW, Zuo QF, et al: Plasma microRNAs, miR-223,
miR-21 and miR-218, as novel potential biomarkers for gastric
cancer detection. PLoS One. 7:e416292012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li J, Yen C, Liaw D, Podsypanina K, Bose
S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, et al:
PTEN, a putative protein tyrosine phosphatase gene mutated in human
brain, breast, and prostate cancer. Science. 275:1943–1947. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tay Y, Kats L, Salmena L, Weiss D, Tan SM,
Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, et al:
Coding-independent regulation of the tumor suppressor PTEN by
competing endogenous mRNAs. Cell. 147:344–357. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu W, Yang Z, Zhou SF and Lu N:
Posttranslational regulation of phosphatase and tensin homolog
(PTEN) and its functional impact on cancer behaviors. Drug Des
Devel Ther. 8:1745–1751. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wu W, Yang J, Feng X, Wang H, Ye S, Yang
P, Tan W, Wei G and Zhou Y: MicroRNA-32 (miR-32) regulates
phosphatase and tensin homologue (PTEN) expression and promotes
growth, migration, and invasion in colorectal carcinoma cells. Mol
Cancer. 12:302013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang ZX, Lu BB, Wang H, Cheng ZX and Yin
YM: MicroRNA-21 modulates chemosensitivity of breast cancer cells
to doxorubicin by targeting PTEN. Arch Med Res. 42:281–290. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ma F, Zhang J, Zhong L, Wang L, Liu Y,
Wang Y, Peng L and Guo B: Upregulated microRNA-301a in breast
cancer promotes tumor metastasis by targeting PTEN and activating
Wnt/β-catenin signaling. Gene. 535:191–197. 2014. View Article : Google Scholar
|
|
33
|
Datta SR, Brunet A and Greenberg ME:
Cellular survival: A play in three Akts. Genes Dev. 13:2905–2927.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lim SC, Parajuli KR and Han SI: The
alkyllysophospholipid edelfosine enhances TRAIL-mediated apoptosis
in gastric cancer cells through death receptor 5 and the
mitochondrial pathway. Tumour Biol. 37:6205–6216. 2016. View Article : Google Scholar
|