Introduction
Colorectal cancer (CRC) is the third most common
cancer and the fourth most common cancer cause of death globally
with ~1.2 million new cases and 600,000 deaths per year (1,2).
Metastatic diseases occur in ~30–50% of CRC patients, either at the
time of initial diagnosis or during follow-up (3,4). CRC
metastasizes predominantly to the liver, and liver metastases
account for at least two thirds of CRC deaths (3,5,6).
Despite advances in research on primary CRC development, the
mechanism for the progression from local disease to metastasis is
not fully understood. Therefore, it is highly required to identify
sensitive biological markers for early detection and the risk of
metastasis in CRC.
MicroRNA (miRNA) is a small non-coding RNA of ~20–24
base in length, and it post-transcriptionally regulates the
expression of multiple target genes by binding to complementary
sequences, mainly in the 3′-untranslated regions (3′-UTR) of the
genes (7,8). miRNA also plays a crucial role in
cancer, and its aberrant expression may cause uncontrolled cell
proliferation, invasion, and metastasis (9–12).
Many miRNAs involved in development and progression of CRC have
been identified, and some of them are reportedly shown to have
potential value as biomarkers for diagnosis, prognosis and
susceptibility in CRC (13,14).
Recently, a few miRNAs are being tested for the clinical practice.
For instance, a phase I trial with miRNA-34 is currently ongoing to
evaluate its safety (15).
However, further studies are needed to comprehend the whole
biological systems of miRNAs in CRC.
We previously performed microarray analyses to
explore miRNAs that showed differential expression among primary
CRC tumors with or without liver metastasis, and liver metastatic
lesions. We reported that miRNA-132 was downregulated in the liver
metastasis-related samples, and that this miRNA was associated with
better prognosis in CRC (16). In
this study, we aimed at identifying another novel miRNA that was
aberrantly expressed in liver metastasis, using the microarray
data. We found that miRNA-487b (miR-487b) decreased in CRC tumors
with liver metastasis and liver metastatic lesions when compared
with CRC tumors without metastasis. Studies show that miR-487b
inhibits WNT/β-catenin signaling pathway and suppresses metastasis
in lung cancer (17,18). Xi et al also reported that
KRAS was a direct target of miR-487b in lung cancer (17). Furthermore, Gattolliat et al
showed that miR-487b could be a favorable prognostic marker in
neuroblastoma (19). These
findings suggest a possibility that miR-487b functions as a tumor
suppressor. However, to our knowledge, no studies have examined
miR-487b expression in CRC. The aim of this study was to reveal the
clinical significance of miR-487b in CRC.
Materials and methods
Collection of human tissue specimens
Tumor tissues were collected from 134 patients who
received surgery between 2003 and 2013 at Osaka University Hospital
and its three related facilities: 96 primary CRC tumors with stage
I (n=18), II (n=38), and III (n=40) disease without metastasis, 22
primary CRC tumors with simultaneous liver metastasis (stage IV),
and 16 liver metastatic lesions of CRC. All samples were
immediately frozen in RNAlater (Ambion, Austin, TX, USA) and stored
at −80°C until RNA extraction. This study was approved by the
institutional review board of each institution, and all subjects
provided written informed consents before participation. The
clinical parameters of the validating cohort are shown in Table I.
 | Table IPatient chracteristics. |
Table I
Patient chracteristics.
| Patient
characteristics | Without metastasis
(stage I/II/III) | With liver
metastasis | Liver metastasis
lesion |
|---|
| No. of
patients | 96 (18/38/40) | 22 | 16 |
| Tumor size |
| ≤35 mm | 21 | 6 | |
| >35 mm | 75 | 16 | |
| Location |
| Colon | 53 | 12 | |
| Rectum | 43 | 10 | |
|
Differentiation |
| tub1, tub2 | 91 | 21 | |
| muc, por | 5 | 1 | |
| Depth |
| T1, T2 | 20 | 0 | |
| T3, T4 | 76 | 22 | |
| Lymph node
metastasis |
| Negative | 56 | 5 | |
| Positive | 40 | 17 | |
| Lymphatic
invasion |
| Negative | 51 | 4 | |
| Positive | 45 | 18 | |
| Venous
invasion |
| Negative | 40 | 2 | |
| Positive | 56 | 20 | |
Cell lines and culture
Human CRC cell lines DLD-1 (KRAS G13D), HCT116 (KRAS
G13D), HT29 (BRAF V600E), and SW480 (KRAS G12V) were purchased from
the American Type Culture Collection (Rockville, MD, USA) in 2001.
Cell lines were maintained in Dulbecco's modified Eagle's medium
(Sigma-Aldrich, St. Louis, MO, USA) with 10% fetal bovine serum in
a humidified 5% CO2 at 37°C.
Transient oligonucleotide
transfection
miR-487b and negative control miRNA were purchased
from Gene Design Inc. (Osaka, Japan). These were transfected into
cells 24 h after seeding with Lipofectamine RNAiMAX reagent
(Invitrogen, Carlsbad, CA, USA) at a final concentration of 30
nmol/l, according to the manufacturer's protocol.
Proliferation assays
Transfected cells were seeded at a density of
2.5–3.0×104 per well in 24-well dishes. After cultured
for 24, 48 and 72 h, cells were trypsinized and stained with trypan
blue solution (Invitrogen). The total number of cells in each well
was determined with Countess Automated Cell Counter
(Invitrogen).
Colony formation assay
Transfected cells were seeded at a density of 500
per well in 6-well plates. After incubation at 37°C for 10 days,
cells were washed with PBS, fixed with formalin and stained with
Giemsa solution. The number of visible colonies was counted with
ImageJ software (National Institutes of Health).
Invasion assay
To measure cell invasion, transwell inserts with
8-µm pores (BD Biosciences, San Jose, CA, USA) were used
according to the manufacturer's protocol. The invading cells were
fixed and stained with Diff-Quik (Sysmex, Hyogo, Japan), and
counted under a light microscope in three random fields (×40
magnification).
Quantitative real-time polymerase chain
reaction (qRT-PCR)
Total RNA was isolated using miRNeasy Mini kit
(Qiagen, Hilden, Germany) for clinical tissues and TRIzol reagent
(Invitrogen) for cell lines following the manufacturer's protocol.
RNA concentration and purity were assessed with NanoDrop ND-1000
spectrophotometer (NanoDrop Technologies, Rockland, DE, USA).
For miRNA quantification, TaqMan microRNA assays
(Applied Biosystems, Foster City, CA, USA) were used: hsa-miR-487b
ID 001285, and RNU6B ID 001093. Total RNA was reverse transcribed
using TaqMan MicroRNA Reverse Transcription kit (Applied
Biosystems). qRT-PCR was performed with ABI PRISM 7900HT Sequence
Detection system (Applied Biosystems) using TaqMan Universal PCR
Master Mix, No AmpErase UNG (Applied Biosystems). Relative
expression was quantified with the ΔΔCq method (20).
For mRNA quantification, total RNA was reverse
transcribed using High Capacity RNA-to-cDNA kit (Applied
Biosystems). To measure KRAS expression level, qRT-PCR was
performed with LightCycler 480 Real-Time PCR system (Roche
Diagnostics, Mannheim, Germany) using the specific primers and
LightCycler-DNA Master SYBR Green I (Roche Diagnostics) (21). The specifically designed primers
were as follows: ACTB forward, 5′-GATGAGATTGGCATGGCTTT-3′; reverse,
5′-CACCTTCACCGTTCCAGTTT-3′. KRAS forward,
5′-ATTCCTTTTATTGAAACATCAGCA-3′; reverse 5′-TCGGATCTCCCTCACCAAT-3′.
For LRP6 expression analysis, qRT-PCR was performed with ABI PRISM
7900HT Sequence Detection system (Applied Biosystems) using TaqMan
Gene Expression Assays (Applied Biosystems) and TaqMan Universal
PCR Master Mix, No AmpErase UNG (Applied Biosystems). The product
numbers of TaqMan Gene Expression assay were as follows: ACTB ID
Hs99999903_m1, LRP6 ID Hs00999795_m1. Relative expression was
quantified using the ΔΔCq method (20).
Western blot analysis
Western blot analysis was performed as described
previously (22). Briefly, cells
were collected and lysed in RIPA buffer, containing phosphatase
inhibitor and protease inhibitor cocktail. The protein lysates (20
µg) from each sample were separated with sodium dodecyl
sulfate-poly-acrylamide gel electrophoresis (SDS-PAGE), and
transferred electronically to a polyvinylidene difluoride membrane.
The membrane was blocked with 5% skimmed milk and incubated with
anti-human polyclonal antibodies against ERK, phosphorylated ERK,
phosphorylated LRP6 (Cell Signaling Technology, Beverly, MA, USA),
ACTB (Sigma-Aldrich), and anti-human monoclonal antibodies against
AKT1, phosphorylated AKT, cleaved PARP, LRP6 (Cell Signaling
Technology), KRAS (Sigma-Aldrich). The membrane was incubated with
secondary antibodies, and visualized with the ECL Detection system
(GE Healthcare, Little Chalfont, UK).
Luciferase reporter assay
For serum response element (SRE) luciferase reporter
assay, the vector (pGL4.33[luc2P/SRE/Hygro]) was obtained from
Promega (Madison, WI, USA). For pmirGLO luciferase reporter assay,
the vector was made as follows. The 3′-UTR of human LRP6 mRNA
containing the putative miR-487b binding site was amplified by
polymerase chain reaction (PCR). The primer sequences were;
forward, 5′-GCTCGCTAGCCTCGAAGCAGGATGGGCGATAGA-3′; reverse
5′-ATGCCTGCAGGTCGATGGACAAGGGCTGACCAA-3′. The amplified DNA product
was cloned into the restriction site downstream of the firefly
luciferase gene in pmirGLO vector (Promega). The vector with mutant
3′-UTR sequence was constructed using the QuikChange Site-Directed
Mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA)
according to the manufacturer's protocol.
Cells were seeded in 96-well plates and transfected
with both reporter vector and miRNA using Lipofectamine 2000
(Invitrogen), and collected 48 h after co-transfection. Luciferase
activity was measured using Dual Luciferase Reporter assay system
(Promega), as described previously (16,23).
Briefly, the cell extracts were prepared by rinsing each plate
twice with PBS and lysing the cells in Passive Lysis buffer
(Promega). The transfection efficiency was evaluated with renilla
luciferase activity, and firefly luciferase activity was
normalized.
Statistical analysis
The data are expressed as the mean ± standard
deviation. Statistical differences were analyzed by the Student's
t-test for continuous variables and by the Chi-square test for the
others. Survival curves were drawn by the Kaplan-Meier method, and
compared using the log-rank test. The Cox proportional hazard
regression model was used to estimate the hazard ratio (HR) and the
95% confidence interval (CI). All statistical analyses were
conducted with JMP ver. 11.0.0 (SAS Institute, Inc., Cary, NC,
USA). P-values of <0.05 were considered to be statistically
significant.
Results
Identification of microRNAs with altered
expression between colorectal and hepatic tissues by the microarray
data
As a testing cohort, we used microarray data that
were previously registered in the NCBI Gene Expression Omnibus
database available through GEO with accession number GSE72199,
consisting of tumor tissues derived from 36 patients: 16 primary
CRC tumors with stage II and III disease without metastasis, 12
primary CRC tumors with simultaneous liver metastasis (stage IV),
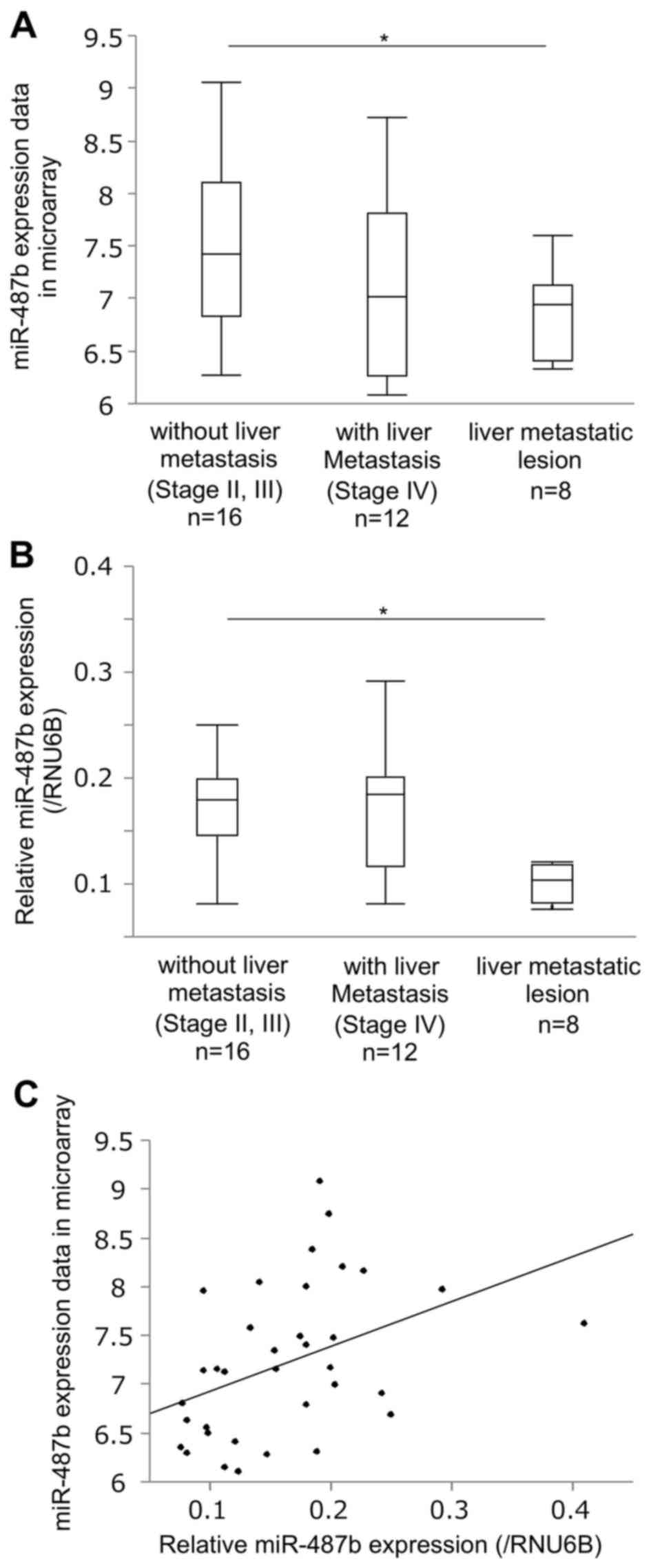
and 8 liver metastatic lesions of CRC (16). Consequently, 38 miRNAs that showed
significantly differential expression with fold change >2.0 in
microarray analyses were identified by comparison between primary
tumors without metastasis and liver metastatic lesions (Table II). Among them, we found that the
expression of miR-487b, which is reportedly shown as an
anti-oncomir in several human malignancies (17–19),
significantly decreased in liver metastatic lesions as compared to
primary tumors without metastasis (P=0.030, Fig. 1A). qRT-PCR validated the result in
the same testing cohort (n=36, P=0.001, Fig. 1B). To confirm the statistical
correlation between the results of microarray and qRT-PCR,
Spearman's rank order correlation coefficient was calculated, which
showed a significant correlation (ρ=0.487, P=0.003, Fig. 1C).
 | Table IIThe result of microarray analysis
(fold change >2.0, P<0.05). |
Table II
The result of microarray analysis
(fold change >2.0, P<0.05).
| 1 | hsa-miR-1 | 20 | hsa-miR-28-3p |
| 2 | hsa-miR-10b | 21 | hsa-miR-3132 |
| 3 | hsa-miR-1273e | 22 | hsa-miR-3154 |
| 4 | hsa-miR-1288 | 23 | hsa-miR-3202 |
| 5 | hsa-miR-132 | 24 | hsa-miR-337-5p |
| 6 | hsa-miR-133a | 25 | hsa-miR-363 |
| 7 | hsa-miR-133b | 26 | hsa-miR-381 |
| 8 | hsa-miR-139-3p | 27 | hsa-miR-3917 |
| 9 | hsa-miR-142-5p | 28 | hsa-miR-409-3p |
| 10 | hsa-miR-143-3p | 29 | hsa-miR-4261 |
| 11 | hsa-miR-143-5p | 30 | hsa-miR-4324 |
| 12 | hsa-miR-145-5p | 31 | hsa-miR-4328 |
| 13 | hsa-miR-145-3p | 32 | hsa-miR-450a |
| 14 | hsa-miR-146a | 33 | hsa-miR-487b |
| 15 | hsa-miR-152 | 34 | hsa-miR-493-5p |
| 16 |
hsa-miR-199a-3p | 35 | hsa-miR-495 |
| 17 |
hsa-miR-199b-5p | 36 | hsa-miR-582-5p |
| 18 | hsa-miR-212 | 37 | hsa-miR-601 |
| 19 | hsa-miR-223 | 38 | hsa-miR-625 |
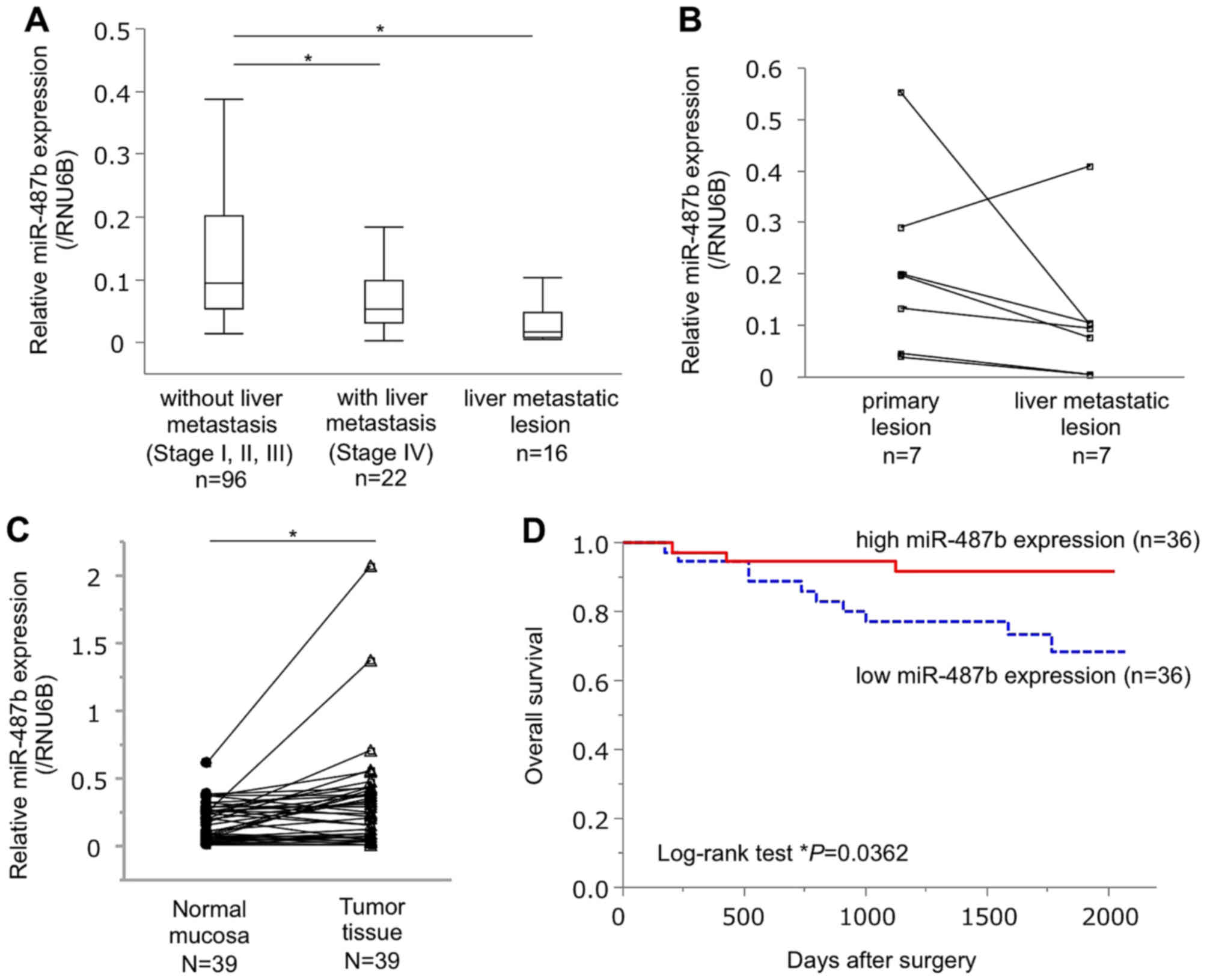
miR-487b is downregulated in CRC liver
metastasis
To confirm the differential expression of miR-487b
between primary tumors without metastasis, and liver metastatic
lesions, miR-487b expression was measured by qRT-PCR in the
validating cohort of 134 patients: primary tumors without
metastasis (n=96), primary tumors with liver metastasis (n=22), and
liver metastatic lesions (n=16). It was confirmed that miR-487b
expression in liver metastatic lesion was significantly lower than
that in primary tumors without metastasis (P=0.041, Fig. 2A). In addition, we found that
miR-487b expression significantly decreased in primary tumors with
liver metastasis, as compared to that without metastasis (P=0.049,
Fig. 2A). When we examined
miR-487b expression in a pair of CRC tissue and its corresponding
synchronous liver metastasis (n=7), metastatic lesions tended to
express lower levels of miR-487b than the primary tumors (P=0.0996,
Fig. 2B). miR-487b expression in
tumor tissues was significantly higher than that in their
pair-matched adjacent normal mucosal tissues (P=0.007, Fig. 2C).
Impact of miR-487b expression on the
patient outcome
To reveal the impact of miR-487b expression on
patient prognosis, 72 CRC patients were divided into two groups
according to the median value of the miR-487b expression.
Kaplan-Meier survival curve showed that the patients with high
expression of miR-487b (n=36) demonstrated better prognosis for
overall survival (OS) than those with low miR-487b expression
(n=36, P=0.036, median follow-up 56.4 months, Fig. 2D). Univariate analysis showed that
tumor depth (P<0.0001), tumor differentiation (P=0.040), lymph
node metastasis (P=0.010), lymphatic invasion (P=0.002), clinical
stage (P<0.0001), and miR-487b expression (P<0.032) were
significant prognostic parameters (Table III). Multivariate analysis
revealed that miR-487b expression was an independent prognostic
factor for 5-year OS, (RR of 4.164, 95% CI of 0.035), in addition
to clinical stage (Table III).
Clinical and pathological survey showed that miR-487b expression
was significantly associated with tumor differentiation (P=0.040,
Table IV).
 | Table IIIUnivariate and multivariate analyses
for 5-year overall survival. |
Table III
Univariate and multivariate analyses
for 5-year overall survival.
| Univariate analysis
| P-value | Multivariate
analysis
| P-value |
|---|
| RR | 95% CI | RR | 95% CI |
|---|
| Gender
(male/female) | 1.069 | 0.356–3.545 | 0.906 | | | |
| Tumor size (≤35
mm/>35 mm) | 0.745 | 0.167–2.438 | 0.647 | | | |
| Location
(colon/rectum) | 0.757 | 0.244–2.279 | 0.616 | | | |
| Differentiation
(tub1, tub2/muc, por) | 0.569 | 0.112–10.383 | 0.617 | | | |
| Depth
(T1-3/T4) | 0.075 | 0.012–0.282 | <0.0001a | 0.316 | 0.045–1.363 | 0.131 |
| Lymph node
metastasis (−/+) | 0.184 | 0.028–0.688 | 0.010a | 0.319 | 0.047–1.311 | 0.120 |
| Lymphatic invasion
(−/+) | 0.09 | 0.005–0.459 | 0.002a | 0.193 | 0.010–1.214 | 0.085 |
| Venous invasion
(−/+) | 0.262 | 0.014–1.331 | 0.120 | 0.890 | 0.196–2.999 | 0.860 |
| Stage
(I–III/IV) | 0.025 | 0.001–0.126 | <0.0001a | 0.099 | 0.005–0.714 | 0.020a |
| miR-487b expression
(low/high) | 3.628 | 1.109–16.185 | 0.032a | 4.164 | 1.097–21.600 | 0.035a |
 | Table IVmiR-487b expression and
clinicopathological characteristics in colorectal cancer
patients. |
Table IV
miR-487b expression and
clinicopathological characteristics in colorectal cancer
patients.
| Patient
characteristics | High miR-487b
expression
(n=36) | Low miR-487b
expression
(n=36) | P-value |
|---|
| Gender |
| Male | 21 | 23 | |
| Female | 15 | 13 | 0.629 |
| Tumor size |
| ≤35 mm | 10 | 10 | |
| >35 mm | 26 | 26 | 1 |
| Location |
| Colon | 16 | 21 | |
| Rectum | 20 | 15 | 0.238 |
|
Differentiation |
| tub1, tub2 | 36 | 32 | |
| muc, por | 0 | 4 | 0.040a |
| Depth |
| T1,T2,T3 | 27 | 20 | |
| T4 | 9 | 16 | 0.083 |
| Lymph node
metastasis |
| Negative | 14 | 18 | |
| Positive | 22 | 18 | 0.343 |
| Lymphatic
invasion |
| Negative | 13 | 18 | |
| Positive | 23 | 18 | 0.234 |
| Venous
invasion |
| Negative | 6 | 11 | |
| Positive | 30 | 25 | 0.165 |
| Stage |
| I, II, III | 29 | 22 | |
| IV | 7 | 14 | 0.07 |
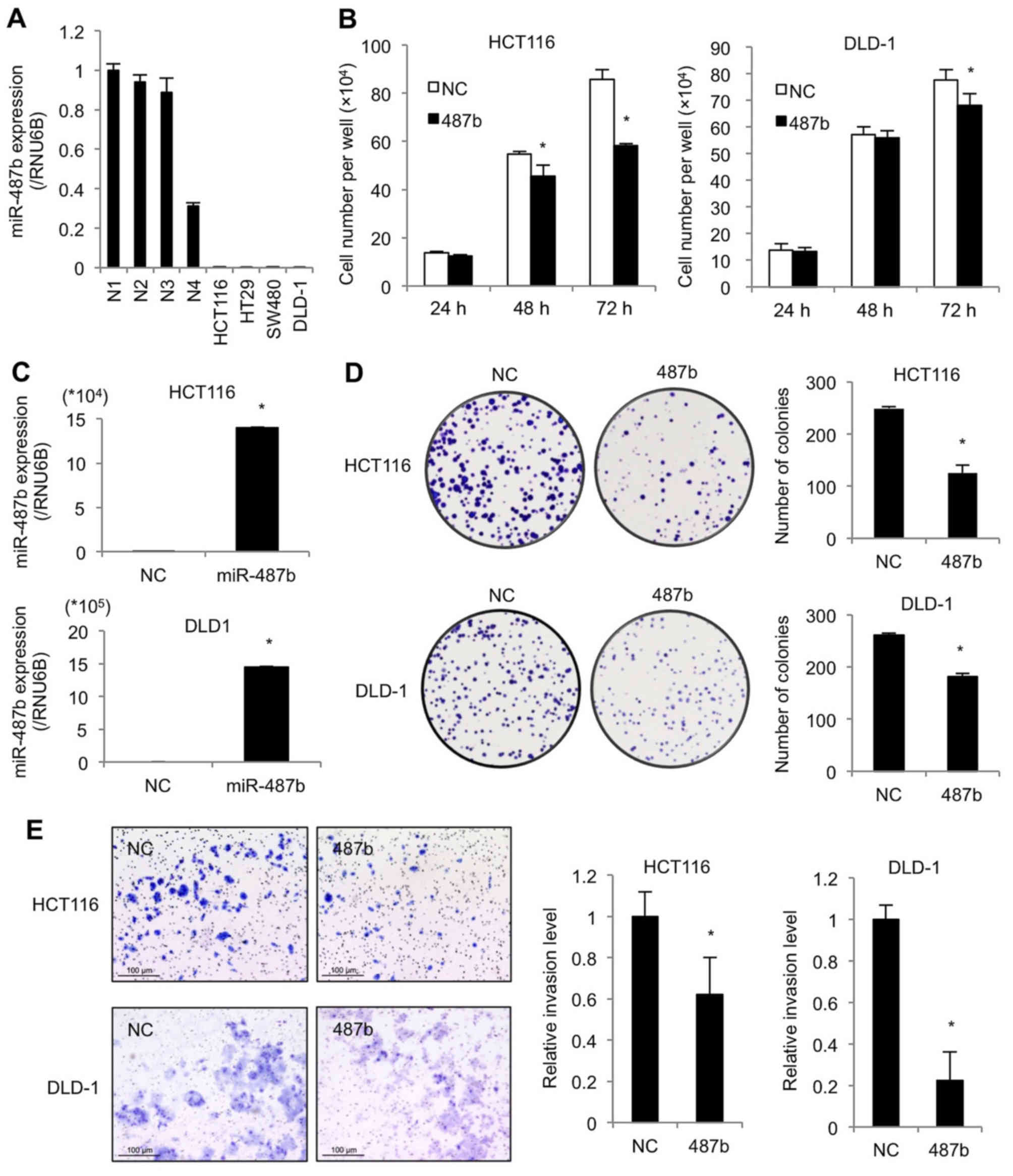
miR-487b inhibits cell proliferation and
invasion ability in CRC cells
We then performed in vitro experiments using
human CRC cell lines. First, qRT-PCR revealed that the miR-487b
levels of representative four CRC cell lines were low compared to
clinical normal tissue samples (Fig.
3A). The ectopic expression of miR-487b resulted in reduction
of cell viability at 48 and 72 h after transfection in HCT116, and
at 72 h in DLD-1 (Fig. 3B).
Increased expression of miR-487b was validated at 48 h by qRT-PCR
in both cell lines (Fig. 3C). In
colony formation assay, the numbers of colonies significantly
decreased in miR-487b transfected cells as compared to negative
control cultures in both cell lines (Fig. 3D). We then assessed cell invasion,
since it is thought to play a crucial role at an initial step of
metastasis. miR-487b transduced cells showed a marked reduction in
the invasion ability in both cell lines (Fig. 3E). These results indicate that
miR-487b suppresses cell proliferation, and more evidently inhibits
invasion ability in CRC.
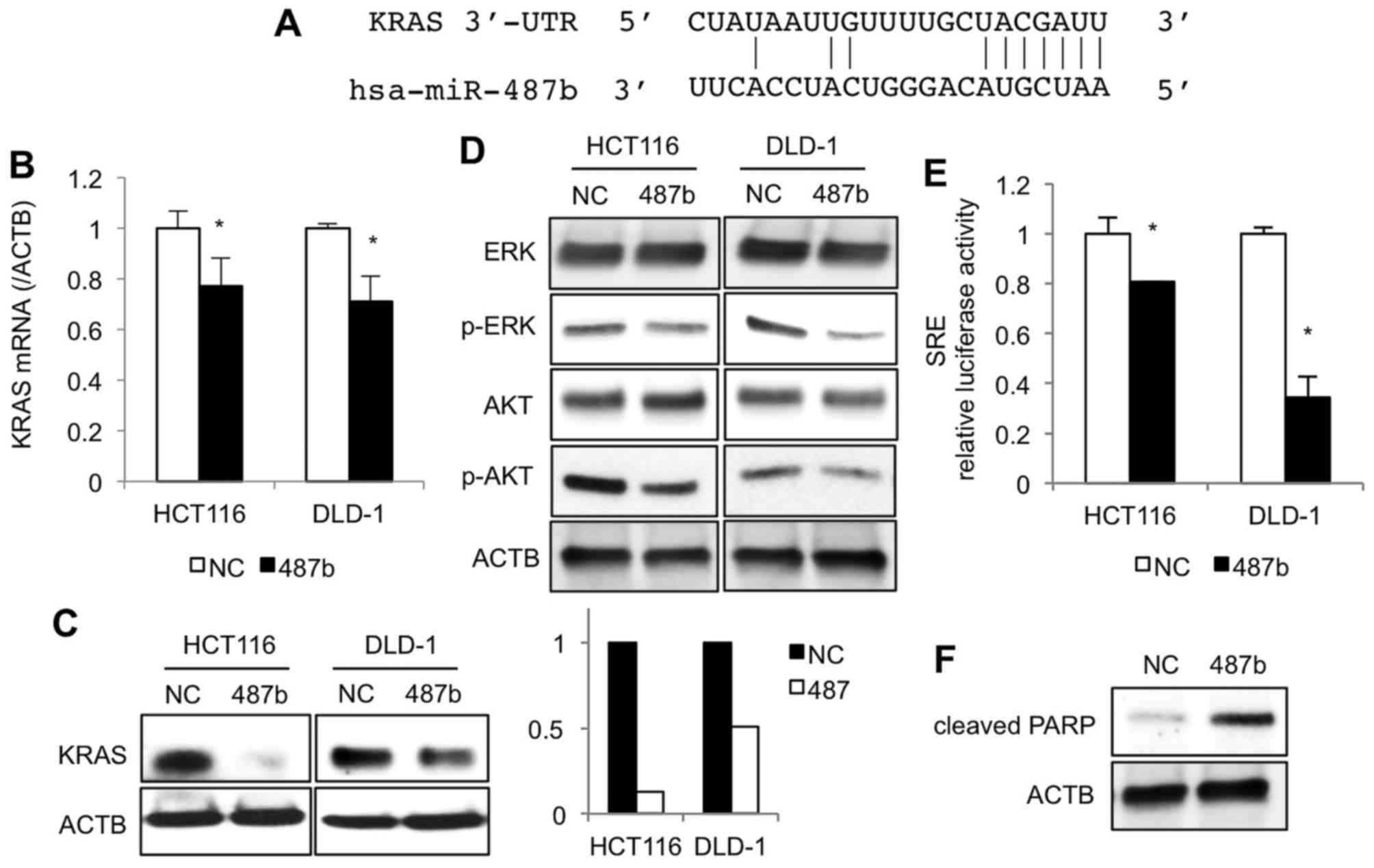
miR-487b downregulates KRAS signaling
pathway in CRC cells
To reveal the underlying mechanisms of how miR-487b
would suppress proliferation and invasion ability, we explored KRAS
signaling pathway because there is evidence that miR-487b targets
3′-UTRs of KRAS mRNA in lung cancer cell lines (17) (Fig.
4A). At 48 h after transfection, KRAS mRNA levels in miR-487b
transfected cells significantly decreased compared with negative
control cultures of HCT116 and DLD-1 cells (Fig. 4B), which was clearly demonstrated
with western blot analysis at a protein level (Fig. 4C). Additional studies were
undertaken to examine the impact of miR-487b on downstream
molecules of the KRAS signaling pathway. Phosphorylation of ERK and
AKT in the miR-487b transfected cells decreased when compared to
negative control cultures (Fig.
4D). SRE reporter assay revealed that miR-487b significantly
inhibited SRE luciferase activities, indicating that the MAPK/ERK
signaling pathway was partially blocked by miR-487b (Fig. 4E). Since miR-487b could also
regulate AKT signaling pathways, we next investigated whether
miR-487b would induce apoptosis. We found that introduction of
miR-487b increased cleaved PARP expression in HCT116 cells
(Fig. 4F), but not in DLD-1 cells
(data not shown).
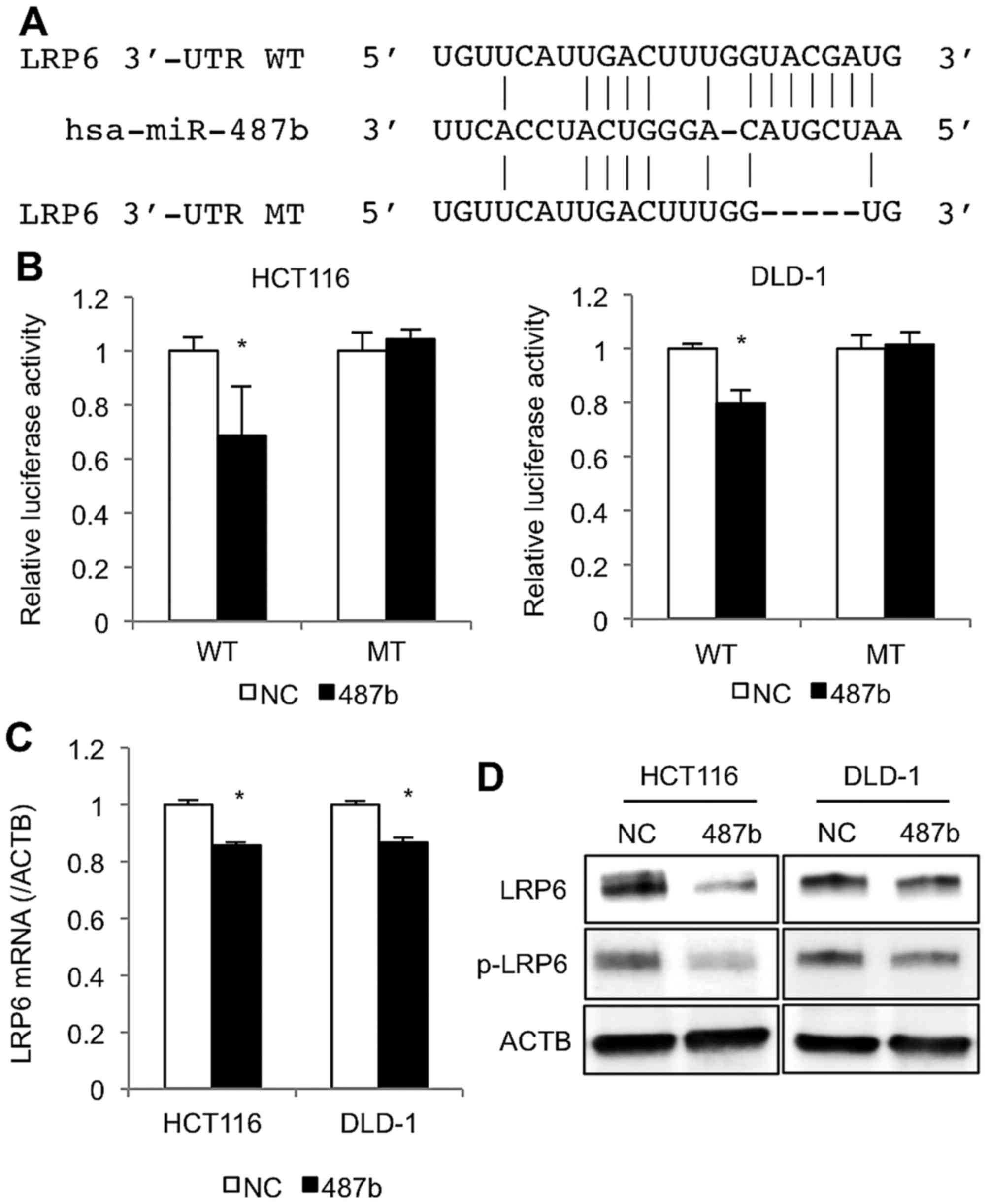
miR-487b directly targets LRP6 in CRC
cells
By using the target prediction tool (TargetScan 7.0,
and microRNA.org.), we found that miR-487b might bind
to the 3′-UTR of LRP6, a Frizzled (FZD) co-receptor for WNT
signaling pathway (Figs. 5A and
6). After miR-487b was
co-transfected with a reporter plasmid harboring LRP6 3′-UTR
sequence, luciferase activities of the reporter were significantly
reduced in HCT116 and DLD-1 cell lines (Fig. 5B), whereas, such decrease was not
observed with mutant LRP6 3′-UTR sequence, indicating that LRP6
could be a direct target of miR-487b. At 48 h after transfection,
LRP6 levels significantly decreased in both cell lines at mRNA and
protein levels by treatment of miR-487b (Fig. 5C and D). Moreover, transfection of
miR-487b reduced the phosphorylated LRP6 protein (Fig. 5D), which was thought to be crucial
for signal transduction in the WNT pathway.
Discussion
Studies have shown that miRNA-181a and miRNA-214
have the potential to suppress liver metastasis through targeting
WNT inhibitory factor 1 (WIF-1) and fibroblast growth factor
receptor 1 (FGFR1), respectively (24,25).
Our group also reported that miRNA-132 was related to liver
metastasis of CRC and targets anoctamin 1 (ANO1) (16). In an effort to identify another
novel miRNA related to liver metastasis, we focused on miR-487b
that significantly decreased in liver metastatic lesions as
compared to primary tumors without metastasis. In this study, we
found that miR-487b level in tumors was even higher than that in
their paired normal tissues. Since miR-487b acts as a tumor
suppressor, this result seems to be somewhat contradictory.
However, such paradox could happen in the process of
carcinogenesis. Weinstein emphasized the existence of feedback
mechanisms to maintain the homeostatic balance in human
malignancies between positive and negative regulators of the cell
cycle, by showing that CDK inhibitors p21Cip1/Waf1,
p27Kip1, or even Rb tumor suppressor gene
products increased in several cancer types (26,27).
Similar scenario is reported in case of miRNA. Thus, anti-onco
miR-132 or -133b rather increased in tumor tissues (28–30).
In addition, some reports showed that miR-34a, a representative
anti-oncomir (31,32), was shown to be upregulated in
cancer tissue rather than in normal mucosa (30,33).
Therefore, we postulate that the present findings of an increase in
miR-487b in tumor tissues may not be unique considering the complex
miRNA networks in cancer development.
Our study revealed that miR-487b inhibited cell
proliferation, colony formation, and cell invasion. These findings
suggest that miR-487b may play a role as an anti-oncomir (i.e.,
tumor suppressor) as demonstrated by other studies; Xi et al
reported that miR-487b targets SUZ12, BMI1, WNT5A, MYC and KRAS and
that its expression was downregulated in lung cancer (17). Gattolliat et al showed that
high miR-487b expression was a marker for improved prognosis in
neuroblastoma (19). In this
study, we focused on RAS and WNT/β-catenin signaling pathway, both
of which are essential for cancer metastasis (34,35).
Consistent with a previous study by Xi et al
(17), we confirmed that miR-487b
clearly downregulated KRAS expression in CRC, which linked to
decrease in phosphorylation of ERK and AKT. miR-487b eventually
decreased the SRE transcription activity. Earlier studies have
established the roles of KRAS-dependent signaling in enhancement of
cell proliferation, invasion and metastasis in CRC (34,36).
The mutation status of KRAS gene is one of the most
essential factors for treatment strategy against CRC in the use of
anti-EGFR antibody. Moreover, recent studies have shown that the
emergence of KRAS mutations is a mediator of acquired
resistance to EGFR blockade (37,38).
In this regard, the ability of miR-487b to suppress MAPK/ERK and
AKT signaling might be an effective approach to CRC with
KRAS mutation.
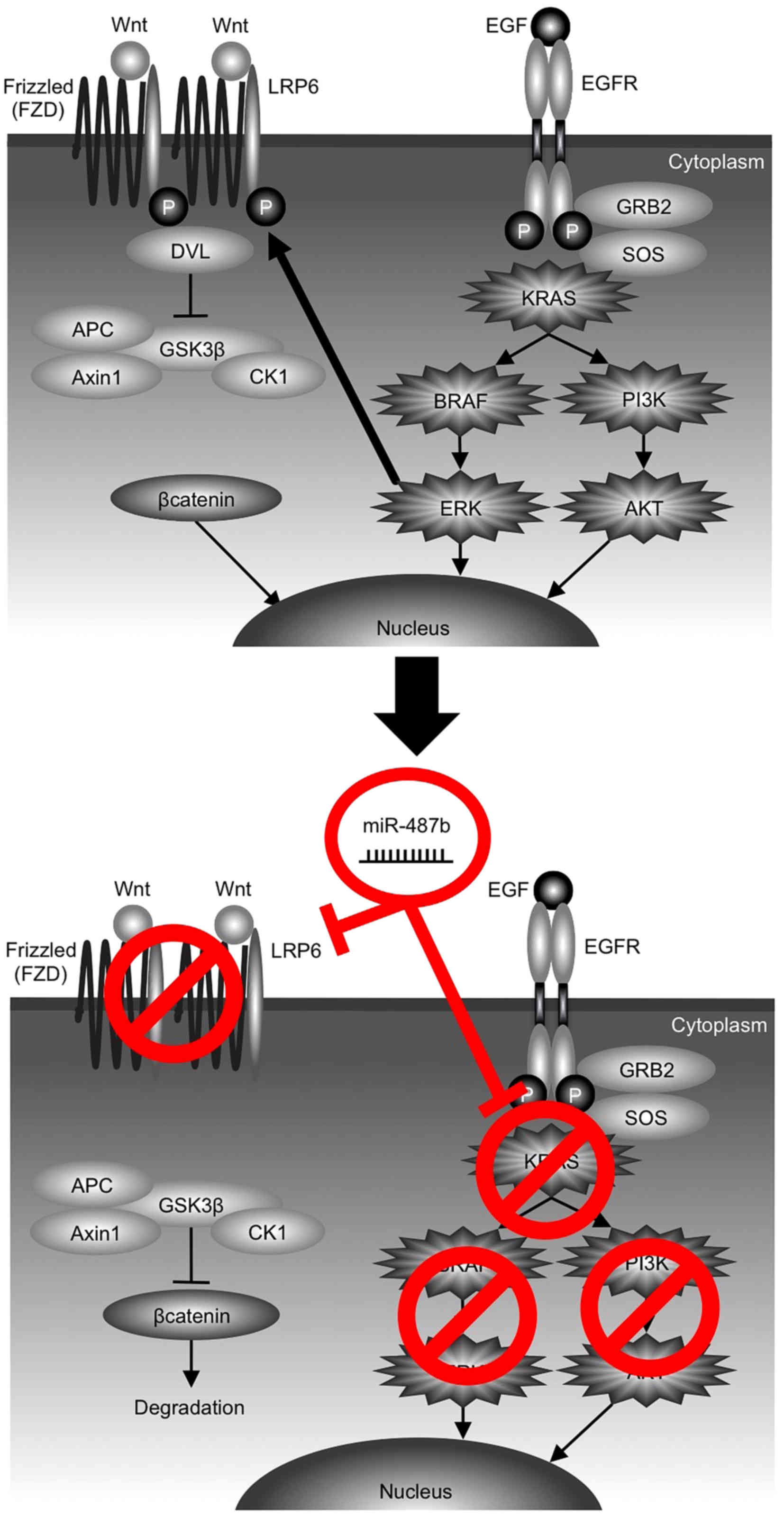
LRP6, a single span transmembrane protein, is a
crucial component of the WNT/β-catenin signaling pathway. We found
that LRP6 harbored miR-487b binding sequence in its 3′-UTR by miRNA
target screening and validated this binding. LRP6 forms a receptor
complex with FZD in presence of WNT ligand, and DVL recruitment by
FZD leads to LRP6 phosphorylation and Axin recruitment, which
allows β-catenin to accumulate in the nucleus where it serves as a
coactivator for TCF to activate WNT-responsive genes (39) (Fig.
6). LRP6 has been shown to function as an oncogene by promoting
cell growth, invasion and migration, and is expected to be a target
for cancer therapy (40–45). Notably, there is evidence that
ERK1/2 in the RAS/MAPK pathway could facilitate WNT/β-catenin
signaling via LRP6 phosphorylation on serine-1490 (S1490) and
threonine-1572 (T1572) at both mature (transmenbrane LRP6) and
immature state during its Golgi network-based maturation process
(46,47). Taken together, it is conceivable
that miR-487b may further contribute to break the synergistic
interaction between ERK and LRP6, as a result of the concurrent
dual targeting of KRAS and LRP6 (Fig.
6). Downstream of LRP6, however, it is known that a majority of
CRC harbors mutation in APC or CTNNB1 gene and the
frequency of gene mutations involved in the canonical WNT/β-catenin
pathway accounted for more than 90% (48–51).
Therefore, it should be carefully assessed to what extent
downregulation of LRP6 would affect the WNT/β-catenin signaling and
the malignant potential of CRC. Further investigation on this issue
is essential as the next step.
In conclusion, this study demonstrated that
decreased expression of miR-487b was associated with liver
metastasis and that miR-487b may play a crucial role in regulating
tumor progression in CRC through targeting KRAS. Our data suggest
that miR-487b could be a sensitive marker for liver metastasis and
prognosis of the patients. Further study is needed to establish a
novel strategy using miR-487b for treatment against CRC.
Acknowledgments
This study was supported by a Grant-in Aid for
Scientific Research (KAKENHI) to Hirofumi Yamamoto (nos. 21390360,
30322184 and 24390315).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brenner H, Kloor M and Pox CP: Colorectal
cancer. Lancet. 383:1490–1502. 2014. View Article : Google Scholar
|
|
3
|
Kopetz S, Chang GJ, Overman MJ, Eng C,
Sargent DJ, Larson DW, Grothey A, Vauthey JN, Nagorney DM and
McWilliams RR: Improved survival in metastatic colorectal cancer is
associated with adoption of hepatic resection and improved
chemotherapy. J Clin Oncol. 27:3677–3683. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Leonard GD, Brenner B and Kemeny NE:
Neoadjuvant chemotherapy before liver resection for patients with
unresectable liver metastases from colorectal carcinoma. J Clin
Oncol. 23:2038–2048. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Amano R, Yamada N, Nakata B, Kimura K,
Yashiro M, Ohira M and Hirakawa K: Prognostic indicator for the
resection of liver metastasis of colorectal cancer. Surg Today.
44:1287–1292. 2014. View Article : Google Scholar
|
|
6
|
Sorski L, Levi B, Shaashua L, Neeman E,
Benish M, Matzner P, Hoffman A and Ben-Eliyahu S: Impact of
surgical extent and sex on the hepatic metastasis of colon cancer.
Surg Today. 44:1925–1934. 2014. View Article : Google Scholar
|
|
7
|
Chen K and Rajewsky N: The evolution of
gene regulation by transcription factors and microRNAs. Nat Rev
Genet. 8:93–103. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang H, Li Y and Lai M: The microRNA
network and tumor metastasis. Oncogene. 29:937–948. 2010.
View Article : Google Scholar
|
|
10
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kent OA and Mendell JT: A small piece in
the cancer puzzle: MicroRNAs as tumor suppressors and oncogenes.
Oncogene. 25:6188–6196. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Stiegelbauer V, Perakis S, Deutsch A, Ling
H, Gerger A and Pichler M: MicroRNAs as novel predictive biomarkers
and therapeutic targets in colorectal cancer. World J
Gastroenterol. 20:11727–11735. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu WK, Law PT, Lee CW, Cho CH, Fan D, Wu
K, Yu J and Sung JJ: MicroRNA in colorectal cancer: From benchtop
to bedside. Carcinogenesis. 32:247–253. 2011. View Article : Google Scholar
|
|
15
|
US National Institutes of Health: A
multicenter phase I study of MRX34, microRNA miR-RX34 liposomal
injection. https://clinicaltrials.gov/ct2/show/NCT01829971.
Accessed May 27, 2016.
|
|
16
|
Mokutani Y, Uemura M, Munakata K, Okuzaki
D, Haraguchi N, Takahashi H, Nishimura J, Hata T, Murata K,
Takemasa I, et al: Down-regulation of microRNA-132 is associated
with poor prognosis of colorectal cancer. Ann Surg Oncol. Feb
11–2016.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xi S, Xu H, Shan J, Tao Y, Hong JA,
Inchauste S, Zhang M, Kunst TF, Mercedes L and Schrump DS:
Cigarette smoke mediates epigenetic repression of miR-487b during
pulmonary carcinogenesis. J Clin Invest. 123:1241–1261. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Stewart DJ: Wnt signaling pathway in
non-small cell lung cancer. J Natl Cancer Inst. 106:djt3562014.
View Article : Google Scholar
|
|
19
|
Gattolliat CH, Thomas L, Ciafrè SA,
Meurice G, Le Teuff G, Job B, Richon C, Combaret V, Dessen P,
Valteau-Couanet D, et al: Expression of miR-487b and miR-410
encoded by 14q32.31 locus is a prognostic marker in neuroblastoma.
Br J Cancer. 105:1352–1361. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
21
|
Yamamoto H, Murata K, Fukunaga M, Ohnishi
T, Noura S, Miyake Y, Kato T, Ohtsuka M, Nakamura Y, Takemasa I, et
al: Micrometastasis volume in lymph nodes determines disease
recurrence rate of stage II colorectal cancer: A prospective
multicenter trial. Clin Cancer Res. 22:3201–3208. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hamabe A, Konno M, Tanuma N, Shima H,
Tsunekuni K, Kawamoto K, Nishida N, Koseki J, Mimori K, Gotoh N, et
al: Role of pyruvate kinase M2 in transcriptional regulation
leading to epithelial-mesenchymal transition. Proc Natl Acad Sci
USA. 111:15526–15531. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hiraki M, Nishimura J, Takahashi H, Wu X,
Takahashi Y, Miyo M, Nishida N, Uemura M, Hata T, Takemasa I, et
al: Concurrent targeting of KRAS and AKT by miR-4689 is a novel
treatment against mutant KRAS colorectal cancer. Mol Ther Nucleic
Acids. 4:e2312015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ji D, Chen Z, Li M, Zhan T, Yao Y, Zhang
Z, Xi J, Yan L and Gu J: MicroRNA-181a promotes tumor growth and
liver metastasis in colorectal cancer by targeting the tumor
suppressor WIF-1. Mol Cancer. 13:862014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen DL, Wang ZQ, Zeng ZL, Wu WJ, Zhang
DS, Luo HY, Wang F, Qiu MZ, Wang DS, Ren C, et al: Identification
of microRNA-214 as a negative regulator of colorectal cancer liver
metastasis by way of regulation of fibroblast growth factor
receptor 1 expression. Hepatology. 60:598–609. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Weinstein IB: Disorders in cell circuitry
during multistage carcinogenesis: The role of homeostasis.
Carcinogenesis. 21:857–864. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Weinstein IB, Begemann M, Zhou P, Han EK,
Sgambato A, Doki Y, Arber N, Ciaparrone M and Yamamoto H: Disorders
in cell circuitry associated with multistage carcinogenesis:
Exploitable targets for cancer prevention and therapy. Clin Cancer
Res. 3:2696–2702. 1997.
|
|
28
|
Zheng YB, Luo HP, Shi Q, Hao ZN, Ding Y,
Wang QS, Li SB, Xiao GC and Tong SL: miR-132 inhibits colorectal
cancer invasion and metastasis via directly targeting ZEB2. World J
Gastroenterol. 20:6515–6522. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin CW, Li XR, Zhang Y, Hu G, Guo YH, Zhou
JY, Du J, Lv L, Gao K, Zhang Y, et al: TAp63 suppress metastasis
via miR-133b in colon cancer cells. Br J Cancer. 110:2310–2320.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kara M, Yumrutas O, Ozcan O, Celik OI,
Bozgeyik E, Bozgeyik I and Tasdemir S: Differential expressions of
cancer-associated genes and their regulatory miRNAs in colorectal
carcinoma. Gene. 567:81–86. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gao J, Li N, Dong Y, Li S, Xu L, Li X, Li
Y, Li Z, Ng SS, Sung JJ, et al: miR-34a-5p suppresses colorectal
cancer metastasis and predicts recurrence in patients with stage
II/III colorectal cancer. Oncogene. 34:4142–4152. 2015. View Article : Google Scholar
|
|
32
|
Raver-Shapira N, Marciano E, Meiri E,
Spector Y, Rosenfeld N, Moskovits N, Bentwich Z and Oren M:
Transcriptional activation of miR-34a contributes to p53-mediated
apoptosis. Mol Cell. 26:731–743. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Krell J, Frampton AE, Mirnezami R, Harding
V, De Giorgio A, Roca Alonso L, Cohen P, Ottaviani S, Colombo T,
Jacob J, et al: Growth arrest-specific transcript 5 associated
snoRNA levels are related to p53 expression and DNA damage in
colorectal cancer. PLoS One. 9:e985612014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Smakman N, Borel Rinkes IH, Voest EE and
Kranenburg O: Control of colorectal metastasis formation by K-Ras.
Biochim Biophys Acta. 1756:103–114. 2005.PubMed/NCBI
|
|
35
|
Anastas JN and Moon RT: WNT signalling
pathways as therapeutic targets in cancer. Nat Rev Cancer.
13:11–26. 2013. View Article : Google Scholar
|
|
36
|
Pretlow TP and Pretlow TG: Mutant KRAS in
aberrant crypt foci (ACF): Initiation of colorectal cancer? Biochim
Biophys Acta. 1756:83–96. 2005.PubMed/NCBI
|
|
37
|
Misale S, Yaeger R, Hobor S, Scala E,
Janakiraman M, Liska D, Valtorta E, Schiavo R, Buscarino M,
Siravegna G, et al: Emergence of KRAS mutations and acquired
resistance to anti-EGFR therapy in colorectal cancer. Nature.
486:532–536. 2012.PubMed/NCBI
|
|
38
|
Diaz LA Jr, Williams RT, Wu J, Kinde I,
Hecht JR, Berlin J, Allen B, Bozic I, Reiter JG, Nowak MA, et al:
The molecular evolution of acquired resistance to targeted EGFR
blockade in colorectal cancers. Nature. 486:537–540.
2012.PubMed/NCBI
|
|
39
|
MacDonald BT, Tamai K and He X:
Wnt/beta-catenin signaling: Components, mechanisms, and diseases.
Dev Cell. 17:9–26. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li Y, Lu W, He X, Schwartz AL and Bu G:
LRP6 expression promotes cancer cell proliferation and
tumorigenesis by altering beta-catenin subcellular distribution.
Oncogene. 23:9129–9135. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu CC, Prior J, Piwnica-Worms D and Bu G:
LRP6 overexpression defines a class of breast cancer subtype and is
a target for therapy. Proc Natl Acad Sci USA. 107:5136–5141. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tung EK, Wong BY, Yau TO and Ng IO:
Upregulation of the Wnt co-receptor LRP6 promotes
hepatocarcinogenesis and enhances cell invasion. PLoS One.
7:e365652012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Du C, Lv Z, Cao L, Ding C, Gyabaah OA, Xie
H, Zhou L, Wu J and Zheng S: MiR-126-3p suppresses tumor metastasis
and angiogenesis of hepatocellular carcinoma by targeting LRP6 and
PIK3R2. J Transl Med. 12:2592014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang Y, Zheng D, Xiong Y, Xue C, Chen G,
Yan B and Ye Q: miR-202 suppresses cell proliferation in human
hepatocellular carcinoma by downregulating LRP6
post-transcriptionally. FEBS Lett. 588:1913–1920. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zeng XC, Liu FQ, Yan R, Yi HM, Zhang T,
Wang GY, Li Y and Jiang N: Downregulation of miR-610 promotes
proliferation and tumorigenicity and activates Wnt/β-catenin
signaling in human hepatocellular carcinoma. Mol Cancer.
13:2612014. View Article : Google Scholar
|
|
46
|
Lemieux E, Cagnol S, Beaudry K, Carrier J
and Rivard N: Oncogenic KRAS signalling promotes the Wnt/β-catenin
pathway through LRP6 in colorectal cancer. Oncogene. 34:4914–4927.
2015. View Article : Google Scholar
|
|
47
|
Krejci P, Aklian A, Kaucka M, Sevcikova E,
Prochazkova J, Masek JK, Mikolka P, Pospisilova T, Spoustova T,
Weis M, et al: Receptor tyrosine kinases activate canonical
WNT/β-catenin signaling via MAP kinase/LRP6 pathway and direct
β-catenin phosphorylation. PLoS One. 7:e358262012. View Article : Google Scholar
|
|
48
|
Morin PJ, Sparks AB, Korinek V, Barker N,
Clevers H, Vogelstein B and Kinzler KW: Activation of
beta-catenin-Tcf signaling in colon cancer by mutations in
beta-catenin or APC. Science. 275:1787–1790. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Bienz M and Clevers H: Linking colorectal
cancer to Wnt signaling. Cell. 103:311–320. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Cancer Genome Atlas N; Cancer Genome Atlas
Network: Comprehensive molecular characterization of human colon
and rectal cancer. Nature. 487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Masuda M, Uno Y, Ohbayashi N, Ohata H,
Mimata A, Kukimoto-Niino M, Moriyama H, Kashimoto S, Inoue T, Goto
N, et al: TNIK inhibition abrogates colorectal cancer stemness. Nat
Commun. 7:125862016. View Article : Google Scholar : PubMed/NCBI
|