Introduction
Prostate cancer (PCa) is the most common solid
cancer entity among men in industrialized countries (1). Annually more than 250,000 men die
from metastasized PCa worldwide. However, many aspects in PCa
development and progression are still unknown.
In many different cancer entities signaling via
extracellular particles has impact on different tumor-related
processes (2,3). Especially extracellular vesicles are
in the focus of current research. They play a role in modulation of
tumor microenvironment (4),
neo-angiogenesis (5), metastatic
niche formation (6) and systemic
inflammation (7). PCa
extracellular vesicles are involved in crosstalk between tumor
cells and the surrounding stroma (8,9).
Exosomes are the best-studied subpopulation of
extracellular vesicles. They have a typical size of 30–100 nm and,
unlike the larger microvesicles which directly originate from the
plasma membrane, are generated from the membranes of endosomes.
These endosomes are termed as multi-vesicular-bodies (MVB). Exosome
generation is mainly mediated by the components of the ESCRT
machinery (endosomal sorting complexes required for transport),
with tetraspanin-rich microdomains and lipids like
sphingosin-1-phosphate also being involved in these processes
(10–12). MVBs release exosomes into the
extracellular space by fusion with the plasma membrane (12). Based on current knowledge, fusion
with the plasma membrane is mainly mediated by the GTPases RAB27A
and RAB27B (13). Further proteins
such as Plectin and Rab11 are also involved in this process
(12). Extracellular vesicles are
rich in proteins involved in membrane transportation and membrane
fusion (GTPases, annexins, flotillins), tetraspanins like CD9, CD63
and CD81, heat shock proteins HSP70 and HSP90 and proteins involved
in MVB formation such as ALIX and TSG101 (14,15).
Compared to microvesicles, the composition of exosomes is more
stringently controlled by specific processes (16–18).
Many studies deal with the function of extracellular
vesicles and their potential use as biomarkers. However, not much
is known about the role of genes involved in vesicular trafficking
and secretion, in cancer. Therefore, it was the aim of this study
to elucidate their relevance in PCa. Their expression was
re-analyzed in existing microarray data with regard to PCa patient
outcome prediction. qRT-PCR profiling of PCa tissue samples at
various stages served as validation. In vitro studies were
done to functionally examine the impact of the genes associated
with patient outcome on proliferation, colony formation and release
of extracellular particles.
Materials and methods
Candidate list generation and in silico
analyses
Based on recent literature a list of genes with
known functions in vesicular trafficking and secretion, like the
ESCRT components TSG101, MVB12, VPS36 and PDCD6IP (ALIX), and of
typical extracellular vesicle markers (e.g., flotillins and the
tetraspanins CD9, CD63 and CD81) was assembled. Furthermore, genes
typically associated with PCa, such as KLK3 (PSA) or PTEN were
added. The full list of genes is given in Table I. RNA expression data and clinical
and pathological parameters of a PCa dataset (19), comprising of 131 primary (mean age,
58.03±6.99 years; mean PSA, 8.48±12.28 ng/ml; T2, 85; T3, 40; T4,
6) and 19 metastatic tumor samples (mean age, 60.51±7.38 years;
mean PSA, 72.99±135.59 ng/ml) were derived from cBioPortal
(20). Gene expression in tumor
samples is compared to healthy controls and adjacent tumor-free
tissue.
 | Table IA list of genes identified from the
literature with known function in exosome biogenesis and secretion
and known PCa biomarkers was assembled for further in silico
analyses.a |
Table I
A list of genes identified from the
literature with known function in exosome biogenesis and secretion
and known PCa biomarkers was assembled for further in silico
analyses.a
| Group | Gene symbol | Gene name |
|---|
| Known PCa
markers | AMACR | α-methylacyl-CoA
racemase |
| AR | Androgen
receptor |
| EZH2 | Enhancer of zeste 2
polycomb repressive complex 2 subunit |
| FOLH1
(PSMA) | Folate hydroxylase
1 (prostate specific membrane antigen) |
| KLK3
(PSA) | Kallikrein 3
(prostate specific antigen) |
| PCA3 | Prostate cancer
antigen 3 |
| PTEN | Phosphatase and
tensin homolog |
| ESCRT-0 | STAM1 | Signal transducing
adaptor molecule 1 |
| STAM2 | Signal transducing
adaptor molecule 2 |
| VPS27 | Vacuolar protein
sorting 27 homolog |
| ESCRT-I | MVB12 | Multivesicular body
subunit 12 |
| PDCD6IP | Programmed cell
death 6 interacting protein |
| TSG101 | Tumor
susceptibility gene 101 |
| UBAP1 | Ubiquitin
associated protein 1 |
| VPS28 | VPS28, ESCRT-I
subunit |
| VPS37 | VPS37, ESCRT-I
subunit |
| ESCR-II | SNF8
(EAP30) | SNF8, ESCRT-II
complex subunit |
| VPS25
(EAP20) | Vacuolar protein
sorting 25 homolog |
| VPS36
(EAP45) | Vacuolar protein
sorting 36 homolog |
| ESCRT-III | CHMP6 | Charged
multivesicular body protein 6 |
| IST1 | IST1, ESCRT-III
associated factor |
| VPS4A | Vacuolar protein
sorting 4 homolog A |
| VTA1
(LIP5) | Vesicle trafficking
1 |
| Rab-kinases | RAB5a | RAB5A, member RAS
oncogene family |
| Rab11 | RAB11, member RAS
oncogene family |
| RAB27A | RAB27A, member RAS
oncogene family |
| RAB27B | RAB27B, member RAS
oncogene family |
| Rab35 | RAB35, member RAS
oncogene family |
| RAB36 | RAB36, member RAS
oncogene family |
| TBC1D10A
(EPI64) | TBC1 domain family
member 10A |
| Exosome-associated
genes | CD9 | CD9 molecule |
| CD63 | CD63 molecule |
| CD81 | CD81 molecule |
| CHMP4C | Charged
multivesicular body protein 4C |
| DCT
(TYRP2) | Dopachrome
tautomerase |
| EPCAM | Epithelial cell
adhesion molecule |
| FLOT1 | Flotillin 1 |
| FLOT2 | Flotillin 2 |
| HSP4A | Heat shock protein
family A (Hsp70) member 4 |
|
HSP90AA1 | Heat shock protein
90 α family class A member 1 |
| MMP2 | Matrix
metallopeptidase 2 |
| MMP9 | Matrix
metallopeptidase 9 |
| MMP14 | Matrix
metallopeptidase 14 |
| STEAP3 | STEAP3
metalloreductase |
The expression z-scores in patients with localized
PCa, obtained from the original data, were used to define
expression cut-off values (high vs. low expression) for the event
of biochemical recurrence (BCR). Calculations were done using the
partition test in SAS JMP 11 (SAS Institute, Cary, NC, USA). These
cut-off values were applied to the cohort and correlated with
BCR-free survival using log-rank test. Only candidates with >20
patients in the high and low expression groups were followed-up.
BCR-free survival was displayed using Kaplan-Meier graphs. In
candidates reaching significance for prediction of BCR, expression
values were stratified according to T-stage and Gleason grade.
Correlation with clinical and pathological parameters was
additionally done using two further microarray datasets (Tomlins
et al and Yu et al) (21,22).
In all three datasets correlations between the expression of the
different candidate genes were done using Spearman correlation.
Cohorts and patient samples
A cDNA array (Origene, Rockville, MD, USA)
consisting of 40 localized PCa and 8 benign control samples was
used as a first step to independently re-test the expression of
RAB27A, RAB27B and VPS36. Patient characteristics of this cohort
are shown in Table II. Expression
of all three candidates was correlated with T-stage and Gleason
grade.
 | Table IIPatient characteristics in the
Origene cDNA array. |
Table II
Patient characteristics in the
Origene cDNA array.
| Patients | Localized PCa
(n=40) | BPH (n=8) |
|---|
| Mean age (SD) | 62.75±8.15 | 64.15±10.9 |
| T1 | – | |
| T2 | 22 | |
| T3 | 12 | |
| T4 | – | |
| Not specified | 6 | |
| Gleason 5 | 2 | |
| Gleason 6 | 8 | |
| Gleason 7a | 14 | |
| Gleason 7b | 8 | |
| Gleason 8 | 3 | |
| Gleason 9 | 4 | |
| Not specified | 1 | |
In a second step a cohort of 41 patients who
underwent transurethral resection of PCa for palliation in the
Department of Urology of the Mannheim Medical Center between May
2005 and September 2015 was retrospectively analyzed. Nine patients
were treated twice with transurethral resection, resulting in a
total of 50 FFPE tumor samples. FFPE prostate tissue specimen of
ten patients who underwent transurethral resection due to benign
prostatic hyperplasia (BPH), histologically negative for PCa,
served as controls. Patient data of this cohort is given in
Table III. All experiments were
in accordance with the institutional ethics review board (Medical
Ethics Committee II of the Medical Faculty Mannheim, ethics
approval 2013-845R-MA). Expression data were correlated with tumor
stage and Gleason grade in both datasets. In tumor samples obtained
from transurethral resection further correlations with castration
resistance were done.
 | Table IIIPatient characteristics in the TUR
cohort. |
Table III
Patient characteristics in the TUR
cohort.
Patients
Samplesa | PCa
(n=41)
PCa (n=50) | BPH
(n=10)
BPH (n=10) |
|---|
| Mean age (SD) | 75.48±6.86 | 66.9±13.56 |
|
Hormone-sensitive | 74.43±6.13 | |
|
Castration-resistant | 75.89±7.15 | |
|
Hormone-sensitive | 14 | |
| Gleason 5: | 2 | |
| Gleason 6 | 3 | |
| Gleason 7 | 4 | |
| Gleason 8 | 4 | |
| Gleason 9 | 1 | |
|
Castration-resistant | 36 | |
RNA-extraction and cDNA-synthesis
Tumor-bearing and tumor-free FFPE prostate tissue
specimen of patients from our institution were sectioned and
stained with hematoxylin and eosin and reviewed microscopically.
Subsequent unstained 10-μm sections were placed on glass
slides. Tissue sections of control patients and those of PCa
patients bearing ≥80% of tumor tissue were used completely. On
sections containing <80% of tumor, non-tumorous areas were
scratched off prior to RNA extraction using a sterile scalpel.
Sections were deparaffinized three times in 2 ml of Neo Clear
(Merck Millipore, Billerica, MA, USA). The deparaffinized tissue
was then scraped off the glass slides into a reaction tube
containing 150 μl of lysis buffer. RNA extraction was
conducted with the XTRACT FFPE kit (Stratifyer, Cologne, Germany)
as suggested by the manufacturer (23,24).
In brief the tubes were incubated 30 min at 80°C with shaking.
After cooling to 65°C 50 μl of Proteinase K (Promega,
Fitchburg, WI, USA) were added and incubated for 30 min with
shaking. Subsequently 800 μl of MagiX-RNA buffer and 40
μl of MagiX-RNA ferromagnetic beads were added and incubated
15 min at room temperature with shaking. The tubes were placed on a
magnetic rack and the supernatant was removed. Three washing steps
were subsequently conducted, after each washing, supernatant was
removed with the tubes on the magnetic rack. The RNA was eluted by
adding 100 μl of elution buffer which was incubated for 15
min at 70°C. With the tube on the magnetic rack the supernatant,
containing the RNA, was transferred to a new reaction tube and
either used immediately or stored at −80°C. RNA-concentration was
determined spectrophotometrically using the NanoDrop 1000 (Thermo
Fisher Scientific, Waltham, MA, USA).
cDNA synthesis was performed using the SuperScript
III reverse transcription kit (Thermo Fisher Scientific) with
sequence specific primers. In brief 2.5 μl of forward
primers of RAB27A, RAB27B, VPS36, androgen receptor (AR) or the
house keeping gene calmodulin 2 (CALM2), 1 μl of 10 mM dNTP
Mix and 3.5 μl of nuclease-free H2O were added to
5 μl of RNA template. This mixture was incubated for 10 min
at 65°C and was subsequently cooled on ice. For cDNA synthesis 4
μl of First Strand synthesis buffer, 1 μl of 0.1 M
DDT, 1 μl of RNAseOut (Thermo Fisher Scientific) and 2
μl of SuperSript III reverse transcriptase were added and
incubated for 120 min at 70°C. cDNA was immediately used for
qRT-PCR or stored at −20°C.
qRT-PCR analyses in patient samples
The expression of RAB27A, RAB27B and VPS36 was
determined in all samples of the cDNA array in relation to the
housekeeping gene CALM2. For all assays intron spanning primer
pairs were designed using Primer-BLAST, based on the primer3
algorithm (25).
In brief, 10 μl of TaqMan Fast Universal PCR
Mastermix (Life Technologies, Darmstadt, Germany), 0.75 μl
of forward and reverse primer, each (300 nM), 0.5 μl of PCR
probe (200 nM) (both MWG Eurofins, Ebersberg, Germany) and 6
μl of nuclease-free H2O were added to 2 μl
of cDNA template. Subsequently 50 cycles of PCR amplification with
3 sec of 95°C and 30 sec of 60°C were conducted in a StepOnePlus™
System (Thermo Fisher Scientific). Relative candidate gene
expression normalized to the expression of CALM2 was calculated in
reference to the average expression in non-tumorous samples using
the 2−ΔΔCT-method (26). Expression of each gene was
correlated with clinical data. Correlations between the expression
of different genes were done using Spearman correlation.
Cell lines, siRNA knockdown and qRT-PCR
of in vitro samples
Human PC3 metastatic PCa cells and MDA-MB-231 breast
cancer cells, which is a well-studied model in case of
extracellular vesicle release, were obtained from ATCC (Wesel,
Germany) and grown under standard conditions either in DMEM (PC3)
or RPMI (MDA-MB-231) (each from Life Technologies) supplemented
with 10% FCS (Sigma-Aldrich, St. Louis, MO, USA) and 2 nM glutamine
(Life Technologies).
Cell lines were transfected with siGenome pooled and
individual siRNAs against RAB27A, RAB27B and VPS36 at a
concentration of 30 nmol using Dharmafect I transfection reagent
(all from Dharmacon, Lafayette, USA). Dharmacon non-targeting siRNA
served as negative control. Transfection was conducted as
recommended by the manufacturer with minor modifications. While
cells were detached, harvested, spun down and diluted to the
desired concentration, siRNA and Dharmafect I were separately
incubated in pure RPMI (Life Technologies) for 10 min at room
temperature and where then mixed 1:1 for subsequent 30 min
incubation at room temperature. Hereafter cell suspension was added
to the transfection mix 3:1 and incubated at 37°C. The supernatant
was replaced with fresh medium after 24 h.
RNA-extraction was performed after further 48 h
using RNeasy Mini kit (Qiagen, Hilden, Germany) according to the
recommendations of the manufacturer. For cNDA-synthesis 40
μl of diluted RNA were mixed with 4 μl of 5 mg/ml
pdN6 random primers, 4 μl of 10 mM dNTP Mix, 16 μl of
5X M-MLV buffer, 8 μl of 0.1 M RNase inhibitor, 4 μl
of 0.1 M DTT and 4 μl of M-MLV reverse transcriptase (all
from Roche Diagnostics). After an incubation for 2 h at 37°C and a
deactivation step of 5 min at 65°C, cDNA was directly used for
qRT-PCR or stored at −20°C. qRT-PCR was performed and analyzed
analogously to patient samples.
qRT-PCR was conducted to validate knockdown
efficiency and to monitor the impact of the knockdown of one
candidated gene on the other two. PCR experiments and functional
assays were conducted in three biological replicates each.
Proliferation assay
PC3 cells were seeded and transfected following the
protocol described above in 96-well plates with a total volume of
100 μl/well. After 24 h the supernatant was replaced by 100
μl of fresh growth medium. After further 24, 48 and 72 h of
incubation, 10 μl of MTT-reagent (Promega) was added per
well and incubated for 3 h at 37°C. Absorption measurement at 570
nm was performed using an Infinite M1000 Pro plate reader (Tecan,
Männerdorf, Switzerland).
Scratch assay
Using the same transfection protocol, PC3 cells were
seeded in 24-well plates with a volume of 1 ml per well. The medium
was changed 24 h after transfection. Again 24 h later a defined
scratch was introduced in the center of the well, using a sterile
200 μl pipette tip. The medium was changed again. At this
point of time and after further 24, 48 and 72 h the scratch was
photographed at a ×20 magnification. The calculation of the
cell-free space in the scratch area was performed with the open
source software tscratch (ETH Zürich, Switzerland) (27). The open area 24 h after scratch was
compared to the initial scratch size.
Colony formation assay
In total, 500 PC3 cells were seeded and transfected
per well in 6-well plates. Medium was changed after 24 h. After 14
days of cultivation, the supernatant was removed and the cells were
stained using 0.05% crystal violet for 15 min. Wells were
photographed at ×20 and the number of colonies was counted.
Nanoparticle tracking analysis of
supernatant of transfected and inhibitor-treated cells
PC3 cells were seeded and transfected in 24-well
plates as described above. Forty-eight hours after transfection the
supernatant was removed and the cells were washed with sterile PBS
two times. Afterwards cells were again incubated for 48 h in
serum-free RPMI medium. Subsequently the supernatant was recovered
and sequentially centrifuged at 300, 2,000 and 12,000 g to remove
cellular debris, larger particles and microvesicles. Each sample of
cleared supernatant was measured five times for 45 sec using
nanoparticle tracking analysis (NTA) on an LM10 (Malvern
Instruments, Malvern, UK) at dilutions between 1:20 and 1:50.
Particle concentration and size were determined. The same was done
for the supernatant of PC3 cells treated with the sphingomyelinase
inhibitor GW4869, known to block exosome release in vitro.
For GW4869 a concentration of 5 ng/μl in 1.7% DMSO was
chosen. The same concentration of DMSO without inhibitor served as
control condition.
Statistics
Statistical calculations and graph design were
performed using Prism 6 (GraphPad Software, Inc., La Jolla, CA,
USA) or JMP 11 (SAS Institute GmbH, Heidelberg, Germany). Testing
for normality showed non-normal data distribution in all tested
datasets thus non-parametric tests were used. Calculation of
inter-group gene expression changes, for both in silico
cohorts and patient samples analyzed with qRT-PCR, were performed
using Mann-Whitney test. Correlation of candidate gene expression
was done using Spearman correlation. Differences in BCR were
calculated using log-rank test. For in vitro assays
parametric t-test was used. P-values ≤0.05 were considered
statistically significant.
Results
Several candidate genes are associated
with BCR in localized PCa in silico
Thirty-seven genes either involved in vesicular
trafficking and secretion or typically present on extracellular
vesicles were identified from recent literature. These and five
genes associated with PCa were tested in the microarray dataset of
localized PCa by Taylor et al (19) for their prediction of BCR. Gene
expression cut-off levels, defined by partition test, divided the
dataset into two groups of patients for each candidate, one group
with candidate gene high expression and the other with candidate
gene low expression. Table IV
summarizes cut-off values, group sizes, number of BCR-events and
p-values of log-rank test for BCR. For 22 genes a significant
correlation between gene expression and BCR was seen. Yet, for many
genes partition test had resulted in very imbalanced group sizes.
With a minimum group size requirement set to 20 patients, six genes
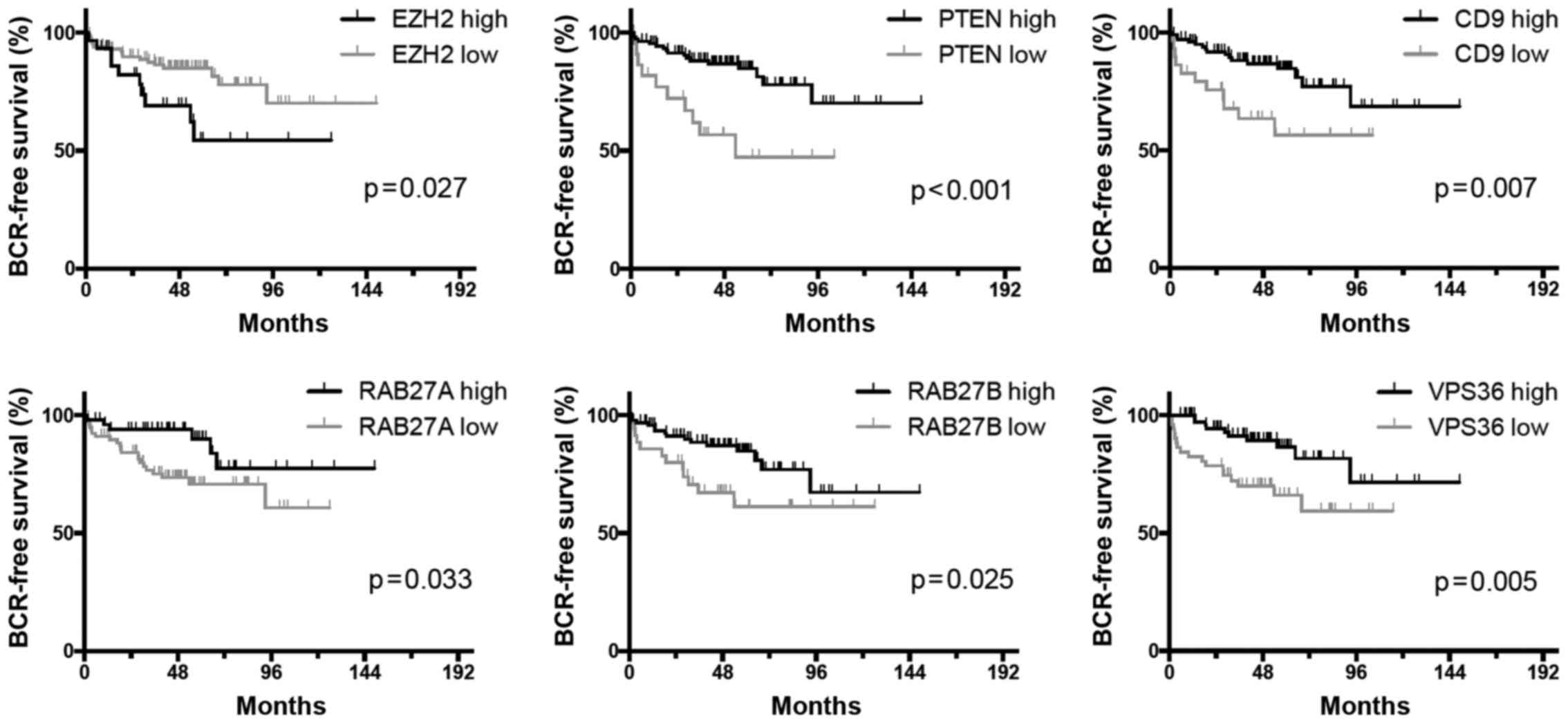
remained. Kaplan-Meier graphs with BCR-free survival as outcome
parameter for these genes are displayed in Fig. 1. Beside genes known to be
associated with PCa outcome, such as EZH2 and PTEN, there were also
the two RABs RAB27A and RAB27B and the ESCRT-II component VPS36
associated with BCR. In case of the latter three candidates a lower
expression was predictive for a worse outcome.
 | Table IVCut-off values, group sizes, number
of BCR-events and p-values of the log-rank test for time to BCR for
37 genes tested in the Taylor et al dataset (19) are displayed.a |
Table IV
Cut-off values, group sizes, number
of BCR-events and p-values of the log-rank test for time to BCR for
37 genes tested in the Taylor et al dataset (19) are displayed.a
| Gene | Cut-off
(z-score) | High
expression
(patients/events) | Low
expression
(patients/events) | p-value
(BCR) |
|---|
| AMACR | 3.390 | 72 (18) | 59 (9) | 0.213 |
| AR | 1.113 | 32 (8) | 99 (19) | 0.424 |
| EZH2 | 3.883 | 30 (10) | 101 (17) | 0.027 |
| FOLH1 | 1.957 | 11 (9) | 120 (18) |
<0.001 |
| KLK3 | 0.683 | 31 (4) | 99 (23) | 0.255 |
| PCA3 | 2.452 | 85 (21) | 46 (6) | 0.147 |
| PTEN | −1.300 | 108 (17) | 23 (10) |
<0.001 |
| PDCD6IP | −1,909 | 126 (24) | 5 (3) | 0.001 |
| TSG101 | −1.672 | 124 (23) | 7 (4) | 0.002 |
| UBAP1 | −1.771 | 115 (19) | 16 (8) |
<0.001 |
| VPS28 | 0.842 | 27 (3) | 104 (24) | 0.912 |
| SNF8 | 1.397 | 10 (0) | 121 (27) | 0.186 |
| VPS25 | −1.651 | 114 (21) | 17 (6) | 0.206 |
| VPS36 | −0.604 | 79 (10) | 52 (17) | 0.005 |
| ST1 | −2.274 | 121 (20) | 10 (7) |
<0.001 |
| VPS4A | −2.468 | 126 (23) | 5 (4) |
<0.001 |
| VTA1 | −1.277 | 120 (22) | 11 (5) |
<0.001 |
| RAB5A | −1.506 | 121 (22) | 10 (5) |
<0.001 |
| RAB27A | 0.607 | 52 (6) | 79 (21) | 0.033 |
| RAB27B | −0.301 | 96 (15) | 35 (12) | 0.025 |
| RAB35 | −0.802 | 79 (12) | 52 (15) | 0.077 |
| TBC1D10A | −0.787 | 61 (9) | 70 (18) | 0.064 |
| CD9 | −0.9227 | 102 (16) | 29 (11) | 0.007 |
| CD63 | 1.814 | 5 (3) | 126 (24) | 0.014 |
| CD81 | 0.531 | 8 (4) | 123 (23) | 0.014 |
| CHMP4C | 3.266 | 7 (4) | 124 (23) | 0.004 |
| DCT | −1.809 | 124 (24) | 7 (3) | 0.083 |
| EPCAM | 1.9265 | 68 (18) | 63 (9) | 0.111 |
| HSPA4 | −1.489 | 120 (22) | 11 (5) |
<0.001 |
| HSP90AA1 | −1.352 | 124 (22) | 7 (5) |
<0.001 |
| MMP2 | 1.583 | 7 (3) | 124 (24) | 0.043 |
| MMP9 | −0.669 | 125 (27) | 6 (0) | 0.233 |
| MMP14 | 1.740 | 5 (3) | 126 (24) |
<0.001 |
| STEAP3 | −2.119 | 120 (22) | 11 (5) | 0.007 |
RAB27A, RAB27B and VPS36 are
overexpressed in metastatic PCa in silico
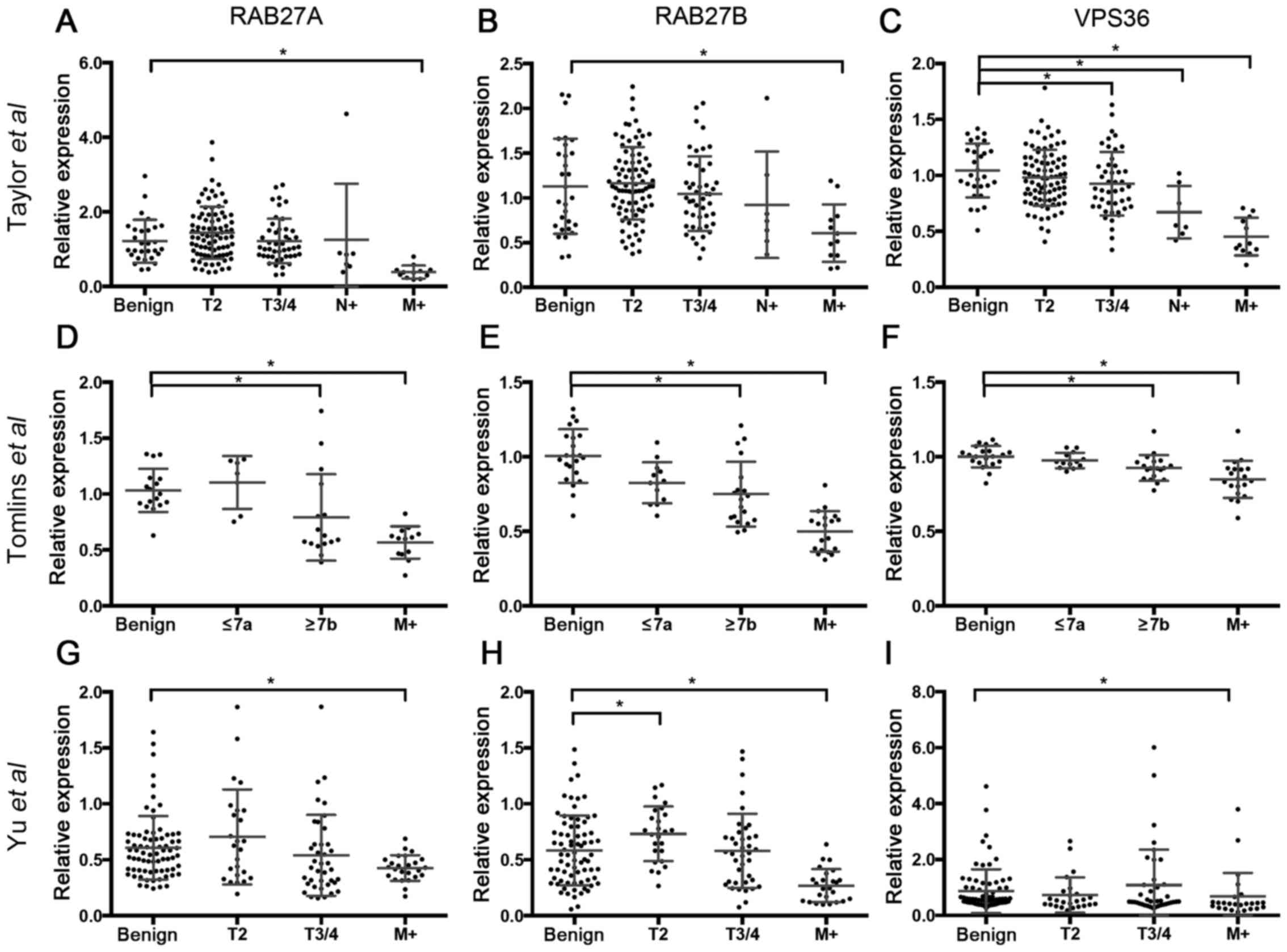
The dataset by Taylor et al (19) was used to stratify gene expression
of RAB27A, RAB27B and VPS36 for tumor stage. Both for RAB27A and
RAB27B a significantly decreased expression in distant metastatic
lesions (RAB27A: p<0.001; RAB27B: p<0.001) but not in primary
tumors and lymph node metastases (Fig.
2A and B) was seen.
The expression of VPS36 was significantly reduced in
locally advanced PCa (p=0.041), as well as in lymph node (p=0.003)
and distant metastases (p<0.001) (Fig. 2C). These findings were re-assessed
in two other publicly available microarray datasets. In the Tomlins
et al dataset (21) the
three candidate genes were significantly reduced in tumors with
Gleason score ≥7b (RAB27A, p=0.007; RAB27B, p=0.003; VPS36,
p=0.001) and metastatic lesions (RAB27A, p<0.001; RAB27B,
p<0.001; VPS36, p<0.001) (Fig.
2D–F). In the Yu et al dataset (22) all three candidates were
underexpressed in metastases (RAB27A, p=0.0389; RAB27B, p<0.001;
VPS36, p=0.001). RAB27B was also higher expressed in stage 2 tumors
(p=0.014) (Fig. 2G–H).
Expression of candidate genes correlates
among each other in silico
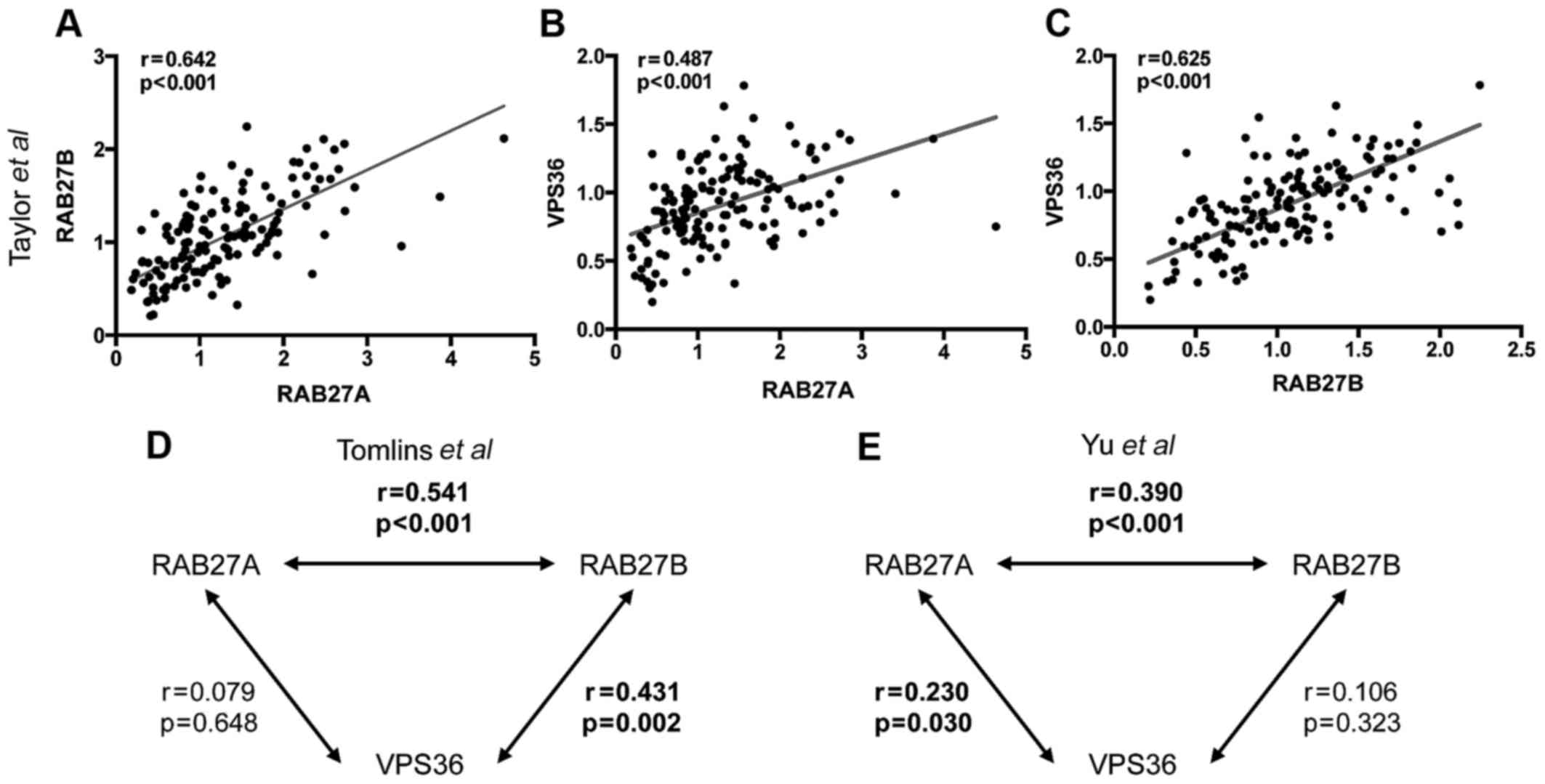
Analysis of microarray datasets furthermore revealed
the expression of RAB27A, RAB27B and VPS36 to be positively
correlated with each other. A low expression of RAB27A was
associated with a low expression of RAB27B in all three datasets.
In the Taylor et al dataset (19) expression of both genes was also
positively correlated with the expression of VPS36. This could
partly be confirmed in the other two datasets. Bivariate
correlation plots are shown in Fig.
3A–C for the Taylor et al dataset (19) exemplarily. For the two other
analyzed datasets correlation coefficients and p-values are shown
as interaction graphs in Fig. 3D and
E.
VPS36 expression is decreased in advanced
primary PCa
To validate these findings in independent patient
samples with a different technique, two cohorts of PCa patients and
benign prostatic tissue controls were analyzed using qRT-PCR. In
the cDNA array consisting of 40 patients with localized PCa and 8
patients with BPH no altered expression of all three candidates,
compared to benign controls, was seen when stratifying for tumor
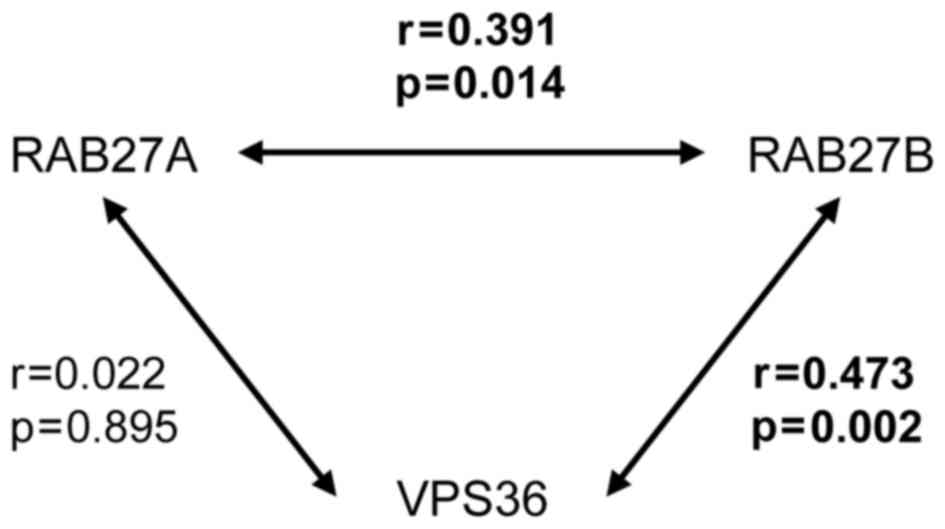
stage and Gleason score (data not shown). Consistent with the in
silico analyses, RAB27A and RAB27 were positively correlated in
this dataset and also a positive correlation between RAB27B and
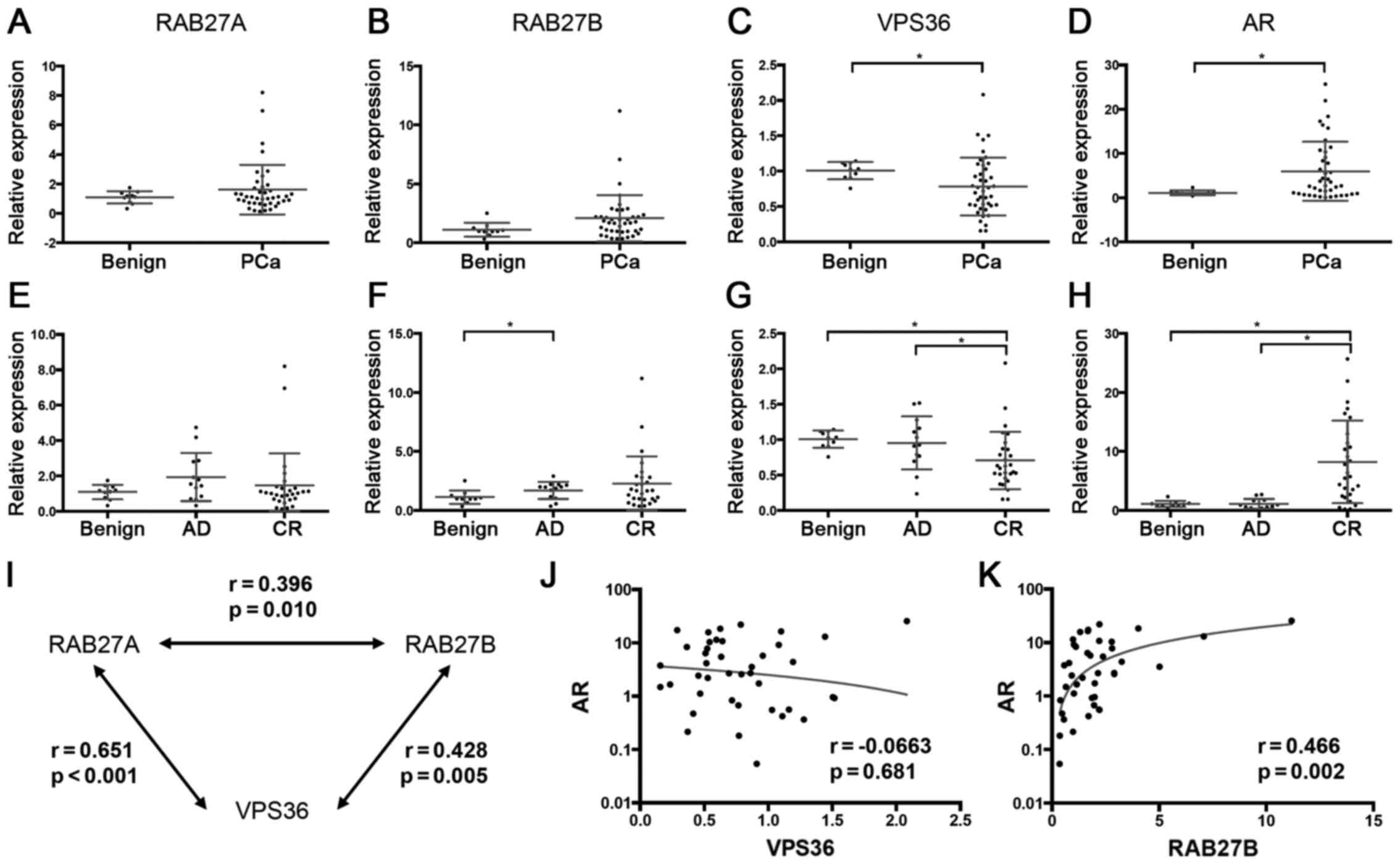
VPS36 was found (Fig. 4).
Since in silico a downregulation of RAB27A,
RAB27B and VPS36 was seen in metastatic PCa, but not in organ
confined PCa, a second dataset consisting of patients undergoing
transurethral resection of PCa for palliation, with many of them
having metastatic and/or castration-resistant disease and therefore
reflecting an advanced disease state, was analyzed.
In this cohort a significant reduction of VPS36
expression in tumor samples (p=0.022) was found, whilst RAB27A and
RAB27B were not reduced in tumor samples, instead showing a slight
but not significant increase in expression (Fig. 5A–C). When stratifying the tumor
samples into androgen-dependent (AD) and castration-resistant (CR)
tumors according to the clinical data, VPS36, but not RAB27A and
RAB27B, was lower expressed in CR tumors, compared to both controls
(p=0.002) and AD tumors (p=0.026) (Fig. 5E–G).
Furthermore, the expression of AR, typically
overexpressed in CR tumors as a sign of aberrant androgen
signaling, was analyzed. As expected a strong overexpression of AR
in CR tumors compared to controls (p<0.001) and to AD tumors
(p<0.001) was seen (Fig. 5D and
H).
The expression of the three candidate genes
positively correlated with each other (Fig. 5I). The expression of VPS36 and
RAB27A did not show a significant correlation with AR (Fig. 5J), whilst for RAB27B a positive
correlation with AR was seen (Fig.
5K).
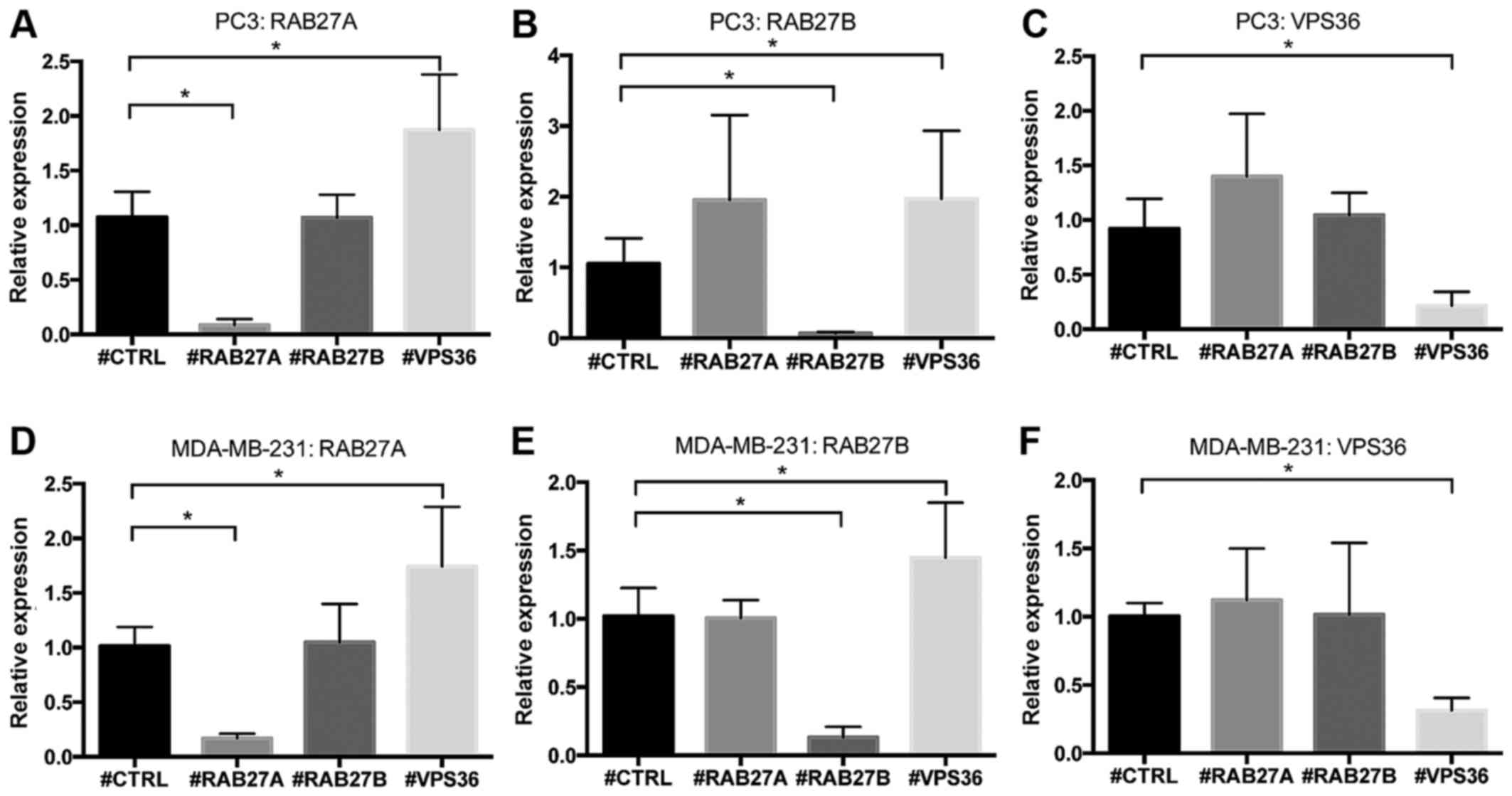
Knockdown experiments confirm
interdependency of candidate genes
To further elucidate the unknown role of RAB27A,
RAB27B and VPS36 in PCa, siRNA-mediated knockdown experiments were
performed in the castration-resistant PC3 cell line. Whilst
knockdown of RAB27A and RAB27B resulted in no significant changes
in the expression of the other genes in PC3 cells, knockdown of
VPS36 resulted in a significant increase in the expression of
RAB27A (p=0.001) and RAB27B (p=0.046), indicating potential
crosstalk (Fig. 6A–C).
Interestingly, in MDA-MB-231 breast cancer cells the same
dependency with an elevated expression of RAB27A (p=0.003) and
RAB27B (p=0.019) upon knockdown of VPS36 was seen (Fig. 6D–F).
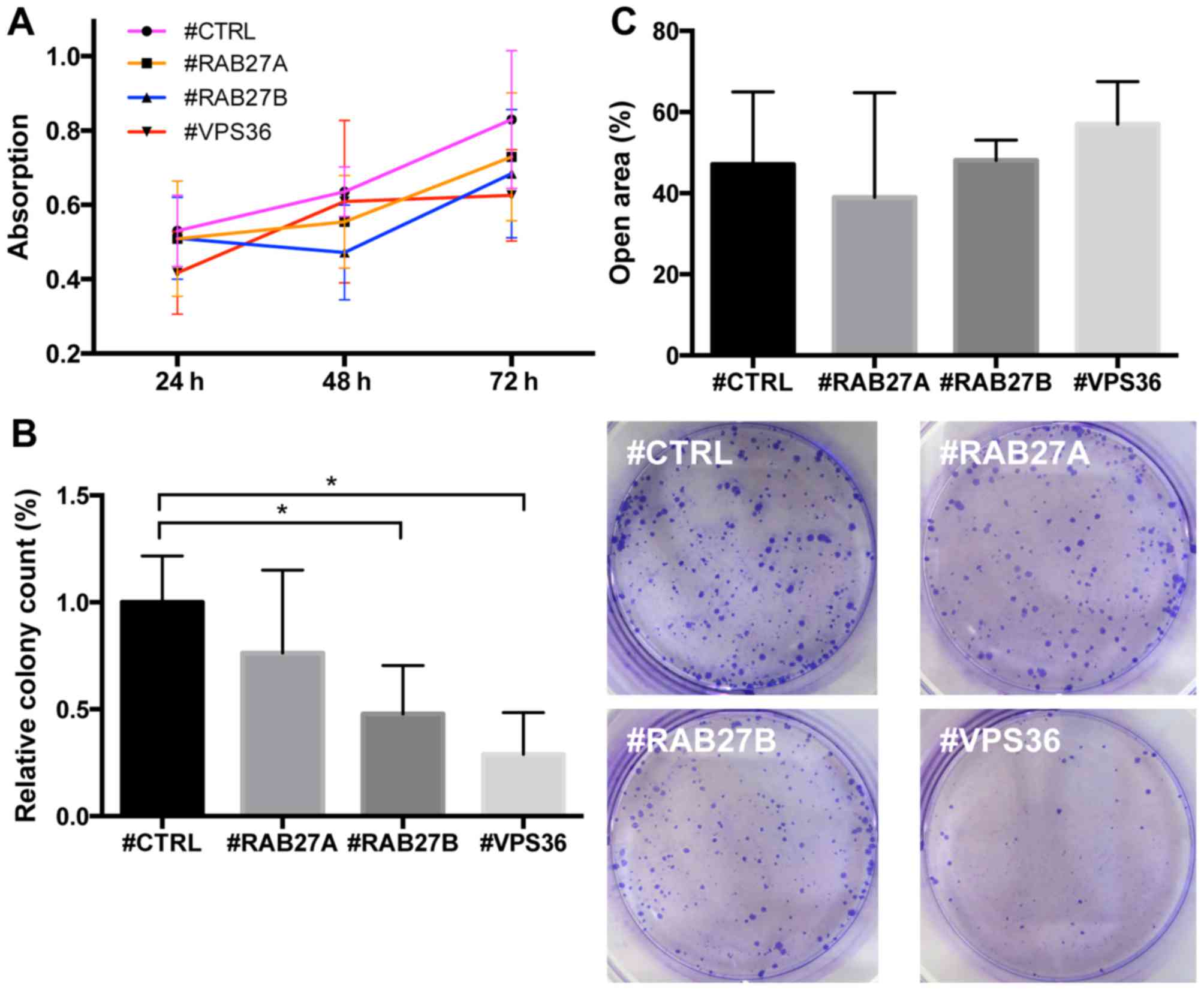
Knockdown of RAB27B and VPS36 reduces
colony formation and increases extracellular particle release
Knockdown of none of the candidates significantly
reduced the growth of PC3 cells, measured with MTT assay under
standard dense 2D cell culture conditions (Fig. 7A). Also under standard conditions,
the migration of PC3 cells, determined by the scratch assay, was
not significantly altered upon knockdown of the candidate genes
(Fig. 7C).
Unlike in the MTT assay, when transfected PC3 cells
were seeded at low density for long-term cultivation, a strong
reduction of colony formation upon knockdown of RAB27B (p<0.001)
and VPS36 (p<0.001), potentially indicating an impairment of
paracrine cell-to-cell signaling, was seen (Fig. 7B).
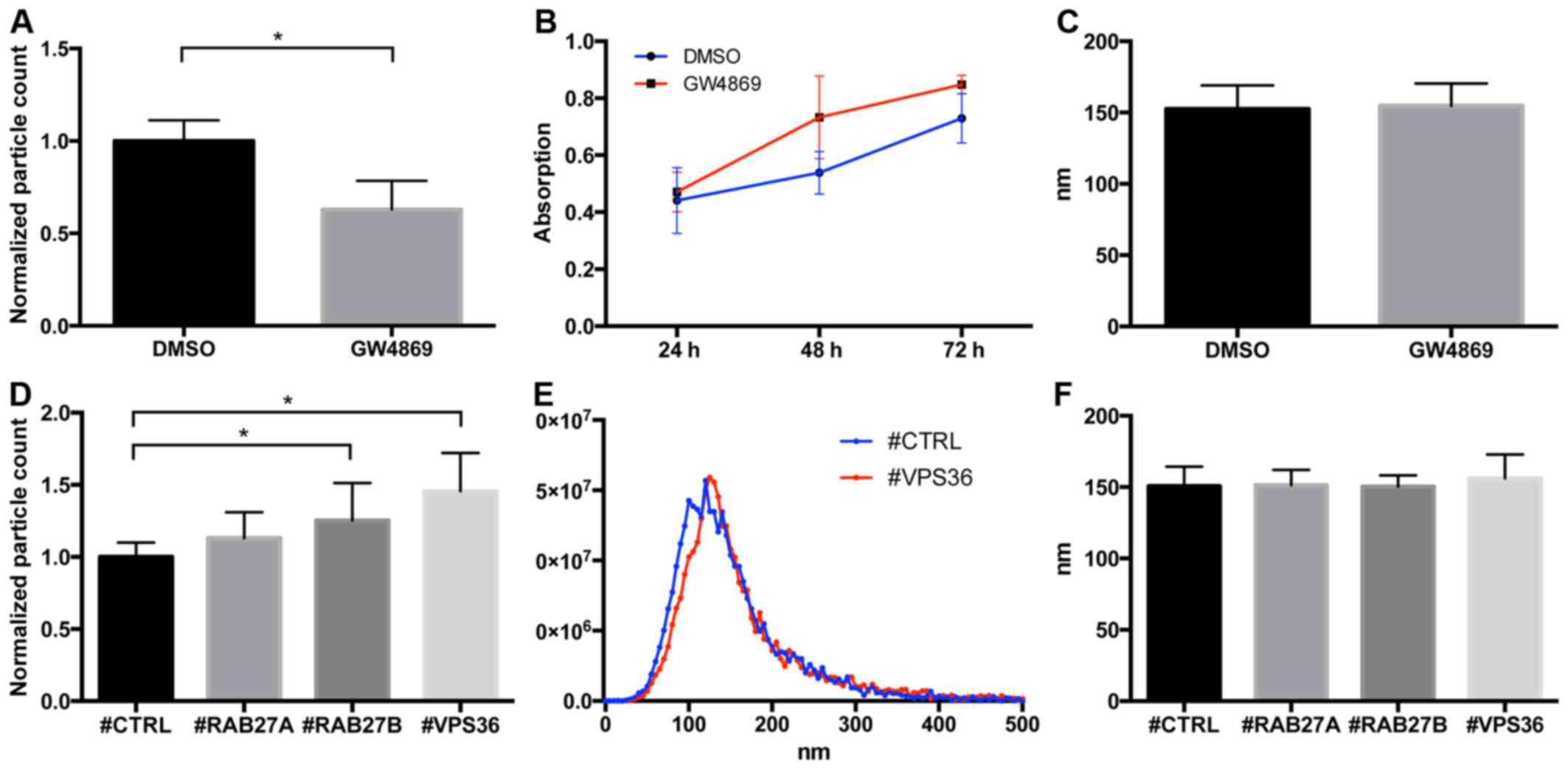
Following this approach, the influence of the
candidate genes on the production of extracellular particles not
pelleting at 12,000 g centrifugation, being a surrogate for small
extracellular vesicles, was investigated. As control PC3 cells were
treated with GW4869, which is a known blocker of the release of
exosomes. GW4869 treatment did not significant increase cellular
growth, but reduced the number of released particles by 37.1%
(p<0.001), indicating that the experimental setup could monitor
modulations in extracellular vesicle release (Fig. 8A and B). Interestingly there was a
significant increase in particle release in PC3 cells upon
knockdown of RAB27B (p=0.014) and VPS36 (p<0.001), again
pointing to an alteration in cell-to-cell signaling dependent on
the candidate genes (Fig. 8D).
Both for inhibitor treatment and RNAi no changes in the size of the
released particles were seen (Fig. 8C
and F). Fig. 8E shows the
typical size distribution of the particles released from PC3 cells
treated with #CTRL and #VPS36.
Discussion
Genes with known function in vesicular trafficking
and secretion and genes coding for typical extracellular vesicle
marker proteins were analyzed in existing microarray datasets, to
investigate their role in PCa, Among these, the expression of
VPS36, RAB27A and RAB27B was inversely correlated with BCR-free
survival. These three genes were underexpressed in metastatic PCa
tissue samples.
RAB27A and RAB27B are deemed to be key regulators of
exosome secretion, since they mediate the transport of MVBs to the
plasma membrane and their fusion with the plasma membrane (13). Several studies have shown their
expression to be correlated with exosome release in vitro
and in vivo in different cancer entities (4,28).
Typically blockage of their expression reduces the number of
exosomes secreted (29). Two
studies suggest RAB27A to be involved in the secretion of typical
PCa markers the PSA and PSAP by modulating PI3K-signaling (30,31).
Furthermore, reduction of RAB27A expression results in a reduction
of stroma-assisted tumor growth by reducing the number of secreted
exosomes from PCa cells (32). It
is known that the regulatory effects of RAB27 GTPases are
cell-specific, depending on expression and function of potential
effector molecules (33). In mast
cells they were shown to regulate granule exocytosis, with RAB27B
being more of a positive regulator and RAB27A more of a negative
regulator of degranulation (34).
This underlines that they can both have concordant and discordant
function and are also involved in immune-modulatory processes
(35).
RAB27B is known to be expressed in various
epithelial tissues in adults, while RAB27B expression is low in
fetal tissue (36). Yet, the
expression of RAB27A and RAB27B has only been studied in some
cancer entities. In breast cancer an elevated expression of RAB27B
is associated with lymph node metastasis and predicts poor
prognosis (37). Also in
colorectal cancer RAB27B has been identified as potential predictor
of metastasis and outcome (38). A
higher expression of RAB27B mRNA was observed in cancer tissue and
was associated with decreased overall survival. Conversely another
study found a lower expression of RAB27A and RAB27B to be
correlated with colorectal cancer progression (39). To date nothing is known about the
expression of RAB27A and RAB27B in PCa. In this study data from
in silico analysis consistently showed the expression of
both genes to be lower in PCa metastases. In the Tomlins et
al dataset (21) both genes
were also under-expressed in localized PCa with a Gleason score of
7b and higher. Also a positive correlation between RAB27B and AR
was seen. A recent study found the expression of RAB27A to be
regulated by AR upon androgen deprivation therapy in PCa (40), but the underlying mechanisms are
yet unknown.
Unlike for RAB27A and RAB27B, the role of VPS36 in
cancer has not been investigated at all. As a member of the
ESCRT-II complex it is mainly associated with MVB biogenesis,
cellular abscission and viral budding (41). Deregulation of JAK/STAT signaling
in tissues with modifications in the ESCRT-II complex resulted in
facilitated tumorigenesis in Drosophila, indicating
crosstalk with known cancer-pathways (42). VPS36, also known as EAP45, is
located on chromosome 13 and transcribes for three different
transcript variants. Crystal structure analyses propose it to
directly interact with ubiquitinated proteins (43). It showed a negative correlation
with advanced tumor stage and worse outcome in silico. This
was confirmed by qRT-PCR analysis. Especially in CR PCa VPS36
expression was significantly reduced, whilst AR expression was
strongly increased. Yet, no significant correlation between these
two genes was seen.
Both in existing microarray datasets and in tumor
samples analyzed with qRT-PCR, the expression of the candidate
genes was consistently correlated among each other. This might be
due to a mechanism simultaneously regulating the candidate gene
expression. In vitro a significant increase of RAB27A and
RAB27B expression was seen upon knockdown of VPS36 in PC3 cells,
but not vice versa. This suggests an additional directional
interdependency of these genes. At least with regard to exosome
release VPS36 is located prior in the signaling cascade, making
this relationship plausible. Re-testing in the MDA MB-231 breast
cancer cell line confirmed these results and suggests this
interdependency not to be PCa-specific.
Knockdown of the candidate genes did not result in a
significant alteration of growth and migration of PC3 cells under
standard culture conditions. The significant reduction of colony
formation upon knockdown of RAB27B and VPS36 seems to be
controversial to the findings in patient samples. Yet, cellular
behavior in this long-term setup is less dependent on the induced
changes in the individual cells and direct cell-to-cell
interaction. Instead it also reflects aspects of paracrine
stimulation via secreted molecules and particles like exosomes.
Again comparable data from other studies in PCa are lacking. In
vivo data from lung cancer mouse models show a reduction of
tumor growth only for metastatic, but not for non-metastatic cell
lines treated with shRNAs targeting RAB27A (4).
To investigate the impact of candidate gene
expression on extracellular vesicle secretion, NTA analyses of the
supernatant of treated cells were conducted. In order to facilitate
measurement of multiple samples under different treatment
conditions, particles in the supernatant of cancer cells not
pelleting at 12,000 g were used as a surrogate for small
extracellular vesicles. Using this system, treatment of PC3 cells
with the sphingomyelinase inhibitor GW4869, a known blocker of
exosome release (44), resulted in
a strong reduction of extracellular particles, indicating the
applicability of this setup. Unexpectedly, knockdown of RAB27B and
VPS36 resulted in a significant increase of extracellular particles
remaining in the PC3 supernatant after centrifugation at 12,000 g.
Again, there is not much comparable data in the literature. One
study has used a similar experimental setup to monitor vesicle
release from MDA MB-231 cells. shRNA-mediated knockout of RAB27A
resulted in a significant decrease of secreted vesicles (29). As in this study, no changes in the
size of secreted particles were observed. RAB27B and VPS36 were not
tested in this study. Ostrowski et al (13) also found no changes in size and
morphology of HeLa exosomes upon shRNA mediated knockdown of RAB27A
and RAB27B. By analyzing ultracentrifugation pellets they saw a
reduction of exosomes upon knockdown, which is controversial to our
findings. These different observations might be attributed to the
different assay and readout conditions. Also a time-dependent
regulation of extracellular particle release upon modulation of
RAB27A and RAB27B expression can be debated. Furthermore, the used
cell lines represent different cancer entities with a potentially
different regulation of RAB27A and RAB27B expression and function.
Also the different cell lines originate from different
microenvironmental conditions. In this context it has been shown
that extracellular vesicles of malignant cells have an ambivalent
role both in immune-evasion (45,46)
and the activation of the immune system against cancer cells
(47), adding a further potential
regulatory element on extracellular vesicle biogenesis and
secretion.
Our study is limited by the heterogeneous assay
platforms used for microarray experiments. Furthermore, sample
handling and data post-processing offer potential source of bias.
The cohorts analyzed by qRT-PCR were heterogeneous with regard to
tumor stage and, mainly for the patients treated with transurethral
resection, also for iatrogenous factors as pretreatment and
castration-resistance. Also the sample material itself, being FFPE
tissue, is not optimal for downstream analyses due to a strong
degradation of nucleic acids. Yet studies relying on antibody-based
protein analyses, being an alternative strategy to analyze
candidate expression, also have specific limitations such as
degradation of epitopes, unspecific antibody binding and a more
difficult quantification.
In conclusion, several genes with known function in
vesicular trafficking and secretion are deregulated in PCa. Of
these, RAB27A, RAB27B and VPS36 are consistently underexpressed in
advanced PCa and predict worse outcome. For VPS36 underexpression
could also be shown in primary tumors undergoing local palliation.
Subgroup analysis indicates a stronger decrease in CR tumors,
whilst no correlation was found with the expression the AR.
In the analyzed datasets the expression of RAB27A,
RAB27B and VPS36 showed a strong positive correlation among each
other, pointing to a common regulatory mechanism. Additionally
in vitro manipulation of the gene expression indicates
RAB27A and RAB27B to be dependent of VPS36. An increase of
extracellular particles and a reduction of colony formation upon
knockdown of RAB27B and VPS36 indicate an involvement in paracrine
cell-cell communication.
Further analyses in larger cohorts are needed to
validate the repressed expression of VPS36, RAB27A and RAB27B in
advanced and metastatic PCa. Additionally, studies dissecting their
specific regulation and function should help to elucidate their
role in vesicular trafficking and secretion and paracrine signaling
in PCa.
Abbreviations:
|
AD
|
androgen dependent
|
|
AR
|
androgen receptor
|
|
BCR
|
biochemical recurrence
|
|
CR
|
castration-resistant
|
|
FFPE
|
formalin-fixed paraffin-embedded
tissue
|
|
PCa
|
prostate cancer
|
|
PSA
|
prostate-specific antigen
|
|
PSMA
|
prostate-specific membrane antigen
|
|
TUR-P
|
transurethral resection of the
prostate
|
Acknowledgments
This study was supported by the Foundation on Cancer
and Scarlet Research of the University of Heidelberg. T.S.W. was
supported by a Ferdinand Eisenberger scholarship of the German
Society of Urology.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vlassov AV, Magdaleno S, Setterquist R and
Conrad R: Exosomes: Current knowledge of their composition,
biological functions, and diagnostic and therapeutic potentials.
Biochim Biophys Acta. 1820:940–948. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Junker K, Heinzelmann J, Beckham C, Ochiya
T and Jenster G: Extracellular vesicles and their role in urologic
malignancies. Eur Urol. 70:323–331. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bobrie A, Krumeich S, Reyal F, Recchi C,
Moita LF, Seabra MC, Ostrowski M and Théry C: Rab27a supports
exosome-dependent and -independent mechanisms that modify the tumor
microenvironment and can promote tumor progression. Cancer Res.
72:4920–4930. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Beckham CJ, Olsen J, Yin P-N, Wu CH, Ting
HJ, Hagen FK, Scosyrev E, Messing EM and Lee YF: Bladder cancer
exosomes contain EDIL-3/Del1 and facilitate cancer progression. J
Urol. 192:583–592. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Alderton GK: Metastasis. Exosomes drive
premetastatic niche formation. Nat Rev Cancer. 12:4472012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peinado H, Alečković M, Lavotshkin S,
Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M,
Williams C, García-Santos G, Ghajar C, et al: Melanoma exosomes
educate bone marrow progenitor cells toward a pro-metastatic
phenotype through MET. Nat Med. 18:883–891. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Webber MM, Trakul N, Thraves PS,
Bello-DeOcampo D, Chu WW, Storto PD, Huard TK, Rhim JS and Williams
DE: A human prostatic stromal myofibroblast cell line WPMY-1: A
model for stromal-epithelial interactions in prostatic neoplasia.
Carcinogenesis. 20:1185–1192. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Webber J, Steadman R, Mason MD, Tabi Z and
Clayton A: Cancer exosomes trigger fibroblast to myofibroblast
differentiation. Cancer Res. 70:9621–9630. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Colombo M, Moita C, van Niel G, Kowal J,
Vigneron J, Benaroch P, Manel N, Moita LF, Théry C and Raposo G:
Analysis of ESCRT functions in exosome biogenesis, composition and
secretion highlights the heterogeneity of extracellular vesicles. J
Cell Sci. 126:5553–5565. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Adell MA and Teis D: Assembly and
disassembly of the ESCRT-III membrane scission complex. FEBS Lett.
585:3191–3196. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Brinton LT, Sloane HS, Kester M and Kelly
KA: Formation and role of exosomes in cancer. Cell Mol Life Sci.
72:659–671. 2015. View Article : Google Scholar
|
|
13
|
Ostrowski M, Carmo NB, Krumeich S, Fanget
I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, et
al: Rab27a and Rab27b control different steps of the exosome
secretion pathway. Nat Cell Biol. 12:19–30. 2010. View Article : Google Scholar
|
|
14
|
Théry C, Amigorena S, Raposo G and Clayton
A: Isolation and characterization of exosomes from cell culture
supernatants and biological fluids. Curr Protoc Cell Biol. Chapter
3: Unit 3.22. 2006. View Article : Google Scholar
|
|
15
|
Théry C, Zitvogel L and Amigorena S:
Exosomes: Composition, biogenesis and function. Nat Rev Immunol.
2:569–579. 2002.PubMed/NCBI
|
|
16
|
Villarroya-Beltri C, Gutiérrez-Vázquez C,
Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N,
Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M and
Sánchez-Madrid F: Sumoylated hnRNPA2B1 controls the sorting of
miRNAs into exosomes through binding to specific motifs. Nat
Commun. 4:29802013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Koppers-Lalic D, Hackenberg M, Bijnsdorp
IV, van Eijndhoven MA, Sadek P, Sie D, Zini N, Middeldorp JM,
Ylstra B, de Menezes RX, et al: Nontemplated nucleotide additions
distinguish the small RNA composition in cells from exosomes. Cell
Rep. 8:1649–1658. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
van der Pol E, Böing AN, Harrison P, Sturk
A and Nieuwland R: Classification, functions, and clinical
relevance of extracellular vesicles. Pharmacol Rev. 64:676–705.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Taylor BS, Schultz N, Hieronymus H,
Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva
B, et al: Integrative genomic profiling of human prostate cancer.
Cancer Cell. 18:11–22. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tomlins SA, Mehra R, Rhodes DR, Cao X,
Wang L, Dhanasekaran SM, Kalyana-Sundaram S, Wei JT, Rubin MA,
Pienta KJ, et al: Integrative molecular concept modeling of
prostate cancer progression. Nat Genet. 39:41–51. 2007. View Article : Google Scholar
|
|
22
|
Yu YP, Landsittel D, Jing L, Nelson J, Ren
B, Liu L, McDonald C, Thomas R, Dhir R, Finkelstein S, et al: Gene
expression alterations in prostate cancer predicting tumor
aggression and preceding development of malignancy. J Clin Oncol.
22:2790–2799. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Specht E, Kaemmerer D, Sänger J, Wirtz RM,
Schulz S and Lupp A: Comparison of immunoreactive score, HER2/neu
score and H score for the immunohistochemical evaluation of
somatostatin receptors in bronchopulmonary neuroendocrine
neoplasms. Histopathology. 67:368–377. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kaemmerer D, Reimann C, Specht E, Wirtz
RM, Sayeg M, Baum RP, Schulz S and Lupp A: Differential expression
and prognostic value of the chemokine receptor CXCR4 in
bronchopulmonary neuroendocrine neoplasms. Oncotarget. 6:3346–3358.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Untergasser A, Cutcutache I, Koressaar T,
Ye J, Faircloth BC, Remm M and Rozen SG: Primer3 - new capabilities
and interfaces. Nucleic Acids Res. 40:e1152012. View Article : Google Scholar
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−∆∆C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
27
|
Ashby WJ and Zijlstra A: Established and
novel methods of interrogating two-dimensional cell migration.
Integr Biol. 4:1338–1350. 2012. View Article : Google Scholar
|
|
28
|
Li W, Hu Y, Jiang T, Han Y, Han G, Chen J
and Li X: Rab27A regulates exosome secretion from lung
adenocarcinoma cells A549: Involvement of EPI64. APMIS.
122:1080–1087. 2014.PubMed/NCBI
|
|
29
|
Zheng Y, Campbell EC, Lucocq J, Riches A
and Powis SJ: Monitoring the Rab27 associated exosome pathway using
nanoparticle tracking analysis. Exp Cell Res. 319:1706–1713. 2013.
View Article : Google Scholar
|
|
30
|
Johnson JL, Ellis BA, Noack D, Seabra MC
and Catz SD: The Rab27a-binding protein, JFC1, regulates
androgen-dependent secretion of prostate-specific antigen and
prostatic-specific acid phosphatase. Biochem J. 391:699–710. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Catz SD: Characterization of Rab27a and
JFC1 as constituents of the secretory machinery of
prostate-specific antigen in prostate carcinoma cells. Methods
Enzymol. 438:25–40. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Webber JP, Spary LK, Sanders AJ, Chowdhury
R, Jiang WG, Steadman R, Wymant J, Jones AT, Kynaston H, Mason MD,
et al: Differentiation of tumour-promoting stromal myofibroblasts
by cancer exosomes. Oncogene. 34:290–302. 2015. View Article : Google Scholar
|
|
33
|
Catz SD: Regulation of vesicular
trafficking and leukocyte function by Rab27 GTPases and their
effectors. J Leukoc Biol. 94:613–622. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Singh RK, Mizuno K, Wasmeier C,
Wavre-Shapton ST, Recchi C, Catz SD, Futter C, Tolmachova T, Hume
AN and Seabra MC: Distinct and opposing roles for Rab27a/Mlph/MyoVa
and Rab27b/Munc13-4 in mast cell secretion. FEBS J. 280:892–903.
2013.PubMed/NCBI
|
|
35
|
Elstak ED, Neeft M, Nehme NT, Voortman J,
Cheung M, Goodarzifard M, Gerritsen HC, van Bergen En Henegouwen
PM, Callebaut I, de Saint Basile G, et al: The munc13-4-rab27
complex is specifically required for tethering secretory lysosomes
at the plasma membrane. Blood. 118:1570–1578. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hendrix A, Lambein K, Westbroek W, Seabra
MC, Cocquyt V, Pauwels P, Bracke M, Gespach C and De Wever O: An
immunohistochemical analysis of Rab27B distribution in fetal and
adult tissue. Int J Dev Biol. 56:363–368. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang J-X, Huang X-X, Cai M-B, Tong ZT,
Chen JW, Qian D, Liao YJ, Deng HX, Liao DZ, Huang MY, et al:
Overexpression of the secretory small GTPase Rab27B in human breast
cancer correlates closely with lymph node metastasis and predicts
poor prognosis. J Transl Med. 10:2422012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bao J, Ni Y, Qin H, Xu L, Ge Z, Zhan F,
Zhu H, Zhao J, Zhou X, Tang X, et al: Rab27b is a potential
predictor for metastasis and prognosis in colorectal cancer.
Gastroenterol Res Pract. 2014:9131062014. View Article : Google Scholar
|
|
39
|
Dong W, Cui J, Yang J, Li W, Wang S, Wang
X, Li X, Lu Y and Xiao W: Decreased expression of Rab27A and Rab27B
correlates with metastasis and poor prognosis in colorectal cancer.
Discov Med. 20:357–367. 2015.
|
|
40
|
Shaw GL, Whitaker H, Corcoran M, Dunning
MJ, Luxton H, Kay J, Massie CE, Miller JL, Lamb AD, Ross-Adams H,
et al: The early effects of rapid androgen deprivation on human
prostate cancer. Eur Urol. 70:214–218. 2016. View Article : Google Scholar :
|
|
41
|
Schuh AL and Audhya A: The ESCRT
machinery: From the plasma membrane to endosomes and back again.
Crit Rev Biochem Mol Biol. 49:242–261. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Woodfield SE, Graves HK, Hernandez JA and
Bergmann A: De-regulation of JNK and JAK/STAT signaling in ESCRT-II
mutant tissues cooperatively contributes to neoplastic
tumorigenesis. PLoS One. 8:e560212013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Alam SL, Langelier C, Whitby FG, Koirala
S, Robinson H, Hill CP and Sundquist WI: Structural basis for
ubiquitin recognition by the human ESCRT-II EAP45 GLUE domain. Nat
Struct Mol Biol. 13:1029–1030. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chairoungdua A, Smith DL, Pochard P, Hull
M and Caplan MJ: Exosome release of β-catenin: A novel mechanism
that antagonizes Wnt signaling. J Cell Biol. 190:1079–1091. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liu Y, Gu Y and Cao X: The exosomes in
tumor immunity. OncoImmunology. 4:e10274722015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lundholm M, Schröder M, Nagaeva O, Baranov
V, Widmark A, Mincheva-Nilsson L and Wikström P: Prostate
tumor-derived exosomes downregulate NKG2D expression on natural
killer cells and CD8+ T cells: Mechanism of immune
evasion. PLoS One. 9:e1089252014. View Article : Google Scholar
|
|
47
|
Li W, Mu D, Tian F, Hu Y, Jiang T, Han Y,
Chen J, Han G and Li X: Exosomes derived from Rab27a-overexpressing
tumor cells elicit efficient induction of antitumor immunity. Mol
Med Rep. 8:1876–1882. 2013.PubMed/NCBI
|