Introduction
Hepatocellular carcinoma (HCC) is the fifth most
common malignant cancer and the second leading cause of cancer
related deaths globally (1,2).
Despite the improvement of surgical techniques and adjuvant
therapies, approximately 20% of HCC patients still suffer
extra-hepatic metastases within 5–10 years of receiving radical
surgical treatment. The long-term survival of patients with
metastases remains low (3).
Therefore, it is critical to discover the mechanisms underlying HCC
metastasis.
C-reactive protein (CRP), a prototypic acute-phase
protein and a member of the ancient and highly conserved proteins
of the pentraxin family, has a cyclic pentameric structure. CRP is
involved directly in a wide range of inflammatory processes and
contributes to innate host immunity (4). Moreover, CRP is a sensitive systemic
marker of inflammation and tissue damage. Elevated levels of CRP
are detected in patients with infections, inflammatory diseases, or
necrosis (5). Upregulation of CRP
expression has been implicated in many types of tumors, including
ovarian (6), lung (7), colon cancer (8), multiple myeloma (9) and lymphoma (10). Previous studies focused mostly on
the expression level of CRP in cancer patients. Several studies
have demonstrated that CRP promotes cell proliferation in
endothelial cells, endothelial progenitor cells, renal tubular
epithelial cells, and protects against apoptosis in myeloma cells
(11,12), which are implicated in
tumorigenesis and the development of HCC. Despite evidence of CRP
being involved in a variety of cancers, the role of CRP in
regulating HCC metastasis remains unclear.
Recently, the use of isobaric tags for relative and
absolute quantitation (iTRAQ) technology has become a particularly
powerful tool and has been recommended by the proteomics community
to enable deeper proteome coverage, since it can facilitate
simultaneous analysis of up to eight samples in one experiment. The
aim of the present study was to use iTRAQ to identify alterations
in the proteome of CRP siRNA treated samples, compared to control
samples, in order to identify proteins participating in the
migration and invasion of HCC.
In the present study, we hypothesized that CRP has
an effect on invasion and metastasis of HCC. To elucidate the
potential mechanism/pathway by which CRP contributes to migration
and invasion, iTRAQ-based MS was performed to analyze
differentially expressed proteins (DEPs) between the supernatants
of CRP siRNA-treated supernatant and the negative siRNA-treated
HepG2 cells.
Materials and methods
Immunohistochemistry (IHC) and tissue
microarrays (TMA)
A commercial tissue microarray (BC03117; Us Biomax,
Inc., Rockville, MD, USA), containing 40 cases of hepatocellular
carcinoma and 40 matched cancer adjacent normal tissues, was used
for IHC evaluation of CRP. Liver sections embedded in paraffin were
deparaffinized in xylene, rehydrated in ethanol and washed in
double-distilled H2O (13). Endogenous peroxidase activity was
quenched by incubating the sections for 10 min in 3%
H2O2, and the liver sections were then
blocked with BSA for 30 min. The sections were incubated with
primary antibodies against CRP (1:100 dilution) overnight at 4°C.
IHC visualization of CRP was performed with an EnVision system with
horseradish peroxidase (Dako Cytomation, Glostrup, Denmark)
(13).
Cell lines
Human HCC cell lines, HepG2 (ATCC, Manassas, VA,
USA) and the BEL7402 (Cell Bank of the Chinese Academy of Medical
Science, Beijing, China), were cultured in an atmosphere of 5.0%
carbon dioxide at 37°C in RPMI-1640 medium that was supplemented
with 10% fetal bovine serum (FBS; Gibco, San Diego, CA, USA) and
100 IU/ml penicillin.
CRP siRNA transfection, Transwell assays
and wound healing
HepG2 and BEL7402 cells were transfected with 100 nM
of CRP specific Stealth Select RNAi™ siRNA (sc-40816) or a negative
control siRNA (12935-400) using Lipofectamine 2000 (Invitrogen,
Carlsbad, CA, USA), following the manufacturer's instructions.
After transfection for 6 h, the culture medium was replaced with
fresh RPMI-1640, supplemented with 10% FBS and penicillin, and the
cells continued in culture for an additional 42 h. To explore the
role of CRP in the in vitro progression of HCC,
wound-healing, cell migration and invasion assays were conducted
two days after transfection. The wound healing assays were
performed in 6-well plates. As the cell reached confluence, a wound
was incised in the cell monolayer using a sterile p200 pipette tip,
followed by three washes with medium. Digital images of the wound
areas were captured after 0 and 24 h, using a phase contrast
microscope. The Transwell invasion assays were performed using a
24-well Cell Invasion Assay kit (Cell Biolabs, Inc., San Diego, CA,
USA). Briefly, HepG2 cells were harvested and re-suspended in
serum-free media until they were transfected with CRP or control
siRNA for 48 h. Approximately 2×105 transfected cells
were loaded into the upper chamber and 500 µl media (1640
plus 10% FBS) was loaded into the lower chambers. Cells were
incubated for 24 h. The non-invasive cells were removed using
cotton swabs, and the number of invading cells on the bottom of the
filters were measured using CyQUANT GR fluorescent dye and
detection at 560 nm. In each case, the silencing of CRP expression
was verified by western blot analysis.
The HepG2 cell secretory proteins
collection
Two days following HepG2 transfected with 100 nM of
CRP specific Stealth Select RNAi™ siRNA (sc-40816) or a negative
control siRNA, the culture medium was again replaced with fresh
RPMI-1640 without FBS or penicillin and continued to culture for 2
days. Then the HepG2 cell culture medium without FBS or penicillin
(secretory proteins) were concentrated and used for iTRAQ-coupled
LC-MS/MS analyses.
ITRAQ labeling
The 8-plex iTRAQ kits were purchased from Applied
Biosystems (Foster City, CA, USA). The secretory proteins was
collected as described above and the protein concentrations were
quantified by 2D Quant kit (Amersham Biosciences). Approximately
100 µg of protein from each sample was precipitated,
dissolved in dissolution buffer, denatured, cysteine blocked,
digested with 2 µg of sequencing grade modified trypsin and
labeled using iTRAQ reagents as follows: negative control siRNA
transfected protein, 113 and 116 tags; pooled CRP siRNA-treated
protein, 115 and 117 tags: pooled negative siRNA-treated protein.
The labeled samples were combined before analysis.
Peptide fractionation
The method of peptide fractionation was
immobilized-pH-gradient isoelectric focusing (IPG-IEF), as
previously described (14,15). Briefly, the pooled iTRAQ-labeled
samples were solubilized in Pharmalyte (Amersham Biosciences) and 8
M urea, rehydrated on 18 cm-long IPG gel strips (pH 3–10; Amersham
Biosciences), and then subjected to IEF focusing at 68 kV/h with an
IPGphor System (GE Healthcare). Peptides were extracted by
incubating the gel pieces in acetonitrile and formic acid. The
pieces were purified and concentrated on a C18 Discovery DSC-18 SPE
column (Sigma-Aldrich), lyophilized and stored at −20°C until
LC-MS/MS analysis.
Mass spectrometry
A QStar Elite mass spectrometer (Applied Biosystems)
coupled with an Dionex UltiMate 3000 liquid chromatography system
(Thermo Fisher Scientific, Amsterdam, The Netherlands) was used for
mass spectrometric analysis (16).
Peptide separation was carried out on a C18 analytical column
(Thermo Fisher Scientific, Beijing, China). Purified peptide
fractions were dissolved in buffer A (98% ACN), loaded onto a C18
trap column and subsequently eluted from the trap column over the
C18 analytical column at a flow rate of 300 nl/min in 125 min
linear gradient ranging from 2 to 100% mobile phase B (0.1% formic
acid, 98% acetonitrile). Data acquisition was done in the positive
ion mode, with a selected mass range of 300–1800 m/z. The two most
abundantly charged ions which exceeded 20 counts were chosen for
MS/MS at a dynamic exclusion of 30 sec (17). Protein identification and
quantification were performed with ProteinPilot v2.0 (AB Sciex).
MS/MS data were processed by searching the International Protein
Index (IPI) human database v3.77. Methyl methane thiosulfate (MMTS)
modified cysteine was specified as a fixed modification. Proteins
having at least two unique peptides with fold-change >1.3 or
<0.77 (P<0.05) between two stages were considered to be
differentially expressed proteins.
Bioinformatics
Gene Ontology analysis was performed using PANTHER
(http://www.pantherdb.org/) to classify
biological processes, protein classes and molecular functions.
RNA extraction and quantitative
RT-PCR
Total RNA was extracted from HepG2 cells with TRIzol
reagent (Gibco-BRL, Gaithersburg, MD, USA), according to the
manufacturer's instructions. First-strand cDNA was synthesized
using a Thermo Scientific RevertiAid First Strand cDNA synthesis
kit (Thermo Fisher Scientific). RT-PCR was performed on an ABI
7900HT system using the KAPA SYBR® FAST Universal 2X
qPCR Master Mix and primers for GAPDH (Hs00486019_CE), GFAP
(Hs00167550_CE), CUL1 (Hs00667710_CE), FAHD1 (Hs00636334_CE), NME2
(Hs00543451_CE), GNPDA2 (Hs00831453_CE), CALM2 (Hs00710519_CE),
DDB1 (Hs00586106_CE), HSPD1 (Hs00830627_CE), CTSZ (Hs00664339_CE),
CDH2 (Hs00805624_CE), IL11 (Hs00545902_CE), (Hs00829008_ CE), NUCB1
(Hs00817382_CE), COL1A1 (Hs00747266_CE), SDF4 (Hs00796825_CE) and
CSRP1 (Hs00723484_CE). The relative changes of gene expression were
calculated according to the 2−ΔΔCT quantification method
(18).
Western blotting
HepG2 cells were lysed with a non-ionic detergent
(NID) lysis buffer. The resulting soluble cell extract was
centrifuged for 30 min at 12,000 × g and the intracellular protein
was collected. Additional, the secretory proteins was concentrated
for 30 min at 3400 × g using an ultra-filtration centrifuge tube,
after which the extracellular protein was collected. The
intracellular and extracellular protein concentrations were
determined using a 2-D Quant kit (GE Healthcare). Protein (40
µg) was separated by SDS-PAGE and transferred to PVDF
membranes (Amersham Biosciences). Membranes were blocked for 1 h
with 5% non-fat powdered milk in TBS-T buffer (pH 7.6, 0.5%
Tween-20), then incubated overnight with primary antibodies at 4°C.
These monoclonal antibodies against CRP, CTSZ, IL11, CTSD, COL1A1,
CUL1, CALM2, HIF-1α, p-AKT, AKT, p-ERK, ERK and actin (Abcam,
Cambridge, MA, USA) were diluted from 1:2,000 to 1:10,000. After
washing three times with TBS-T buffer, the membranes were incubated
with a horseradish peroxidease-conjugated (HRP) goat anti-rabbit
IgG or goat anti-mouse IgG (Santa Cruz Biotechnology) as the
secondary antibody (1:5,000 dilution) for 1 h at room temperature.
The membranes were washed again three times with TBS-T buffer and
visualized with the ChemiDoc MP Imaging system (Bio-Rad
Laboratories, Hercules, CA, USA).
The detection of HIF-1α luciferase
activity
HepG2 cells transfected with either CRP siRNA or
control siRNA were plated into 24-well plates. After 24 h of
transfection, the HepG2 cells were co-transfected with 500 ng of
plasmid pGL3 and 15 ng of pRL-SV40 using Lipofectamine 2000. After
transfection for additional 24 h, the Luciferase activity of HIF-1α
was determined on a GloMax 20/20 using the Dual-luciferase assay
kit (Promega GmbH, Mannheim, Germany). Firefly luciferase units
were normalized with Renilla luciferase carried by pRL-SV40
plasmid. Each experiment was performed in triplicate.
Statistical analysis
The experimental data are presented as the mean ±
standard deviation (SD), and differences between the two groups
were analyzed using the Student's t-test. P<0.05 was considered
statistically significant. SPSS software v16.0 (SPSS, Inc.,
Chicago, IL, USA) was used for the analyses.
Results
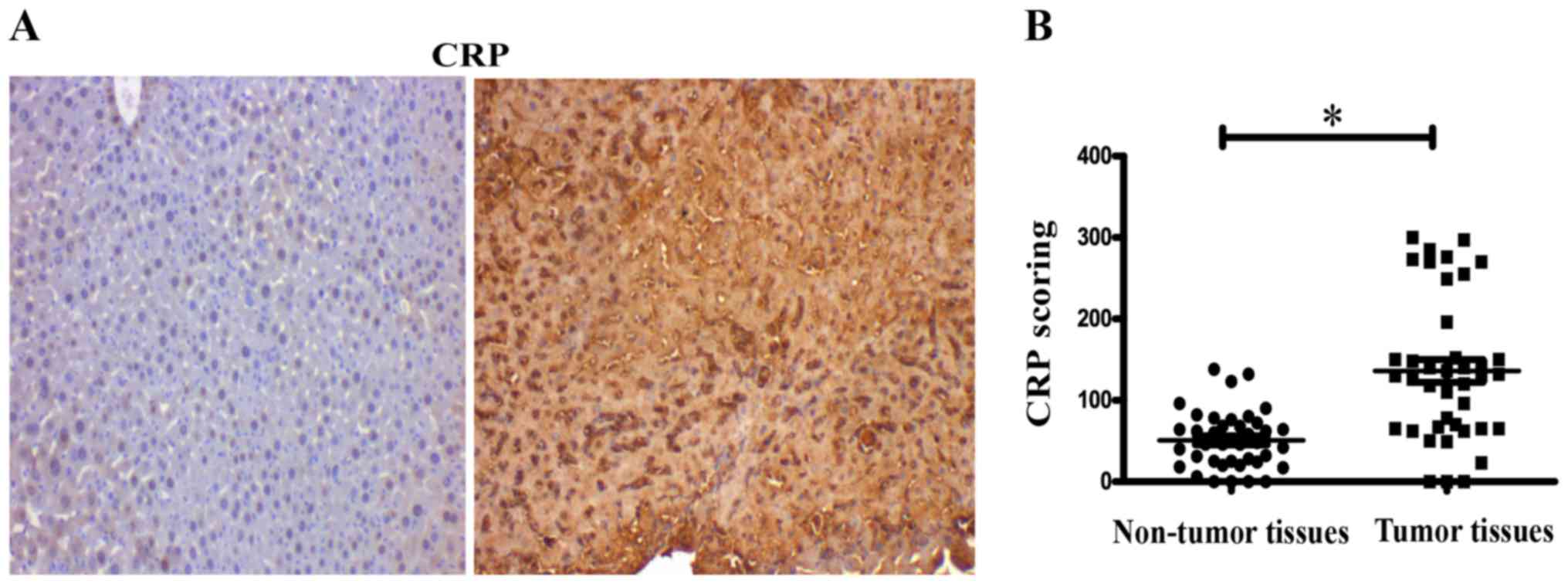
Overexpression of CRP in hepatocellular
carcinoma tissues
The expression of CRP was assessed in tissue
microarrays, containing 40 cases of hepatocellular carcinoma and 40
matched adjacent normal tissue by IHC. IHC evaluation of tissue
microarrays showed that expression of CRP was significantly
stronger in tumor tissues than in normal tissues (P<0.05)
(Fig. 1A). IHC score values of CRP
were significantly higher in the HCC tissues than in the HCC
adjacent normal tissues (P<0.05) (Fig. 1B).
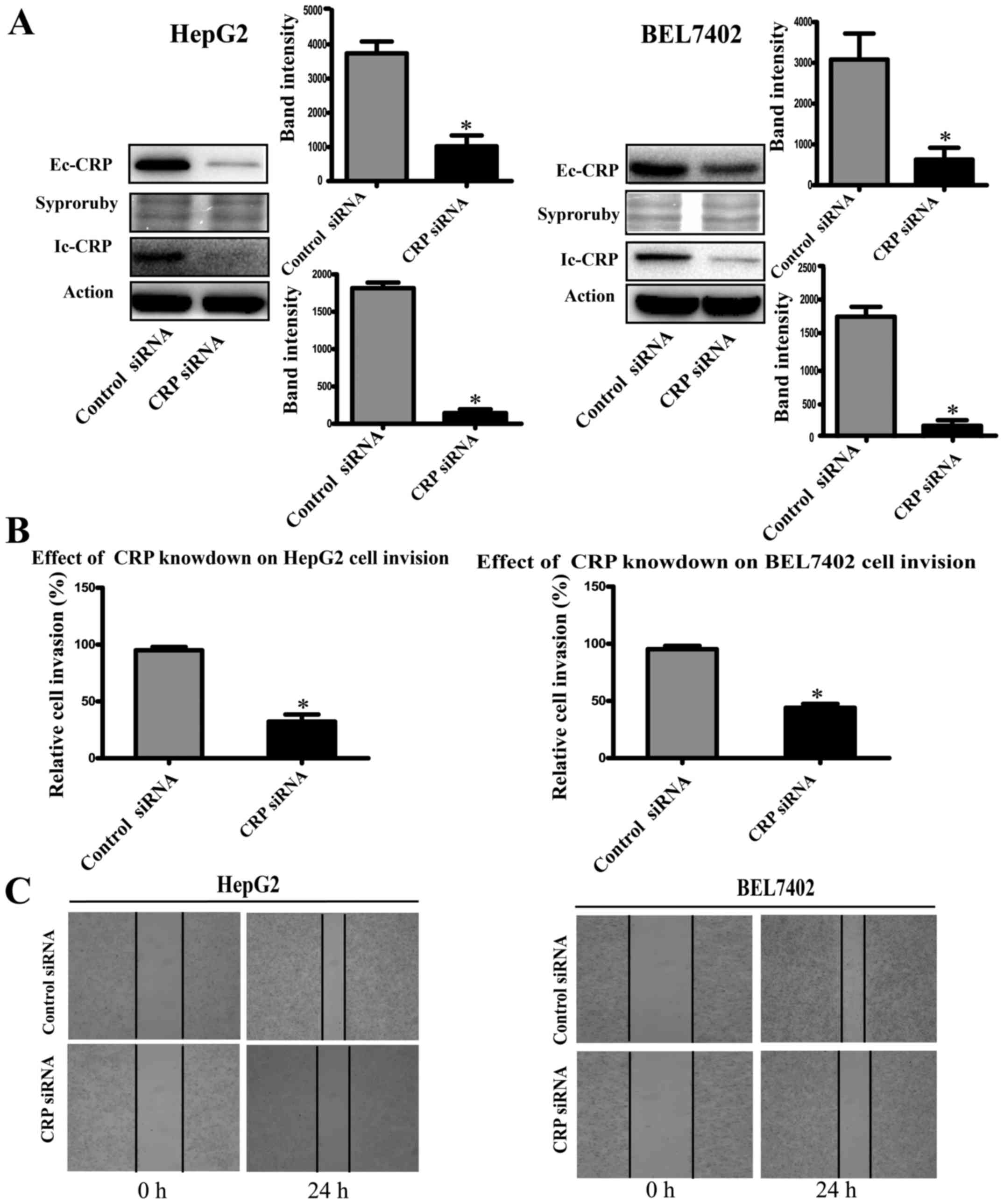
CRP inhibits HCC cell migration, invasion
and wound healing
To study the role of CRP in tumor cell motility, we
used CRP-specific siRNA to silence CRP expression in HCC cell lines
(HepG2 and BEL7402). Western blot analysis showed that CRP
siRNA-treated cell lines downregulated CRP expression significantly
(Fig. 2A). The invasion assay
demonstrated that the downregulation of CRP markedly weakened the
migration and invasion capabilities of HepG2 and BEL7402 cells by
63 and 50%, respectively (P<0.05) (Fig. 2B). Similarly, the ability to close
scratch wounds was decreased in HepG2 and BEL7402 cells (Fig. 2C).
Analysis of iTRAQ data of aberrantly
expressed proteins
To investigate the molecular mechanism of CRP in the
suppression of HCC migration and invasion, we conducted iTRAQ-based
MS to analyze secretory proteins from CRP siRNA-treated and
negative control siRNA-treated HepG2 cells. The ratio of 115:113
and 117:116 expressed the relative protein expression in the CRP
siRNA-treated and negative control siRNA-treated secretory
proteins. Hundreds of proteins were identified by ProteinPilot 2.0
software. The protein threshold was set to achieve 95% confidence
at 5% FDR (false discovery rate). To define the differentially
expressed proteins (DEPs), we introduced an additional ±1.3-fold
cut-off for all iTRAQ ratios (19,20).
Using this value, the overall data from technical replicate
analyses produces <30% variation. A total of 401 unique proteins
were confidently identified and quantified, regardless of whether
the P-value was <0.05 in the iTRAQ ratios. Of the 109 proteins
expressed differentially between the CRP siRNA-treated samples and
the negative control siRNA-treated samples, 45 proteins were
overexpressed and 64 were downregulated. The top 30 upregulated and
downregulated proteins are shown in Table I.
 | Table IThe top 30 upregulated and
downregulated differentially expressed proteins, as identified
using iTRAQ technology. |
Table I
The top 30 upregulated and
downregulated differentially expressed proteins, as identified
using iTRAQ technology.
| N | Accession | Gene symbol | Protein name | Peptides −95% | CRP knockdown:
control (115:113) | PVal 115:113 | CRP knockdown:
control (117:116) | PVal 117:116 |
|---|
| Top 30 proteins
upregulated in CRP siRNA secretory protein |
| 1 |
sp|P14136|GFAP_HUMAN | GFAP | Glial fibrillary
acidic protein | 15 | 2.697659 | 0.009843 | 2.729757 | 0.010676 |
| 2 |
tr|J3QR68|J3QR68_HUMAN | HP | Haptoglobin | 11 | 2.262996 | 0.002992 | 2.545902 | 0.002731 |
| 3 |
sp|P51884|LUM_HUMAN | BLMH | Bleomycin
hydrolase | 3 | 1.987454 | 0.039766 | 1.995493 | 0.046258 |
| 4 |
tr|A8K3S1|A8K3S1_HUMAN | GNPDA2 |
Glucosamine-6-phosphate isomerase | 10 | 1.874345 | 0.006277 | 1.693967 | 0.030001 |
| 5 |
sp|P10155|RO60_HUMAN | TROVE2 | 60 kDa SS-A/Ro
ribonucleoprotein isoform | 8 | 1.810782 | 0.000753 | 1.685377 | 0.00063 |
| 6 |
tr|Q53GX6|Q53GX6_HUMAN | GSTO1 | Highly similar to
Homo sapiens glutathione S-transferase omega 1 | 4 | 1.794569 | 0.016479 | 1.71704 | 0.015401 |
| 7 |
tr|H0Y8C6|H0Y8C6_HUMAN | IPO5 | Importin-5 | 27 | 1.661198 | 0.00088 | 1.677587 | 0.000784 |
| 8 |
tr|Q32Q12|Q32Q12_HUMAN | NME2 | Nucleoside
diphosphate kinase | 54 | 1.660392 | 0.014814 | 1.723658 | 0.004326 |
| 9 |
tr|H0Y7A7|H0Y7A7_HUMAN | CALM2 | Calmodulin | 23 | 1.618516 | 0.014581 | 1.497814 | 0.048883 |
| 10 |
sp|Q6P587|FAHD1_HUMAN | FAHD1 | Acylpyruvase
FAHD1 | 6 | 1.597314 | 0.048898 | 1.745514 | 0.006089 |
| 11 |
tr|Q6LET3|Q6LET3_HUMAN | HPRT1 | HPRT1 protein | 12 | 1.561261 | 0.003935 | 1.6111 | 0.000739 |
| 12 |
tr|K7EKD8|K7EKD8_HUMAN | CAPNS1 | Calpain small
subunit 1 | 2 | 1.550796 | 0.050152 | 1.564854 | 0.057883 |
| 13 | tr|A0A024R462
_HUMAN | FN1 | Fibronectin 1 | 129 | 1.507385 | 4.96E-11 | 1.516702 | 3.37E-09 |
| 14 |
sp|Q13616|CUL1_HUMAN | CUL1 | Cullin-1 | 4 | 1.468003 | 0.029943 | 1.863864 | 0.026399 |
| 15 |
sp|Q04760|LGUL_HUMAN | GLO1 | Lactoylglutathione
lyase | 13 | 1.452267 | 0.018025 | 1.434937 | 0.010939 |
| 16 |
sp|P30041|PRDX6_HUMAN | PRDX6 |
Peroxiredoxin-6 | 28 | 1.438182 | 0.038665 | 1.523574 | 0.027196 |
| 17 |
sp|P26641|EF1G_HUMAN | EEF1G | Elongation factor
1-gamma | 14 | 1.434626 | 0.025754 | 1.47196 | 0.020297 |
| 18 |
tr|B0YIW6|B0YIW6_HUMAN | ARCN1 | Archain 1 | 7 | 1.432327 | 0.035547 | 1.374012 | 0.10264 |
| 19 |
sp|P55060|XPO2_HUMAN | CSE1L | Exportin-2 | 18 | 1.413718 | 0.009854 | 1.407874 | 0.025418 |
| 20 |
sp|O75436|VP26A_HUMAN | VPS26A | Vacuolar protein
sorting-associated protein 26A | 8 | 1.424428 | 0.042069 | 1.388839 | 0.022327 |
| 21 |
sp|P27695|APEX1_HUMAN | APEX1 | DNA-(apurinic or
apyrimidinic site) lyase | 9 | 1.388838 | 0.031594 | 1.508206 | 0.008443 |
| 22 |
sp|P48637|GSHB_HUMAN | GSS | Glutathione
synthetase | 16 | 1.386073 | 0.020581 | 1.319895 | 0.000501 |
| 23 |
sp|Q16531|DDB1_HUMAN | DDB1 | DNA damage-binding
protein 1 | 35 | 1.370885 | 0.002909 | 1.491143 | 0.000105 |
| 24 |
tr|H7C2I1|H7C2I1_HUMAN | PRMT1 | Protein arginine
N-methyltransferase 1 | 10 | 1.369152 | 0.013057 | 1.446224 | 0.026285 |
| 25 |
sp|P60900|PSA6_HUMAN | PSMA6 | Proteasome subunit
alpha type-6 | 22 | 1.359674 | 0.007907 | 1.347712 | 0.034374 |
| 26 |
sp|Q13907|IDI1_HUMAN | IDI1 |
Isopentenyl-diphosphate Delta-isomerase
1 | 4 | 1.359607 | 0.030252 | 1.374364 | 0.105732 |
| 27 |
sp|Q8N543|OGFD1_HUMAN | OGFOD1 | Prolyl
3-hydroxylase OGFOD1 | 5 | 1.351501 | 0.050466 | 1.353389 | 0.038133 |
| 28 |
sp|Q93009|UBP7_HUMAN | USP7 | Ubiquitin
carboxyl-terminal hydrolase 7 | 10 | 1.33261 | 0.020238 | 1.417985 | 0.135236 |
| 29 |
sp|P13639|EF2_HUMAN | EEF2 | Elongation factor
2 | 48 | 1.324262 | 0.041668 | 1.352563 | 0.000136 |
| 30 |
tr|H0UID3|H0UID3_HUMAN | AP2B1 | Adaptor-related
protein complex 2 | 16 | 1.311697 | 0.003873 | 1.434811 | 0.014619 |
| Top 30 proteins
downregulated in CRP siRNA secretory protein |
| 1 |
tr|B4E3Q1|B4E3Q1_HUMAN | BMP2 | Bone morphogenetic
protein 2 | 3 | 0.317011 | 0.10559 | 0.278341 | 0.051668 |
| 2 |
tr|K7EKD8|K7EKD8_HUMAN | SDF4 | 45 kDa
calcium-binding protein | 3 | 0.385897 | 0.051998 | 0.349369 | 0.049074 |
| 3 |
tr|Q8WUV3|Q8WUV3_HUMAN | CSRP1 | Cysteine and
glycine-rich protein 1 | 4 | 0.403368 | 0.067666 | 0.32749 | 0.047143 |
| 4 |
tr|H0Y7A7|H0Y7A7_HUMAN | NUCB1 | Nucleobindin 1
variant | 18 | 0.493801 | 8.39E-05 | 0.480406 | 0.000151 |
| 5 |
sp|Q9HAV7|GRPE1_HUMAN | CLU | Clusterin | 63 | 0.518533 | 0.001495 | 0.480786 | 0.000236 |
| 6 |
sp|Q12841|FSTL1_HUMAN | FSTL1 | Follistatin-related
protein 1 | 11 | 0.526944 | 0.005066 | 0.591959 | 0.002263 |
| 7 |
sp|O75787|RENR_HUMAN | ATP6AP2 | Renin receptor | 8 | 0.564742 | 0.00759 | 0.616903 | 0.009215 |
| 8 |
sp|Q16270|IBP7_HUMAN | IGFBP7 | Insulin-like growth
factor-binding protein 7 | 36 | 0.568981 | 0.005411 | 0.525776 | 0.002737 |
| 9 |
sp|P20809|IL11_HUMAN | IL-11 | Interleukin-11 | 3 | 0.579813 | 0.077419 | 0.579813 | 0.077419 |
| 10 |
sp|P04406|G3P_HUMAN | GAPDH |
Glyceraldehyde-3-phosphate
dehydrogenase | 108 | 0.581235 | 0.001442 | 0.49832 | 2.85E-05 |
| 11 |
sp|P07339|CATD_HUMAN | CTSD | Cathepsin D | 23 | 0.596307 | 0.011429 | 0.556281 | 0.005984 |
| 12 |
sp|P19022|CADH2_HUMAN | CDH2 | Cadherin-2 | 10 | 0.597 | 0.029602 | 0.583355 | 0.045716 |
| 13 |
tr|Q5U000|Q5U000_HUMAN | CTSZ | Cathepsin Z | 11 | 0.614062 | 0.04132 | 0.588521 | 0.004804 |
| 14 |
sp|Q13421|MSLN_HUMAN | MSLN | Mesothelin | 63 | 0.619085 | 0.031075 | 0.643571 | 0.01251 |
| 15 |
tr|Q7Z3Z9|Q7Z3Z9_HUMAN | L1CAM | L1 cell adhesion
molecule | 42 | 0.620206 | 0.000147 | 0.604773 | 0.000235 |
| 16 |
sp|Q8NES3|LFNG_HUMAN | LFNG |
β-1,3-N-acetylglucosaminyltransferase
lunatic fringe | 3 | 0.630394 | 0.003585 | 0.512136 | 0.00982 |
| 17 |
sp|P20827|EFNA1_HUMAN | EFNA1 | Ephrin-A1 | 8 | 0.641523 | 0.038208 | 0.654235 | 0.0573 |
| 18 |
sp|P00966|ASSY_HUMAN | ASS1 | Argininosuccinate
synthase | 62 | 0.652695 | 0.015085 | 0.635661 | 0.016378 |
| 19 |
sp|P35555|FBN1_HUMAN | FBN1 | Fibrillin-1 | 23 | 0.676557 | 0.006903 | 0.736246 | 0.016766 |
| 20 |
sp|P17936|IBP3_HUMAN | IGFBP3 | Insulin-like growth
factor-binding protein 3 | 9 | 0.703906 | 0.005939 | 0.758705 | 0.01324 |
| 21 |
sp|P51884|LUM_HUMAN | LUM | Lumican | 21 | 0.707534 | 0.007503 | 0.71794 | 5.66E-05 |
| 22 |
sp|P16035|TIMP2_HUMAN | TIMP2 | Metalloproteinase
inhibitor 2 | 15 | 0.717572 | 0.036572 | 0.683828 | 0.037238 |
| 23 |
tr|Q2M1J3|Q2M1J3_HUMAN | ROBO1 | ROBO1 protein | 16 | 0.717682 | 0.005528 | 0.71049 | 0.014984 |
| 24 |
sp|P28799|GRN_HUMAN | GRN | Granulins | 13 | 0.718265 | 0.042939 | 0.644782 | 0.013296 |
| 25 |
sp|Q9H4F8|SMOC1_HUMAN | SMOC1 | SPARC-related
modular calcium-binding protein 1 | 16 | 0.727941 | 0.035403 | 0.708092 | 0.011316 |
| 26 |
sp|P24752|THIL_HUMAN | ACAT1 | Acetyl-CoA
acetyltransferase, mitochondrial | 6 | 0.732777 | 0.045963 | 0.709778 | 0.042653 |
| 27 |
sp|P10809|CH60_HUMAN | HSPD1 | 60 kDa heat shock
protein, mitochondrial | 24 | 0.73754 | 0.007491 | 0.636544 | 0.000373 |
| 28 |
sp|P05067|A4_HUMAN | APP | Amyloid β A4
protein | 18 | 0.741021 | 0.01009 | 0.716255 | 0.020473 |
| 29 |
sp|Q92626|PXDN_HUMAN | PXDN | Peroxidasin
homolog | 51 | 0.746291 | 0.000592 | 0.747293 | 0.001493 |
| 30 |
sp|Q9BRK5|CAB45_HUMAN | COL1A1 | Collagen α-1(I)
chain | 5 | 0.753276 | 0.020072 | 0.447282 | 0.011558 |
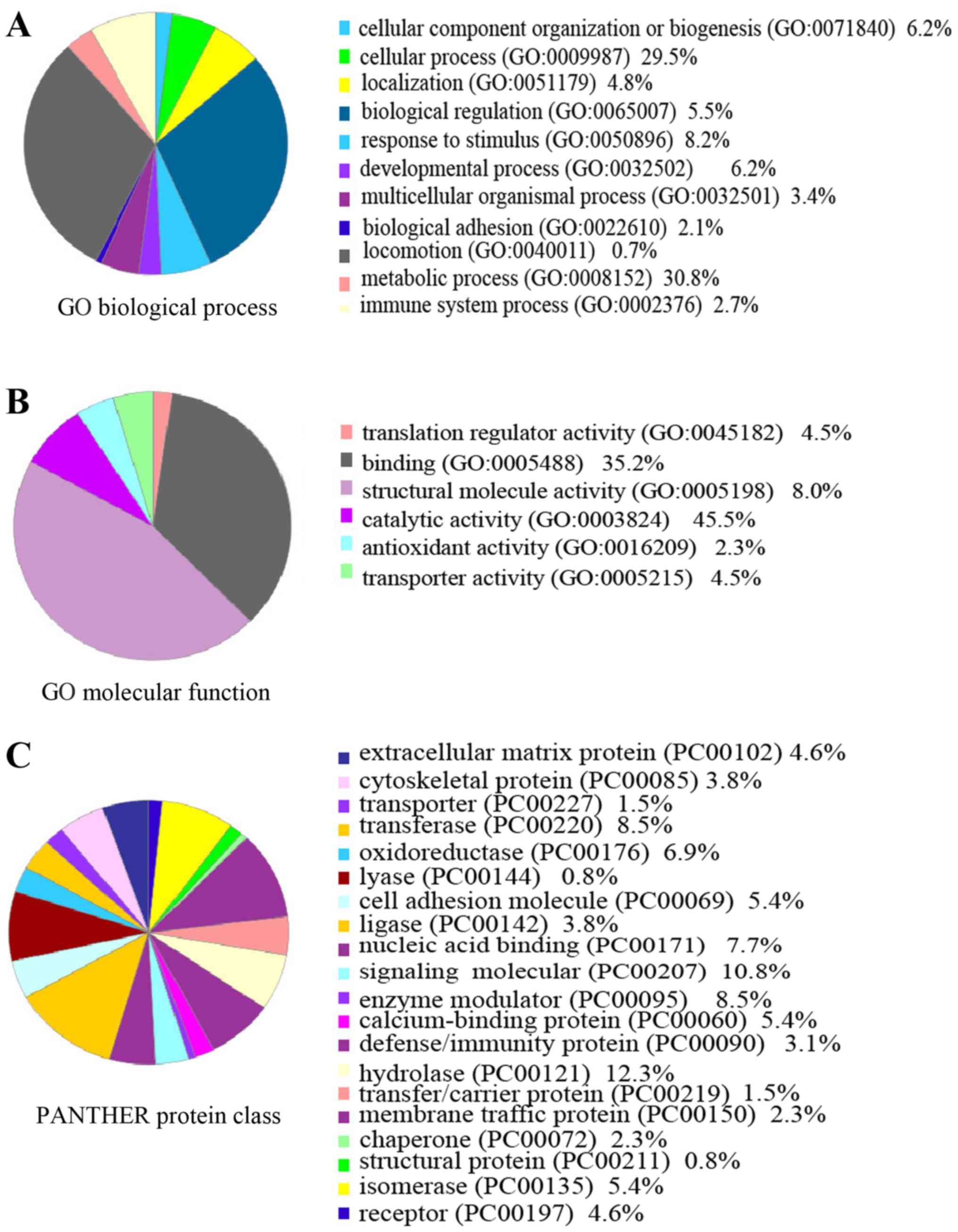
Cellular and molecular functional
characteristics of the differentially expressed proteins
To better identify the functional characteristics of
the 109 DEPs, these proteins were grouped by PANTHER Classification
System according to their reported biological process, protein
class and molecular functions. Gene Ontology analysis with PANTHER
suggested that the DEPs was found to represent a total of 11
biological processes, 20 protein classes and 6 molecular functions
(Fig. 3). Metabolic, cellular and
developmental processes were the most common biological processes
reported.
Validation of differentially expressed
proteins
To validate the reliability of the iTRAQ analysis
data, we chose samples used in the iTRAQ assays and conducted
western blotting and RT-PCR to detect the extracellular levels of
several DEPs. Fig. 4A shows the
relative mRNA expression levels of CTSZ, CDH2, IL11, CTSD, NUCB1,
COL1A1, SDF4, CSRP1, HSPD1, GFAP, CUL1, FAHD1, NME2, CALM2, DDB1,
and GNPDA2 as normalized to GADPH. RT-PCR showed the mRNA levels of
CTSZ, CDH2, IL11, CTSD, NUCB1, COL1A1, SDF4, CSRP1 and HSPD1 were
downregulated, whereas the mRNA levels of GFAP, CUL1, FAHD1, NME2,
CALM2, DDB1 and GNPDA2 were upregulated in the CRP siRNA-treated
samples, compared to the negative control siRNA-treated samples.
The trend was consistent with the results of the iTRAQ approach. In
order to validate the levels of several proteins, western blot
analyses were performed. Fig. 4B
shows the western blot analysis results of CTSZ, IL11, CTSD, COL1A1
CUL1 and CALM2 expression in intracellular and extracellular
samples. Supernatant protein from CRP siRNA-treated cells had
obviously decreased expression levels of CTSZ, IL11, CTSD, COL1A1
and increased expression levels of CUL1 and CALM2, compared to
negative control siRNA-treated cells. IL11 and CTSD are,
intracellularly, low expression proteins, thus, these were not
detected in the western blot analyses. In addition to these two
proteins, other proteins are similarly expressed
intracellularly.
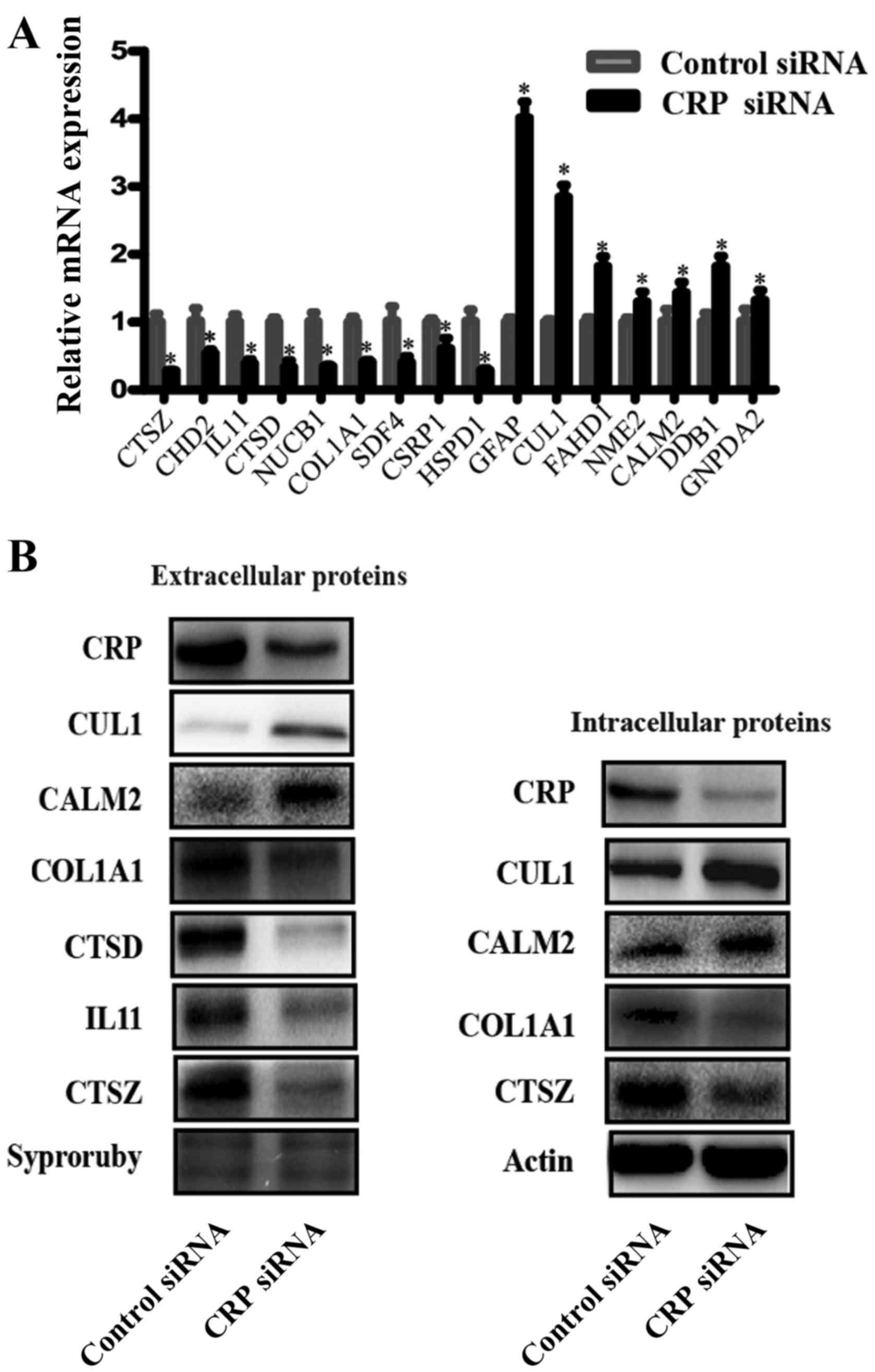 | Figure 4Verification of the differentially
expressed proteins. (A) Real-time RT-PCR detected the relative mRNA
expression levels of CTSZ, CDH2, IL11, CTSD, NUCB1, COL1A1, SDF4,
CSRP1, HSPD1, GFAP, CUL1, FAHD1, NME2, CALM2, DDB1 and GNPDA2, as
normalized to GADPH (*P<0.05). (B) A representative
western blot analysis for CTSZ, IL11, CTSD, COL1A1 CUL1 and CALM2
expression in intracellular and extracellular samples from HepG2
lines. (Bars indicate SD, *P<0.05). Actin was used as
the normalization standard. |
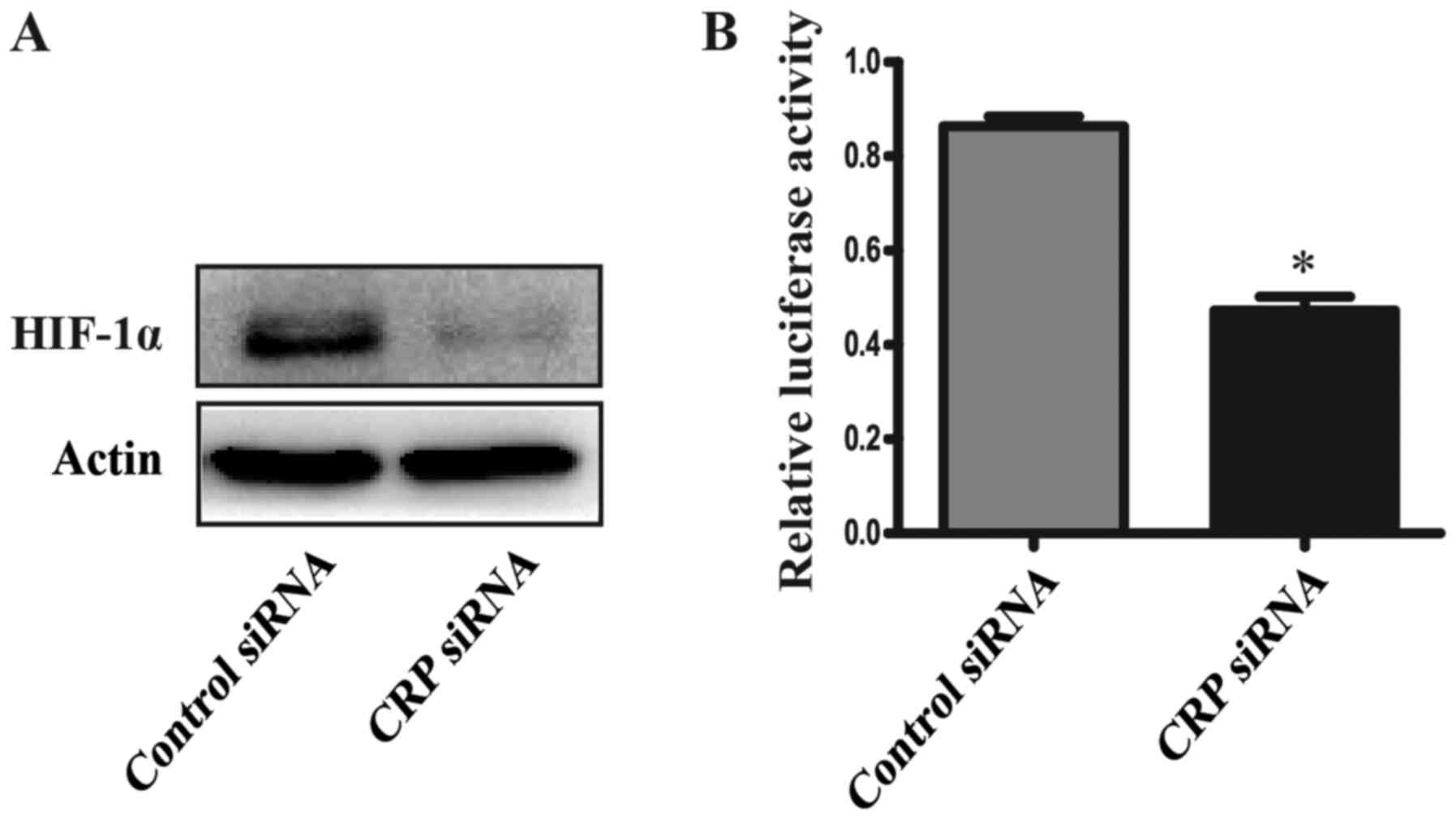
CRP knockdown downregulates HIF-1α
expression and the luciferase activity of HIF-1α in HepG2
cells
Because CRP could affect the activity of HIF-1α,
which has also been shown to induce expression of CTSD (21). CTSD accociated with the growth,
proliferation and metastasis of tumors (22). The expression and luciferase
activity of HIF-1α was determined using the western blot analyses
and Dual-luciferase reporter assay system. Our results showed that
the expression and luciferase activity of HIF-1α was significantly
reduced in CRP siRNA treated HepG2 cells, compared to the control
group (Fig. 5).
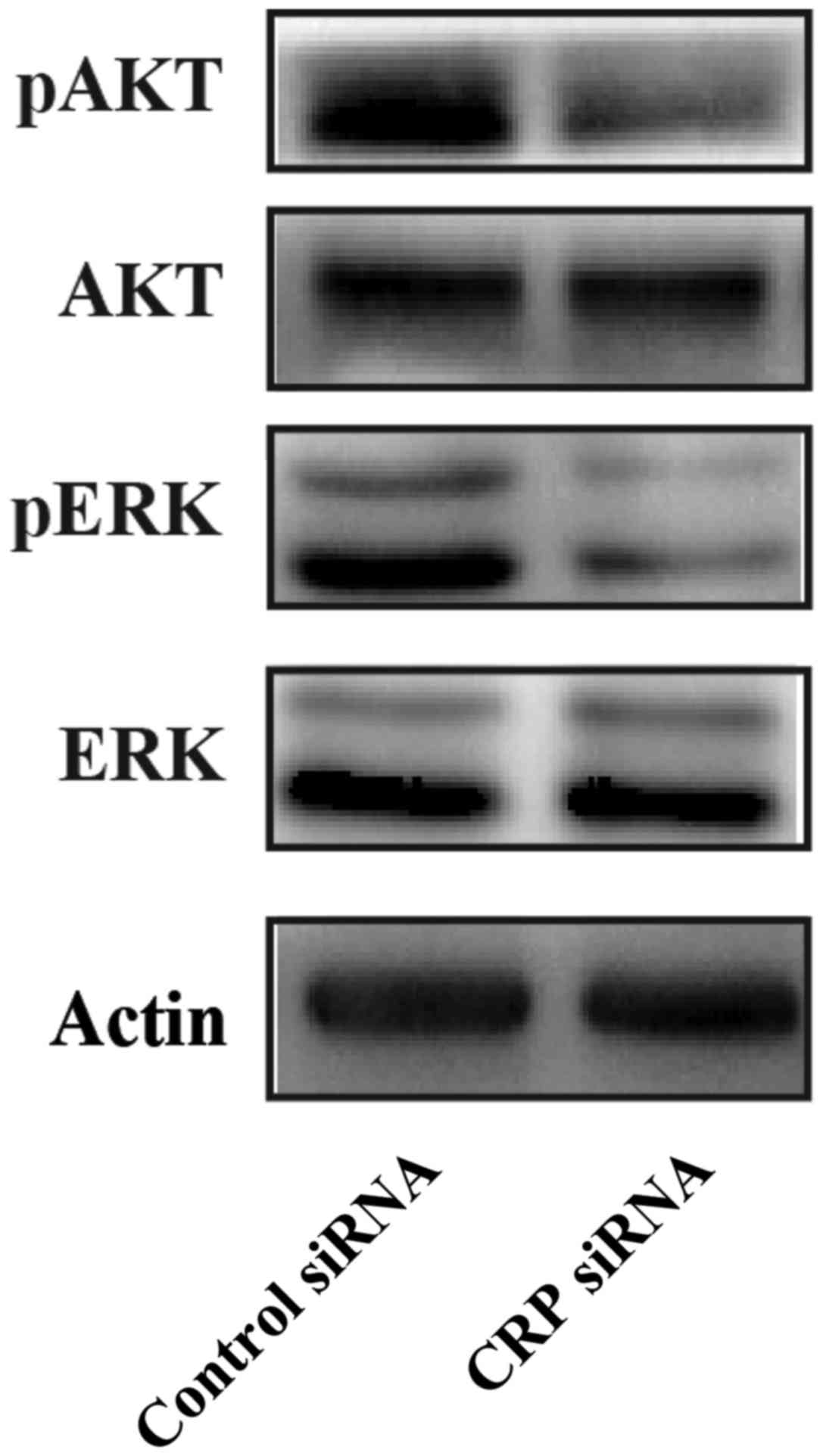
CRP knockdown suppresses ERK and Akt
phosphorylation in HepG2 cells
MEK/ERK and PI3K/AKT signaling pathways play
important roles in migration, invasion and metastasis of cancer
(23). CRP could upregulate VEGF-A
expression via the MEK/ERK and PI3K/AKT signaling pathways in
adipose-derived stem cells (24).
We hypothesize that CRP promotes migration, invasion, and
metastasis of HCC through MEK/ERK and PI3K/AKT signaling pathways.
We observed that the downregulation of CRP remarkably inhibits ERK
and Akt phosphorylation at the protein level when compared to
control siRNA in HepG2 cells (Fig.
6). These results indicate that CRP may induce cell migration,
and invasion through MEK/ERK and PI3K/AKT signaling pathways.
Discussion
Using proteomic strategies, a growing body of
evidence has identified proteins specifically upregulated or
downregulated in HCC tissues that can be considered as early
diagnostic markers, prognostic markers and therapeutic targets
(25,26). CRP is such a protein overexpressed
in various types of tumors (6–10),
and is a promising biomarker of HBV-related HCC (27). However, little is known about the
function of CRP in HCC cells.
The present study demonstrates that CRP was
significantly overexpressed in HCC tissues compared to
non-cancerous tissues. Extra-hepatic metastases of HCC, which is
the main cause of cancer-related death, depend largely on the
migratory and invasive capabilities of HCC cells. Several studies
have demonstrated that CRP promoted cell proliferation in
endothelial cells, endothelial progenitor cells, renal tubular
epithelial cells and provided protection from apoptosis in myeloma
cells, in vitro and in vivo (11,12).
Here, we demonstrated that CRP knockdown in HepG2 and BEL7402 cells
significantly suppressed cell growth, migration and invasion in
vitro, as shown in wound assays and Transwell assays. Our
findings provide the first piece of evidence that CRP silencing
inhibited migration and invasion, suggesting a carcinogenic role
for CRP in HCC.
To investigate the potential molecular mechanism by
which CRP contributes to migration and invasion, iTRAQ-based MS was
performed to analyze secretory DEPs between CRP siRNA-treated and
negative siRNA-treated HepG2 cells. Our iTRAQ analysis identified
109 aberrantly expressed proteins in CRP siRNA-treated samples.
Many of them, including CTSZ, IL11, CTSD, COL1A1, CUL1 and CALM2,
were identified using western blot analysis and RT-PCR analyses.
The data indicated that the iTRAQ technology is both reliable and
powerful for protein quantification. Among these proteins, we
focused on cathepsin D (CTSD) because the expression of CTSD was
obviously downregulated in CRP siRNA-treated samples, compared to
control negative siRNA-treated samples, and is closely associated
with invasion and metastasis of cancer cells.
Cathepsin D, a member of the aspartic proteinase
super-family in the lysosomes of eukaryotic cells (28), degrades the extracellular matrix
(ECM) and is overexpressed and hyper-secreted by carcinoma cells
(29). Accumulated data show that
CTSD is secrected in breast, prostate, ovaria, and lung cancer cell
lines, and acts as a autocrine cancer cell growth factor involving
cancer development (30). In
addition, CTSD correlates with poor prognoses, invasion and
metastasis in many malignancies (22,31,32).
Several possible mechanisms have been proposed. For instance, CTSD
promotes angiogenesis by releasing basic fibroblast growth factor
(33). Additionally, CTSD can
degrade anti-angiogenesis growth factors, such as angiogenesis
inhibitor 16K prolactin and endostatin (34). HIF1, a transcription factor, is one
of the important players in modulation of cell metabolism and plays
an essential role in cellular and systemic homeostatic responses to
hypoxia. HIF-1α in itself induces expression of several glycolytic
enzymes, as well as inhibits entry into the TCA-cycle (35). Several studies have shown that high
expression of HIF-1α correlated with a short survival in non-small
cell lung cancer (NSCLC) (36–38).
HIF-1α has also been shown to induce expression ofCTSD (21). Based on the close relationship
between HIF-1α and CTSD, and the observation that CTSD was
significantly decreased when CRP was silenced. We further measured
expression and activity of HIF-1α. Our result showed that
expression and luciferase activity of HIF-1α was decreased in CRP
siRNA treated HepG2 cells. Thus, we hypothesized that when
silencing CRP, the decrease in CTSD may be through this
pathway.
It has been reported that activation of MEK/ERK and
PI3K/AKT signaling pathways play important roles in migration,
invasion and metastasis of cancer (23). CRP could upregulate VEGF-A
expression by activating HIF-1α via the MEK/ERK and PI3K/AKT
signaling pathways in adipose-derived stem cells (ADSCs) (24). However, few studies have reported
on the relationship of CRP and MEK/ERK and PI3K/AKT signaling
pathways in HCC. Therefore, we investigated whether CRP was capable
of promoting migration, invasion via the MEK/ERK and PI3K/AKT
pathway in HCC cells. Our results showed that CRP knockdown
remarkably inhibits ERK and Akt phosphorylation at the protein
level when compared to control siRNA in HepG2 cells. Therefore, our
data support that activation of MEK/ERK and PI3K/AKT signaling
pathways may required for CRP-stimulated cell migration and
invasion of HCC cells.
In conclusion, we have demonstrated that CRP is
highly expressed in tumor tissues and promotes invasion and
metastases in HCC cell lines. In addition, we have performed a
quantitative proteomic profiling of supernatant proteins from CRP
siRNA-treated and negative control siRNA-treated HepG2 cells. We
observed 109 aberrantly expressed proteins in CRP siRNA-treated
samples. Moreover, silencing of CRP abrogates HIF-1α expression
levels, the luciferase activity of HIF-1α, and ERK and Akt
phosphorylation in HepG2 cells. The present study provides a novel
mechanism by which CRP promotes the proliferation, migration,
invasion, metastasis of hepatocellular carcinoma cells. Inhibition
of CRP could suppress migration, invasion and healing of hepatoma
carcinoma cells by decreasing HIF-1α activity and CTSD.
Acknowledgments
The present study was supported by the National
Natural Science Foundation of China (81171560), the National Key
Technology Support Program (2012BAI35B03), the 'Par-Eu Scholars
Program' of Chongqing City, the National Science and Technology
Major Project of China (2012ZX10002007001), the Chongqing Natural
Science Foundation, the Chongqing Municipal Science and Technology
(no. cstc2012jjA10064), and the Natural Science Foundation Project
of CQ CSTC (2013jcyjA10060).
References
|
1
|
El-Serag HB: Epidemiology of viral
hepatitis and hepatocellular carcinoma. Gastroenterology.
142:1264–1273.e1. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
El-Serag HB and Kanwal F: Epidemiology of
hepatocellular carcinoma in the United States: Where are we? Where
do we go? Hepatology. 60:1767–1775. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ren W, Qi X, Jia J, Yang M and Han G:
Hepatocellular carcinoma. Lancet (London, England). 380:469author
reply 470–461. 2012. View Article : Google Scholar
|
|
4
|
Stein MP, Edberg JC, Kimberly RP, Mangan
EK, Bharadwaj D, Mold C and Du Clos TW: C-reactive protein binding
to FcgammaRIIa on human monocytes and neutrophils is
allele-specific. J Clin Invest. 105:369–376. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pepys MB: C-reactive protein: The role of
an ancient protein in modern rheumatology. Clin Exp Rheumatol.
1:3–7. 1983.PubMed/NCBI
|
|
6
|
Hefler LA, Concin N, Hofstetter G, Marth
C, Mustea A, Sehouli J, Zeillinger R, Leipold H, Lass H, Grimm C,
et al: Serum C-reactive protein as independent prognostic variable
in patients with ovarian cancer. Clin Cancer Res. 14:710–714. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu M, Zhu M, Du Y, Yan B, Wang Q, Wang C
and Zhao J: Serum C-reactive protein and risk of lung cancer: A
case-control study. Med Oncol. 30:3192013. View Article : Google Scholar
|
|
8
|
Chung YC and Chang YF: Serum C-reactive
protein correlates with survival in colorectal cancer patients but
is not an independent prognostic indicator. Eur J Gastroenterol
Hepatol. 15:369–373. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bataille R, Boccadoro M, Klein B, Durie B
and Pileri A: C-reactive protein and beta-2 microglobulin produce a
simple and powerful myeloma staging system. Blood. 80:733–737.
1992.PubMed/NCBI
|
|
10
|
Legouffe E, Rodriguez C, Picot MC, Richard
B, Klein B, Rossi JF and Commes T: C-reactive protein serum level
is a valuable and simple prognostic marker in non Hodgkin's
lymphoma. Leuk Lymphoma. 31:351–357. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang J, Wezeman M, Zhang X, Lin P, Wang M,
Qian J, Wan B, Kwak LW, Yu L and Yi Q: Human C-reactive protein
binds activating Fcgamma receptors and protects myeloma tumor cells
from apoptosis. Cancer Cell. 12:252–265. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu F, Chen HY, Huang XR, Chung AC, Zhou
L, Fu P, Szalai AJ and Lan HY: C-reactive protein promotes diabetic
kidney disease in a mouse model of type 1 diabetes. Diabetologia.
54:2713–2723. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ho J, Kong JW, Choong LY, Loh MC, Toy W,
Chong PK, Wong CH, Wong CY, Shah N and Lim YP: Novel breast cancer
metastasis-associated proteins. J Proteome Res. 8:583–594. 2009.
View Article : Google Scholar
|
|
14
|
Hu H, Ding X, Yang Y, Zhang H, Li H, Tong
S, An X, Zhong Q, Liu X, Ma L, et al: Changes in
glucose-6-phosphate dehydrogenase expression results in altered
behavior of HBV-associated liver cancer cells. Am J Physiol
Gastrointest Liver Physiol. 307:G611–G622. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ran X, Xu X, Yang Y, She S, Yang M, Li S,
Peng H, Ding X, Hu H, Hu P, et al: A quantitative proteomics study
on olfactomedin 4 in the development of gastric cancer. Int J
Oncol. 47:1932–1944. 2015.PubMed/NCBI
|
|
16
|
Wang LN, Tong SW, Hu HD, Ye F, Li SL, Ren
H, Zhang DZ, Xiang R and Yang YX: Quantitative proteome analysis of
ovarian cancer tissues using a iTRAQ approach. J Cell Biochem.
113:3762–3772. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang Y, Toy W, Choong LY, Hou P, Ashktorab
H, Smoot DT, Yeoh KG and Lim YP: Discovery of SLC3A2 cell membrane
protein as a potential gastric cancer biomarker: Implications in
molecular imaging. J Proteome Res. 11:5736–5747. 2012.PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
19
|
Gan CS, Chong PK, Pham TK and Wright PC:
Technical, experimental, and biological variations in isobaric tags
for relative and absolute quantitation (iTRAQ). J Proteome Res.
6:821–827. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou C, Simpson KL, Lancashire LJ, Walker
MJ, Dawson MJ, Unwin RD, Rembielak A, Price P, West C, Dive C, et
al: Statistical considerations of optimal study design for human
plasma proteomics and biomarker discovery. J Proteome Res.
11:2103–2113. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Krishnamachary B, Berg-Dixon S, Kelly B,
Agani F, Feldser D, Ferreira G, Iyer N, LaRusch J, Pak B, Taghavi
P, et al: Regulation of colon carcinoma cell invasion by
hypoxia-inducible factor 1. Cancer Res. 63:1138–1143.
2003.PubMed/NCBI
|
|
22
|
Dian D, Heublein S, Wiest I, Barthell L,
Friese K and Jeschke U: Significance of the tumor protease
cathepsin D for the biology of breast cancer. Histol Histopathol.
29:433–438. 2014.
|
|
23
|
Yoo YA, Kang MH, Lee HJ, Kim BH, Park JK,
Kim HK, Kim JS and Oh SC: Sonic hedgehog pathway promotes
metastasis and lymphangiogenesis via activation of Akt, EMT, and
MMP-9 pathway in gastric cancer. Cancer Res. 71:7061–7070. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen J, Gu Z, Wu M, Yang Y, Zhang J, Ou J,
Zuo Z, Wang J and Chen Y: C-reactive protein can upregulate VEGF
expression to promote ADSC-induced angiogenesis by activating
HIF-1α via CD64/PI3k/Akt and MAPK/ERK signaling pathways. Stem Cell
Res Ther. 7:1142016. View Article : Google Scholar
|
|
25
|
Lee IN, Chen CH, Sheu JC, Lee HS, Huang
GT, Chen DS, Yu CY, Wen CL, Lu FJ and Chow LP: Identification of
complement C3a as a candidate biomarker in human chronic hepatitis
C and HCV-related hepatocellular carcinoma using a proteomics
approach. Proteomics. 6:2865–2873. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Feng JT, Liu YK, Song HY, Dai Z, Qin LX,
Almofti MR, Fang CY, Lu HJ, Yang PY and Tang ZY: Heat-shock protein
27: A potential biomarker for hepatocellular carcinoma identified
by serum proteome analysis. Proteomics. 5:4581–4588. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
She S, Xiang Y, Yang M, Ding X, Liu X, Ma
L, Liu Q, Liu B, Lu Z, Li S, et al: C-reactive protein is a
biomarker of AFP-negative HBV-related hepatocellular carcinoma. Int
J Oncol. 47:543–554. 2015.PubMed/NCBI
|
|
28
|
Westley B and Rochefort H: A secreted
glycoprotein induced by estrogen in human breast cancer cell lines.
Cell. 20:353–362. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rochefort H, Capony F, Garcia M, Cavaillès
V, Freiss G, Chambon M, Morisset M and Vignon F: Estrogen-induced
lysosomal proteases secreted by breast cancer cells: A role in
carcinogenesis? J Cell Biochem. 35:17–29. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Berchem G, Glondu M, Gleizes M, Brouillet
JP, Vignon F, Garcia M and Liaudet-Coopman E: Cathepsin-D affects
multiple tumor progression steps in vivo: Proliferation,
angiogenesis and apoptosis. Oncogene. 21:5951–5955. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Vashishta A, Ohri SS, Proctor M, Fusek M
and Vetvicka V: Ribozyme-targeting procathepsin D and its effect on
invasion and growth of breast cancer cells: An implication in
breast cancer therapy. Int J Oncol. 30:1223–1230. 2007.PubMed/NCBI
|
|
32
|
Glondu M, Liaudet-Coopman E, Derocq D,
Platet N, Rochefort H and Garcia M: Down-regulation of cathepsin-D
expression by antisense gene transfer inhibits tumor growth and
experimental lung metastasis of human breast cancer cells.
Oncogene. 21:5127–5134. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Briozzo P, Badet J, Capony F, Pieri I,
Montcourrier P, Barritault D and Rochefort H: MCF7 mammary cancer
cells respond to bFGF and internalize it following its release from
extracellular matrix: A permissive role of cathepsin D. Exp Cell
Res. 194:252–259. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Piwnica D, Fernandez I, Binart N, Touraine
P, Kelly PA and Goffin V: A new mechanism for prolactin processing
into 16K PRL by secreted cathepsin D. Mol Endocrinol. 20:3263–3278.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Keith B, Johnson RS and Simon MC: HIF1α
and HIF2α: Sibling rivalry in hypoxic tumour growth and
progression. Nat Rev Cancer. 12:9–22. 2011.PubMed/NCBI
|
|
36
|
Hung JJ, Yang MH, Hsu HS, Hsu WH, Liu JS
and Wu KJ: Prognostic significance of hypoxia-inducible
factor-1alpha, TWIST1 and Snail expression in resectable non-small
cell lung cancer. Thorax. 64:1082–1089. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kim SJ, Rabbani ZN, Dewhirst MW,
Vujaskovic Z, Vollmer RT, Schreiber EG, Oosterwijk E and Kelley MJ:
Expression of HIF-1alpha, CA IX, VEGF, and MMP-9 in surgically
resected non-small cell lung cancer. Lung Cancer. 49:325–335. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Giatromanolaki A, Koukourakis MI, Sivridis
E, Turley H, Talks K, Pezzella F, Gatter KC and Harris AL: Relation
of hypoxia inducible factor 1 alpha and 2 alpha in operable
non-small cell lung cancer to angiogenic/molecular profile of
tumours and survival. Br J Cancer. 85:881–890. 2001. View Article : Google Scholar : PubMed/NCBI
|