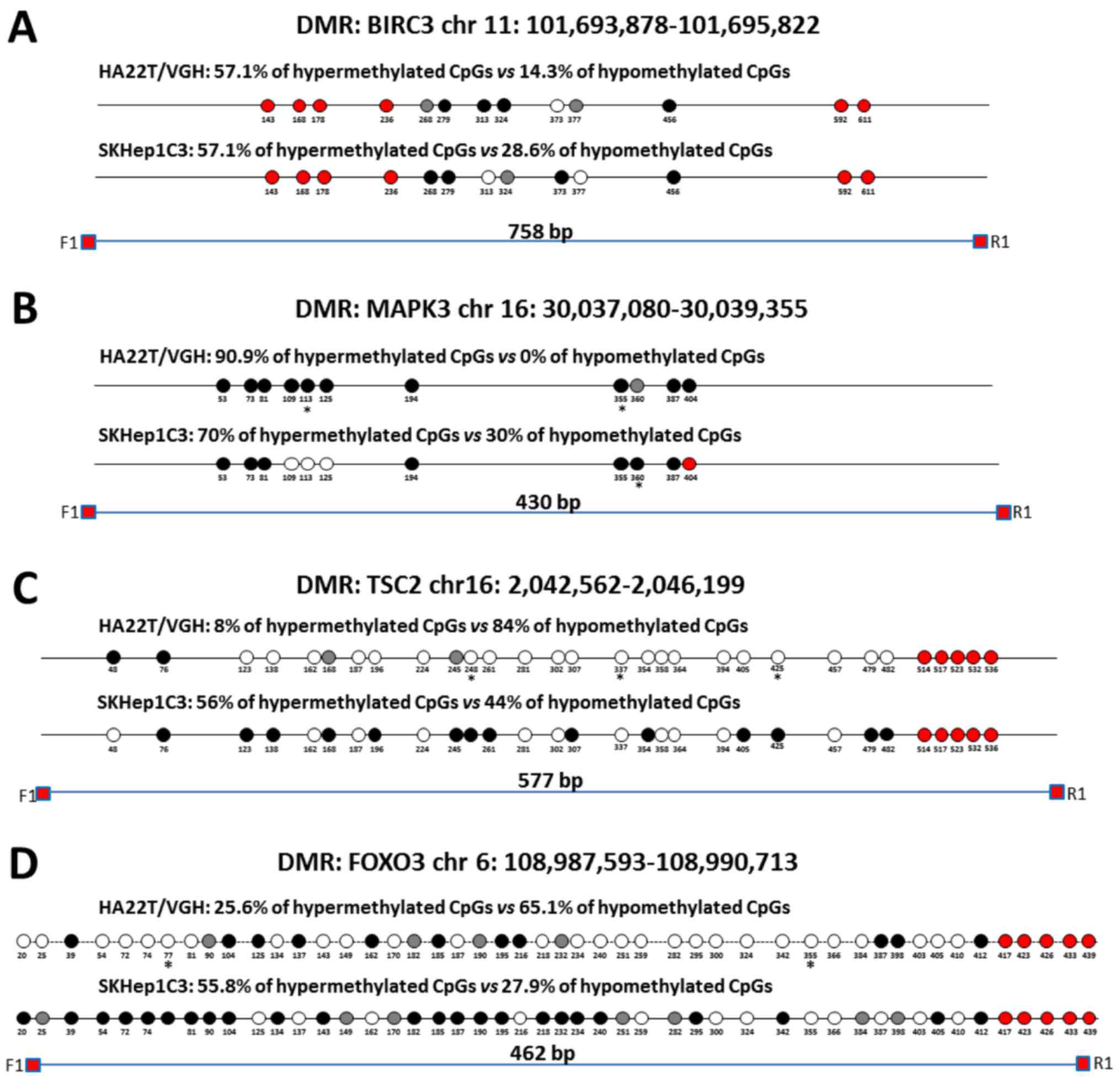
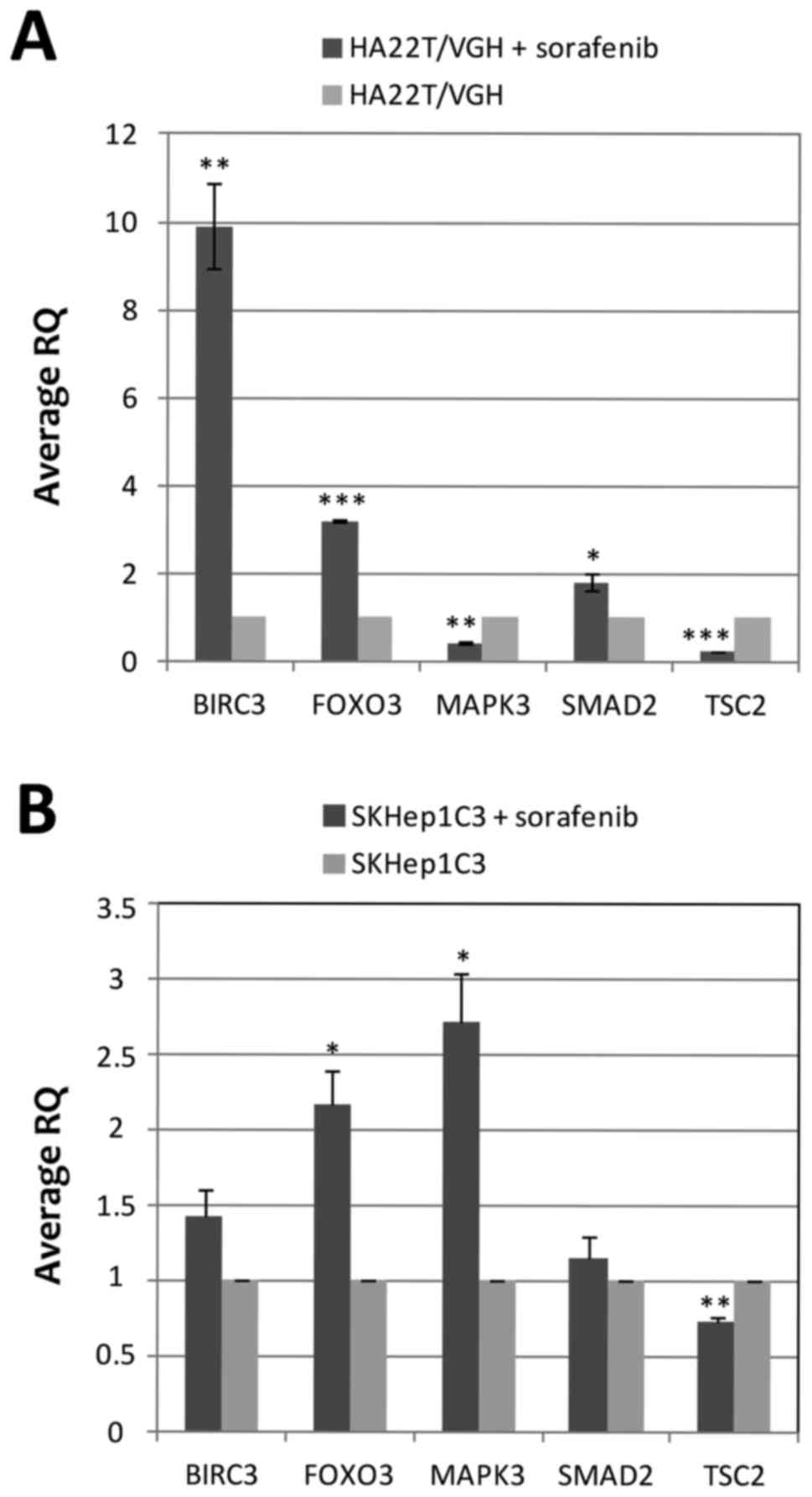
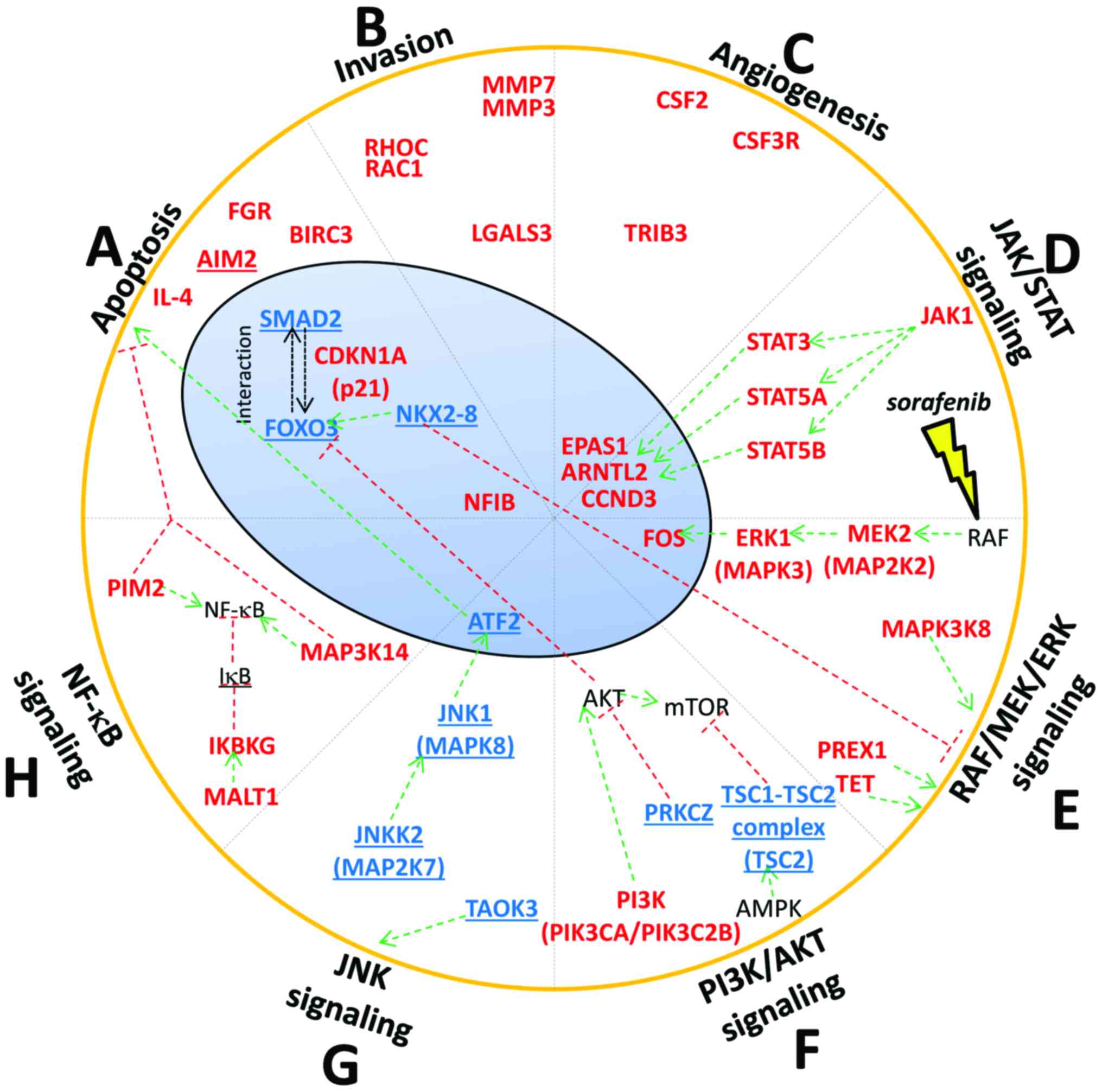
|
1
|
Mittal S and El-Serag HB: Epidemiology of
hepatocellular carcinoma: Consider the population. J Clin
Gastroenterol. 47(Suppl): S2–S6. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Llovet JM, Burroughs A and Bruix J:
Hepatocellular carcinoma. Lancet. 362:1907–1917. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Llovet JM, Ricci S, Mazzaferro V, Hilgard
P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A,
et al SHARP Investigators Study Group: Sorafenib in advanced
hepatocellular carcinoma. N Engl J Med. 359:378–390. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wilhelm SM, Carter C, Tang L, Wilkie D,
McNabola A, Rong H, Chen C, Zhang X, Vincent P, McHugh M, et al:
BAY 43-9006 exhibits broad spectrum oral antitumor activity and
targets the RAF/MEK/ERK pathway and receptor tyrosine kinases
involved in tumor progression and angiogenesis. Cancer Res.
64:7099–7109. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Carlomagno F, Anaganti S, Guida T,
Salvatore G, Troncone G, Wilhelm SM and Santoro M: BAY 43-9006
inhibition of oncogenic RET mutants. J Natl Cancer Inst.
98:326–334. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu L, Cao Y, Chen C, Zhang X, McNabola A,
Wilkie D, Wilhelm S, Lynch M and Carter C: Sorafenib blocks the
RAF/MEK/ERK pathway, inhibits tumor angiogenesis, and induces tumor
cell apoptosis in hepatocellular carcinoma model PLC/PRF/5. Cancer
Res. 66:11851–11858. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gauthier A and Ho M: Role of sorafenib in
the treatment of advanced hepatocellular carcinoma: An update.
Hepatol Res. 43:147–154. 2013. View Article : Google Scholar
|
|
8
|
Pogribny IP and Rusyn I: Role of
epigenetic aberrations in the development and progression of human
hepatocellular carcinoma. Cancer Lett. 342:223–230. 2014.
View Article : Google Scholar :
|
|
9
|
Zhang J, Chen YL, Ji G, Fang W, Gao Z, Liu
Y, Wang J, Ding X and Gao F: Sorafenib inhibits
epithelial-mesenchymal transition through an epigenetic-based
mechanism in human lung epithelial cells. PLoS One. 8:e649542013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang S, Zhu Y, He H, Liu J, Xu L, Zhang H,
Liu H, Liu W, Liu Y, Pan D, et al: Sorafenib suppresses growth and
survival of hepatoma cells by accelerating degradation of enhancer
of zeste homolog 2. Cancer Sci. 104:750–759. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Salvi A, Conde I, Abeni E, Arici B, Grossi
I, Specchia C, Portolani N, Barlati S and De Petro G: Effects of
miR-193a and sorafenib on hepatocellular carcinoma cells. Mol
Cancer. 12:1622013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Grossi I, Arici B, Portolani N, De Petro G
and Salvi A: Clinical and biological significance of miR-23b and
miR-193a in human hepatocellular carcinoma. Oncotarget.
8:6955–6969. 2017.
|
|
13
|
Weber M, Hellmann I, Stadler MB, Ramos L,
Pääbo S, Rebhan M and Schübeler D: Distribution, silencing
potential and evolutionary impact of promoter DNA methylation in
the human genome. Nat Genet. 39:457–466. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Johnson WE, Li W, Meyer CA, Gottardo R,
Carroll JS, Brown M and Liu XS: Model-based analysis of
tiling-arrays for ChIP-chip. Proc Natl Acad Sci USA.
103:12457–12462. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Parrish RR, Day JJ and Lubin FD: Direct
bisulfite sequencing for examination of DNA methylation with gene
and nucleotide resolution from brain tissues. Curr Protoc Neurosci
Chapter 7. Unit 7.24. 2012. View Article : Google Scholar
|
|
16
|
Ferreira E and Cronjé MJ: Selection of
suitable reference genes for quantitative real-time PCR in
apoptosis-induced MCF-7 breast cancer cells. Mol Biotechnol.
50:121–128. 2012. View Article : Google Scholar
|
|
17
|
Lu M, Ma J, Xue W, Cheng C, Wang Y, Zhao
Y, Ke Q, Liu H, Liu Y, Li P, et al: The expression and prognosis of
FOXO3a and Skp2 in human hepatocellular carcinoma. Pathol Oncol
Res. 15:679–687. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wehler TC, Hamdi S, Maderer A, Graf C,
Gockel I, Schmidtmann I, Hainz M, Berger MR, Theobald M, Galle PR,
et al: Single-agent therapy with sorafenib or 5-FU is equally
effective in human colorectal cancer xenograft - no benefit of
combination therapy. Int J Colorectal Dis. 28:385–398. 2013.
View Article : Google Scholar
|
|
19
|
Seoane J, Le HV, Shen L, Anderson SA and
Massagué J: Integration of Smad and forkhead pathways in the
control of neuroepithelial and glioblastoma cell proliferation.
Cell. 117:211–223. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wettersten HI, Hee Hwang S, Li C, Shiu EY,
Wecksler AT, Hammock BD and Weiss RH: A novel p21 attenuator which
is structurally related to sorafenib. Cancer Biol Ther. 14:278–285.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ha TY, Hwang S, Moon KM, Won YJ, Song GW,
Kim N, Tak E, Ryoo BY and Hong HN: Sorafenib inhibits migration and
invasion of hepatocellular carcinoma cells through suppression of
matrix metalloproteinase expression. Anticancer Res. 35:1967–1976.
2015.PubMed/NCBI
|
|
22
|
Chiang IT, Liu YC, Wang WH, Hsu FT, Chen
HW, Lin WJ, Chang WY and Hwang JJ: Sorafenib inhibits TPA-induced
MMP-9 and VEGF expression via suppression of ERK/NF-κB pathway in
hepatocellular carcinoma cells. In Vivo. 26:671–681.
2012.PubMed/NCBI
|
|
23
|
Yang W, Lv S, Liu X, Liu H, Yang W and Hu
F: Up-regulation of Tiam1 and Rac1 correlates with poor prognosis
in hepatocellular carcinoma. Jpn J Clin Oncol. 40:1053–1059. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang W, Yang LY, Yang ZL, Huang GW and Lu
WQ: Expression and significance of RhoC gene in hepatocellular
carcinoma. World J Gastroenterol. 9:1950–1953. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zheng D, Hu Z, He F, Gao C, Xu L, Zou H,
Wu Z, Jiang X and Wang J: Downregulation of galectin-3 causes a
decrease in uPAR levels and inhibits the proliferation, migration
and invasion of hepatocellular carcinoma cells. Oncol Rep.
32:411–418. 2014.PubMed/NCBI
|
|
26
|
Bangoura G, Liu ZS, Qian Q, Jiang CQ, Yang
GF and Jing S: Prognostic significance of HIF-2alpha/EPAS1
expression in hepatocellular carcinoma. World J Gastroenterol.
13:3176–3182. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cervello M, Bachvarov D, Lampiasi N,
Cusimano A, Azzolina A, McCubrey JA and Montalto G: Molecular
mechanisms of sorafenib action in liver cancer cells. Cell Cycle.
11:2843–2855. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Calvisi DF, Ladu S, Gorden A, Farina M,
Conner EA, Lee JS, Factor VM and Thorgeirsson SS: Ubiquitous
activation of Ras and Jak/Stat pathways in human HCC.
Gastroenterology. 130:1117–1128. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang F, Brown C, Buettner R, Hedvat M,
Starr R, Scuto A, Schroeder A, Jensen M and Jove R: Sorafenib
induces growth arrest and apoptosis of human glioblastoma cells
through the dephosphorylation of signal transducers and activators
of transcription 3. Mol Cancer Ther. 9:953–962. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Walter I, Wolfesberger B, Miller I, Mair
G, Burger S, Gallè B and Steinborn R: Human osteosarcoma cells
respond to sorafenib chemotherapy by downregulation of the tumor
progression factors S100A4, CXCR4 and the oncogene FOS. Oncol Rep.
31:1147–1156. 2014.PubMed/NCBI
|
|
31
|
Gedaly R, Angulo P, Chen C, Creasy KT,
Spear BT, Hundley J, Daily MF, Shah M and Evers BM: The role of
I3K/mTOR inhibition in combination with sorafenib in hepatocellular
carcinoma treatment. Anticancer Res. 32:2531–2536. 2012.PubMed/NCBI
|
|
32
|
Zhang CZ, Wang XD, Wang HW, Cai Y and Chao
LQ: Sorafenib inhibits liver cancer growth by decreasing mTOR, AKT,
and I3K expression. J BUON. 20:218–222. 2015.PubMed/NCBI
|
|
33
|
Kulis M, Queirós AC, Beekman R and
Martín-Subero JI: Intragenic DNA methylation in transcriptional
regulation, normal differentiation and cancer. Biochim Biophys
Acta. 1829:1161–1174. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Irizarry RA, Ladd-Acosta C, Wen B, Wu Z,
Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al:
The human colon cancer methylome shows similar hypo- and
hypermethylation at conserved tissue-specific CpG island shores.
Nat Genet. 41:178–186. 2009. View
Article : Google Scholar : PubMed/NCBI
|