Introduction
Prostate cancer (PCa) is the most frequently
diagnosed cancer among men in developed countries (1). Although PCa is initially responsive
to androgen-deprivation therapy (ADT), most patients experience
disease relapse and develop castration-resistant prostate cancer
(CRPC) (2). The survival rate of
patients with CRPC is poor owing to the occurrence of metastasis
(3) and patients with metastatic
CRPC cannot be effectively treated using recently developed
molecular-targeted therapies (2,3).
Therefore, new treatment strategies are needed for these patients.
The molecular mechanisms underlying the aggressiveness of CRPC
cells and the acquisition of treatment resistance are still
unclear.
MicroRNAs (miRNAs) are noncoding RNAs that act as
sequence-specific fine tuners for regulating the expression levels
of proteins and RNAs (4,5). Notably, a single miRNA can regulate a
large number of RNA transcripts in human cells (6). Accordingly, dysregulation of miRNAs
can disrupt the tightly regulated RNA networks in cancer cells,
leading to cancer cell initiation, development, metastasis, and
drug resistance (7,8). The discovery of miRNAs has
complicated the analysis of intracellular RNA networks in
cancer.
We recently constructed an RNA-sequence based miRNA
expression signature using autopsy specimens from patients with
CRPC (9). To elucidate the
miRNA-mediated RNA networks in metastatic CRPC, we have
sequentially identified antitumor miRNAs and oncogenic genes
regulated by these miRNAs based on our miRNA expression signatures
(9,10). Analysis of our miRNA signatures,
including that in CRPC, has shown that both strands of
pre-miR-150, i.e., miR-150-5p (the guide strand) and
miR-150-3p (the passenger strand), are significantly reduced
in cancer tissues (9).
In miRNA biogenesis, pre-miRNA is cleaved by Dicer
into the miRNA duplex, containing the guide and passenger strands.
The mature guide strand miRNA is incorporated into the RNA-induced
silencing complex (RISC) and represses mRNA translation or cleaves
mRNA (11). In contrast, the
passenger strand of miRNA was previously thought to be degraded and
to have no function (12–14). However, our expression data have
contradicted this established miRNA theory. We hypothesized that
the passenger strand of miR-150-3p may have functions in
naïve PCa and CRPC cells by targeting novel oncogenic genes.
Accordingly, in this study, we aimed to investigate the functional
significance of miR-150-5p and miR-150-3p in naïve
PCa and CRPC cells and to identify oncogenic genes involved in the
pathogenesis of the disease.
Materials and methods
Clinical prostate specimens
Clinical prostate specimens were obtained from
patients admitted to Teikyo University Chiba Medical Center from
2014 to 2015. All patients had elevated prostate-specific antigen
(PSA) and had undergone trans-rectal biopsies for definitive cancer
diagnoses. Particularly, in patients with CRPC, neuroendocrine
differentiation was excluded. The patient backgrounds are
summarized in Table I. Samples
were staged according to the UICC TNM classification (15).
 | Table ICharacteristics of patients with
non-PCa and naïve PCa and CRPC. |
Table I
Characteristics of patients with
non-PCa and naïve PCa and CRPC.
| No. | Diagnosis | Age (years) | PSA (ng/ml) | Gleason score | T | N | M | Stage |
|---|
| 1 | Non-PCa | 57 | 5.71 | – | – | – | – | – |
| 2 | Non-PCa | 74 | 9.45 | – | – | – | – | – |
| 3 | Non-PCa | 70 | 8.58 | – | – | – | – | – |
| 4 | Non-PCa | 73 | 4.8 | – | – | – | – | – |
| 5 | Non-PCa | 67 | 6.91 | – | – | – | – | – |
| 6 | Non-PCa | 50 | 7.05 | – | – | – | – | – |
| 7 | Non-PCa | 74 | 9.91 | – | – | – | – | – |
| 8 | Non-PCa | 76 | 20.9 | – | – | – | – | – |
| 9 | Non-PCa | 59 | 4.5 | – | – | – | – | – |
| 10 | Non-PCa | 75 | 1.1 | – | – | – | – | – |
| 11 | Non-PCa | 60 | 7.29 | – | – | – | – | – |
| 12 | Non-PCa | 73 | 38.7 | – | – | – | – | – |
| 13 | Non-PCa | 69 | 11.9 | – | – | – | – | – |
| 14 | Non-PCa | 77 | 23.3 | – | – | – | – | – |
| 15 | Non-PCa | 61 | 4.57 | – | – | – | – | – |
| 16 | Non-PCa | 59 | 7.37 | – | – | – | – | – |
| 17 | Non-PCa | 65 | 5.06 | – | – | – | – | – |
| 18 | PCa | 70 | 75.7 | 4+5 | 4 | 1 | 1 | D2 |
| 19 | PCa | 78 | 1,800 | 4+5 | 4 | 1 | 1 | D2 |
| 20 | PCa | 75 | 68.4 | 5+4 | 4 | 1 | 0 | D1 |
| 21 | PCa | 62 | 38.7 | 4+5 | 2b | 1 | 0 | D1 |
| 22 | PCa | 70 | 25.5 | 4+5 | 3b | 0 | 0 | C |
| 23 | PCa | 75 | 1,260 | 4+5 | 3b | 1 | 1 | D2 |
| 24 | PCa | 88 | 888 | 4+5 | 3b | 1 | 1 | D2 |
| 25 | PCa | 69 | 33.9 | 4+5 | 4 | 0 | 1 | D2 |
| 26 | PCa | 62 | 62.3 | 4+5 | 3b | 1 | 0 | D1 |
| 27 | PCa | 78 | 5 | 4+5 | 2c | 0 | 1b | D2 |
| 28 | PCa | 64 | 449 | 4+5 | 3b | 1 | 1 | D2 |
| 29 | PCa | 81 | 365 | 4+5 | 4 | 1 | 1 | D2 |
| 30 | PCa | 76 | 715 | 5+4 | 4 | 1 | 1 | D2 |
| 31 | PCa | 79 | 555 | 4+5 | 3 | 1 | 1 | D2 |
| 32 | PCa | 63 | 1,120 | 4+5 | 2c | 0 | 1b | D2 |
| 33 | PCa | 67 | 4.95 | 4+5 | 4 | 1 | 1b | D2 |
| 34 | PCa | 70 | 19.5 | 5+5 | 4 | 1 | 1c | D2 |
| 35 | CRPC | 69 | 15.8 | 5+4 | 3b | 1 | 1 | D2 |
| 36 | CRPC | 72 | 212 | 5+4 | 4 | 1 | 1 | D2 |
| 37 | CRPC | 71 | 4.4 | 4+5 | 4 | 1 | 1 | D2 |
| 38 | CRPC | 66 | 295 | 4+5 | 4 | 1 | 1 | D2 |
| 39 | CRPC | 68 | 7.54 | 4+5 | 4 | 1 | 1b | D2 |
All patients in this study provided written informed
consent for tissue donation for research purposes. The protocol was
approved by the Institutional Review Boards of Chiba University and
Teikyo University.
Tissue collection and cell culture
Prostate tissues were immersed in RNAlater (Thermo
Fisher Scientific, Waltham, MA, USA) and stored at 4°C until RNA
extraction. Human prostate cancer cells (PC3 and PC3M cells) were
obtained from the American Type Culture Collection (Manassas, VA,
USA).
Quantitative real-time reverse
transcription polymerase chain reaction (RT-q-PCR)
Stem-loop RT-PCR (TaqMan MicroRNA assays; product
ID: 000473 for miR-150-5p and 002637 for miR-150-3p;
Applied Biosystems, Foster City, CA, USA) was used in this assays.
TaqMan probes and primers for SPOCK1 (product ID:
Hs00270274_m1; Applied Biosystems) were assay-on-demand gene
expression products. We used GUSB (product ID:
Hs00939627_m1; Applied Biosystems), GAPDH (product ID:
Hs02758991_g1; Applied Biosystems), and RNU48 (product ID:
001006; Applied Biosystems) as internal controls.
Cell proliferation, migration, and
invasion assays
Cell proliferation, migration, and invasion assays
were carried out as previously described (9,10).
Argonaute 2 (AGO2)-bound miRNA isolation
by immunoprecipitation
PC3 cells were transfected with 10 nM miRNA by
reverse transfection and plated in 10-cm plates at 1×105
cells/ml. After 48 h, immunoprecipitation was performed using a
microRNA Isolation kit, Human Ago2 (Wako, Osaka, Japan) according
to the manufacturer's protocol. Expression levels of miRNAs bound
to Ago2 were measured by TaqMan RT-qPCR. miRNA expression data were
normalized to the expression of miR-26a (product ID: 000404;
Applied Biosystems), which was not affected by miR-150-5p
and miR-150-3p.
Western blot analysis
Immunoblotting was conducted with diluted monoclonal
anti-SPOCK1 antibodies (1:100 dilution; sc-398782; Santa Cruz
Biotechnology, Santa Cruz, CA, USA) and with diluted
anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibodies
(1:1,000 dilution; ab8245; Abcam, Cambridge, UK) as a loading
control. The procedures were performed as previously described
(16).
Selection of putative target genes
regulated by miR-150-5p and miR-150-3p in PCa cells
To identify miR-150-5p and miR-150-3p
target genes, we analyzed gene expression profile (Gene Expression
Omnibus database; accession no. GSE29079). Gene expression data
were obtained from miR-150-5p and miR-150-3p
transfectant PC3 cells (A SurePrint G3 Human 60K, Agilent
Technologies, Santa Clara, CA, USA). We merged these datasets and
selected putative miR-150-5p and miR-150-3p target
genes using the TargetScan database (Release 7.1; http://www.targetscan.org/vert_71/).
Plasmid construction and dual-luciferase
assays
The partial wide-type sequences of the
3′-untranslated region (UTR) of SPOCK1 or those with a
deleted miR-150-5p or miR-150-3p target site were
inserted in the psiCHECK-2 vector (C8021; Promega, Madison, WI,
USA). Alternatively, we used sequences that were missing the
miR-150-5p target site (position 182–188) or
miR-150-3p target sites (position 1477–1483, position
1749–1756, or position 2593–2599). The procedure for
dual-luciferase reporter assays was described previously (17).
Immunohistochemistry using tissue
microarrays
We used a tissue microarray of prostate cancer
samples obtained from Provitro (Berlin, Germany; cat. no. 401–2209,
Lot no. 146.1P), which contained a total of 78 prostate samples
(PCa specimens: n=58, prostatic intraepithelial neoplasia: n=10,
and normal prostate samples: n=10). Detailed information on these
samples is summarized in Table
II. The tissue microarray was immunostained following the
manufacturer's protocol, as described previously (10,18).
 | Table IICharacteristics of patients, as
evaluated using immunohistochemistry. |
Table II
Characteristics of patients, as
evaluated using immunohistochemistry.
| No. | Diagnosis | Age (years) | Gleason score | T | N | IHC score of
SPOCK1 |
|---|
| 1 | PCa | 64 | 4+3 | 3b | 0 | 5 |
| 2 | PCa | 67 | 3+4 | 2b | 0 | 3 |
| 3 | PCa | 58 | 3+4 | 2b | 0 | 5 |
| 4 | PCa | 63 | 7 | 3b | 0 | 6 |
| 5 | PCa | 65 | 3+3 | 2b | 0 | 5 |
| 6 | PCa | 61 | 4+4 | 3b | × | 4 |
| 7 | PCa | 62 | 3+4 | 2b | × | 2 |
| 8 | PCa | 66 | 4+4 | 2b | × | 4 |
| 9 | PCa | 61 | 3+4 | 3a | × | 4 |
| 10 | PCa | 74 | 4+3 | 2b | × | 5 |
| 11 | PCa | 59 | 2+3 | 2b | × | 4 |
| 12 | PCa | 69 | 3+4 | 3a | 0 | 4 |
| 13 | PCa | 54 | 3+4 | 2c | × | 6 |
| 14 | PCa | 68 | 3+4 | 3a | 0 | 5 |
| 15 | PCa | 58 | 3+4 | 3a | 0 | 5 |
| 16 | PCa | 67 | 3+4 | 3a | 0 | 4 |
| 17 | PCa | 65 | 3+3 | 2a | 0 | 4 |
| 18 | PCa | 77 | 3+4 | 4 | 0 | 5 |
| 19 | PCa | 64 | 4+3 | 2b | 0 | 4 |
| 20 | PCa | 58 | 3+4 | 3a | 0 | 6 |
| 21 | PCa | 50 | 4+3 | 2b | 0 | 6 |
| 22 | PCa | 53 | 3+3 | 2b | 0 | 3 |
| 23 | PCa | 59 | 4+5 | 3a | 0 | 5 |
| 24 | PCa | 70 | 2+3 | 2b | 0 | 5 |
| 25 | PCa | 65 | 5+4 | 3a | 0 | 4 |
| 26 | PCa | 57 | 3+5 | 2b | 0 | 5 |
| 27 | PCa | 68 | 4+4 | 2b | 0 | 6 |
| 28 | PCa | 58 | 3+3 | 2b | 0 | 5 |
| 29 | PCa | 66 | 3+3 | 2b | 0 | 3 |
| 30 | PCa | 63 | 3+4 | 2b | 0 | 6 |
| 31 | PCa | 56 | 3+4 | 2b | 0 | 4 |
| 32 | PCa | 63 | 5+3 | 3a | 0 | 3 |
| 33 | PCa | 64 | 3+5 | 3a | 0 | 5 |
| 34 | PCa | 60 | 3+4 | 2b | 0 | 5 |
| 35 | PCa | 60 | 3+3 | 3a | 0 | 5 |
| 36 | PCa | 57 | 3+2 | 2b | 0 | 4 |
| 37 | PCa | 50 | 3+3 | 2a | 0 | 6 |
| 38 | PCa | 68 | 3+3 | 3a | 0 | 5 |
| 39 | PCa | 65 | 3+4 | 3b | 1 | 4 |
| 40 | PCa | 69 | 5+5 | 3a | 1 | 5 |
| 41 | PCa | 63 | 3+4 | 2b | 0 | 5 |
| 42 | PCa | 51 | 2+3 | 2b | 0 | 4 |
| 43 | PCa | 62 | 3+3 | 3a | 0 | 5 |
| 44 | PCa | 61 | 3+4 | 3a | 0 | 5 |
| 45 | PCa | 53 | 4+4 | 3b | 1 | 4 |
| 46 | PCa | 56 | 4+3 | 2b | 0 | 5 |
| 47 | PCa | 59 | 2+3 | 2b | 0 | 5 |
| 48 | PCa | 61 | 3+4 | 2b | 0 | 5 |
| 49 | PCa | 51 | 3+4 | 3a | 0 | 6 |
| 50 | PCa | 62 | 3+4 | 3b | 1 | 5 |
| 51 | PCa | 66 | 3+3 | 3a | 0 | 5 |
| 52 | PCa | 62 | 3+3 | 2b | 0 | 4 |
| 53 | PCa | 56 | 3+3 | 2b | 0 | 5 |
| 54 | PCa | 58 | 3+3 | 3a | 0 | 5 |
| 55 | PCa | 66 | 5+4 | 3a | 0 | 6 |
| 56 | PCa | 55 | 3+4 | 3a | 0 | 5 |
| 57 | PCa | 67 | 2+3 | 2b | 0 | 5 |
| 58 | PCa | 61 | 3+5 | 2b | 0 | 6 |
| 59 | PIN | 59 | – | – | – | 3 |
| 60 | PIN | 58 | – | – | – | 5 |
| 61 | PIN | 62 | – | – | – | 4 |
| 62 | PIN | 51 | – | – | – | 6 |
| 63 | PIN | 58 | – | – | – | 3 |
| 64 | PIN | 68 | – | – | – | 4 |
| 65 | PIN | 64 | – | – | – | 5 |
| 66 | PIN | 56 | – | – | – | 5 |
| 67 | PIN | 61 | – | – | – | 3 |
| 68 | PIN | 51 | – | – | – | 5 |
| 69 | Normal | 70 | – | – | – | 4 |
| 70 | Normal | 63 | – | – | – | 3 |
| 71 | Normal | 62 | – | – | – | 4 |
| 72 | Normal | 81 | – | – | – | 0 |
| 73 | Normal | 67 | – | – | – | 3 |
| 74 | Normal | 76 | – | – | – | 4 |
| 75 | Normal | 66 | – | – | – | 4 |
| 76 | Normal | 69 | – | – | – | 3 |
| 77 | Normal | 63 | – | – | – | 5 |
| 78 | Normal | 71 | – | – | – | 4 |
Statistical analysis
The relationships between 2 groups and expression
values obtained by RT-PCR were analyzed using Mann-Whitney U tests.
The correlations between miR-150-5p and miR-150-3p
expression were evaluated using Spearman's rank test. The
relationships among more than 3 variables and numerical values were
analyzed using Bonferroni-adjusted Mann-Whitney U tests. We used
Expert StatView software (version 5.0 SAS Institute Inc., Cary, NC,
USA) for these analyses.
Results
The expression levels of miR-150-5p and
miR-150-3p in naïve PCa and CRPC specimens and cell lines
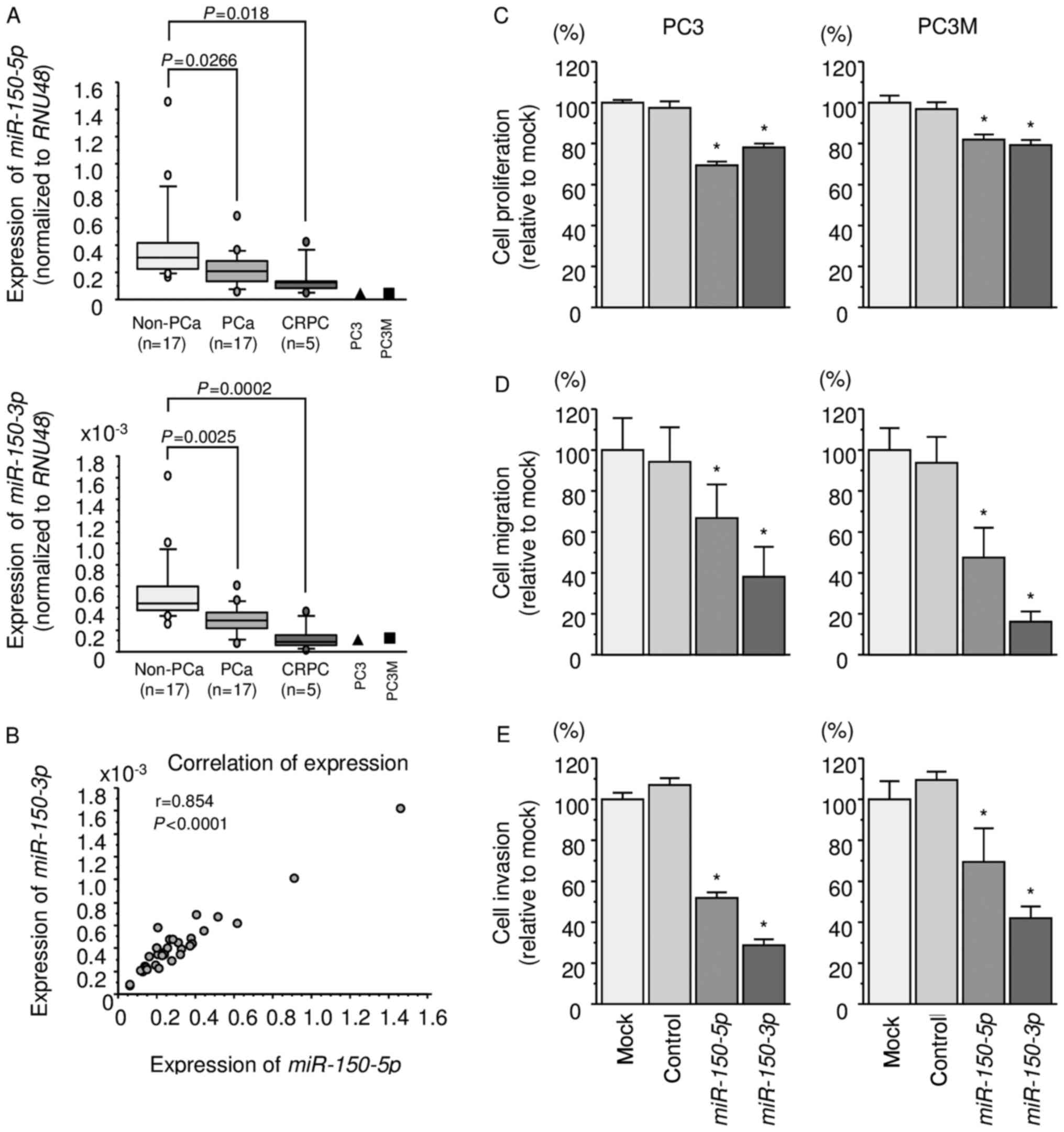
We evaluated the expression levels of
miR-150-5p and miR-150-3p in PCa tissues (PCa: n=17,
CRPC: n=5), normal tissues (non-PCa: n=17), and PCa cell lines (PC3
and PC3M cells). The patient backgrounds are summarized in Table I. The expression levels of
miR-150-5p and miR-150-3p were significantly lower in
PCa and CRPC tissues than normal tissues (miR-150-5p:
P=0.0266 and P=0.018, respectively; miR-150-3p: P=0.0025 and
P=0.0002, respectively; Fig. 1A).
There was a positive correlation between expression levels of
miR-150-5p and miR-150-3p (r=0.854, P<0.0001;
Fig. 1B).
Effects of ectopic expression of
miR-150-5p and miR-150-3p on cell proliferation, migration, and
invasion assays in PCa cell lines
To examine the functional roles of miR-150-5p
and miR-150-3p, we performed gain-of-function studies by
using PC3 and PC3M cells transfected with mature miRNAs.
XTT assays revealed that proliferation was
significantly inhibited in PC3 and PC3M cells transfected with
miR-150-5p and miR-150-3p in comparison with that of
mock or miR-control-transfected cells (P<0.0001; Fig. 1C). Wound-healing and Matrigel
invasion assays demonstrated significant inhibition of cell
migration and invasion in both miR-150-5p and
miR-150-3p transfectants (P<0.0001; Fig. 1D and E).
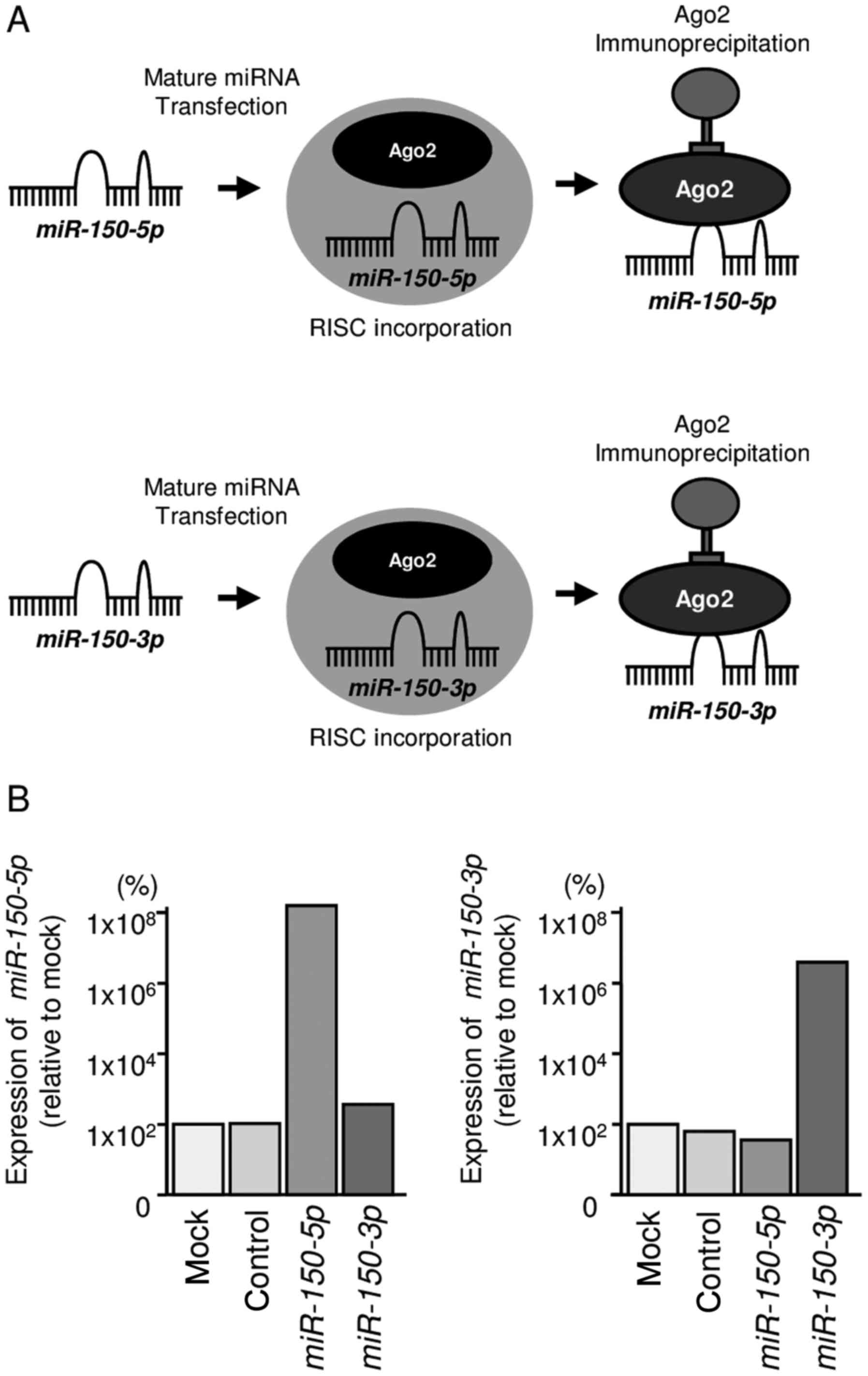
Both miR-150-5p and miR-150-3p bound to
Ago2
We hypothesized that both miR-150-5p and
miR-150-3p may be incorporated into and function as part of
the RISC. To test this hypothesis, we performed immunoprecipitation
with antibodies targeting Ago2, which plays a central role in the
RISC. After transfection with miR-150-5p or
miR-150-3p, Ago2-bound miRNAs were isolated, and RT-qPCR was
carried out to determine whether miR-150-5p and
miR-150-3p bound to Ago2 (Fig.
2A).
After transfection with miR-150-5p and
immunoprecipitation by anti-Ago2 antibodies, miR-150-5p
levels were significantly higher than those of mock- or miR
control-transfected cells and those of
miR-150-3p-transfected PC3 cells (P<0.0001; Fig. 2B). Similarly, after transfection
with miR-150-3p and immunoprecipitation by anti-Ago2
antibodies, miR-150-3p levels were significantly higher than
those of mock- or miR control-transfected cells and those of
miR-150-5p-transfected PC3 cells (P<0.0001; Fig. 2B).
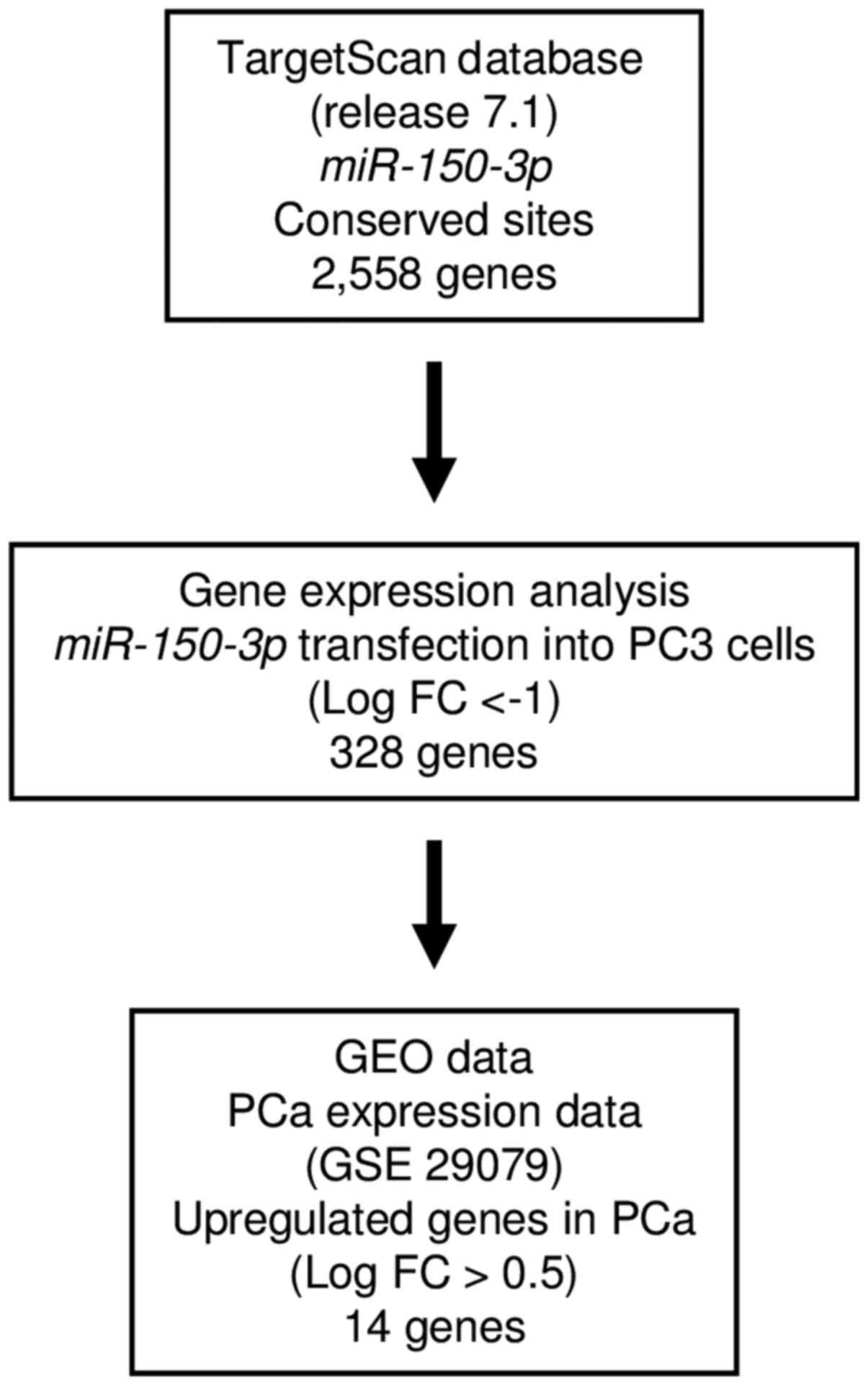
Screening of target genes regulated by
miR-150-3p in PCa cells
To obtain further insights into the molecular
mechanisms regulated by antitumor miR-150-3p in PCa cells,
we screened these miRNA-regulated genes by using in silico
and genome-wide gene expression analyses. First, we undertook
genome-wide gene expression analysis using PC3 cells.
Analysis of the TargetScan database showed that
2,558 genes had putative target sites for miR-150-3p in
their 3′-UTRs. Next, we screened 328 of these genes using
genome-wide gene expression analysis. Finally, we found 14 genes
that were upregulated (fold-change log2 >0.5) in
cancer tissues by GEO database analyses (GEO accession no.
GSE29079). Our strategy for analysis is shown in Fig. 3. Putative target genes of
miR-150-3p are summarized in Table III. Among these candidate genes,
SPOCK1 had a putative binding site for miR-150-5p
according to the TargetScan database. Therefore, we focused on
SPOCK1 as a candidate target gene of the dual strands of
pre-miR-150 and performed further investigations of this target in
PCa.
 | Table IIIPutative target genes regulated by
miR-150-3p in PCa cells. |
Table III
Putative target genes regulated by
miR-150-3p in PCa cells.
| Entrez gene ID | Gene symbol | Gene name | PC3
miR-150-3p transfectant (log2 ratio) | Site counts | GEO Fold
change |
|---|
| 55771 | SPOCK1 | Sparc/osteonectin,
cwcv and kazal-like domains proteoglycan (testican) 1 | −2.718465 | 3 | 1.112207 |
| 11113 | LIG3 | Ligase III, DNA,
ATP-dependent | −1.013958 | 2 | 0.833472 |
| 9448 | MLEC | Malectin | −1.05939 | 1 | 0.817763 |
| 1017 | NETO2 | Neuropilin (NRP)
and tolloid (TLL)-like 2 | −1.639355 | 2 | 0.808991 |
| 51400 |
HIST1H3B | Histone cluster 1,
H3b | −1.070237 | 1 | 0.803539 |
| 22877 | PHF12 | PHD finger protein
12 | −1.531983 | 1 | 0.779377 |
| 4615 | NCALD | Neurocalcin δ | −1.345623 | 2 | 0.772963 |
| 6695 | SIM2 | Single-minded
family bHLH transcription factor 2 | −2.060616 | 1 | 0.707018 |
| 79071 | TAB3 | TGF-β activated
kinase 1/MAP3K7 binding protein 3 | −1.341698 | 1 | 0.642641 |
| 8358 | MAP4K4 | Mitogen-activated
protein kinase kinase kinase kinase 4 | −1.043328 | 1 | 0.580977 |
| 10186 | HNRNPAB | Heterogeneous
nuclear ribonucleoprotein A/B | −1.501292 | 1 | 0.571322 |
| 11113 | FARP1 | FERM, RhoGEF
(ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) | −1.379558 | 1 | 0.536826 |
| 54475 | SPECC1L | Sperm antigen with
calponin homology and coiled-coil domains 1-like | −1.168633 | 1 | 0.506925 |
| 6548 | NFIB | Nuclear factor
I/B | −1.189804 | 1 | 0.505127 |
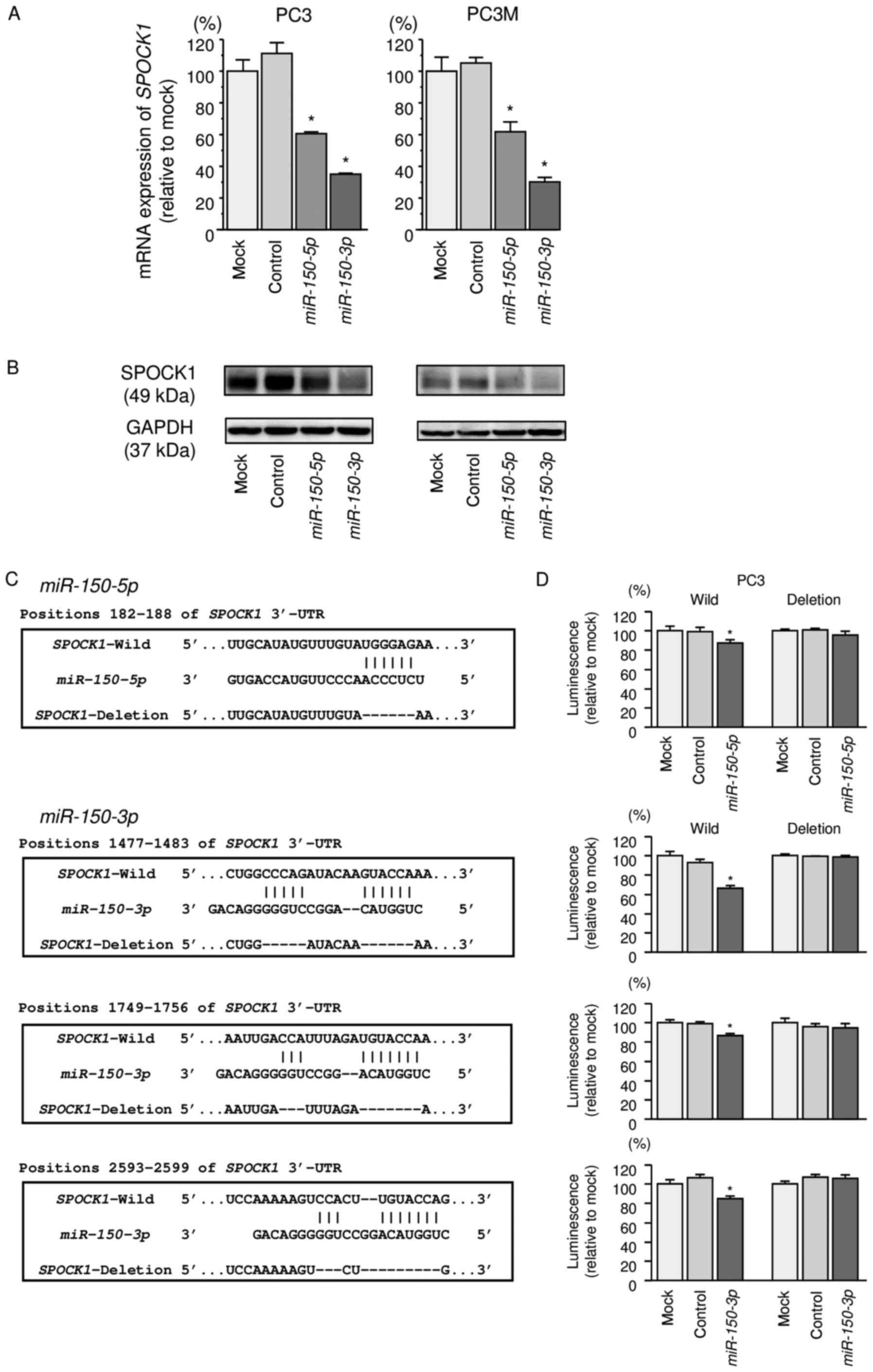
Regulation of SPOCK1 expression by
miR-150-5p and miR-150-3p in PCa cells
Our studies revealed that SPOCK1 mRNA was
significantly reduced in both miR-150-5p and
miR-150-3p transfectants in comparison with those in mock or
miR-control transfectants (P<0.0001 and P<0.0001; Fig. 4A). Expression of SPOCK1 protein was
also repressed in these miRNAs transfectants (Fig. 4B).
Target prediction databases indicated that
miR-150-5p had one putative target site in the 3′-UTR of
SPOCK1 (Fig. 4C). Likewise,
miR-150-3p had three putative target sites (Fig. 4C). To determine whether
SPOCK1 mRNA contained functional target sites, we performed
a dual-luciferase reporter assay.
The TargetScan database identified one putative
target site in the 3′-UTR of SPOCK1 for miR-150-5p
(position 182-188) and three target sites of SPOCK1 for
miR-150-3p (positions 1477-1483, 1749-1756, and 2593-2599).
We used vectors encoding a partial wild-type sequence of the 3′-UTR
of SPOCK1 mRNA, including the predicted miR-150-5p
and miR-150-3p target site, or a vector lacking the
miR-150-5p and miR-150-3p target sites. We found that
the luminescence intensity was significantly reduced by
co-transfection with miR-150-5p or miR-150-3p and the
vector carrying the wild-type 3′-UTR of SPOCK1 (P<0.05;
Fig. 4D).
Effects of silencing SPOCK1 on cell
proliferation, migration, and invasion in PCa cells
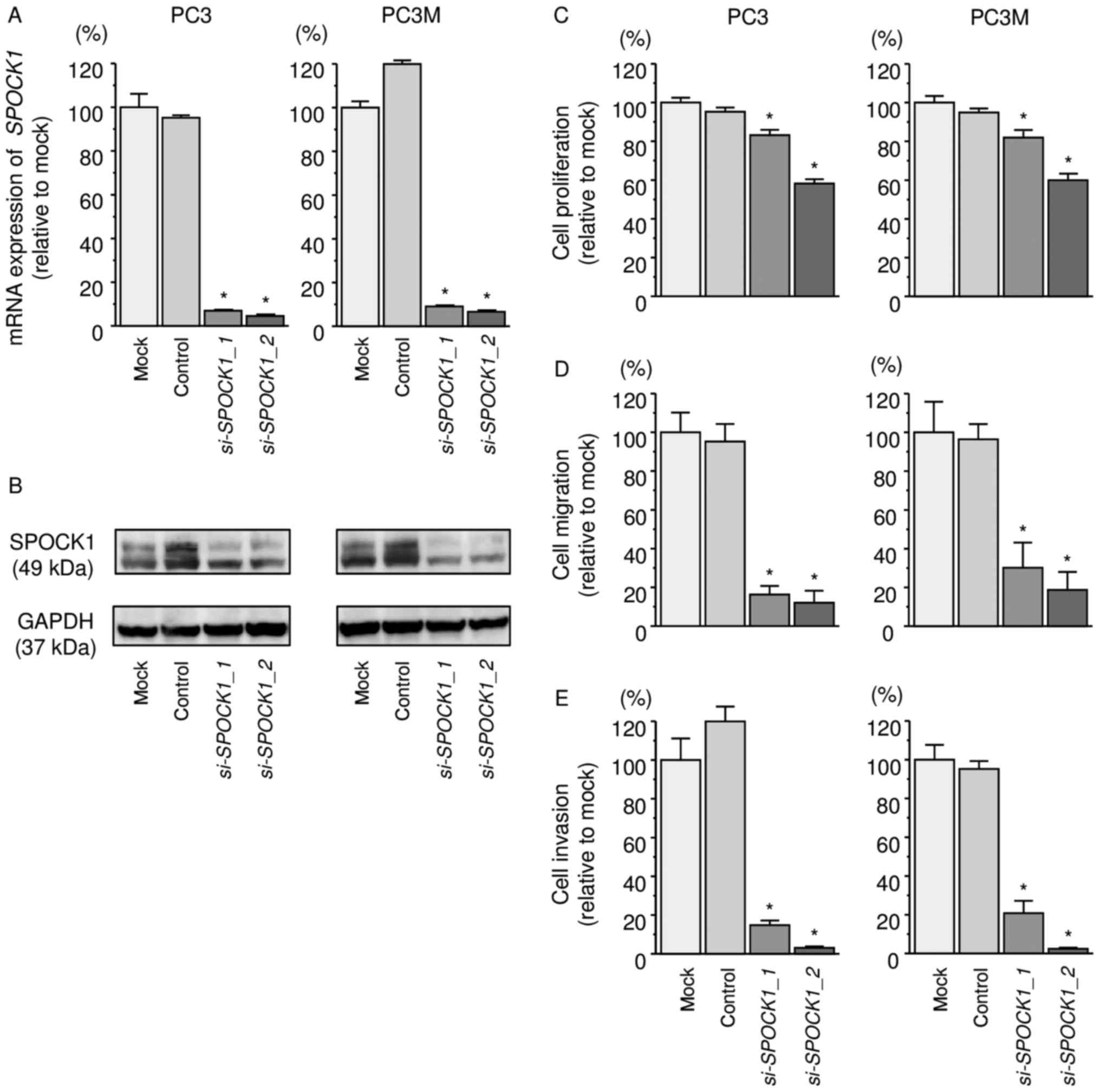
We evaluated the knockdown efficiency of
si-SPOCK1 transfection in PC3 cells. Our present data showed
that si-SPOCK1 transfection effectively downregulated
SPOCK1 expression in PC3 and PC3M cells (Fig. 5A and B).
Functional assays demonstrated that cell
proliferation, migration, and invasion were inhibited in
si-SPOCK1 transfectants compared with those in mock- or miR
control-transfected cells (P<0.0001, Fig. 5C–E).
Analysis of SPOCK1 expression in naïve
PCa and CRPC clinical specimens by immunohistochemistry
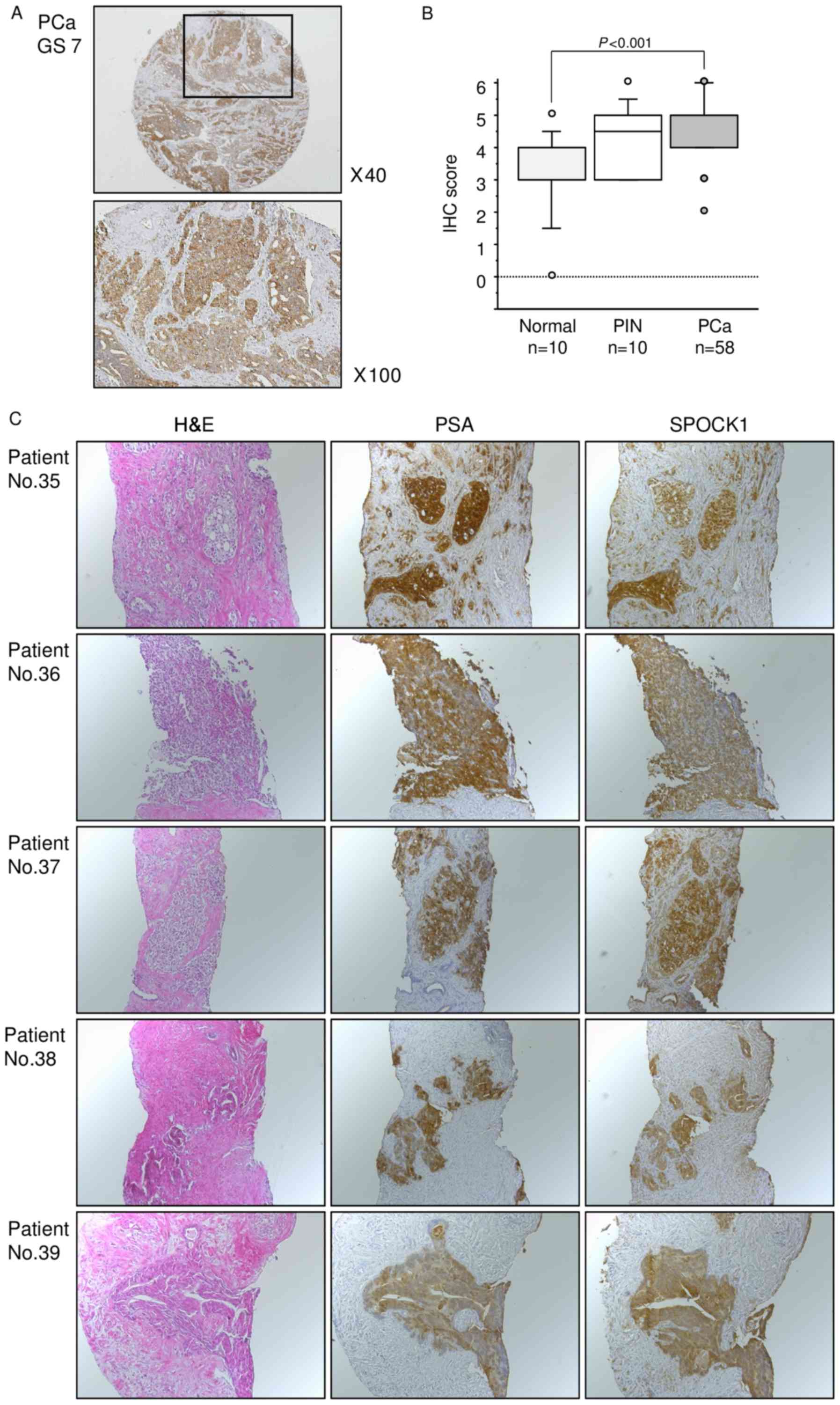
Next, we examined the expression levels of SPOCK1 in
naïve PCa specimens by immunohistochemical staining. SPOCK1 was
strongly expressed in several cancer tissues, while low expression
was observed in normal tissues (Fig.
6A). Moreover, the expression score for SPOCK1 protein was
significantly higher in PCa tissues than in normal tissues
(P<0.001, Fig. 6B). The patient
backgrounds and clinicopathological characteristics are summarized
in Table II.
To analyze SPOCK1 protein expression,
immunohistochemistry was performed with CRPC specimens.
Immunohistochemical staining demonstrated high expression of SPOCK1
in CRPC tissues. SPOCK1 was strongly expressed in the cytoplasm of
the PCa cells almost in the same area where PSA was expressed
(Fig. 6C).
Discussion
Androgen-dependent PCa initially responds to ADT,
which can result in disease control. However, most PCa cells
eventually acquire ADT-resistance mechanisms. Moreover, there are
no curative treatments for patients with metastatic CRPC (19). One of the main challenges in
treating CRPC is controlling aggressive, lethal metastatic PCa
cells. We believe identification of the genes and pathways
responsible for metastasis may lead to the development of new
therapeutic strategies. Accordingly, we have focused on identifying
antitumor miRNAs and oncogenic RNA networks mediated by these
miRNAs in naïve PCa and CRPC cells (9,20,21).
For example, antitumor miR-223 inhibits cancer cell migration and
invasion by targeting ITGA3 and ITGB1 (18). Antitumor miR-218 suppresses
migration and invasion by regulating LASP1 (22). Moreover, antitumor miRNAs
(miR-26a/b, miR-29a/b/c and miR-218) function
cooperatively to suppress metastasis-promoting LOXL
(23).
In this study, we demonstrated that the dual strands
of pre-miR-150, i.e., miR-150-5p and
miR-150-3p, acted as antitumor miRNAs in naïve PCa and CRPC
cells. According to the miRNA database (miRBase: http://www.mirbase.org/), miR-150-5p is a guide
strand of pre-miR-150, and miR-150-3p is the
corresponding passenger strand. Previous studies have shown that
miR-150-5p (the guide strand) is frequently downregulated in
cancer tissues and functions as an antitumor miRNA in several types
of cancer (24–26). In this study, we focused on
miR-150-3p (the passenger strand) and investigated the
antitumor roles of this miRNA in naïve PCa and CRPC cells because
no prior studies had evaluated the functions of miR-150-3p
in cancer cells.
Passenger strands of miRNA are thought to be
degraded and are not expected to be incorporated into the RISC
(4). However, our data showed that
the passenger strand of miR-150 was incorporated into the
RISC in PCa cells, and this is the first report of the antitumor
function of miR-150-3p in cancer cells. Our recent studies
demonstrated that miR-145-3p (the passenger strand of
pre-miR-145) acted as an antitumor miRNA targeting oncogenic
UHRF1 and MTDH in bladder and lung cancer,
respectively (16,27). Similarly, we confirmed the
antitumor function of miR-139-3p (a passenger strand of
pre-miR-139) in bladder cancer (17). These findings suggested that the
passenger strands of miRNAs may have some biological functions in
human cells, similar to the guide strands of miRNAs. The
involvement of passenger strand miRNAs in the regulation of
cellular processes is a novel concept in RNA research.
One of the main challenges in miRNA studies is to
seek out miRNA targeting genes and RNA networks mediated by these
miRNAs in cancer cells. We revealed that SPOCK1 was a direct
target of dual strands of miR-150-5p and miR-150-3p
in PCa cells. Moreover, we demonstrated the overexpression of
SPOCK1 in naïve PCa and CRPC clinical specimens.
SPOCK1/testican-1 belongs to the Ca2+-binding
proteoglycan family, which includes SPARC, testican-2, and
testican-3 (28). Overexpression
of SPOCK1 was observed in several cancers and has been shown
to play pivotal roles in cancer cell progression, metastasis, and
drug resistance (29–31). SPOCK1 is upregulated in lung
cancer and is associated with metastasis and survival (32). Interestingly, ectopic expression of
SPOCK1 induces the epithelial-mesenchymal transition (EMT)
in lung cancer cells (33).
Another study demonstrated that SPOCK1 induces MET-dependent
EMT signaling in lapatinib-resistant gastric cancer (34). Several reports have indicated that
tyrosine kinase receptor inhibitors (TKIs) can frequently cause the
acquisition of TKI resistance in cells and that the EMT is deeply
involved in these events (35,36).
These findings suggest that SPOCK1 mediates the EMT
signaling to regulate cancer cell aggressiveness and drug
resistance.
Most patients with PCa exhibit ADT failure and
progress to CRPC with metastasis. Moreover, no curative treatments
are available for advanced CRPC with metastasis (37). In CRPC cells, the EMT is associated
with metastatic processes and is involved in drug resistance
(38). Interestingly, androgens
and androgen receptor-mediated signaling enhance the EMT and cancer
cell aggressiveness (39).
Transforming growth factor β (TGFβ) is a pivotal player that
induces EMT in cancer cells (40).
Increased expression of TGFβ and EMT-related proteins has been
observed in CRPC bone metastasis (41). Interestingly, expression of SPOCK1
is elevated by TGFβ treatment in lung cancer cells, suggesting that
SPOCK1 mediates downstream TGFβ signaling (33). We suggest that SPOCK1
expression may be related to induction of TGFβ signaling and
epigenetic regulation of miR-150-5p and miR-150-3p in
naïve PCa and CRPC cells. A recent study showed that overexpression
of SPOCK1 in RWPE-1 cells (non-neoplastic adult human
prostatic epithelial cells) promotes cell viability and cell
migration and invasion abilities (42). These findings were supported by our
present data. Thus, the expression of SPOCK1 may be involved
in the pathogenesis of naïve PCa and CRPC, and
SPOCK1-mediated signaling may be a promising therapeutic
target in this disease.
In conclusiom, the dual strands of pre-miR-150,
i.e., miR-150-5p and miR-150-3p, were significantly
reduced in naïve PCa and CRPC tissues and acted as antitumor mRNAs.
The passenger strand of miR-150-3p was found to have a
specific function in cancer cells. SPOCK1 was directly
regulated by dual strands of miR-150-5p and
miR-150-3p in PCa cells. Overexpression of SPOCK1 was
confirmed in naïve PCa and CRPC tissues and acted as an oncogene in
this disease. Elucidation of miR-150/SPOCK1-mediated
molecular networks may lead to a better understanding of the
pathogenesis of naïve PCa and CRPC and facilitate the development
of new treatment strategies.
Acknowledgments
This study was supported by KAKENHI grants
15K10801(C), 15K20071(C), 16K20125, and 16H05462(B).
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sridhar SS, Freedland SJ, Gleave ME,
Higano C, Mulders P, Parker C, Sartor O and Saad F:
Castration-resistant prostate cancer: From new pathophysiology to
new treatment. Eur Urol. 65:289–299. 2014. View Article : Google Scholar
|
|
3
|
Sturge J, Caley MP and Waxman J: Bone
metastasis in prostate cancer: Emerging therapeutic strategies. Nat
Rev Clin Oncol. 8:357–368. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Carthew RW and Sontheimer EJ: Origins and
mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Y, Wang Z and Gemeinhart RA:
Progress in microRNA delivery. J Control Release. 172:962–974.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nelson KM and Weiss GJ: MicroRNAs and
cancer: Past, present, and potential future. Mol Cancer Ther.
7:3655–3660. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Goto Y, Kojima S, Nishikawa R, Kurozumi A,
Kato M, Enokida H, Matsushita R, Yamazaki K, Ishida Y, Nakagawa M,
et al: MicroRNA expression signature of castration-resistant
prostate cancer: The microRNA-221/222 cluster functions as a tumour
suppressor and disease progression marker. Br J Cancer.
113:1055–1065. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Okato A, Goto Y, Kurozumi A, Kato M,
Kojima S, Matsushita R, Yonemori M, Miyamoto K, Ichikawa T and Seki
N: Direct regulation of LAMP1 by tumor-suppressive microRNA-320a in
prostate cancer. Int J Oncol. 49:111–122. 2016.PubMed/NCBI
|
|
11
|
Gregory RI, Chendrimada TP, Cooch N and
Shiekhattar R: Human RISC couples microRNA biogenesis and
posttranscriptional gene silencing. Cell. 123:631–640. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chendrimada TP, Gregory RI, Kumaraswamy E,
Norman J, Cooch N, Nishikura K and Shiekhattar R: TRBP recruits the
Dicer complex to Ago2 for microRNA processing and gene silencing.
Nature. 436:740–744. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hutvágner G and Zamore PD: A microRNA in a
multiple-turnover RNAi enzyme complex. Science. 297:2056–2060.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Matranga C, Tomari Y, Shin C, Bartel DP
and Zamore PD: Passenger-strand cleavage facilitates assembly of
siRNA into Ago2-containing RNAi enzyme complexes. Cell.
123:607–620. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sobin LH, Gospodarowicz MK and Wittekind
Ch: TNM Classification of Malignant Tumours. 7th edition.
Wiley-Blackwell; Chichester: 2009
|
|
16
|
Mataki H, Seki N, Mizuno K, Nohata N,
Kamikawaji K, Kumamoto T, Koshizuka K, Goto Y and Inoue H:
Dual-strand tumor-suppressor microRNA-145 (miR-145-5p and
miR-145-3p) coordinately targeted MTDH in lung squamous cell
carcinoma. Oncotarget. 7:72084–72098. 2016.PubMed/NCBI
|
|
17
|
Yonemori M, Seki N, Yoshino H, Matsushita
R, Miyamoto K, Nakagawa M and Enokida H: Dual tumor-suppressors
miR-139-5p and miR-139-3p targeting matrix metalloprotease 11 in
bladder cancer. Cancer Sci. 107:1233–1242. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kurozumi A, Goto Y, Matsushita R, Fukumoto
I, Kato M, Nishikawa R, Sakamoto S, Enokida H, Nakagawa M, Ichikawa
T, et al: Tumor-suppressive microRNA-223 inhibits cancer cell
migration and invasion by targeting ITGA3/ITGB1 signaling in
prostate cancer. Cancer Sci. 107:84–94. 2016. View Article : Google Scholar
|
|
19
|
Semenas J, Allegrucci C, Boorjian SA,
Mongan NP and Persson JL: Overcoming drug resistance and treating
advanced prostate cancer. Curr Drug Targets. 13:1308–1323. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fuse M, Kojima S, Enokida H, Chiyomaru T,
Yoshino H, Nohata N, Kinoshita T, Sakamoto S, Naya Y, Nakagawa M,
et al: Tumor suppressive microRNAs (miR-222 and miR-31) regulate
molecular pathways based on microRNA expression signature in
prostate cancer. J Hum Genet. 57:691–699. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Goto Y, Kurozumi A, Enokida H, Ichikawa T
and Seki N: Functional significance of aberrantly expressed
microRNAs in prostate cancer. Int J Urol. 22:242–252. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nishikawa R, Goto Y, Sakamoto S, Chiyomaru
T, Enokida H, Kojima S, Kinoshita T, Yamamoto N, Nakagawa M, Naya
Y, et al: Tumor-suppressive microRNA-218 inhibits cancer cell
migration and invasion via targeting of LASP1 in prostate cancer.
Cancer Sci. 105:802–811. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kato M, Kurozumi A, Goto Y, Matsushita R,
Okato A, Nishikawa R, Fukumoto I, Koshizuka K, Ichikawa T and Seki
N: Regulation of metastasis-promoting LOXL2 gene expression by
antitumor microRNAs in prostate cancer. J Hum Genet. 62:123–132.
2017. View Article : Google Scholar
|
|
24
|
Abe F, Kitadate A, Ikeda S, Yamashita J,
Nakanishi H, Takahashi N, Asaka C, Teshima K, Miyagaki T, Sugaya M,
et al: Histone deacetylase inhibitors inhibit metastasis by
restoring a tumor suppressive microRNA-150 in advanced cutaneous
T-cell lymphoma. Oncotarget. 8:7572–7585. 2017.
|
|
25
|
Srivastava SK, Bhardwaj A, Singh S, Arora
S, Wang B, Grizzle WE and Singh AP: MicroRNA-150 directly targets
MUC4 and suppresses growth and malignant behavior of pancreatic
cancer cells. Carcinogenesis. 32:1832–1839. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Qu Y, Pan S, Kang M, Dong R and Zhao J:
MicroRNA-150 functions as a tumor suppressor in osteosarcoma by
targeting IGF2BP1. Tumour Biol. 37:5275–5284. 2016. View Article : Google Scholar
|
|
27
|
Matsushita R, Yoshino H, Enokida H, Goto
Y, Miyamoto K, Yonemori M, Inoguchi S, Nakagawa M and Seki N:
Regulation of UHRF1 by dual-strand tumor-suppressor microRNA-145
(miR-145-5p and miR-145-3p): Inhibition of bladder cancer cell
aggressiveness. Oncotarget. 7:28460–28487. 2016.PubMed/NCBI
|
|
28
|
Bradshaw AD: Diverse biological functions
of the SPARC family of proteins. Int J Biochem Cell Biol.
44:480–488. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li Y, Chen L, Chan TH, Liu M, Kong KL, Qiu
JL, Li Y, Yuan YF and Guan XY: SPOCK1 is regulated by CHD1L and
blocks apoptosis and promotes HCC cell invasiveness and metastasis
in mice. Gastroenterology. 144:179–191.e4. 2013. View Article : Google Scholar
|
|
30
|
Ma LJ, Wu WJ, Wang YH, Wu TF, Liang PI,
Chang IW, He HL and Li CF: SPOCK1 Overexpression confers a poor
prognosis in urothelial carcinoma. J Cancer. 7:467–476. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shu YJ, Weng H, Ye YY, Hu YP, Bao RF, Cao
Y, Wang XA, Zhang F, Xiang SS, Li HF, et al: SPOCK1 as a potential
cancer prognostic marker promotes the proliferation and metastasis
of gallbladder cancer cells by activating the PI3K/AKT pathway. Mol
Cancer. 14:122015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kusakabe M, Kutomi T, Watanabe K, Emoto N,
Aki N, Kage H, Hamano E, Kitagawa H, Nagase T, Sano A, et al:
Identification of G0S2 as a gene frequently methylated in squamous
lung cancer by combination of in silico and experimental
approaches. Int J Cancer. 126:1895–1902. 2010.
|
|
33
|
Miao L, Wang Y, Xia H, Yao C, Cai H and
Song Y: SPOCK1 is a novel transforming growth factor-β target gene
that regulates lung cancer cell epithelial-mesenchymal transition.
Biochem Biophys Res Commun. 440:792–797. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kim HP, Han SW, Song SH, Jeong EG, Lee MY,
Hwang D, Im SA, Bang YJ and Kim TY: Testican-1-mediated
epithelial-mesenchymal transition signaling confers acquired
resistance to lapatinib in HER2-positive gastric cancer. Oncogene.
33:3334–3341. 2014. View Article : Google Scholar
|
|
35
|
Uramoto H, Iwata T, Onitsuka T, Shimokawa
H, Hanagiri T and Oyama T: Epithelial-mesenchymal transition in
EGFR-TKI acquired resistant lung adenocarcinoma. Anticancer Res.
30:2513–2517. 2010.PubMed/NCBI
|
|
36
|
Choe C, Shin YS, Kim C, Choi SJ, Lee J,
Kim SY, Cho YB and Kim J: Crosstalk with cancer-associated
fibroblasts induces resistance of non-small cell lung cancer cells
to epidermal growth factor receptor tyrosine kinase inhibition.
Onco Targets Ther. 8:3665–3678. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Crawford ED, Higano CS, Shore ND, Hussain
M and Petrylak DP: Treating patients with metastatic castration
resistant prostate xancer: A comprehensive review of available
therapies. J Urol. 194:1537–1547. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Martin SK, Pu H, Penticuff JC, Cao Z,
Horbinski C and Kyprianou N: Multinucleation and
mesenchymal-to-epithelial transition alleviate resistance to
combined cabazitaxel and anti-androgen therapy in advanced prostate
cancer. Cancer Res. 76:912–926. 2016. View Article : Google Scholar
|
|
39
|
Nakazawa M and Kyprianou N:
Epithelial-mesenchymal-transition regulators in prostate cancer:
Androgens and beyond. J Steroid Biochem Mol Biol. 166:84–90. 2017.
View Article : Google Scholar
|
|
40
|
Xu J, Lamouille S and Derynck R:
TGF-beta-induced epithelial to mesenchymal transition. Cell Res.
19:156–172. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Haider M, Zhang X, Coleman I, Ericson N,
True LD, Lam HM, Brown LG, Ketchanji M, Nghiem B, Lakely B, et al:
Epithelial mesenchymal-like transition occurs in a subset of cells
in castration resistant prostate cancer bone metastases. Clin Exp
Metastasis. 33:239–248. 2016. View Article : Google Scholar :
|
|
42
|
Chen Q, Yao YT, Xu H, Chen YB, Gu M, Cai
ZK and Wang Z: SPOCK1 promotes tumor growth and metastasis in human
prostate cancer. Drug Des Devel Ther. 10:2311–2321. 2016.
View Article : Google Scholar : PubMed/NCBI
|