Introduction
Liver cancer is one of the most common malignant
tumors of the digestive system (1). In China, the incidence of liver
cancer has increased annually due to the persistent high infection
rate of hepatitis B virus in the population (2). The major forms of liver cancer are
hepatocellular carcinoma (HCC) and intrahepatic cholangiocarcinoma.
In addition, there are two rare malignant liver tumors occurring in
adolescence, hepatoblastoma and fibrolamellar carcinoma, both
developing in a non-cirrhotic liver (3). At present, the treatment options for
liver cancer and its prognosis depend on a number of factors,
including tumor size, staging and the extent of liver injury
(4). Surgical resection offers the
optimal prognosis for long-term survival; however, only 10–15% of
patients are considered suitable for surgical resection (5). The overall recurrence rate following
resection is 50–60% (6). If the
tumor cannot be removed completely, the disease is usually fatal
within 3–6 months (5). Although
radiotherapy and chemotherapy are often employed, the treatment
effects are extremely limited (7).
An emerging theory in recent years is that the
existence of cancer stem cells is the basic cause of the
post-operative recurrence of liver tumors (8). The main obstacle is that these cancer
stem cells are often in a state of dormancy which impairs the
treatment effects of chemotherapeutic drugs (9). Some recent studies have shown that
the herpes simplex virus thymidine kinase (HSVtk) gene and the
complementary treatment with ganciclovir (GCV) exert marked
antitumor effects against HCC by promoting apoptosis and inhibiting
angiogenesis (10,11). However, it is still difficult to
prevent the recurrence of liver tumors (10–12).
It is well known that normal gap junction
intercellular communication (GJIC) between tumor cells is crucial
for HSVtk/GCV to exert its therapeutic effects (10). However, there are few studies on
the structure and function of GJIC between liver cancer stem cells,
although some studies have reported that their GJIC is obviously
defective in liver tumors (13).
Whether GJIC deficiency also occurs in liver cancer stem cells, and
whether this feature promotes cancer stem cell resistance to
chemotherapeutic drugs needs to be explored further.
Small ubiquitin-like modifier (SUMO)ylation is an
essential post-translational modification that regulates a number
of physiological and pathological events in cells, such as those
involved in the stress response, nuclear translocation and the
stabilization of protein structures (14). Although enhanced SUMOylation and
the accumulation of SUMO-conjugated proteins have been widely
observed in patients with various disorders, including cancers
(15), their roles remain largely
unknown. Recently, it has been reported that connexin 43 (Cx43), a
protein involved in GJIC, can be covalently modified and regulated
by SUMOylation in HeLa cells (16). However, to date, at least to the
best of our knowledge, there are no studies available on the role
of Cx43 SUMOylation in liver cancer stem cells, as well as no
studies on whether Cx43 SUMOylation is triggered and plays roles in
certain molecular events, such as stemness maintenance, immune
escape and drug resistance, when cancer stem cells are exposed to
different niches, or whether artificial Cx43 SUMOylation can be a
clinical therapy for liver cancer stem cells.
Based on the above-mentioned issues, in this study,
we detected the expression status of Cx43 and SUMO1 in cancer stem
cells of HCC and hepatoblastoma origin. To reveal the function of
Cx43 SUMOylation in liver cancer stem cells, two exogenous
plasmids, Cx43-wild-type (wt) and green fluorescent protein
(GFP)-SUMO1, were co-transfected into hepatoblastoma-derived HepG2
cells, which are significantly less responsive to chemotherapy
(17), and GJIC structure and
function were examined. To further examine the effects of Cx43
SUMOylation on the effectiveness of chemotherapeutic drugs in liver
cancer stem cells, we used HSVtk/GCV both in vitro and in
vivo.
Materials and methods
Tissue specimens
Fresh surgical specimens (tumor and adjacent) were
collected from 8 patients with liver cancer at the Department of
General Surgery, the Fifth Central Hospital of Tianjin (Tianjin,
China) between January, 2015 and December, 2015. The initial
diagnosis of all frozen samples was completed by a senior
pathologist, and their paraffin-embedded sections were re-examined
by another pathologist to confirm the initial diagnosis. This study
was approved by the Ethics Committee of the Fifth Central Hospital
of Tianjin. Written informed consent was obtained from all
patients.
Cell culture
Two HCC cell lines (SNU182 and SNU449) and one
hepatoblastoma-derived cell line (HepG2) were purchased from the
American Type Culture Collection (Rockville, MD, USA) and cultured
in Dulbecco's modified Eagle's medium supplemented with 10% fetal
bovine serum (both from Invitrogen, Carlsbad, CA, USA), 100 U/ml
penicillin and 100 µg/ml streptomycin (both from Sigma, St.
Louis, MO, USA). Primary human normal liver cells were obtained
from the left liver tissue of a 51-year-old male patient who
underwent surgical resection for hepatic duct stones, which were
cultured in Roswell Park Memorial Institute (RPMI)-1640 medium
(Invitrogen) with 10% fetal bovine serum. All the cells were
incubated at 37°C in a humidified 5% CO2 atmosphere.
Isolation of liver cancer stem cells
CD133+ and CD133− HepG2 cells
were obtained by fluorescence-activated cell sorting using
fluorescence-activated cell sorting. These CD133+ and
CD133− cells were stained for Cx43 and SUMO1 proteins by
immunofluorescence. The CD133+ cells were cultured in
neurobasal medium containing 1X B27 (both from Invitrogen/Thermo
Fisher Scientific, Inc., Carlsbad, CA, USA), 20 ng/ml basic
fibroblast growth factor (Miltenyi Biotec, Bergisch Gladbach,
Germany) and 20 ng/ml epidermal growth factor (Provitro
Biosciences, Mt. Vernon, WA, USA), and incubated at 37°C with 5%
CO2. After ~3 weeks, cell spheres were collected, and
immunofluorescence staining was performed to detect the expression
of Cx43 and SUMO1 proteins. The detailed method of
immunofluorescence is described below.
Plasmids, transfection and treatment
The Ad-CMV-TK plasmid containing the HSVtk gene was
provided by the Institute of Life Science, Nankai University
(Tianjin, China). Lentiviral plasmids (pWPXLD-Cx43-wt,
pWPXLD-GFP-SUMO1, pWPXLD-Cx43-K144R, pWPXLD-Cx43-K237R and
pWPXLD-SENP1) were synthesized by Biogot Technology, Co. Ltd.
(Nanjing, China). All constructs were verified by nucleic acid
sequencing. The plasmids were then transfected into the
CD133+ HepG2 cells according to the experimental design,
following the transfection protocols provided with the lentiviral
plasmids. Finally, stably transfected cells were selected and
cultured in stem cell culture medium until the formation of new
clonal spheres. When the diameter of these spheres was at least 100
µm, GCV was added to the culture medium at 1 mg/ml.
Immunoprecipitation
Total protein was extracted from the cells. For
immunoprecipitation, ~1 mg of proteins was diluted 10-fold with
Triton X-100 lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1
mM EDTA, 0.5% Triton X-100, 1 mM PMSF, 10 mM iodoacetamide and
protease inhibitors), pre-cleared with protein-agarose beads for 1
h at 4°C, followed by the addition of anti-Cx43 antibody (sc-59949,
2 µl per 400 µg of total protein; Santa Cruz
Biotechnology, Dallas, TX, USA). Following incubation at 4°C
overnight, immunoprecipitates were washed 3 times with 1 ml Triton
X-100 lysis buffer and then diluted in 2X SDS sample buffer. After
boiling for 10 min, the samples were evaluated by western blot
analysis.
Western blot analysis
Total protein was extracted from fresh tissues or
cells. Western blot analysis was performed using NuPAGE™ 4-12%
Bis-Tris Protein Gels (NP0336PK2; Invitrogen). Following gel
electrophoresis, protein transfer and blocking, the membranes were
incubated with anti-SUMO1 (ab11672, 1:1,000; Abcam Trading Company
Ltd., Shanghai, China) or anti-Cx43 (sc-59949, 1:2,000; Santa Cruz
Biotechnology) antibodies overnight at 4°C. The membranes were then
incubated with the secondary antibodies [chicken anti-rabbit IgG
conjugated with horseradish peroxidase (HRP) (sc-2955, 1:2,000) or
chicken anti-goat IgG-HRP (sc-2953, 1:2,000) (both from Santa Cruz
Biotechnology)] for 1 h. A Super Signal protein detection kit
(Pierce; Thermo Fisher Scientific, Inc.) was used to detect protein
signals. The membranes were then stripped and re-probed with a goat
anti-human β-actin polyclonal primary antibody (1:1,000; Santa Cruz
Biotechnology). All experiments were repeated 3 times with similar
results.
Immunohistochemistry
The tumor tissues were fixed in 4% paraformaldehyde,
washed with PBS, transferred to 70% ethanol and then embedded in
paraffin in accordance with standard procedures (22). The antibodies used in this study
were a goat anti-human Cx43 monoclonal antibody (ab87645, 1:2,000;
Abcam Trading Company Ltd.) and mouse anti-human SUMO1 monoclonal
antibody (sc-5308, 1:400; Santa Cruz Biotechnology).
Immunofluorescence staining
Cx43 and SUMO1 protein expression was detected by
immunofluorescence staining. Briefly, the cells were fixed with 4%
paraformaldehyde (Sigma), treated with 3%
H2O2 for 10 min, and then incubated overnight
at 4°C with primary antibodies (anti-Cx43, sc-59949, 1:500; Santa
Cruz Biotechnology; anti-SUMO1, ab11672, 1:400; Abcam Trading
Company Ltd.) overnight at 4°C, followed by incubation with
Texas-Red or fluorescein isothiocyanate-labeled secondary
antibodies (sc-2787, 1:500 dilution; Santa Cruz Biotechnology).
Image-Pro Plus 6.0 software (Media Cybernetics, Rockville, MD, USA)
was used for image analysis.
Lactate dehydrogenase (LDH) activity
detection
The LDH content in the conditioned medium was
measured by an enzyme-linked immunosorbent assay using an LDH
Activity assay kit (BioVision, Inc., Milpitas, CA, USA) in
accordance with the manufacturer's instructions. Concisely, the
culture medium was centrifuged at 400 × g at room temperature for 5
min, and 20 µl supernatant was then collected and mixed with
20 µl 2,4-dinitrophenylhydrazine. Following incubation at
37°C for 15 min, 250 µl NaOH (0.4 M) was added to the
mixture followed by incubation for a further 15 min at 37°C. The
mixture was maintained at room temperature for 5 min, and the OD
was subsequently recorded using a microplate reader at 450 nm. The
activity of LDH was derived from the OD values and expressed as
U/l.
Apoptosis assay
Apoptosis was analyzed using an Annexin V/FITC
apoptosis detection kit (BD Biosciences, Franklin Lakes, NJ, USA)
according to the manufacturer's instructions. The cells were washed
twice with cold PBS and then resuspended in 1X binding buffer at a
concentration of 1×106 cells/ml. Subsequently, 100
µl of the solution (1×105 cells) was transferred
to a 5 ml culture tube, followed by the addition of 5 µl of
FITC Annexin V and 5 µl PI into the solution. The cells were
gently vortexed and incubated for 15 min at room temperature (25°C)
in the dark. This was followed by the addition of 400 µl of
1X binding buffer to each tube and analysis by flow cytometry
within 1 h.
Xenograft tumor assay
A total of 40 female nude mice (4 weeks old,
weighing 14–16 g) were purchased from the Animal Center of the
Academy of Military Medical Sciences (Beijing, China) and housed at
the Experimental Animal Center of Tianjin Medical University under
controlled temperature (22–24°C) conditions with a 12 h light/dark
cycle. All experimental procedures were carried out according to
the regulations and internal biosafety and bioethics guidelines of
the Tianjin Municipal Science and Technology Commission and were
approved by the Animal Ethics Committee of the Fifth Central
Hospital of Tianjin. The in vivo cancer model was
established as previously described (18). The 40 nude mice were randomly
divided into 5 groups as follows: i) The control group, in which
HepG2 cell spheres obtained by the stable transfection of the HSVtk
gene were transplanted into the subcutaneous tissues of the left
shoulders of the nude mice, and the mice were then treated with PBS
after 15 days; ii) the HSVtk group, in which HepG2 cell spheres
obtained by the stable transfection of the HSVtk gene were
transplanted, and after 15 days the mice were treated with 15 mg/kg
GCV every 2 days for 15 days; iii) the HSVtk + Cx43 group, in which
HepG2 cell spheres obtained by stable transfection with both HSVtk
and Cx43 genes were transplanted, and the mice were then treated
with GCV in the same manner as described above after 15 days; iv)
the HSVtk + SUMO1 group, in which HepG2 cell spheres obtained by
the stable transfection with both HSVtk and SUMO1 genes were
transplanted, and the mice were then treated with GCV in the same
manner as described above after 15 days; v) the HSVtk + Cx43 +
SUMO1 group, in which the HepG2 cell spheres obtained by the stable
transfection of the HSVtk, SUMO1 and Cx43 genes were transplanted,
and the mice were then treated with GCV in the same manner as
described above after 15 days. Tumor growth was monitored by
caliper measurements every 5 days for 30 days. Tumor volume (V) was
calculated as follows: V = L × W2 × 0.5 (L, length; W,
width). Following observation, the mice were anesthetized by an
intraperitoneal injection of chloral hydrate, according to the dose
of 400 mg/kg/animal body weight, and were then placed in sealed
chambers where a flow rate of 25% volume/min of carbon dioxide gas
are introduced for euthanasia. The weight of the mice upon
sacrifice and of the tumors upon anatomy was detected separately.
All tumor tissues blocks were prepared as paraffin-embedded
sections for in situ apoptosis and immunohistochemical
analyses. Terminal deoxynucleotidyl transferase (TdT)-mediated dUTP
nick-end labeling (TUNEL) assay using the FLOWTAC kit (Trevigen,
Gaithersburg, MD, USA) was used to examine cell apoptosis. The
cells (107 cells) were exposed to 2 mM hydrogen peroxide
(H2O2) or 0.5 µg/ml triclosan for 4 h
at 37°C. The cells were then fixed in 1 ml 3.7% formaldehyde for 10
min followed by permeabilization with Cytonin™ for 30 min. The
cells were then washed and resuspended in labeling reaction mix
(TdT dNTP mix, TdT enzyme, 1X Mn2+, 1X TdT labeling
buffer) at 37°C for 1 h. The reaction was terminated by the
addition of 1X StopBuffer followed by staining with 25 ml
Strep-Fluorescein for 10 min in the dark. The cells were then
stained with propidium iodide and RNase and analyzed using a
FACSCanto cytometer (BD Biosciences).
Statistical analysis
All experiments were repeated at least 3 times. When
the averages of the 2 groups was compared, the results were
analyzed by the Student's t-test. The comparison of caverages among
3 or >3 groups were analyzed by one-way analysis of variance.
Bonferroni correction was used to control the type I error. Data
are expressed as the means ± standard deviation (SD). All tests
were two-tailed, and the level of statistical significance was set
at P≤0.05. GraphPad Prism 6 software (GraphPad Software, Inc., La
Jolla, CA, USA) was used for all statistical tests.
Results
The expression of Cx43 and SUMO1 differs
markedly between liver cancer cells and normal liver cells
Previous studies have reported a relatively high
frequency of Cx43 expression loss in many types of tumors, such as
glioma and breast, colorectal, prostate, lung and pancreatic
cancers (19–21). Therefore, in this study, we first
examined the expression of Cx43 in liver cancer tissues and
adjacent tissues from 8 patients who underwent surgical resection.
The results of western blot analysis revealed that the expression
of Cx43 in the liver cancer tissue was significantly lower than
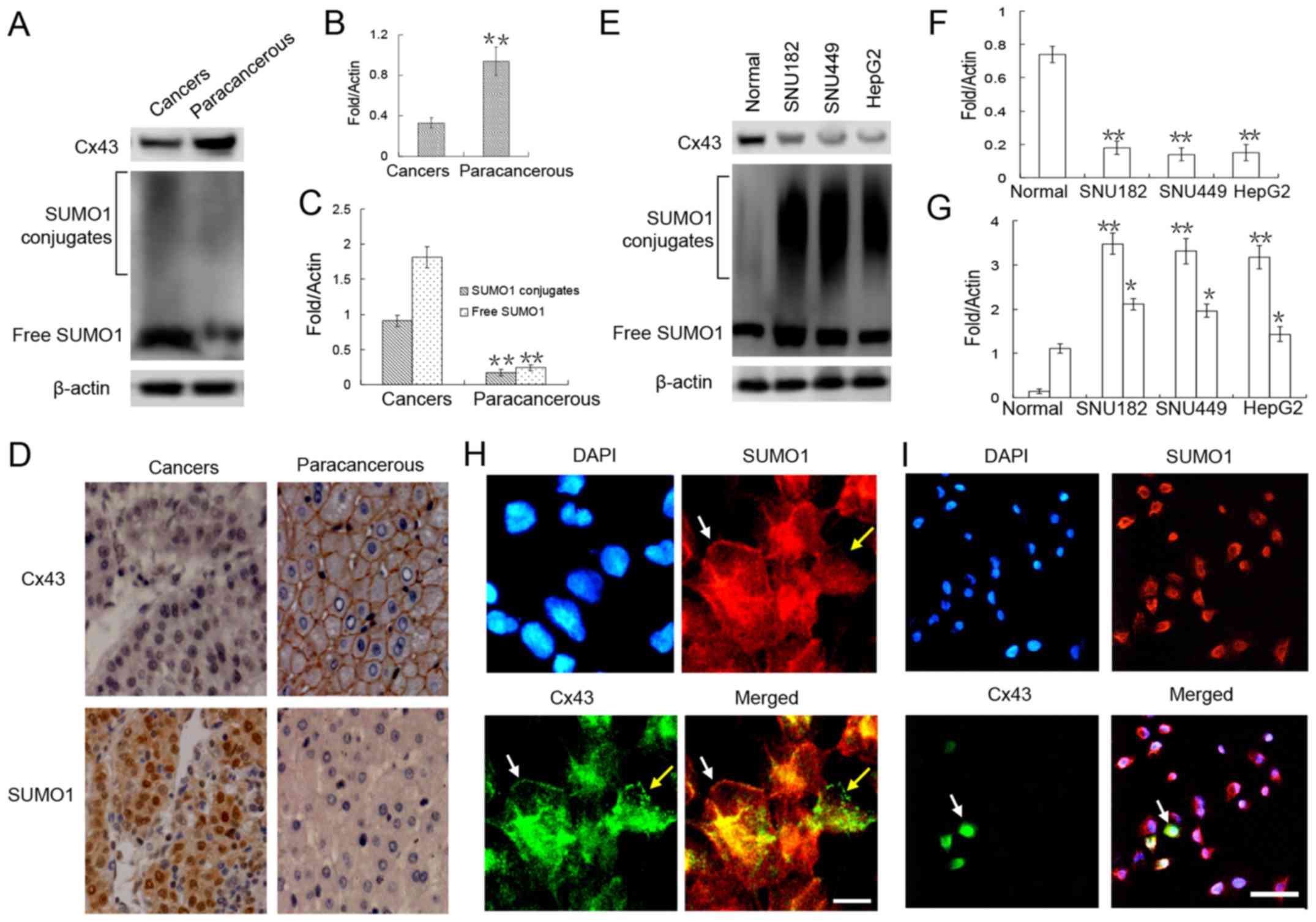
that in the adjacent (paracancerous) tissue by ~0.3-fold (Fig. 1A and B). Further
immunohistochemical analyses revealed that Cx43 was mainly
expressed in the liver cell membranes of the normal tissues
adjacent to the cancer tissues (Fig.
1D). Furthermore, in the cancer nest, Cx43 expression was
obviously weak (Fig. 1D).
Similar trends were detected in the liver cancer
cell lines. Compared with the normal liver cells, there was a
marked downregulation of Cx43 expression in these 3 liver cancer
cell lines (Fig. 1E and F).
Immunofluorescence staining revealed that Cx43 was localized in the
cell membranes of the normal hepatocytes (Fig. 1H). Compared with the normal liver
cells, only a small number (<30%) of HepG2 liver cancer cells
expressed Cx43, and the GJIC structure in the cell membrane was
almost unseen (Fig. 1I).
To verify the correlation between SUMO1 and Cx43
expression, SUMO1 expression was detected in the liver tumors and
liver cancer cell lines. Our results revealed that the expression
level of SUMO1 in both free and conjugated states was significantly
higher in the liver cancer tissues and liver cancer cell lines than
in the adjacent tissues and normal liver cells (Fig. 1A, C, E and G). Of note, we found no
obvious correlation between the expression of SUMO1 and Cx43 in
both normal liver cells and liver cancer cells, as Cx43 and SUMO1
expression was co-localized in the cell membrane (Fig. 1H, white arrow) but was also
expressed separately (Fig. 1H,
yellow arrow).
Cx43 and SUMO1 expression patterns are
distinctly different between cancer stem cells and non-cancer stem
cells
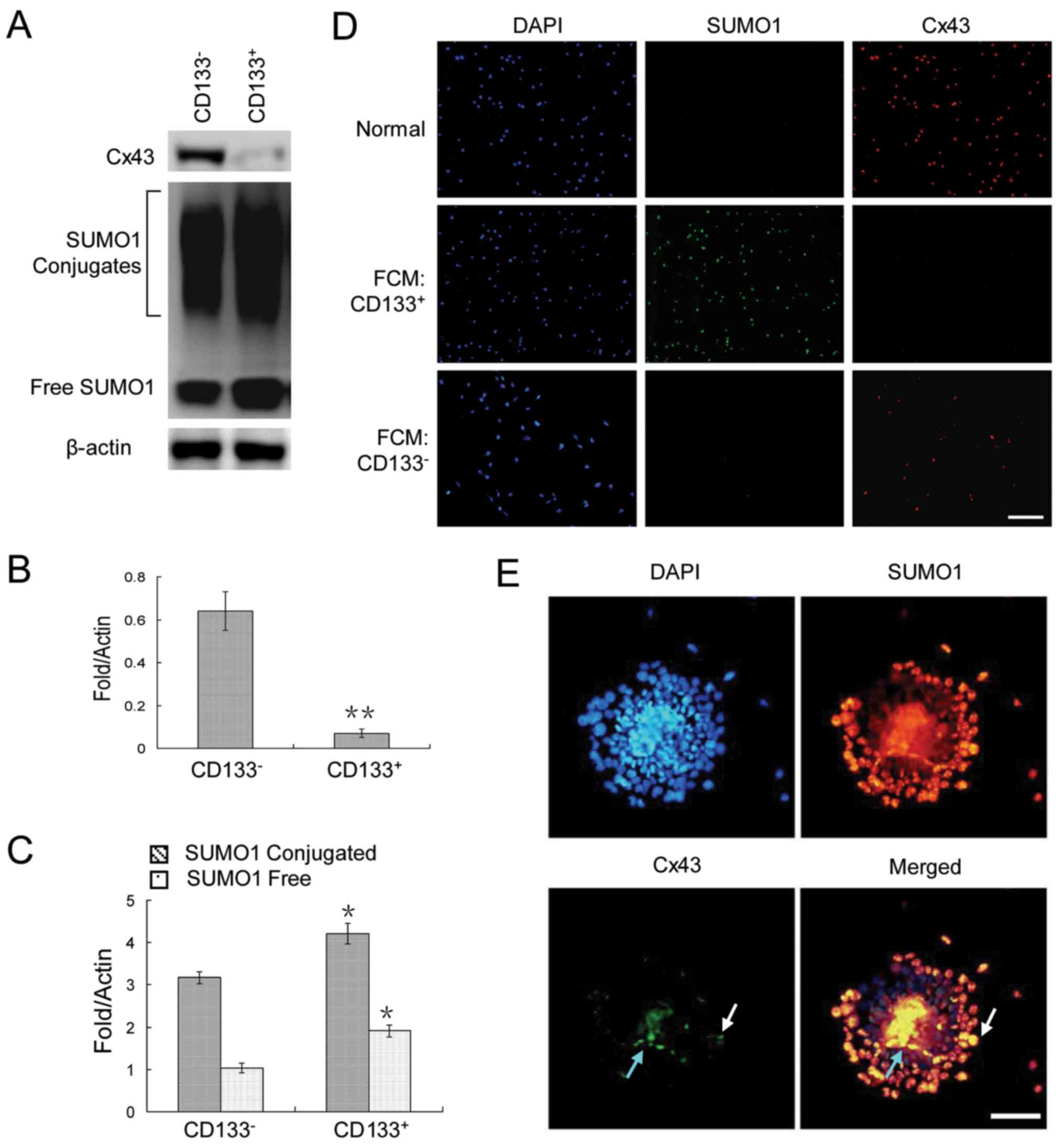
To analyze the expression patterns of Cx43 and SUMO1
in cancer stem cells and non-cancer stem cells, CD133+
and CD133− HepG2 cells were assessed by flow cytometry.
The results revealed a very weak Cx43 expression, but a strong
SUMO1 expression in the CD133+ HepG2 cells (Fig. 2A–C). The same pattern was observed
by immunofluorescence staining of the cells sorted by
fluorescence-activated cell sorting (Fig. 2D). Finally, we examined their
expression in HepG2 cell spheres. The results revealed that Cx43
was expressed in only a very small number of cells in clonal
spheres (Fig. 2E).
Cx43 is modified by SUMO1 in liver cancer
stem cells
Although Kjenseth et al (16) reported that Cx43 was covalently
modified and regulated by SUMOylation in HeLa cells, it is not
known whether the same mechanism exists in cancer stem cells.
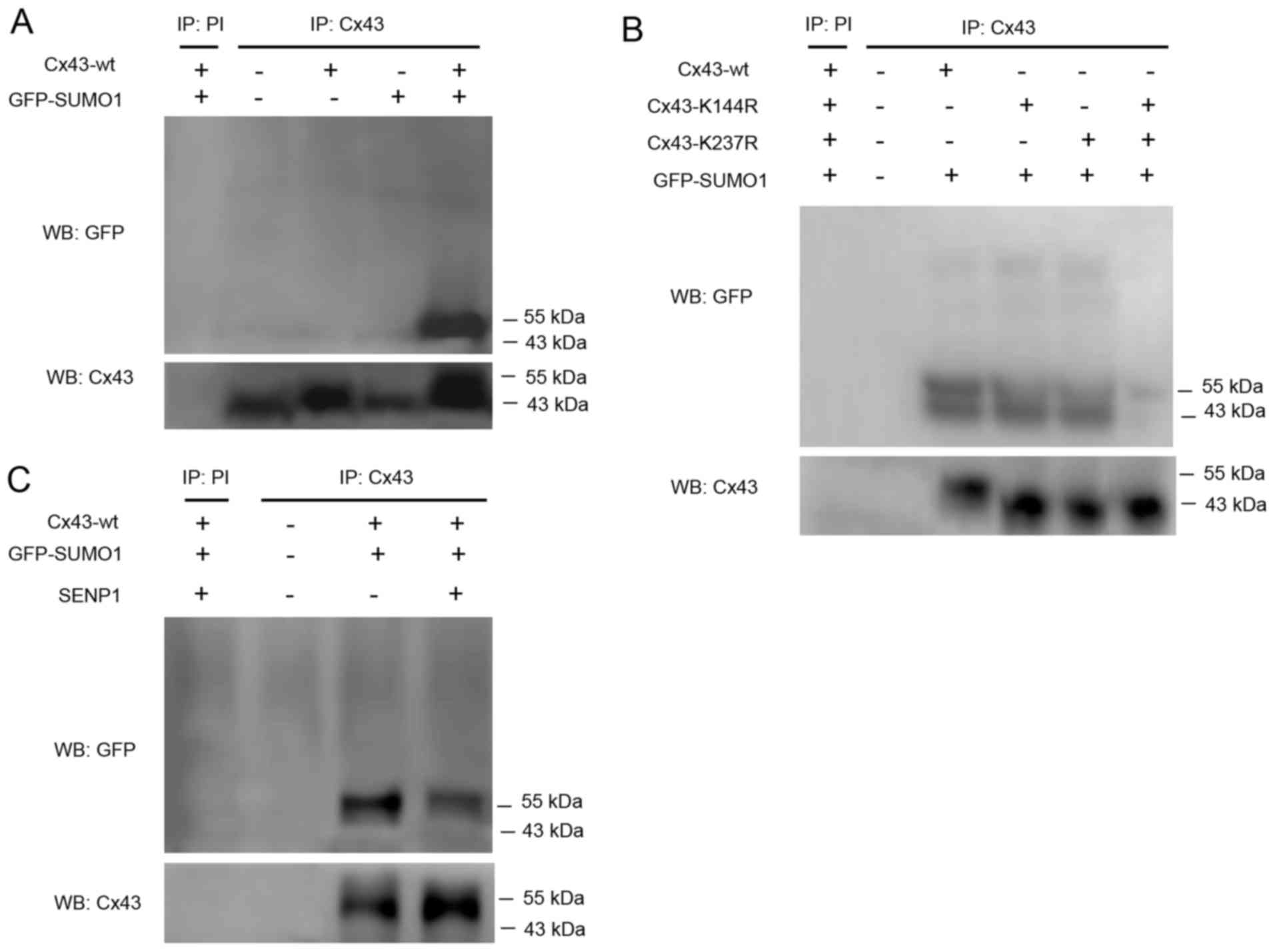
Therefore, in this study, CD133+ HepG2 cells were
transfected with Cx43-wt alone or in combination with GFP-SUMO1.
Cell lysates were subjected to immunoprecipitation using an
anti-Cx43 antibody or preimmune serum as a negative control.
SUMOylated Cx43 was detected by western blot analysis using an
anti-GFP antibody. The results revealed that only Cx43-wt and
GFP-SUMO1 co-transfection led to the appearance of a clear band
near 55 kDa indicating SUMOylated Cx43 (Fig. 3A).
Kjenseth et al (16) also reported that the covalent
binding site of Cx43 modified by SUMO1 was located at Lys-144 and
Lys-237. Based on the high conservation of these two loci among
species, we hypothesized that the covalent binding of Cx43 to SUMO1
may occur at the same site in liver cancer stem cells. Therefore,
two mutants, Cx43-K144R and Cx43-K237R, were co-transfected with
GFP-SUMO1. The results confirmed that the two mutations were indeed
able to abolish the covalent binding of SUMO1 to Cx43 (Fig. 3B).
SENP1 induces the de-conjugation of SUMOs,
particularly SUMO1, from the substrate protein, which was detected
to reveal whether it is capable of dissociation of conjugated SUMO1
from Cx43. The results confirmed that SENP1 partially, but not
completely, counteracted the conjugation of SUMO1 to Cx43 (Fig. 3C). However, the in-depth reasons
require further investigation.
Cx43 SUMOylation increases GJIC among
liver cancer stem cells
To further investigate whether Cx43 SUMOylation
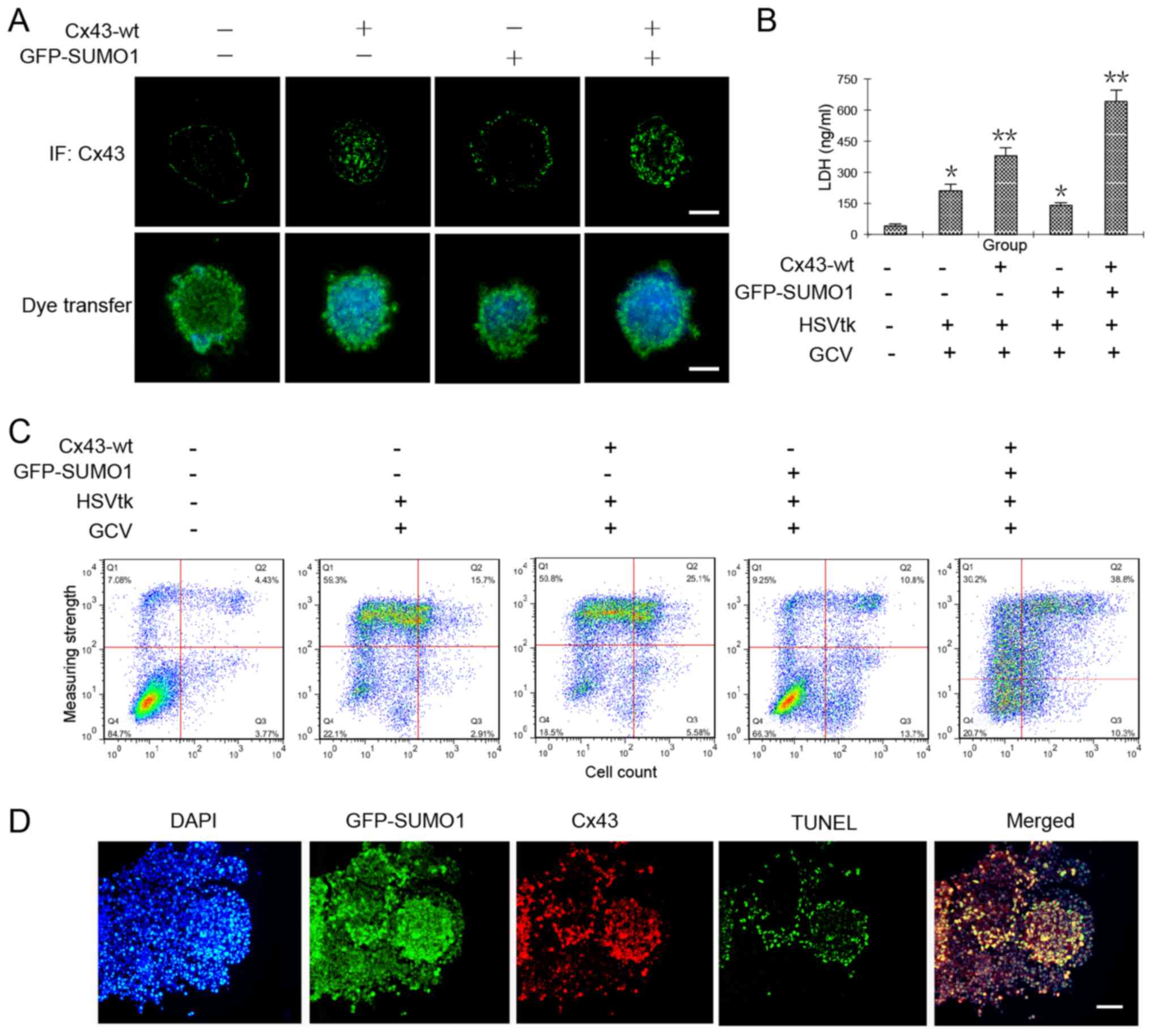
increases GJIC in liver cancer stem cells, we observed the
transmission of Hoechst 33342 in HepG2 cell spheres that were
co-transfected with Cx43-wt alone or in combination with GFP-SUMO1.
In theory, cancer stem cells are able to efflux the fluorescent
dye, Hoechst 33342 (22).
Therefore, fluorescence microscopic observations or flow cytometry
should detect no staining. Our results revealed that when the
cancer stem cells underwent Cx43 and SUMO1 gene transfection, they
were able to efflux most of the dye. However, when they were
transfected with the Cx43 or SUMO1 genes alone, the amount of dye
in the clonal spheres increased significantly. We hypothesized that
SUMO1 transfection alone may induce some unknown events, so that
the degree of dye diffusion among the cells was increased. The
specific mechanisms require further investigation. We found that
when the cells were co-transfected with SUMO1 and Cx43, the spread
of the dye in clonal spheres was increased significantly, which was
the highest in all test groups (Fig.
4A).
Cx43 SUMOylation increases the
sensitivity of liver cancer stem cells to HSVtk/GCV
We then applied this mechanism of increased GJIC by
Cx43 SUMOylation to enhance the sensitivity of cancer stem cells to
a conventional drug system, HSVtk/GCV, for which there no reports
available to date to prove its effectiveness for cancer stem cells;
however, it has been shown to be effective for most cancer cells
(12). The results from the LDH
content and flow cytometric assays both confirmed the obvious
damage and apoptosis of the tumor cells by co-transfection of the
Cx43 and SUMO1 genes (Fig. 4B and
C). Subsequently, the cell spheres were prepared as frozen
sections and subjected immunofluorescence staining or in
situ apoptosis detection. The results revealed that cellular
damage mainly occurred in the Cx43 and SUMO1 co-expression sites
(Fig. 4D).
Cx43 SUMOylation enhances the sensitivity
of subcutaneous tumors in immunodeficient mice to HSVtk/GCV
To verify whether the enhanced sensitivity to
HSVtk/GCV therapy in liver cancer stem cells also occurs in
vivo, immunodeficient mice were employed in this study.
Considering the changes in the growth state of liver cancer stem
cells induced by Cx43 or SUMO1 alone in vitro, measurements
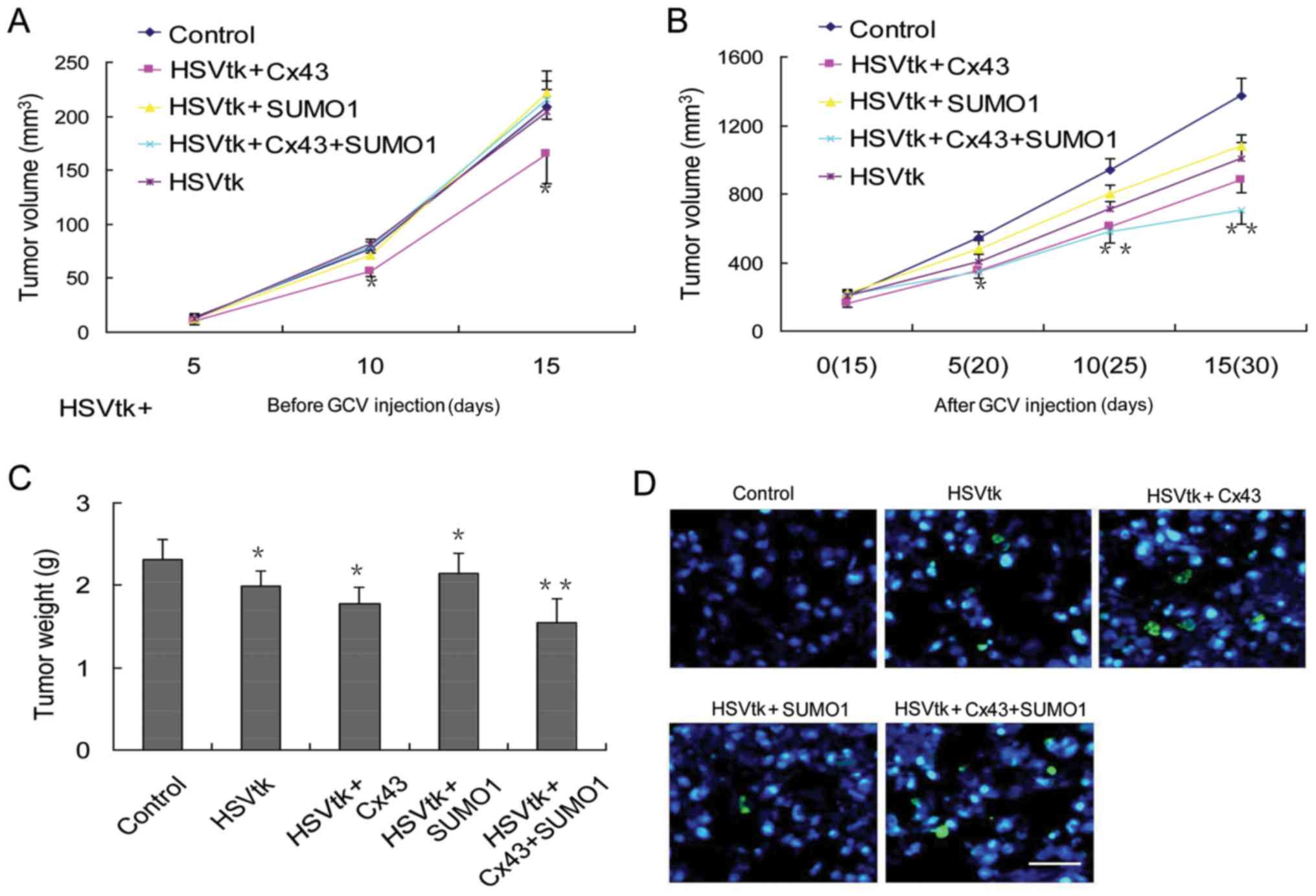
of subcutaneous tumor volumes were performed at 2 time periods.
During the first 15 days, the animals were not administered GCV,
and the tumor volume was recorded. The results revealed that
transfection of the HSVtk or SUMO1 genes alone did not induce
significant differences in tumor volumes compared with the control
group. However, transfection with Cx43 alone delayed tumor growth
(Fig. 5A). Next, GCV was
administered on day 15 following tumor sphere injection. The
results revealed that the tumor stem cells transfected with the
different genes exhibited different therapeutic responses to GCV.
The degree of their sensitivity to GCV treatment was as follows:
The control group < SUMO1 group < HSVtk group < Cx43 group
< Cx43 + SUMO1 group (Fig. 5B).
The results were consistent with the results of tumor weight
measurements (Fig. 5C) and the
rate of apoptosis in tumor tissues (Fig. 5D).
Discussion
The expression of Cx43 is reduced or even absent in
a variety of tumors, including liver cancer (13,23,24).
On the other hand, various tumors highly express SUMO1, and this
trend is increased with the degree of malignancy (24–26).
However, to date, at least to the best of our knowledge, limited
research has been performed into the association between Cx43 and
SUMO1 in liver cancer. In this study, we first examined the
expression of Cx43 and SUMO1 in liver tumor and adjacent tissues.
The results revealed a negative correlation between Cx43 and SUMO1
expression. Further results from 3 liver cancer cell lines
confirmed this conclusion. However, the results from
immunocytochemistry revealed that Cx43 and SUMO1 proteins were not
always co-located in the cell membrane, suggesting that the binding
of the two proteins may be temporary, and their interaction is
produced only when the function is performed jointly.
We then examined the protein expression of Cx43 and
SUMO1 in liver cancer stem cells. The results revealed that Cx43
protein was almost completely absent in the liver cancer stem
cells, which implies that intercellular communication is obviously
absent between liver cancer stem cells. However, the expression of
SUMO1 in liver cancer stem cells was obviously much higher than
that in non-cancer stem cells. It has been reported that several
proteins, such as octamer-binding transcription factor-4 (Oct-4),
hypoxia-inducible factor-1α (HIF-1α) and AKT1, which play key roles
in maintaining the stemness of cancer stem cells, are regulated by
SUMO1 (27–30). This may partly explain the reasons
for the high expression level of SUMO1 in liver cancer stem
cells.
Kjenseth et al reported that Cx43 is a target
protein modified by SUMO1 in HeLa cells (16). Therefore, in this study, we
confirmed the above finding in liver cancer stem cells, and that
SUMO binding sites in the Cx43 protein were Lys-144 and Lys-237.
This result implies that Cx43 SUMOylation is highly conserved among
species and may have a wide range of applications in several areas
in the future.
The main purpose of this study was to observe the
effect of Cx43 SUMOylation on the recovery of GJIC in liver cancer
stem cells and use this feature to restore the sensitivity of
cancer stem cells to chemotherapeutic drugs. Therefore, HepG2 cells
were selected as the research object due to its more pronounced
resistance to chemotherapeutic drugs than other two HCC cell lines.
The results revealed that Cx43 SUMOylation effectively improved
GJIC in liver cancer stem cells and increased their sensitivity to
the chemotherapeutic drug system, HSVtk/GCV. This finding provides
us with a new strategy for the treatment of liver tumors.
In addition to the above-mentioned findings, we
observed that Cx43 gene transfection alone partially inhibited
tumor growth in mice. This result is consistent with previous
reports that considered Cx43 as a tumor suppressor gene (31–33).
However, it is clear that the inhibitory effect of Cx43 alone on
tumor growth is extremely limited (31,33).
Similarly, the killing effect of HSVtk/GCV alone on liver cancer
cells is limited, particularly in CD133+ cell spheres
that were cultured in vitro. However, the induction of Cx43
SUMOylation significantly enhanced the sensitivity of the liver
cancer stem cells to HSVtk/GCV.
In conclusion, we found a novel method which may be
used to restore GJIC in liver cancer stem cells, which
significantly enhances their sensitivity to chemotherapeutic drugs.
Based on the high conservation of Cx43 SUMOylation among species,
it may be extended to more therapeutic areas in the future.
Acknowledgments
This study was supported by the National Natural
Science Foundation of China (grant nos. 81471175 and 1671246), and
the Tianjin Health Bureau Science and Technology Projects (grant
nos. 2014KY23 and 2015KZ018).
Notes
[1] Competing
interests
The authors declare that they have no competing
interests.
References
|
1
|
Kudo M, Trevisani F, Abou-Alfa GK and
Rimassa L: Hepatocellular carcinoma: Therapeutic guidelines and
medical treatment. Liver Cancer. 6:16–26. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang Z, Li Z, Ye Y, Xie L and Li W:
Oxidative stress and liver cancer: Etiology and therapeutic
targets. Oxid Med Cell Longev. 2016:78915742016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Castelli G, Pelosi E and Testa U: Liver
cancer: Molecular characterization, clonal evolution and cancer
stem cells. Cancers (Basel). 9:E1272017. View Article : Google Scholar
|
|
4
|
Salem AI and Winslow ER: Current technical
aspects of oncological hepatic surgery. Hepatobiliary Pancreat Dis
Int. 16:147–154. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim PT, Jang J, Fischer S, Greig PD,
Gallinger S, Wei AC, McGilivray IM, Cattral MS and Cleary SP: Liver
resection for multifocal hepatocellular carcinoma. J Clin Oncol.
30(Suppl 4): 3552012. View Article : Google Scholar
|
|
6
|
Cong WM and Wu MC: New insights into
molecular diagnostic pathology of primary liver cancer: Advances
and challenges. Cancer Lett. 368:14–19. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Büttner N, Schmidt N and Thimme R:
Perspectives of immunotherapy in hepatocellular carcinoma (HCC). Z
Gastroenterol. 54:1334–1342. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nishida N, Kitano M, Sakurai T and Kudo M:
Molecular mechanism and prediction of sorafenib chemoresistance in
human hepatocellular carcinoma. Dig Dis. 33:771–779. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Watson ME, Diepeveen LA, Stubbs KA and
Yeoh GC: Glycosylation-related diagnostic and therapeutic drug
target markers in hepatocellular carcinoma. J Gastrointestin Liver
Dis. 24:349–357. 2015.PubMed/NCBI
|
|
10
|
Wu K, Yang L, Huang Z, Zhao H, Wang J and
Xu S: A double suicide gene system driven by vascular endothelial
growth factor promoter selectively kills human hepatocellular
carcinoma cells. Oncol Lett. 11:3152–3160. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu X, Wang S, Guo X, Wei F, Yin J, Zang
Y, Li N and Chen D: Exogenous p53 and ASPP2 expression enhances
rAdV-TK/GCV-induced death in hepatocellular carcinoma cells lacking
functional p53. Oncotarget. 7:18896–18905. 2016.PubMed/NCBI
|
|
12
|
Park JH, Kim KI, Lee KC, Lee YJ, Lee TS,
Chung WS, Lim SM and Kang JH: Assessment of α-fetoprotein targeted
HSV1-tk expression in hepatocellular carcinoma with in vivo
imaging. Cancer Biother Radiopharm. 30:8–15. 2015. View Article : Google Scholar :
|
|
13
|
Yang Y, Qin SK, Wu Q, Wang ZS, Zheng RS,
Tong XH, Liu H, Tao L and He XD: Connexin-dependent gap junction
enhancement is involved in the synergistic effect of sorafenib and
all-trans retinoic acid on HCC growth inhibition. Oncol Rep.
31:540–550. 2014. View Article : Google Scholar :
|
|
14
|
Da Silva-Ferrada E, Ribeiro-Rodrigues TM,
Rodríguez MS and Girão H: Proteostasis and SUMO in the heart. Int J
Biochem Cell Biol. 79:443–450. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bettermann K, Benesch M, Weis S and
Haybaeck J: SUMOylation in carcinogenesis. Cancer Lett.
316:113–125. 2012. View Article : Google Scholar
|
|
16
|
Kjenseth A, Fykerud TA, Sirnes S, Bruun J,
Yohannes Z, Kolberg M, Omori Y, Rivedal E and Leithe E: The gap
junction channel protein connexin 43 is covalently modified and
regulated by SUMOylation. J Biol Chem. 287:15851–15861. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
López-Terrada D, Cheung SW, Finegold MJ
and Knowles BB: Hep G2 is a hepatoblastoma-derived cell line. Hum
Pathol. 40:1512–1515. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu Z, Jiang Z, Huang J, Huang S, Li Y, Yu
S, Yu S and Liu X: miR-7 inhibits glioblastoma growth by
simultaneously interfering with the PI3K/ATK and Raf/MEK/ERK
pathways. Int J Oncol. 44:1571–1580. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sin WC, Crespin S and Mesnil M: Opposing
roles of connexin43 in glioma progression. Biochim Biophys Acta.
1818:2058–2067. 2012. View Article : Google Scholar
|
|
20
|
Grek CL, Rhett JM, Bruce JS, Ghatnekar GS
and Yeh ES: Connexin 43, breast cancer tumor suppressor: Missed
connections? Cancer Lett. 374:117–126. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Leithe E: Regulation of connexins by the
ubiquitin system: Implications for intercellular communication and
cancer. Biochim Biophys Acta. 1865:133–146. 2016.PubMed/NCBI
|
|
22
|
Khan IS and Ehtesham M: Isolation and
characterization of stem cells from human central nervous system
malignancies. Adv Exp Med Biol. 853:33–47. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mattoscio D and Chiocca S: SUMO pathway
components as possible cancer biomarkers. Future Oncol.
11:1599–1610. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Amelio I, Landré V, Knight RA, Lisitsa A,
Melino G and Antonov AV: Polypharmacology of small molecules
targeting the ubiquitin-proteasome and ubiquitin-like systems.
Oncotarget. 6:9646–9656. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang XJ and Chiang CM: Sumoylation in gene
regulation, human disease, and therapeutic action. F1000Prime Rep.
5:452013. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bettermann K, Benesch M, Weis S and
Haybaeck J: SUMOylation in carcinogenesis. Cancer Lett.
316:113–125. 2012. View Article : Google Scholar
|
|
27
|
Wu Y, Guo Z, Wu H, Wang X, Yang L, Shi X,
Du J, Tang B, Li W, Yang L, et al: SUMOylation represses Nanog
expression via modulating transcription factors Oct4 and Sox2. PLoS
One. 7:e396062012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wei F, Schöler HR and Atchison ML:
Sumoylation of Oct4 enhances its stability, DNA binding, and
transactivation. J Biol Chem. 282:21551–21560. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Han X, Wang XL, Li Q, Dong XX, Zhang JS
and Yan QC: HIF-1α SUMOylation affects the stability and
transcriptional activity of HIF-1α in human lens epithelial cells.
Graefes Arch Clin Exp Ophthalmol. 253:1279–1290. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gu J, Fan Y, Liu X, Zhou L, Cheng J, Cai R
and Xue S: SENP1 protects against myocardial ischaemia/reperfusion
injury via a HIF1α-dependent pathway. Cardiovasc Res. 104:83–92.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Maqbool R, Rashid R, Ismail R, Niaz S,
Chowdri NA and Hussain MU: The carboxy-terminal domain of connexin
43 (CT-Cx43) modulates the expression of p53 by altering miR-125b
expression in low-grade human breast cancers. Cell Oncol (Dordr).
38:443–451. 2015. View Article : Google Scholar
|
|
32
|
Fu Y, Shao ZM, He QZ, Jiang BQ, Wu Y and
Zhuang ZG: Hsa-miR-206 represses the proliferation and invasion of
breast cancer cells by targeting Cx43. Eur Rev Med Pharmacol Sci.
19:2091–2104. 2015.PubMed/NCBI
|
|
33
|
Mauro V, Carette D, Pontier-Bres R,
Dompierre J, Czerucka D, Segretain D, Gilleron J and Pointis G: The
anti-mitotic drug griseofulvin induces apoptosis of human germ cell
tumor cells through a connexin 43-dependent molecular mechanism.
Apoptosis. 18:480–491. 2013. View Article : Google Scholar : PubMed/NCBI
|