Introduction
As the world's second most common type of
gynecological cancer, cervical cancer causes ~12,820 newly
diagnosed cases and 4,210 mortalities each year in the United
States (1). Although radical
surgical treatment and radiotherapy are effective treatments, more
than a third of these patients will develop progressive or
recurrent tumors (2,3). Currently, the International
Federation of Obstetricians and Gynecologists (FIGO) staging
system, depth of invasion and lymph node (LN) status are recognized
prognosis factors in patients with cervical cancer (4–6).
Among these factors, the adverse impact of LN metastasis on
patients with cervical cancer was confirmed in certain recent
clinical studies (4,7). Currently, there are some studies that
have investigated the mechanism of cervical LN metastasis (8,9);
however, a there is still no valid biomarker for LN metastasis.
Therefore, a comprehensive understanding of the pathways and genes
contributing to the development and LN metastasis of cervical
cancer is required.
The peroxisome proliferator-activated receptor
(PPAR) signaling pathway was first reported in 1990, and
demonstrated to be involved in glucose and lipid metabolism
(10). With further investigation
of the association between cell metabolism and cancer, the role of
PPARs in tumors has been continuously explored. In general,
deregulation of PPARγ, a subtype of PPARs, is detected in
peripheral tissues associated with lipid metabolism (11), whereas Mandard and Patsouris
(12) reported that PPARγ was also
deregulated in inflammation and cancer. Although evidence has
suggested that PPAR ligands may inhibit tumor angiogenesis during
tumor formation (13,14), the importance of PPAR signaling
pathway in tumors remains unclear, particularly the role in the
carcinogenesis and metastasis of cervical cancer.
As members of the zinc-dependent protease family,
matrix metalloproteinases (MMPs) are important factors involved in
the degradation of extracellular matrix and proteolysis (15). As an interstitial collagenase,
abnormal expression of MMP1 has been reported to be associated with
progression of human cancer. The increased expression of MMP1
detected in prostate cancer (16),
bladder cancer (17) and gastric
cancer (18) has been demonstrated
to be closely associated with prognosis. In addition, MMP1 has been
reported to promote angiogenesis by activating the endothelial
protease-activated receptor-1 (19). Anand et al (20) reported that glioma cell invasion
can be promoted by MMP1 through the mitogen-activated protein
kinase pathway (20). Whether MMP1
has a role in LN metastasis of cervical cancer is remains
unknown.
The Cancer Genome Atlas (TCGA) is a publicly funded
project that aims to catalogue and discover major cancer-causing
genomic alterations. The present study was based on high-throughput
RNA-sequencing data from TCGA. Gene profiling, molecular signatures
and functional pathway information with Gene Set Enrichment
Analysis (GSEA) were incorporated to identify aberrant pathways and
genes between patients with cervical cancer that were LN-negative
(N0) and LN-positive (N1). The combined approach revealed that MMP1
in the PPAR signaling pathway served a role in cervical cancer LN
metastasis. The effect of MMP1 on tumor metastasis phenotype was
validated by cervical cancer cell line experiments in vitro.
The clinical data in the TCGA database also confirmed the effect of
high expression of MMP1 on clinical prognosis. Critically, it was
demonstrated that MMP1 had key role in the regulation of metastasis
by promoting epithelial-mesenchymal transition (EMT) in cervical
cancer and was an independent prognostic factor for cervical
cancer. The mechanism of how upregulated MMP1 promotes LN
metastasis in cervical cancer requires further validation.
Materials and methods
Datasets
Transcriptome profiling data and prognostic data of
N0 and N1 cervical cancer (portal.gdc.cancer.gov/projects/TCGA-CESC) were
accessed from the TCGA (cancergenome.nih.gov) consortium. In total, 132 N0 and
60 N1 cervical cancer samples were obtained. The gene expression
profiles of GSE9750 were downloaded from Gene Expression Omnibus
database (ncbi.nlm.nih.gov/geo). GSE9750, which
was based on Agilent GPL96 platform (Agilent GeneChip Affymetrix
Human Genome U133A Array; Thermo Fisher Scientific, Inc., Waltham,
MA, USA), was submitted by Scotto et al (21). The GSE9750 dataset contained 66
samples, including 33 cervical cancer samples, 24 normal cervix
epitheliums, and 9 cervical cancer cell lines.
Identification of differentially
expressed genes (DEGs)
The transcriptome profiling data files obtained from
TCGA for analysis were systemized and transferred into a .txt file
which included expression and prognosis data using a Perl order
line. Then, package 'edgeR' (22,23)
of Bioconductor (24) (version
3.4) was applied in RStudio (RStudio, Inc., Boston, MA, USA;
version 3.3.2) to screen out the DEGs. The raw gene expression
profiles of GSE9750 were read using package 'affyPLM' (23), and probe quality control was
determined by relative logarithmic expression. Following
preprocessing of the data by log scale robust multi-array analysis
and filling the missing values using the k-Nearest Neighbor method,
package 'limma' (25) was applied
to obtain the DEGs. Exact test was used to search for the
non-random association between the expression of genes and group
according to the read counts of genes. In order to optimize the
probability of random association and the number of DEGs, fold
change (FC) ^2 and P<0.05 was considered to indicate a
statistically significant difference.
Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathway enrichment analysis of DEGs
KEGG (kegg.jp) is a knowledge base for systematic
and comprehensive analysis of gene functions in pathways and to
link genomic information with higher-level function information.
Database for Annotation, Visualization and Integrated Discovery
(DAVID; david.ncifcrf.gov; version 6.8) is an
important foundation for any high-throughput gene functional
analysis. ClueGO (version 2.2.3) is a plug-in app of Cytoscape
(cytoscape.org; version 3.5.0) where KEGG pathway
enrichment analysis can also be performed based on a different
database from DAVID. In order to analyze the DEGs at the functional
level, KEGG pathway analysis was applied by ClueGo and verified by
DAVID. P<0.05 was considered to indicate statistical
significance, pathways including four or more DEGs are presented in
the ClueGo-KEGG figures.
Pathway gene signatures analyzed using
GSEA
GSEA (version 6.0) (26,27)
is a computational method for exploring whether a given gene set is
significantly enriched in a group of gene markers ranked by their
relevance with a phenotype of interest. The curated KEGG pathway
V6.0 dataset was used to compare the impaired pathways in N0 and N1
cervical cancer samples. Additionally, the gene sets of <15
genes or >500 genes were excluded. The phenotype label was set
as N1 vs. N0. The t-statistic mean of the genes was computed in
each KEGG pathway using a permutation test with 1,000 replications.
The upregulated pathways were defined by a normalized enrichment
score (NES) >0 and the downregulated pathways were defined by an
NES <0. Pathways with a false discovery rate P≤0.1 were
considered significantly enriched.
Cell culture
Human cervical cancer cell lines HeLa and SiHa were
purchased from (American Type Culture Collection, Manassas, VA,
USA). Cells were cultured in Dulbecco's modified Eagle's medium
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) supplemented with
10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.)
at 37°C with 5% CO2 in a cell culture incubator.
Knockdown of MMP1
For designing the single guide RNA (sgRNA), MIT
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)
design software was used (crispr.mit.edu). The MMP1 sgRNA sequence was as
follows: TCACTGAGGGGAACCCTCGC (exon 2; score, 89). The lentiviral
particles were produced by transient transfection of letiX-293
cells cultured in a 10-cm Petri dishes with 10 μg vector DNA
pFgh1tUTG (a gift from Dr Marco Herold; plasmid no. 70183, Addgene,
Inc., Cambridge, MA, USA) (28)
with the target sgRNA (10 μg) inserted and the
pFUCas9mCherry vector (10 μg), along with the packaging
constructs pMDL (5 μg), pRSV-rev (2.5 μg), and pVSV-G
(3 μg; gifts from Dr Dakang Xu; Hudson Institute of Medical
Research) using standard calcium phosphate precipitation method.
Virus-containing supernatants were collected at 48 h after
transfection and passed through a 0.45 μm filter. To
establish MMP1 knockdown cervical cancer cells, HeLa and SiHa cells
were treated with 8 ng/ml polybrene in the viral supernatant,
incubated for 30 min at 37°C, and then centrifuged at 500 × g for 2
h at 32°C. Doxycycline hyclate (1 μg/ml; Sigma-Aldrich;
Merck KGaA) was used for treatment of cell lines to induce
expression of the sgRNA for 3 days. Cells without doxycycline
hyclate treatment were used as the knockdown control. Western
blotting was performed to assess the efficacy of the knockdown.
Western blotting
Cells were washed twice with cold PBS, followed by
lysis buffer [Tris-HCl (3.03 g), SDS (0.5 g), NaCl (4.35 g),
NaN3 (0.1 g), deoxysodium cholate (2.5 g), 1% NP-40 (5
ml) in 1 l ddH2O, plus 100 mM phenylmethane sulfonyl
fluoride (1:100) and inhibitor cocktail (1:50; cat. no. P8340;
Sigma-Aldrich; Merck KGaA) added before use]. The concentration of
the protein was determined by bicinchoninic acid protein assay kit
(Merck KGaA). Protein lysate (2 μg per well) was separated
on 10% SDS-PAGE gels and transferred onto nitrocellulose membranes
(PerkinElmer, Inc., Waltham, MA, USA) using standard western
blotting protocols. After blocking in Odyssey blocking buffer (cat.
no. 927-40000; LI-COR Biosciences, Lincoln, NE, USA) for 50 min,
primary antibody against MMP1 (1:500 dilution; cat. no. sc-21731;
Santa Cruz Biotechnology, Inc., Dallas, TX, USA), β-actin (1:10,000
dilution; cat. no. ab8227; Abcam, Cambridge, MA, USA), E-cadherin
(1:1,000 dilution; cat. no. 3195; Cell Signaling Technology, Inc.,
Danvers, MA, USA) and vimentin (1:1,000 dilution; cat. no. 5741;
Cell Signaling Technology, Inc.) were incubated overnight at 4°C
and the proteins were detected using secondary antibodies
conjugated to IRdye680 (cat. no. 18-4416-32/18-4417-32; Rockland
Immunochemicals Inc., Limerick, PA, USA) or IRdye800 (cat. no.
18-4516-32/18-4517-32; Rockland Immunochemicals Inc.) were used to
image the proteins at 700 or 800 nm under a LI-COR-Odyssey imaging
system (LI-COR Biosciences) scanner.
Proliferation assay
Cell viability was measured over a period of 6 days
using the PrestoBlue kit (Invitrogen; Thermo Fisher Scientific,
Inc.). Cells in the log growth phase were seeded into a 96-well
plate (2,000 cells/wells) and adhered overnight. Following the
manufacturer's instructions, 10 μl PrestoBlue reagent and 90
μl fresh medium were added to each well. After 30 min
incubation at 37°C, the fluorescence at a wavelength of 570 nm was
read using a FLUOstar Optima microplate spectrophotometer from BMG
Labtech GmbH (Ortenberg, Germany). The reagent was replaced with
fresh medium, and repeated on the next day. All experiments were
performed three times independently in biological triplicate.
Migration and invasion assays
Cell migration activity was assessed by a wound
healing assay. Briefly, 106 cells were seeded into
6-well plate at 90% confluency and adhered overnight. The cells
were wounded by scratching lines with a 200-μl pipette tip.
Floating cells were washed off with 1X PBS, then the cells were
observed and imaged under a microscope at different time-points.
The distance of the uncovered wound gap was quantified using ImageJ
software (version: 1.8.0; National Institutes of Health, Bethesda,
MD, USA), the relative wound closure was assessed at 36 h relative
to 0 h. A cell invasion assay was performed using Transwell
chambers (24-well, 8 μm pore size; Corning Life Sciences,
Corning, NJ, USA). In this assay, 600 μl RPMI medium
containing 25% FBS was used as the attractant in the lower chamber,
and 100 μl RPMI medium containing 5% FBS with 105
cells was added to the upper chamber; the cells were allowed to
migrate through the pores for 36 h. The cells on the lower side of
the filter were fixed with 4% paraformaldehyde for 10 min and were
stained with 0.5% crystal violet for 15 min at room temperature.
The number of the cells on the underside of the filter from three
randomly selected microscopic views were counted and digitally
imaged. All experiments were performed three times independently in
biological triplicate.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells using TRIzol
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. RT of cDNA was synthesized using the
Thermo Scientific RevertAid First Strand cDNA Synthesis kit (cat.
no. K1622; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. The mixture of template and primer was
incubated at 65°C for 5 min, then the following substances were
added in sequence and incubated at 42°C for 30 min: Reaction
buffer, 4 μl; RiboLock™ RNA, 1 μl; dNTP mix, 2
μl; RevertAid™ M-MuLV, 1 μl. The reaction was
terminated by 70°C for 5 min. The mRNA levels were determined using
Maxima™ SYBR Green/ROX qPCR Master Mix (2X; cat. no. K0222 Thermo
Fisher Scientific, Inc.) on ABI 7500 Real-Time PCR Systems. The PCR
conditions were as follows: Preheating at 50°C for 2 min; initial
denaturation at 95°C for 10 min; 40 denaturation cycles at 95°C for
15 sec; and primer annealing/elongation at 60°C for 1 min. Melting
curves were analyzed in each run to confirm specificity of
amplification. 18S rRNA were used as internal normalization
controls. Primers specific to E-cadherin and vimentin were as
follows: E-cadherin, forward 5′-GAACGCATTGCCACATACAC-3′, reverse
5′-AGCACCTTCCATGACAGACC-3′; vimentin, forward
5′-CCCTCACCTGTGAAGTGGAT-3′, reverse 5′-GACGAGCCATTTCCTCCTTC-3′; and
18S rRNA, forward 5′-CAAGCCCGACTTTGCAGA-3′, reverse
5′-CGCACGATTCGTCAAGTTATC-3′. The 2−∆∆Cq method was used
to represent fold change (29,30).
In each experiment, E-cadherin/18S and vimentin/18S levels in MMP1
knockdown cells were normalized to E-cadherin/18S and vimentin/18S
levels in MMP1-expressing cells. All experiments were performed in
biological triplicate. Data are presented as the mean ± standard
error of the mean (SEM).
Survival analysis of N1 vs
N0 cervical cancer patients and DEGs. Survival
analysis of N1 vs. N0 cervical cancer patients and DEGs in
overlapped pathway was conducted. Package 'survival' (version 2.38;
CRAN.R-project.org/package=survival) in RStudio was
applied and Kaplan-Meier curves were mapped based on the follow-up
data from TCGA. A log-rank test was used to calculate the
statistical significance of the difference in survival between the
two groups and the cut-off of P<0.05 was consider to be a
statistically significant difference.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism software (version 7.0; GraphPad Software, Inc., La Jolla, CA,
USA) and displayed as the mean ± SEM. SPSS (version 22.0; IBM
Corp., Armonk, NY, USA) was utilized to perform receiver operating
characteristic (ROC) curve analysis and Cox's proportional hazards
regression model. A significant difference between two groups was
analyzed using unpaired Student's t-test with Welch's correction.
Survival curves were constructed with the Kaplan-Meier method and
compared using the log-rank test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Work flow for the identification of key
pathways and genes in LN metastasis of cervical cancer
N1 and N0 cervical cancer samples were compared to
identify significant signatures in cervical cancer (Fig. 1A). In total, 1,287 DEGs, including
406 upregulated and 881 downregulated genes (N1 vs. N0), were
obtained based on 132 N0 and 60 N1 cervical cancer samples from
TCGA. The top 25 of up and downregulated DEGs are presented in
Fig. 1B. PPAR signaling pathway
was identified to be the top distinct signature upregulated pathway
by different enrichment methods (KEGG pathway enrichment and GSAE).
To explore further, GSE9750 datasets were analyzed and 770 DEGs
were obtained. There were 11 mutual DEGs selected following
overlapping DEGs from two different cohorts. Among 11 mutual DEGs,
MMP1 in PPAR signaling pathway was the unique gene with abnormal
high expression. Finally, two cervical cell lines were chosen to
verify the significance of MMP1 on the invasion and metastasis, and
clinical significance of MMP1 was investigated (Fig. 1).
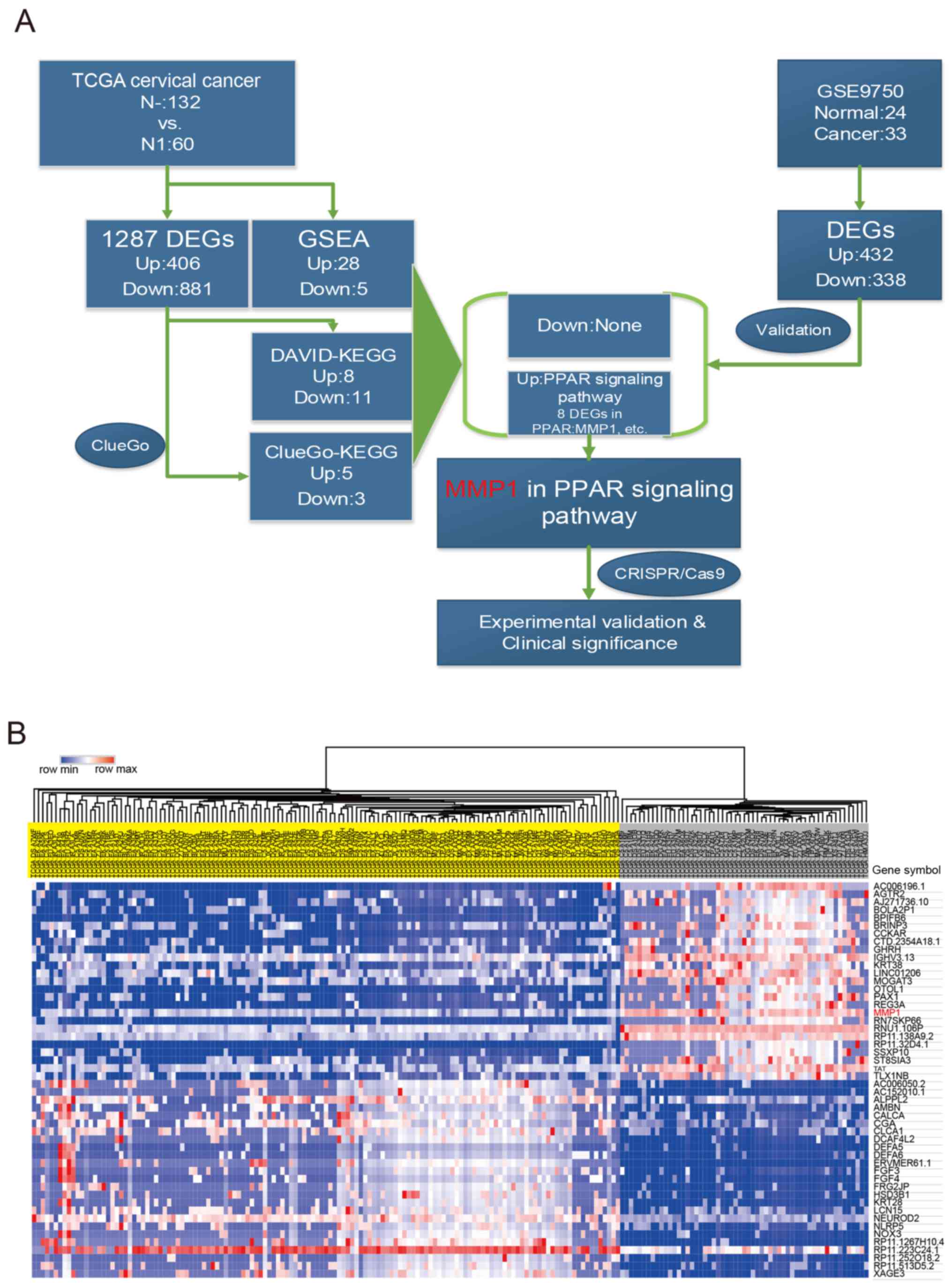 | Figure 1Work flow and heatmap of the top 25
DEGs. (A) Work-flow of the current study. (B) Top 25 up- and
downregulated DEGs in a heatmap. The upper section represents the
hierarchical clustering results, with yellow and grey bars
indicating N0 and N1 samples respectively. Genes are listed on the
right and red grids represented high expression while blue grids
represented low expression in N1 vs. N0. TCGA, The Cancer Genome
Atlas; DEG, differentially expressed gene; GSEA, Gene Set
Enrichment Analysis; DAVID, Database for Annotation, Visualization
and Integrated Discovery; KEGG, Kyoto Encyclopedia of Genes and
Genomes; PPAR, peroxisome proliferator-activated receptor; MMP1,
matrix metalloproteinase 1; CRISPR, Clustered Regularly Interspaced
Short Palindromic Repeats. |
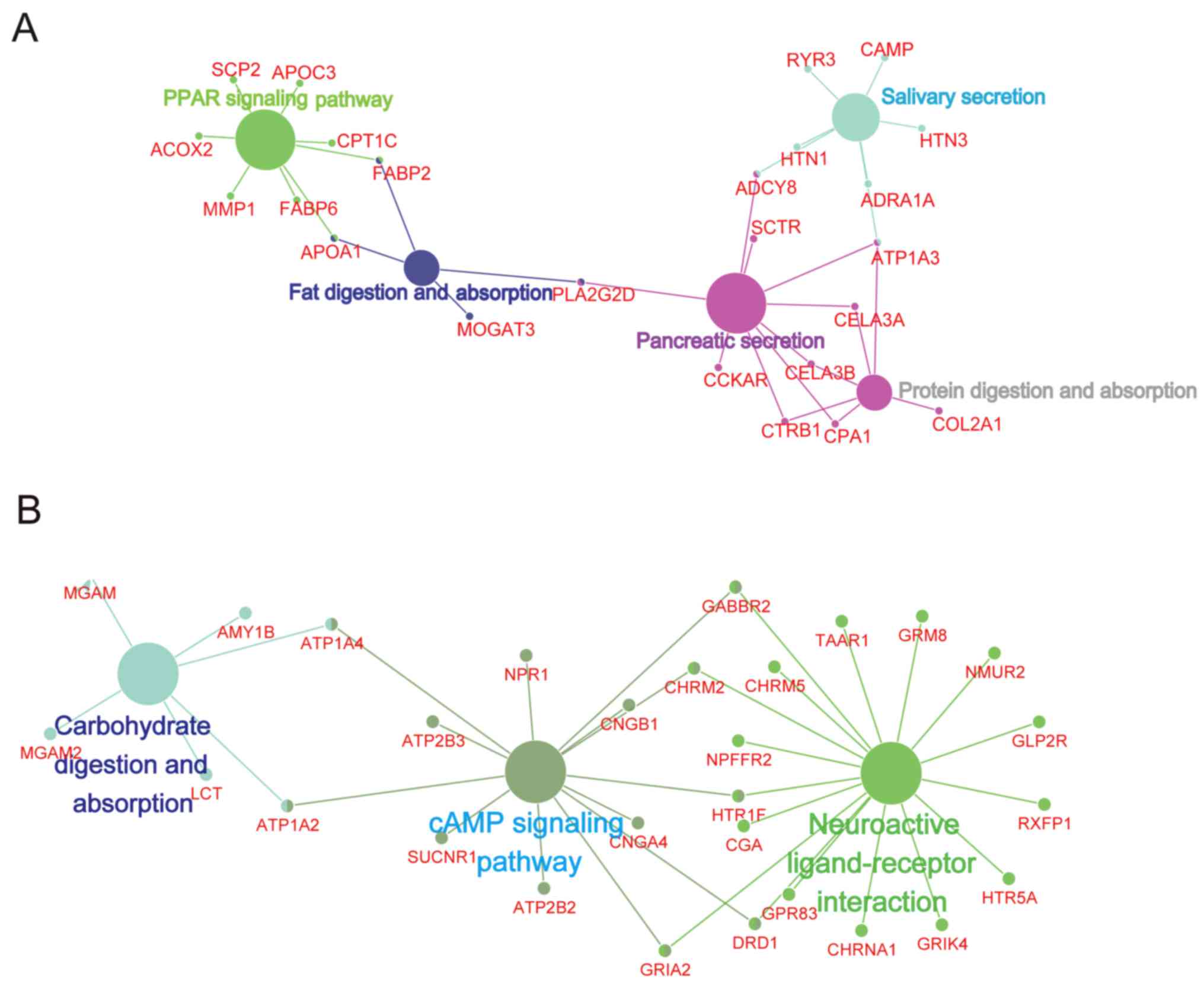
Identification of distinct pathways for
in cervical cancer
Up and downregulated DEGs between N0 and N1 were
analyzed to determine the most significantly enriched pathways
based on ClueGO and DAVID. The ClueGO result demonstrated that
'PPAR signaling pathway', 'Fat digestion and absorption', 'Protein
digestion and absorption' and others were significantly enriched
(Fig. 2A), while the most
significant pathways enriched in the downregulated DEGs were
'Carbohydrate digestion and absorption', 'cAMP signaling pathway'
and 'Neuroactive ligand-receptor interaction' based on the ClueGO
database (Fig. 2B). All the five
upregulated and three downregulated pathways enriched by ClueGO
were validated by DAVID.
GSEA was conducted to investigate the differential
pathways between N0 and N1. Finally, 28 upregulated and 5
downregulated pathways were identified (data not shown).
Identification of key gene signatures in
cervical cancer
Overlapping the GSEA results with the KEGG pathways
produced one upregulated pathway, namely 'PPAR signaling pathway'
and no downregulated pathways were mutual in KEGG and GSEA. The
peak in the GSEA result of the PPAR signaling pathway was shifted
left, with NES=1.412, indicating that most of the genes in PPAR
signaling pathway were upregulated (Fig. 3A and B). Fig. 3C There were 11 mutual genes in the
N0 vs. N1 cohort (1,287 DEGs) and normal vs. cancer cohort (770
DEGs). Among the 11 mutual genes, MMP1 in the 'PPAR signaling
pathway' was the only gene with high expression in cervical cancer
vs. normal cervix and N1 vs. N0 cervical cancer (logFC 4.02 and
1.22, respectively).
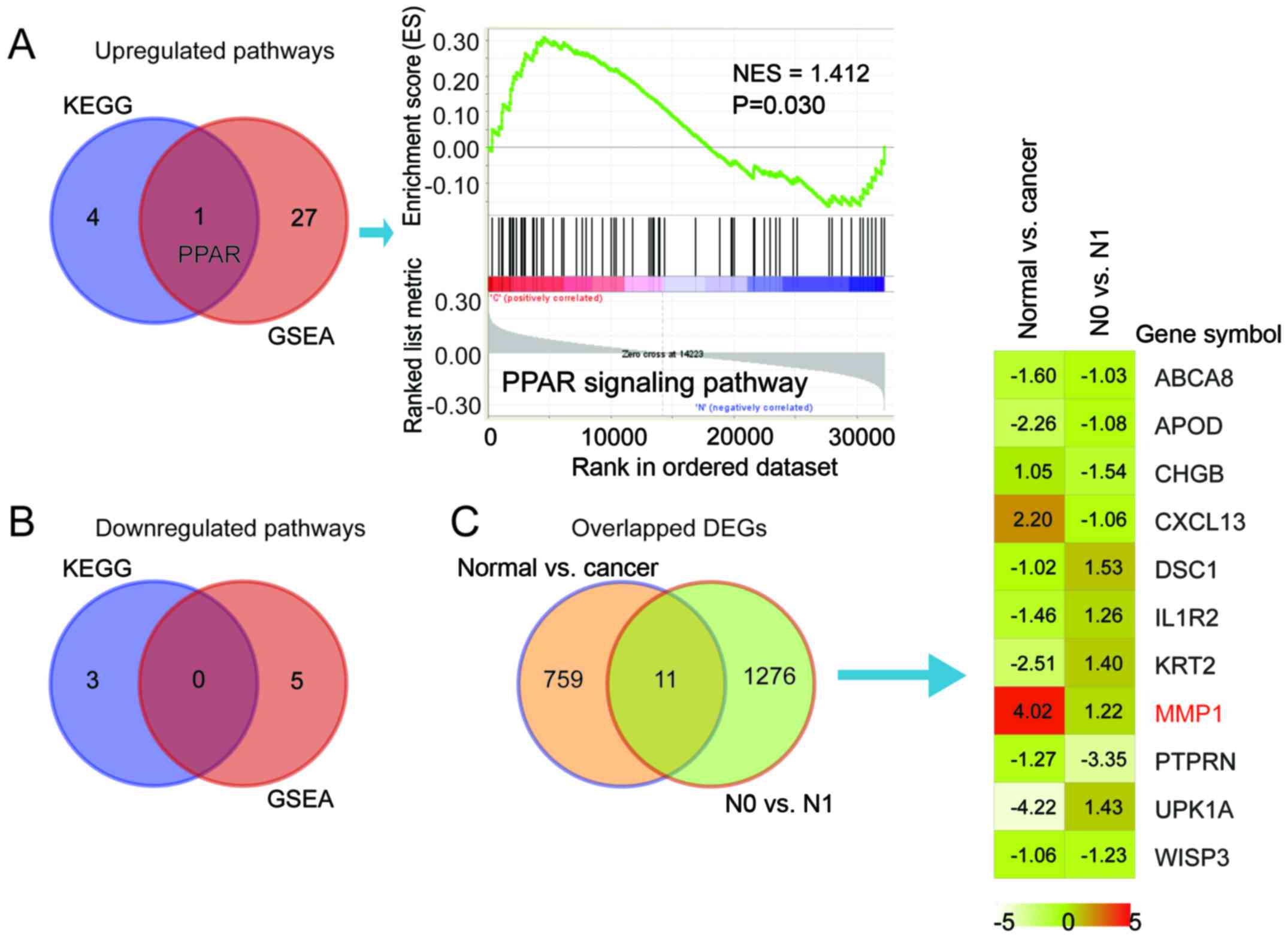 | Figure 3Identification of key gene signatures
and key genes in cervical cancer. (A) Venn diagram illustrating the
overlapped pathways among KEGG and GSEA (left) and the
representative GSEA curve of mutual upregulated pathway PPAR
signaling (right) with NES=1.413 and P=0.030 when comparing N1 and
N0. (B) Venn diagram illustrating the overlapped pathways between
KEGG and GSEA for downregulated pathways. (C) Venn-diagram
illustrating the overlapped DEGs between another GEO cohort (normal
vs. cancer, GSE 9750; N0 vs. N1, TCGA; left) and a heatmap
illustrating the log fold change of the 11 overlapped genes in the
two groups (right). MMP1 is marked as red for its elevation in both
groups. KEGG, Kyoto Encyclopedia of Genes and Genomes; PPAR,
peroxisome proliferator-activated receptor; GSEA, Gene Set
Enrichment Analysis; NES, normalized enrichment score; DEGs,
differentially expressed genes; ABCA8, ATP binding cassette
subfamily A member 8; APOD, apolipoprotein D; CHGB, chromogranin B;
CXCL13, C-X-C motif chemokine ligand 13; DSC1, desmocollin 1;
IL1R2, interleukin 1 receptor type 2; KRT2, keratin 2; MMP1, matrix
metalloproteinase 1; PTPRN, protein tyrosine phosphatase, receptor
type N; UPK1A, uroplakin 1A; WISP3, WNT1 inducible signaling
pathway protein 3. |
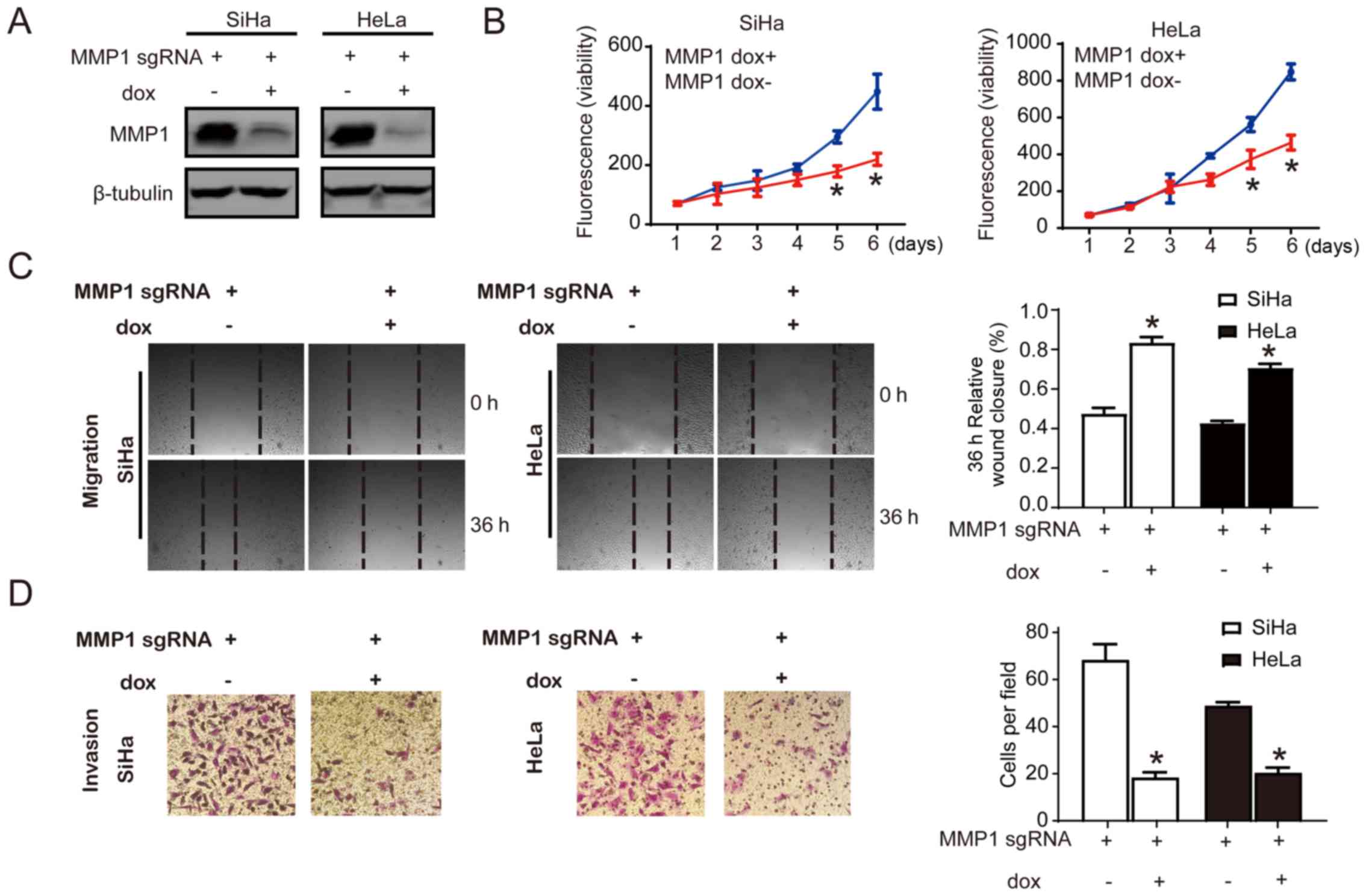
Knockdown of MMP1 alters malignant
behavior in vitro
Cell proliferation, migration and invasion have
important roles in the carcinogenesis. Different functional assays
were performed to explore the functional significance of MMP1.
Inducible CRISPR gene editing of MMP1 was conducted, and the
knockdown efficiency was displayed by immunoblotting (Fig. 4A). To determine the contribution of
MMP1 in cervical cancer cell growth, cell viability was determined.
Growth of MMP1 knockdown cells was significantly reduced compared
with the control cells by day 5 and 6, while there was no
significant difference between MMP1 knockdown cells and control
cells before day 4 (Fig. 4B). Cell
motility was determined by wound healing assay and Transwell assay.
The migration of MMP1 knockdown cell lines was assessed by
measuring the scratch wound closure capacity. There was a
significant delay in wound closure in the MMP1 knockdown cells at
36 h compared with the control cells (P<0.05; percentage of
wound closure) with 70-80% of the wound remaining open; in control
cells, ~40% of the wound remained open (Fig. 4C). The Transwell assay demonstrated
that ~20 cells per field were observed in MMP1 knockdown cells, and
the control cells had 70-80 cells per field, which demonstrated the
reduced invasion ability in MMP1 knockdown cells compared with the
control (P<0.05; Fig. 4D).
Taken together, the data suggested that knockdown of MMP1 decreased
the proliferation, invasion and migration ability in SiHa and HeLa
cervical cancer cells lines.
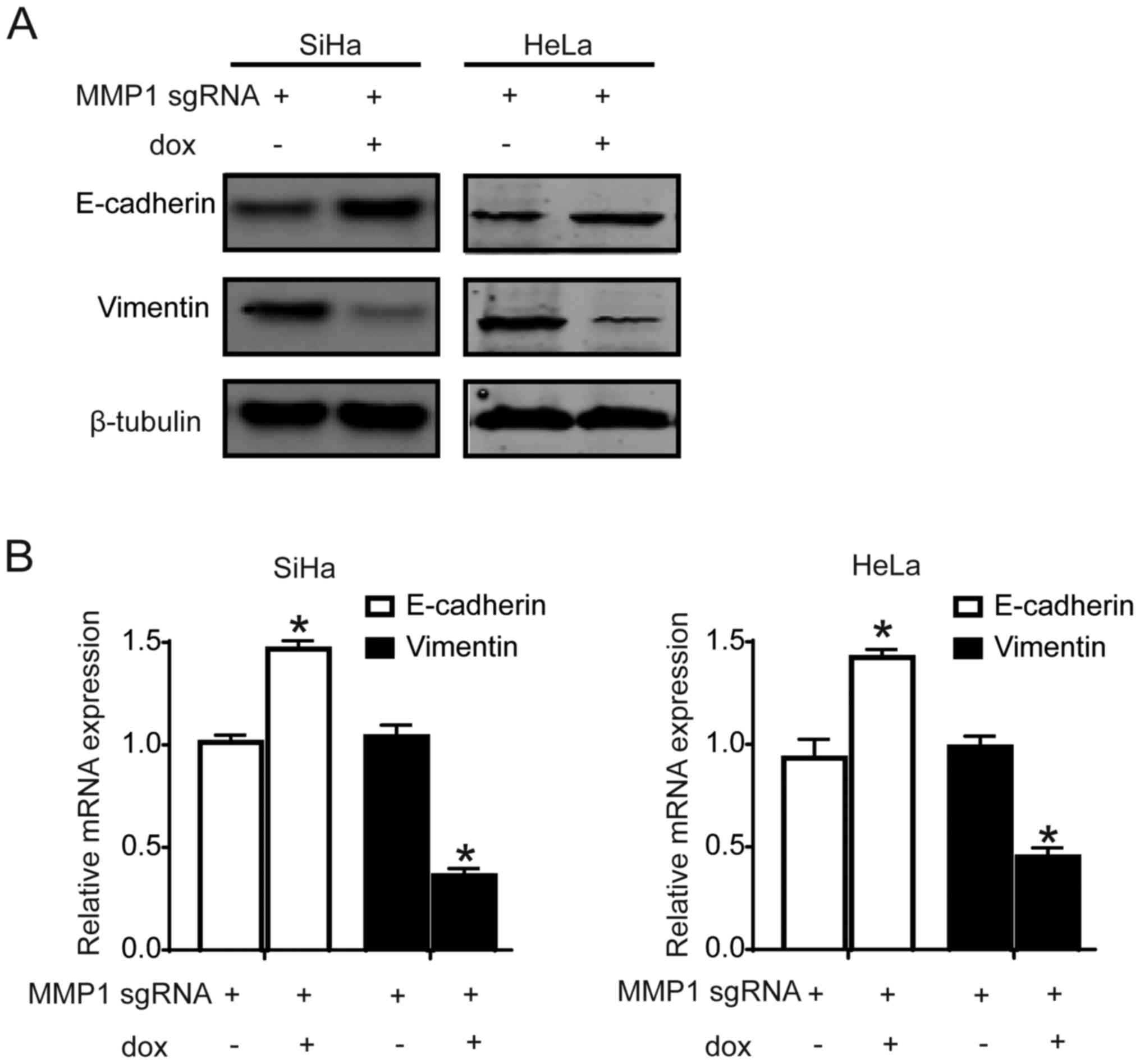
Knockdown of MMP1 inhibited EMT of
cervical cancer cells
To explore the mechanism of MMP1 in promoting
metastasis, EMT markers were detected. The expression of epithelial
marker, E-cadherin, was increased significantly (P<0.05), while
the metastasis-associated gene, vimentin, was decreased
significantly (P<0.05) in MMP1 knockdown cell lines comparing
with control cervical cancer cell lines (Fig. 5A and B). The results suggested that
knockdown of MMP1 may decrease cell migration and invasion ability
by inhibiting EMT in cervical cancer cells.
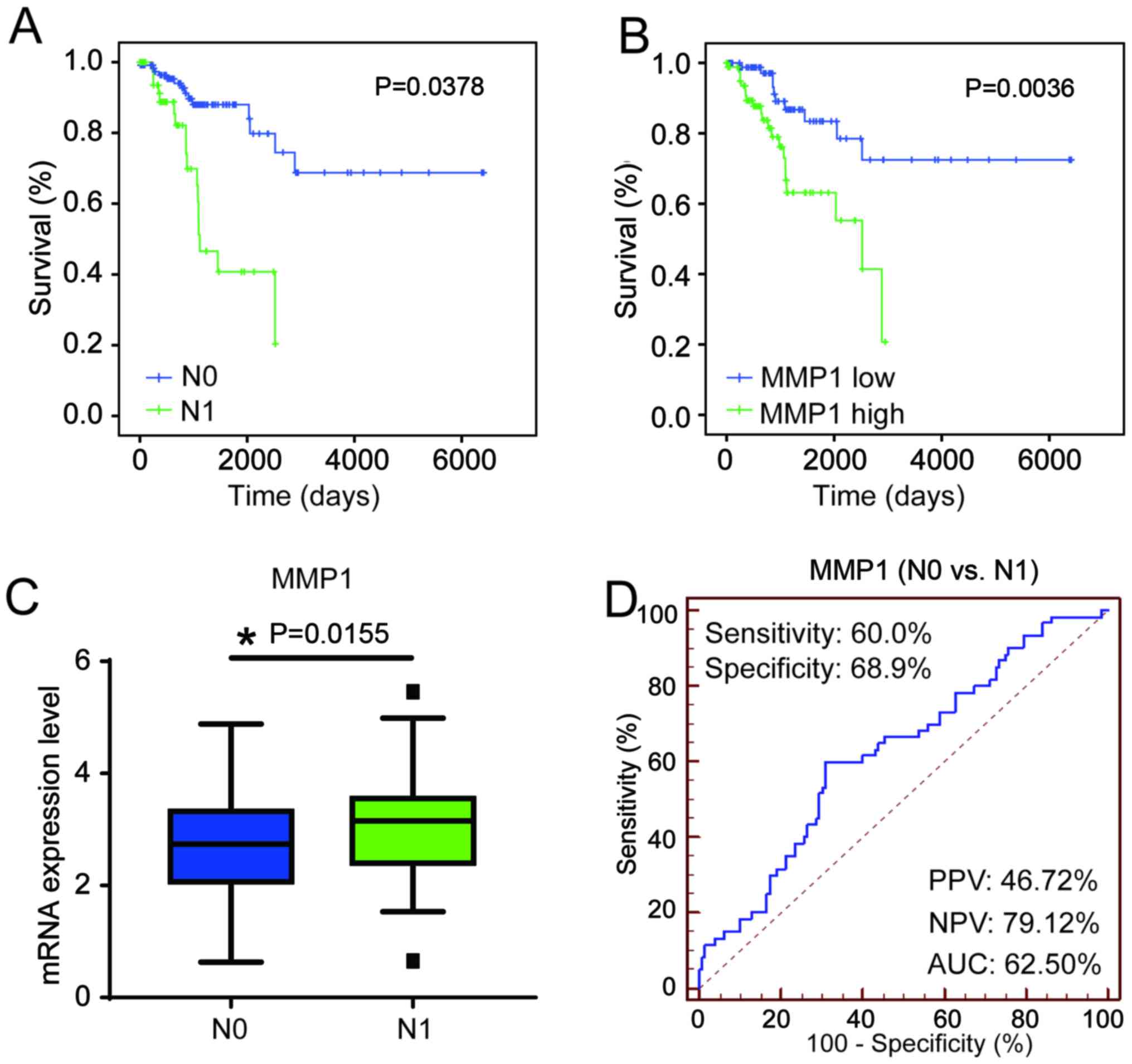
Clinical significance of MMP1 in patients
with cervical cancer
Kaplan-Meier curve analysis TCGA data from patients
with N0 and N1 cervical cancer demonstrated that N1 patients had
poor prognosis (P=0.03789) compared with N0 patients, which
demonstrated the crucial clinical value of predicting LN metastasis
(Fig. 6A). The expression level of
MMP1 was divided into high expression group and low expression
group according to the median expression, and was significantly
associated the overall survival of patients with cervical cancer,
and patients with MMP1 high expression had poor prognosis
(P=0.0036; Fig. 6B). Additionally,
MMP1 expression was significantly increased in N1 cervical cancer
samples compared with that in N0 (P=0.0434; Fig. 6C). The performance of MMP1 for
discriminating between N0 and N1 in patients with cervical cancer
was determined using an ROC curve. The sensitivity and specificity
of MMP1 were >60% (60.00% and 68.90%, respectively; Fig. 6D). Cox multivariate analysis
indicated that FIGO stage [hazard ratio (HR)=1.451, 95% confidence
interval (CI) 1.028-2.413, P=0.040), LN status (HR=2.560, 95% CI
1.177-5.568, P=0.018) and MMP1 (HR=1.699, 95% CI 1.133-2.548,
P=0.010) were significant independent favorable prognostic factors
of OS for patients with cervical cancer (Table I).
 | Table IUnivariate and multivariate Cox
proportional hazards analysis of MMP1 expression and overall
survival of patients with cervical cancer in The Cancer Genome
Atlas cohort. |
Table I
Univariate and multivariate Cox
proportional hazards analysis of MMP1 expression and overall
survival of patients with cervical cancer in The Cancer Genome
Atlas cohort.
| Factor | Univariate analysis
| Multivariate
analysis
|
|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age | 0.992
(0.978–1.006) | 0.272 | | |
| Stage | | | | |
| I–IIA vs.
IIB–IV | 1.536
(1.040–2.269) | 0.031a | 1.451
(1.028–2.413) | 0.040a |
| Grade | | | | |
| G1 vs. G2 + G3 +
G4 | 1.304
(0.940–1.810) | 0.112 | | |
| Lymph node | | | | |
| N0 vs. N1 | 3.158
(1.544–6.458) | 0.002b | 2.560
(1.177–5.568) | 0.018a |
| T | | | | |
| T1 vs. T2 + T3 +
T4 | 1.015
(0.452–2.277) | 0.972 | | |
| MMP1 | | | | |
| High vs. low | 2.014
(1.357–2.989) | 0.001a | 1.699
(1.133–2.548) | 0.010a |
Discussion
LN metastasis is considered as one of the strongest
prognostic markers for patients with cervical cancer; however, the
molecular mechanisms involved remain unclear. In the current study,
integrative bioinformatics methods and databases identified that
the PPAR signaling pathway and MMP1 were vital aberrant signatures
during LN metastasis of cervical cancer.
Currently, the association between metabolism and
cancer is attracting more and more attention. The PPAR signaling
pathway is predominantly involved in lipid and glucose metabolism.
Michalik et al (31)
reported that the increase in PPARβ/δ expression was associated
with a decrease in lipid accumulation in cardiac cells, whereas
overexpression in the intestine was associated with the development
of colon cancer. Disordered metabolism will further lead to
producing high quantities of inflammatory cytokines, predominantly
leptin, and also interleukin (IL)-8, IL-6 and angiopoietin like 4
(32-34). IL-6, a known upstream activator of
signal transducer and activator of transcription 3 (STAT3), has
been demonstrated to be crucial in the metastasis of various types
of human cancer (35,36). Additionally, other studies have
also directly confirmed the association between PPARs and tumors.
Trombetta et al (37)
reported that activation of PPARγ in cancer cells affected tumor
development. PPARγ ligand may be effective in inhibiting tumor
angiogenesis (13,38), and was recognized as important in
the carcinogenesis and metastasis of tumors. In the present study,
PPAR signaling was identified as an essential pathway in LN
metastasis based on a cohort containing 192 patients with cervical
cancer. MMP1 in the PPAR signaling pathway was a DEG with increased
expression in the cancer vs. normal and the N1 vs. N0 datasets.
Considering the anomalies of inflammatory factors and tumor
microenvironment caused by aberrant PPARs and MMP1, it requires
further investigation to understand the crucial role of the PPAR
signaling pathway and MMP1 during the development of cervical
cancer and LN metastasis.
To further validate the role of MMP1 in cervical
cancer, CRISPR/Cas9 genome editing was performed to knockdown MMP1
in SiHa and HeLa cervical cancer cell lines and revealed that cell
proliferation, migration and invasion abilities were reduced
significantly by MMP1 knockdown. Mechanistic studies demonstrated
that knockdown of MMP1 may decrease cell invasion and migration
ability by inhibiting the EMT process. MMP1 has been reported to
have an important role in various types of human tumor. George
et al (39) detected
abnormal expression of MMP1 in oral squamous cell carcinoma with
high histopathological grade. Langenskiöld et al (40) demonstrated that MMP1 was an
independent prognostic factor for colon cancer. Currently, certain
studies suggested that MMP1 promoted tumor cell migration by
degrading specific substrates that regulate cell adhesion and
cell-matrix adhesion (41), and
the subtle balance between MMPs and tissue inhibitor of
metalloproteinases contributed to the tumor microenvironment
(42), which is one of the most
important aspects of tumor invasion and metastasis (43). MMP1 was reported to be involved in
tumor LN metastasis (44), and
also to facilitate peritoneal dissemination (45) and nerve tract migration (46). The results of the current study
suggested that knockdown of MMP1 decreased the invasion and
migration ability of SiHa and HeLa cell lines, which partially
supports by the findings of Sun et al (47). Furthermore, the present study
demonstrated that knockdown of MMP1 inhibited EMT in SiHa and HeLa
cell lines, suggesting that MMP1 may manifest its malignant ability
via EMT. Currently, there are several lines of evidence that a
large number of growth factors, such as transforming growth
factor-β (TGF-β), Wnt and epidermal growth factor (EGF), cytokines,
such as tumor necrosis factor-α, nuclear factor-κB and Akt,
transcription factors, such as Snail, Twist and Slug, and other
markers are involved in the EMT process (48-50).
However, the mechanism of the cooperative regulation between these
factors in the EMT process remains unclear. EMT had been considered
to be an important process in stimulating the metastasis of
cervical cancer (51). Shostak
et al (52) confirmed that
HPV infection promoted EGF-mediated EMT in cervical cancer. In the
current study, MMP1 may exert its activity through the PPAR pathway
to alter the tumor microenvironment, thus inducing the EMT process
by affecting the IL-6/STAT3 axis. Additionally, the stable
expression of the active form of MMP1 was demonstrated to induce
EMT by promoting the generation of TGF-β (53), and certain EMT transcription
factors, including Twist and Slug, have been reported to regulate
MMP1 expression (54,55). Relevant research is required to
determine the detailed mechanism by which MMP1 promotes EMT during
metastasis of cervical cancer.
The carcinogenic properties of MMP1 adversely affect
the prognosis of patients with cervical cancer. Highly expressed
MMP1 was significantly inversely associated with poor overall
survival in a cohort of 306 patients based on TCGA data. MMP1 was
an independent prognostic biomarker of cervical cancer. The
clinical value of MMP1 has been reported in certain other types of
human cancer (15,31,45,56);
however, the importance for cervical cancer remains unclear.
Certain studies have noted the overexpression of MMP1 in cervical
cancer (47,57,58),
while the current study suggested that MMP1 was an independent
prognostic factor for cervical cancer. These findings will have a
guiding role in future clinical work.
The research of the present study still has some
shortcomings. Independent cervical cancer samples were not
available to validate the function of MMP1, and also the relevant
animal experiments have not been performed. Although MMP1 is highly
expressed in cervical cancer from several independent databases,
related MMP1 overexpression experiments were not conducted. The use
of stable cell lines with MMP1 overexpression will be required for
further research. The detailed mechanism by which MMP1 promotes
cervical cancer metastasis required further study. These are the
future research directions, some of which have already been
performed.
In summary, the data of the current study indicated
that the PPAR signaling pathway has important role during LN
metastasis of cervical cancer, and that MMP1 in the PPAR signaling
pathway acted as a key role in the regulation of tumor growth and
LN metastasis via EMT process in cervical cancer. MMP1 was
suggested to be a biomarker for LN metastasis and an independent
prognostic factor in cervical cancer that requires further
validation.
Glossary
Abbreviations
Abbreviations:
|
LN
|
lymph node
|
|
DEGs
|
differently expressed genes
|
|
N0/N1
|
LN-negative/LN-positive
|
|
TCGA
|
The Cancer Genome Atlas
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
GSEA
|
Gene Set Enrichment Analysis
|
|
GEO
|
Gene Expression Omnibus
|
|
PPAR
|
peroxisome proliferator-activated
receptor
|
|
MMP1
|
matrix metalloproteinase 1
|
|
EMT
|
epithelial-mesenchymal transition
|
|
FIGO
|
International Federation of
Obstetricians and Gynecologists
|
Acknowledgments
The authors thank Professor Xu Dakang (Monash
University, Clayton, Australia) for advice on the manuscript and Dr
Henry Clarke (Monash University) for work on the language of this
manuscript.
Notes
[1]
Funding
No funding was received.
[2] Availability
of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
[3] Authors'
contributions
RT analyzed the bioinformatic data. RT and XFL
performed the cell assays and were major contributors in writing
the manuscript. YG, YL, PY and KZW gave advice on the experiments
and writing of the manuscript. RT and KZW designed the study. All
authors read and approved the final manuscript.
[4] Ethics
approval and consent to participate
Not applicable.
[5] Consent for
publication
Not applicable.
[6] Competing
interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Munro A, Codde J, Spilsbury K, Steel N,
Stewart CJ, Salfinger SG, Tan J, Mohan GR, Leung Y, Semmens JB, et
al: Risk of persistent and recurrent cervical neoplasia following
incidentally detected adenocarcinoma in situ. Am J Obstet Gynecol.
216:272.e1–272.e7. 2017. View Article : Google Scholar
|
|
3
|
Kim S-W, Chun M, Ryu H-S, Chang SJ, Kong
TW, Lee EJ, Lee YH and Oh YT: Salvage radiotherapy with or without
concurrent chemotherapy for pelvic recurrence after hysterectomy
alone for early-stage uterine cervical cancer. Strahlenther Onkol.
193:534–542. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li C, Liu W and Cheng Y: Prognostic
significance of metastatic lymph node ratio in squamous cell
carcinoma of the cervix. Onco Targets Ther. 9:3791–3797.
2016.PubMed/NCBI
|
|
5
|
Wang XJ, Xiong Y, Ma ZB, Xia JC and Li YF:
The expression and prognostic value of protein tyrosine kinase 6 in
early-stage cervical squamous cell cancer. Chin J Cancer.
35:542016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li J, Wu MF, Lu HW, Chen Q, Lin ZQ and
Wang LJ: Pretreatment serum lactate dehydrogenase is an independent
prognostic factor for patients receiving neoadjuvant chemotherapy
for locally advanced cervical cancer. Cancer Med. 5:1863–1872.
2016. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fleming ND, Frumovitz M, Schmeler KM, dos
Reis R, Munsell MF, Eifel PJ, Soliman PT, Nick AM, Westin SN and
Ramirez PT: Significance of lymph node ratio in defining risk
category in node-positive early stage cervical cancer. Gynecol
Oncol. 136:48–53. 2015. View Article : Google Scholar :
|
|
8
|
Zhang WN, Li W, Wang XL, Hu Z, Zhu D, Ding
WC, Liu D, Li KZ, Ma D and Wang H: CLDN1 expression in cervical
cancer cells is related to tumor invasion and metastasis.
Oncotarget. 7:87449–87461. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chung IH, Wu TI, Liao CJ, Hu JY, Lin YH,
Tai PJ, Lai CH and Lin KH: Overexpression of lipocalin 2 in human
cervical cancer enhances tumor invasion. Oncotarget. 7:11113–11126.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Issemann I and Green S: Activation of a
member of the steroid hormone receptor superfamily by peroxisome
proliferators. Nature. 347:645–650. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Berger J and Moller DE: The mechanisms of
action of PPARs. Annu Rev Med. 53:409–435. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mandard S and Patsouris D: Nuclear control
of the inflammatory response in mammals by peroxisome
proliferator-activated receptors. PPAR Res. 2013:6138642013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xin X, Yang S, Kowalski J and Gerritsen
ME: Peroxisome proliferator-activated receptor gamma ligands are
potent inhibitors of angiogenesis in vitro and in vivo. J Biol
Chem. 274:9116–9121. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sato M: Peroxisome proliferator activated
receptor ligands and angiogenesis. Nihon Rinsho. 63:603–608.
2005.In Japanese. PubMed/NCBI
|
|
15
|
Liu M, Hu Y, Zhang MF, Luo KJ, Xie XY, Wen
J, Fu JH and Yang H: MMP1 promotes tumor growth and metastasis in
esophageal squamous cell carcinoma. Cancer Lett. 377:97–104. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ozden F, Saygin C, Uzunaslan D, Onal B,
Durak H and Aki H: Expression of MMP-1, MMP-9 and TIMP-2 in
prostate carcinoma and their influence on prognosis and survival. J
Cancer Res Clin Oncol. 139:1373–1382. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shin DH, Dier U, Melendez JA and Hempel N:
Regulation of MMP-1 expression in response to hypoxia is dependent
on the intracellular redox status of metastatic bladder cancer
cells. Biochim Biophys Acta. 1852:2593–2602. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cai QW, Li J, Li XQ, Wang JQ and Huang Y:
Expression of STAT3, MMP-1 and TIMP-1 in gastric cancer and
correlation with pathological features. Mol Med Rep. 5:1438–1442.
2012.PubMed/NCBI
|
|
19
|
Juncker-Jensen A, Deryugina EI, Rimann I,
Zajac E, Kupriyanova TA, Engelholm LH and Quigley JP: Tumor MMP-1
activates endothelial PAR1 to facilitate vascular intravasation and
metastatic dissemination. Cancer Res. 73:4196–4211. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Anand M, Van Meter TE and Fillmore HL:
Epidermal growth factor induces matrix metalloproteinase-1 (MMP-1)
expression and invasion in glioma cell lines via the MAPK pathway.
J Neurooncol. 104:679–687. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Scotto L, Narayan G, Nandula SV,
Arias-Pulido H, Subramaniyam S, Schneider A, Kaufmann AM, Wright
JD, Pothuri B, Mansukhani M, et al: Identification of copy number
gain and overexpressed genes on chromosome arm 20q by an
integrative genomic approach in cervical cancer: Potential role in
progression. Genes Chromosomes Cancer. 47:755–765. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar
|
|
23
|
McCarthy DJ, Chen Y and Smyth GK:
Differential expression analysis of multifactor RNA-Seq experiments
with respect to biological variation. Nucleic Acids Res.
40:4288–4297. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huber W, Carey VJ, Gentleman R and Anders
S: Orchestrating high-throughput genomic analysis with
Bioconductor. Nat Methods. 12:115–121. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mootha VK, Lindgren CM, Eriksson KF,
Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E,
Ridderstråle M, Laurila E, et al: PGC-1alpha-responsive genes
involved in oxidative phosphorylation are coordinately
downregulated in human diabetes. Nat Genet. 34:267–273. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES, et al: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Aubrey BJ, Kelly GL, Kueh AJ, Brennan MS,
O'Connor L, Milla L, Wilcox S, Tai L, Strasser A and Herold MJ: An
inducible lentiviral guide RNA platform enables the identification
of tumor-essential genes and tumor-promoting mutations in vivo.
Cell Reports. 10:1422–1432. 2015. View Article : Google Scholar
|
|
29
|
Li X, Tian R, Gao H, Yang Y, Williams BRG,
Gantier MP, McMillan NAJ, Xu D, Hu Y and Gao Y: Identification of a
histone family gene signature for predicting the prognosis of
cervical cancer patients. Sci Rep. 7:164952017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
31
|
Michalik L, Desvergne B and Wahli W:
Peroxisome-proliferator-activated receptors and cancers: Complex
stories. Nat Rev Cancer. 4:61–70. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cangemi A, Fanale D, Rinaldi G, Bazan V,
Galvano A, Perez A, Barraco N, Massihnia D, Castiglia M, Vieni S,
et al: Dietary restriction: Could it be considered as speed bump on
tumor progression road. Tumour Biol. 37:7109–7118. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
O'Sullivan KE, Reynolds JV, O'Hanlon C,
O'Sullivan JN and Lysaght J: Could signal transducer and activator
of transcription 3 be a therapeutic target in obesity-related
gastrointestinal malignancy. J Gastrointest Cancer. 45:1–11. 2014.
View Article : Google Scholar
|
|
34
|
Zhu P, Goh YY, Chin HF, Kersten S and Tan
NS: Angiopoietin-like 4: A decade of research. Biosci Rep.
32:211–219. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pencik J, Schlederer M, Gruber W, Unger C,
Walker SM, Chalaris A, Marié IJ, Hassler MR, Javaheri T, Aksoy O,
et al: STAT3 regulated ARF expression suppresses prostate cancer
metastasis. Nat Commun. 6:77362015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zheng T, Hong X, Wang J, Pei T, Liang Y,
Yin D, Song R, Song X, Lu Z, Qi S, et al: Gankyrin promotes tumor
growth and metastasis through activation of IL-6/STAT3 signaling in
human cholangiocarcinoma. Hepatology. 59:935–946. 2014. View Article : Google Scholar
|
|
37
|
Trombetta A, Maggiora M, Martinasso G,
Cotogni P, Canuto RA and Muzio G: Arachidonic and docosahexaenoic
acids reduce the growth of A549 human lung-tumor cells increasing
lipid peroxidation and PPARs. Chem Biol Interact. 165:239–250.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Margeli A, Kouraklis G and Theocharis S:
Peroxisome proliferator activated receptor-gamma (PPAR-gamma)
ligands and angiogenesis. Angiogenesis. 6:165–169. 2003. View Article : Google Scholar
|
|
39
|
George A, Ranganathan K and Rao UK:
Expression of MMP-1 in histopathological different grades of oral
squamous cell carcinoma and in normal buccal mucosa - an
immunohistochemical study. Cancer Biomark. 7:275–283. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Langenskiöld M, Ivarsson ML, Holmdahl L,
Falk P, Kåbjörn-Gustafsson C and Angenete E: Intestinal mucosal
MMP-1 - a prognostic factor in colon cancer. Scand J Gastroenterol.
48:563–569. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hua H, Li M, Luo T, Yin Y and Jiang Y:
Matrix metalloproteinases in tumorigenesis: An evolving paradigm.
Cell Mol Life Sci. 68:3853–3868. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang Z, Tao D, Zhang P, Liu X, Zhang Y,
Cheng J, Yan H, Liu L and Jiang H: Hyaluronan synthase 2 expressed
by cancer-associated fibroblasts promotes oral cancer invasion. Exp
Clin Cancer Res. 35:1812016. View Article : Google Scholar
|
|
43
|
Panagopoulos V, Leach DA, Zinonos I,
Ponomarev V, Licari G, Liapis V, Ingman WV, Anderson P, DeNichilo
MO and Evdokiou A: Inflammatory peroxidases promote breast cancer
progression in mice via regulation of the tumour microenvironment.
Int J Oncol. 50:1191–1200. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kuasne H, Barros-Filho MC, Busso-Lopes A,
Marchi FA, Pinheiro M, Muñoz JJ, Scapulatempo-Neto C, Faria EF,
Guimarães GC, Lopes A, et al: Integrative miRNA and mRNA analysis
in penile carcinomas reveals markers and pathways with potential
clinical impact. Oncotarget. 8:15294–15306. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yokoi A, Yoshioka Y, Yamamoto Y, Ishikawa
M, Ikeda SI, Kato T, Kiyono T, Takeshita F, Kajiyama H, Kikkawa F,
et al: Malignant extracellular vesicles carrying MMP1 mRNA
facilitate peritoneal dissemination in ovarian cancer. Nat Commun.
8:144702017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ismail TM, Bennett D, Platt-Higgins AM,
Al-Medhity M, Barraclough R and Rudland PS: S100A4 elevation
empowers expression of metastasis effector molecules in human
breast cancer. Cancer Res. 77:780–789. 2017. View Article : Google Scholar :
|
|
47
|
Sun N-x, Zhao Q, Ye C, Ma Y and Li W: Role
of matrix metal-loproteinase-1 (MMP-1)/protease-activated
receptor-1 (PAR-1) signaling pathway in the cervical cancer
invasion. J Reprod Contracept. 25:18–25. 2014.
|
|
48
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yang L, Han S and Sun Y: An IL6-STAT3 loop
mediates resistance to PI3K inhibitors by inducing
epithelial-mesenchymal transition and cancer stem cell expansion in
human breast cancer cells. Biochem Biophys Res Commun. 453:582–587.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Tse JC and Kalluri R: Mechanisms of
metastasis: Epithelial-to-mesenchymal transition and contribution
of tumor microenvironment. J Cell Biochem. 101:816–829. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Huang L, Huang Z, Fan Y, He L, Ye M, Shi
K, Ji B, Huang J, Wang Y and Li Q: FOXC1 promotes proliferation and
epithelial-mesenchymal transition in cervical carcinoma through the
PI3K-AKT signal pathway. Am J Transl Res. 9:1297–1306.
2017.PubMed/NCBI
|
|
52
|
Shostak K, Zhang X, Hubert P, Göktuna SI,
Jiang Z, Klevernic I, Hildebrand J, Roncarati P, Hennuy B, Ladang
A, et al: NF-κB-induced KIAA1199 promotes survival through EGFR
signalling. Nat Commun. 5:52322014. View Article : Google Scholar
|
|
53
|
Iida J and McCarthy JB: Expression of
collagenase-1 (MMP-1) promotes melanoma growth through the
generation of active transforming growth factor-beta. Melanoma Res.
17:205–213. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Weiss MB, Abel EV, Mayberry MM, Basile KJ,
Berger AC and Aplin AE: TWIST1 is an ERK1/2 effector that promotes
invasion and regulates MMP-1 expression in human melanoma cells.
Cancer Res. 72:6382–6392. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Shen CJ, Kuo YL, Chen CC, Chen MJ and
Cheng YM: MMP1 expression is activated by Slug and enhances
multi-drug resistance (MDR) in breast cancer. PLoS One.
12:e01744872017. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Mueller E, Sarraf P, Tontonoz P, Evans RM,
Martin KJ, Zhang M, Fletcher C, Singer S and Spiegelman BM:
Terminal differentiation of human breast cancer through PPAR gamma.
Mol Cell. 1:465–470. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhang YX and Zhao YL: Pathogenic network
analysis predicts candidate genes for cervical cancer. Comput Math
Methods Med. 2016:31860512016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Rajkumar T, Sabitha K, Vijayalakshmi N,
Shirley S, Bose MV, Gopal G and Selvaluxmy G: Identification and
validation of genes involved in cervical tumourigenesis. BMC
Cancer. 11:802011. View Article : Google Scholar : PubMed/NCBI
|