Introduction
Cancer metastasis is the principal cause of
cancer-related mortality (1). The
processes of metastasis are highly complex and involve a series of
sequential steps that cancer cells must successfully complete,
including the shedding of cells from the primary tumor into the
blood circulation, survival of the circulating tumor cells in the
circulation, initial seeding at a new site, extravasation into the
surrounding tissue, initiation and maintenance of growth, and
vascularization of the metastatic tumor (2,3).
Although large primary tumors are able to shed millions of cancer
cells into the circulatory system every day, very few cells succeed
at establishing a metastatic site at a distant organ (2,4).
These data indicated that the overall process of metastasis is an
inefficient process. To survive in the blood or lymph and arrive at
the specific organs, the escaped cells must exhibit metabolic
flexibility to endure the nutrient-poor and acutely unfavorable
environments (5,6).
Extensive reprogramming of cellular energy
metabolism is a hallmark of cancer (7,8). The
deregulation of cellular energetics (also called metabolic
reprogramming) involves tumor cells ‘rewiring’ their metabolic
pathways to support rapid proliferation, continuous growth,
metastasis, survival and therapy resistance (9-11).
Oxidative phosphorylation (OXPHOS) and glycolysis are the two
principal energy production pathways in the cell (12). Most cancer cells exhibit a high
rate of glycolysis, which not only compensates for the increased
demand for adenosine 5'-triphosphate (ATP), but also contributes to
cell proliferation and survival by affecting signaling pathways and
enhancing the production of macromolecules, including proteins,
nucleic acids and lipids (9,13,14).
Furthermore, tumor cells undergo considerable metabolic
reprogramming to survive and metastasize within the hostile
environment and the limited nutrient supply of solid tumors
(15). Although it has been
suggested that the extensive reprogramming of the cellular energy
metabolism is a hallmark of cancer (14,16),
the potential molecular mechanisms by which metabolic reprogramming
mediates cancer cells invasion and metastasis have yet to be
elucidated. Differing from primary tumor cells, metastatic tumor
cells exhibit specific metabolic characteristics that interact with
the tissue microenvironment and are required for forming metastases
in distant organs (8). However,
metabolic changes associated with metastasis between the different
environments of primary tumor cells and metastatic tumor cells
remain unclear.
Epithelial-mesenchymal transition (EMT) is an
important cellular process, during which cells lose epithelial
characteristics and acquire mesenchymal invasive properties and
stem cell-like features (17,18).
For example, the expression of the epithelial cell marker
Epithelial cadherin (CDH1) is lost, whereas expression of the
mesenchymal cell markers Neural cadherin (CDH2), fibronectin (FN)
and Vimentin (VIM), and the stem cell marker CD133 are induced. The
EMT process is very complex and controlled by different EMT
regulators such as Zinc-finger protein SNAI1, Zinc-finger
E-box-binding homeobox (ZEB), hypoxia-inducible factor (HIF)-1α and
Twist (19,20). During the progression to metastatic
competence, cancer cells acquire mesenchymal gene expression
patterns and properties that confer migratory and invasive
abilities, alter adhesive properties, prevent apoptosis and
senescence, and activate proteolysis and motility (19,21).
In the present study, two cell lines, SW480 and
SW620, were selected. The cells were originally derived from the
same colon cancer patient; SW480 cells were obtained from a primary
tumor and SW620 cells from a lymph node metastasis (22). ATP levels and the metastatic
ability and of the two cell lines were investigated to determine
the different bioenergetic adaptations in the same circumstances.
In addition, the expression levels of the glycolysis regulatory
proteins hexokinase (HK)1 and HK2, the cell cycle regulators
cyclin-dependent kinase (CDK)-2 and CDK4, as well as EMT-related
mRNAs and metastasis-associated proteins, cell apoptosis,
mitochondrial membrane potential (ΔΨm) and the sensitivity to fetal
bovine serum (FBS) of the two cell lines were determined, to reveal
the adaptive mechanisms exhibited by SW620 cells that may confer a
higher metastatic ability when confronted with hostile
environments. The present study provided a novel framework for
understanding the metastatic cancer cells adaptations to hostile
microenvironments and the formation of metastases.
Materials and methods
Antibodies and reagents
Phycoerythrin (PE)-conjugated mouse anti-human CD29
(cat. no. 557332), allophyco-cyanin-conjugated CD44 (cat. no.
559942), fluorescein isothiocyanate (FITC)-conjugated CD47 (cat.
no. 556045), CD54-PE (cat. no. 555511), VIM-PE (cat. no. 562337),
CD49d-FITC (cat. no. 560840), CD49e-PE (cat. no. 555617),
CD51/61-PE (cat. no. 550037), Alexa Fluor 647-conjugated FN (cat.
no. 563098), CD133-PE (cat. no. 566593), CD166-PE (cat. no.
559263), CD324-PE (cat. no. 562870), CD325-Alexa Fluor 488 (cat.
no. 562119) and CD326-PE (cat. no. 347198) and rat anti-human
CD49f-PE (cat. no. 555736) antibodies were purchased from BD
Pharmingen (BD Biosciences, Franklin Lakes, NJ, USA). Rabbit
anti-human p21 (cat. no. 2947), p27 (cat. no. 3686), CDK2 (cat. no.
2546), CDK4 (cat. no. 12790), Cyclin D1 (cat. no. 2978),
phosphorylated (p)-cell-division control protein 2 (CDC2; also
known as CDK1; cat. no. 4539), β-actin (cat. no. 4970), mouse
anti-human CDK6 (cat. no. 3136), Cyclin D3 (cat. no. 2936) and
β-actin (cat. no. 3700) antibodies were purchased from Cell
Signaling Technology, Inc. (Danvers, MA, USA). Mouse anti-HK1 (cat.
no. BM1910) and mouse anti-human HK2 (cat. no. BM1911) antibodies
were purchased from Boster Biological Technology (Pleasanton, CA,
USA). Rabbit anti-human HIF-1α (cat. no. WL01607) and glucose
transporter type (GLUT)-1 (cat. no. WL01163) antibodies and an
Annexin V-Light 650/propidium iodide (PI) Reagent kit (cat. no.
WLA002a) were from Wanleibio Co., Ltd (Shanghai, China).
Horseradish peroxidase (HRP)-conjugated goat anti-mouse (cat. no.
70-GAM007) and goat anti-rabbit (cat. no. 70-GAR007) secondary
antibodies were purchased from Hangzhou MultiSciences (Lianke)
Biotech Co., Ltd. (Hangzhou, China). Oligomycin A (Oligo A) was
purchased from Selleck Chemicals (Houston, TX, USA). An ATP assay
kit was purchased from PerkinElmer, Inc. (Waltham, MA, USA). FBS,
L-15 medium and TRIzol® reagent were obtained from
Thermo Fisher Scientific, Inc. (Waltham, MA, USA). PI, McCoy's 5A
media, 2-Deoxy-D-glucose (2-DG), dimethyl sulfoxide, Triton X-100,
RNase, 4% paraformaldehyde, 3,3'-dihexyloxacarbocyanine iodide
[DiOC6(3)] and a Bicinchoninic
Acid (BCA) Protein assay kit were purchased from Sigma-Aldrich
(Merck KGaA, Darmstadt, Germany). Trypsin-EDTA (0.25%),
penicillin/streptomycin (100X), tetramethylethylenediamine,
ammonium persulfate, tris(hydroxymethyl)aminomethane, glycine,
Tween-20, sodium dodecyl sulfate (SDS), an Efficient
Chemiluminescence kit, radioimmunoprecipitation assay (RIPA) lysis
buffer and bovine serum albumin were obtained from Gen-view
Scientific, Inc. (El Monte, FL, USA). A PrimeScript RT Reagent kit
and SYBR Premix Ex Taq PCR kit were purchased from Takara
Bio, Inc. (Otsu, Japan). Polyvinylidene difluoride (PVDF)
membranes, 30% polyacrylamide and dithiothreitol were purchased
from Bio-Rad Laboratories, Inc. (Hercules, CA, USA).
Cell culture
The human colon cancer cell lines SW480 and SW620
were purchased from the Type Culture Collection of the Chinese
Academy of Sciences (Shanghai, China) and cultured in L-15 medium
containing 10% FBS and 1% penicillin/streptomycin. All cells were
maintained at 37°C in a humidified atmosphere of 5% CO2,
and harvested with 0.25% trypsin before use.
ATP levels assay
Intracellular ATP concentrations were determined by
luminescence assay. SW480 and SW620 cells (1×104
cells/well) were sorted and collected in 96-well plates using a BD
FACSAria III flow cytometer (BD Biosciences). To determine the
effects of Oligo A on the intracellular ATP levels over time, cells
were cultured in L-15 containing 1 µM Oligo A, 1% FBS and 3
mg/ml glucose at 37°C for 0, 0.5, 1, 2, 4, 6, 8, 12, 24, 36, 48 and
60 h. To evaluate the effects of Oligo A or 2-DG on the
intracellular ATP levels over time, SW620 cells (1×104
cells/well) were sorted and collected in 96-well plates as
aforementioned. Cells were cultured in L-15 containing 1 µM
Oligo A, 1% FBS and 3 mg/ml glucose or L-15 containing 1% FBS and 3
mg/ml 2-DG at 37°C for 0, 0.5, 1, 2, 4, 6, 8, 12, 24, 36, 48 and 60
h.
To detect the ATP synthesis ability of the two cell
lines in different microenvironment, cells were cultured for 24 h
at 37°C in different media conditions, as follows: i) NOR, in which
cells were cultured in L-15 containing 1% FBS and 3 mg/ml glucose,
and cells metabolized through OXPHOS and glycolysis; ii) OXPHOS, in
which cells were cultured in L-15 containing 1% FBS without
glucose, and cells metabolized through OXPHOS; iii) GLY, in which
cells were cultured in L-15 containing 1% FBS, 3 mg/ml glucose and
1 µM Oligo A, and cells metabolized through glycolysis; and
iv) UN-(O+G), L-15 containing 1% FBS and 1 µM Oligo A, in
which cells did not metabolize through either OXPHOS or
glycolysis.
ATP production was determined with the ATPlite
1-step Luminescence ATP Detection Assay system (PerkinElmer, Inc.),
according to the manufacturer's protocol. Briefly, following
culture in different media in 96-well plate, ATP assay solution
(100 µl) was added to each well and shaken at 400 rpm, 37°C
for 5 min. Subsequently, cells were incubated in the dark for 10
min at 37°C and luminescence was measured with a TECAN M200 PRO
microplate luminometer at the wavelength of 540-600 nm (Tecan
Group, Ltd., Männedorf, Switzerland).
Cell surface proteins and intracellular
proteins analysis
Expression of the cell surface proteins CD29, CD44,
CD47, CD54, CD49d, CD49e, CD49f, CD51/61, CD133, CD166, CD324,
CD325 and CD326, and the intracellular proteins VIM and FN1 were
detected by BD FACSAria III flow cytometry. Briefly, SW480 and
SW620 cells (5x105 cells/well) were seeded in 6-well
plate and cultured with L-15 media containing 10% FBS and 1%
penicillin/streptomycin at 37°C for 24 h. Subsequently, cells were
collected and washed with phosphate-buffered saline (PBS) three
times. The cells were centrifuged and resuspended with 200
µl staining buffer (PBS + 1% FBS). Anti-cell surface protein
antibodies were added (all 1:10) and incubated at 4°C in the dark
for 20 min. For intracellular proteins, cells were fixed with
Fixation Buffer (BD Biosciences) for 20 min at 4°C. The cells were
washed with PBS and permeabilized with Perm Buffer III (BD
Biosciences) for 30 min at room temperature. Subsequently, cells
were washed with Perm/Wash Buffer I (BD Biosciences) and stained
with mouse anti-human VIM-PE and mouse anti-human FN-Alexa Fluor
647 antibodies. Background staining was determined by staining with
the respective isotype control antibodies. Following staining,
cells were washed with staining buffer and resuspended in 500
µl of the staining buffer. Flow cytometric analysis was
performed on the BD FACSAria III. Data were acquired with BD
FACSDiva software 6.0, and the obtained data were analyzed with
FlowJo software 7.6 (FlowJo LLC, Ashland, OR, USA).
Cell apoptosis analysis
Apoptosis was examined using the Annexin V-Light
650/PI Reagent kit, according to the manufacturer's protocol. SW480
and SW620 cells (5×105 cells/well) were seeded in 6-well
plates and cultured at 37°C for 24 h in the media conditions
aforementioned: i) NOR; ii) OXPHOS; iii) GLY; iv) UN-(O+G).
Following incubation, cells were collected and washed twice with
PBS. Subsequently, cells were stained with 5 µl
Annexin-V-Light 650 and 5 µl PI at room temperature for 15
min in the dark. Cells were analyzed with a BD FACSAria III flow
cytometer and the obtained data were analyzed with FlowJo software
7.6.
Mitochondrial membrane potential (∆Ψm)
measurement
∆Ψm was detected by flow cytometry using
DiOC6(3), as described in our
previous study (23). Briefly,
SW480 and SW620 cells were seeded (5×105 cells/well) in
6-well plates and cultivated in the media conditions
aforementioned: i) UN-(O+G) and ii) GLY. Cells were cultured for 0,
1, 4, 6, 8 h and 24 h at 37°C. Cells were harvested and incubated
with 3 µM DiOC6(3) at room
temperature for 20 min in the dark and subsequently analyzed by the
BD FACSAria III. Data were acquired with BD FACSDiva software 6.0,
and the obtained data were analyzed with FlowJo software 7.6.
Western blot analysis
To evaluate the expression levels of
glycolysis-regulated proteins HIF-1α, HK1, HK2 and GLUT1 in
different microenvironments, western blotting was performed as
previously described (24). SW480
and SW620 cells (5×105 cells/well) were seeded in 6-well
plates and cultivated in three different media conditions, as
aforementioned, at 37°C for 24 h: i) OXPHOS; ii) NOR: and iii) GLY.
To investigate the expression of associated proteins in condition
of glycolysis, ‘OXPHOS’ group served as the control group. To
investigate the expression of glycolysis regulatory proteins (HK1
and HK2) and cell cycle regulators (CDK2, CDK4, CDK6, p21 and p27)
over time when OXPHOS was suppressed, SW480 and SW620 cells
(5×105 cells/well) were seeded in 6-well plates and
cultured in L-15 supplemented with 1 µM Oligo A, 1% FBS and
3 mg/ml glucose at 37°C for 0, 1, 4, 8, 24 and 48 h. To determine
the expression of cell cycle regulators (CDK6, Cyclin D3, Cyclin
D1, pCDC2 and CDK4) in different microenvironment, cells were
cultured in three different media named above as OXPHOS, GLY and
NOR.
Following incubations, cells (1×106
cells/well) were rinsed with PBS and lysed with RIPA lysis buffer
on the ice for 5 min. The lysates were centrifuged at 18,000 × g at
4°C for 15 min. Protein concentrations were determined with the BCA
protein assay kit. The samples were denatured with SDS running
buffer [50 mM Tris-Cl (pH 6.8), 2% (w/v) SDS, 0.1% (w/v)
bromophenol blue, 20% (v/v) glycerol and 200 mM dithiothreitol] at
100°C for 5 min. Proteins (1 mg/ml) were separated by 10% SDS-PAGE
and transferred onto PVDF membranes. The membranes were blocked in
5% skim milk for 1 h at room temperature. The membranes were
incubated with the following primary antibodies overnight at 4°C:
Anti-HK1, anti-HK2 and anti-HIF-1α (all 1:200 dilution);
anti-GLUT1, anti-CDK2, anti-CDK4, anti-CDK6, anti-pCDC2,
anti-Cyclin D1, anti-Cyclin D3, p21, p27 and the loading control
mouse anti-human and rabbit anti-human β-actin (all 1:1,000).
Following washing with TBS + Tween-20, the membranes were incubated
with an HRP-conjugated goat anti-rabbit (1:10,000 dilution) or goat
anti-mouse (1:10,000) secondary antibody for 1 h at room
temperature. Visualization was performed on ChemiDoc XPS system
(Bio-Rad Laboratories, Inc.) using the Efficient Chemiluminescence
kit. Densitometric analysis was performed with Image Lab 5.2.1
(Bio-Rad Laboratories, Inc.); relative protein expression levels
were normalized to β-actin.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
SW480 and SW620 (5×105 cells/ well) were seeded in
6-well plates and cultured at 37°C for 24 h in three different
media conditions, as aforementioned: i) OXPHOS; ii) NOR; and iii)
GLY. RNA was extracted using TRIzol reagent, according to the
manufacturer's protocol, and resuspended in 20 µl RNase-free
water. Following determination of the RNA concentration with
Quawell Q5000 Micro-volume Spectrophotometer (Quawell Technology,
Inc., San Jose, CA, USA), RNA was reverse transcribed into cDNA
using the PrimeScript RT Reagent kit. cDNA was used for each of
three replicates of qPCR using the SYBR Premix Ex Taq PCR
kit and a CFX96 Real-Time PCR Detection system (Bio-Rad
Laboratories, Inc.) with the following thermocycling conditions:
Initial denaturation at 95°C for 30 sec; followed by 40 cycles of
95°C (5 sec) and 60°C (30 sec). Target mRNA expression levels were
normalized to β-actin and calculated by the 2−ΔΔCq
method (25). Results are
presented as the mean ± standard deviation (SD) of three
independent experiments. mRNA expression levels were analyzed for
HK1, HK2, HIF-1α, CDH1, CDH2, ZEB1, SNAI1, SNAI2, FN1, VIM, CD133,
TWIST and β-actin; all primer sequences for PCR analysis are
presented in Table I.
 | Table IPrimer sequences used for reverse
transcription-quantitative polymerase chain reaction analyses of
human colorectal cancer cells. |
Table I
Primer sequences used for reverse
transcription-quantitative polymerase chain reaction analyses of
human colorectal cancer cells.
| Gene | Primer sequence
(5'→3') |
|---|
| HK1 | F:
AATGCTGGGAAACAAAGGT |
| R:
AGAGGAATCCCTTCTTGGG |
| HK2 | F:
CCTCGGTTTCCCAACTCTGC |
| R:
GCTCCAAGCCCTTTCTCCAT |
| HIF-1α | F:
CGTTCCTTCGATCAGTTGTC |
| R:
TCAGTGGTGGCAGTGGTAGT |
| CDH1 | F:
CCGAGGACTTTGGCGTGGGC |
| R:
TCCCTGTCCAGCTCAGCCCG |
| CDH2 | F:
GACCCAAACAGCAACGACGGGT |
| R:
CTGAGGCGGGTGCTGAATTCCC |
| ZEB1 | F:
ATCCTGGGGCCTGAAGCTCAGG |
| R:
TGGTGTGCCCTGCCTCTGGT |
| SNAI1 | F:
GCTGCTACAAGGCCATGTCCGG |
| R:
CTTGGTGCTTGTGGAGCAGGGAC |
| SNAI2 | F:
GATGCCGCGCTCCTTCCTGG |
| R:
TGGAGCAGCGGTAGTCCACAC |
| FN1 | F:
TGCAAGGCCTCAGACCGGGT |
| R:
GCGCTCAGGCTTGTGGGTGT |
| VIM | F:
TTCCAAGCCTGACCTCACGGCTG |
| R:
TTCCGGTTGGCAGCCTCAGAGA |
| CD133 | F:
CAGAGTACAACGCCAAACCA |
| R:
AAATCACGATGAGGGTCAGC |
| TWIST | F:
TCCGCGTCCCACTAGCAGGC |
| R:
CGCCCCACGCCCTGTTTCTT |
| β-actin | F:
CAGAGTACAACGCCAAACCA |
| R:
AAATCACGATGAGGGTCAGC |
In addition, SW480 and SW620 cells (5×105
cells/well) were seeded in 6-well plates and cultivated in L-15
supplemented with 1 µM Oligo A, 1% FBS and 3 mg/ml glucose
at 37°C for 0, 1, 4, 8, 24 or 48 h, and the mRNA expression levels
of the aforementioned genes were detected by RT-qPCR.
FBS-free tolerance and sensitivity
analysis
The tolerance and sensitivity of SW480 and SW620
cells to FBS deprivation was determined by starvation culture.
Cells (5x105 cells/well) were seeded in 6-well plates
cultivated in FBS-free L-15 medium. The FBS-free medium was
replaced every 2 days. Cell morphology and viability were observed
under Zeiss Observer A1 microscope (Zeiss AG, Oberkochen,
Germany).
Wound-healing migration assay
A wound-healing assay was performed to assess the
migratory ability of SW480 and SW620 cells. Cells
(2.5×104 cells/well) were seeded onto a 24-well plate
and cultured for 24 h. Subsequently, a scratch was made across the
cell monolayer with a pipette tip. Cells were washed three times
with PBS and L-15 medium containing 0.5% FBS was added to each
well. Cells were cultured in incubator at 37°C and cell migration
within the scrape line was recorded at 0, 8, 24 and 48 h using a
light microscope (Zeiss AG); migration distance was used to
evaluate migratory ability.
In vivo metastasis analysis
BALB/c nude mice (n=12 females; age, 6-8 weeks;
weight, 20 g) were purchased from the Shanghai SLAC Laboratory
Animal Co., Ltd. (Shanghai, China). Mice were housed in clean,
pathogen-free rooms in an environment with controlled temperature
(26°C), filtered atmosphere and humidity (55%), with a 12-h
light/dark cycle, and provided free access to pellet food and water
in microinsulator cages. All animals used in investigations were
handled in accordance with the Guide for the Care and Use of
Laboratory Animals (National Research Council, 1996). The
Institutional Animal Care and Use Committee of Fuzhou University
(Fuzhou, China) reviewed and approved all animal use procedures
based on the above regulations, including the use of appropriate
species, quality and number of animals, avoidance or minimization
of discomfort, distress and pain of animals with a good scientific
basis, and use of appropriate anesthesia and euthanasia. BALB/c
nude mice were randomly allocated to two groups (n=6/group), which
received an intravenous injection of PBS (200 µl; pH 7.4)
containing either SW480 or SW620 cells (1.0x106 cells)
into the tail vein. At 30 days post-injection, mice were sacrificed
and the lungs were excised and stained with Bouin's solution at
room temperature for 24 h for pulmonary nodule counting.
Statistical analysis
Statistical analyses were performed with the SPSS
statistical software package 22 (IBM Corp., Armonk, NY, USA).
Results are expressed as the mean ± SD. Statistical significance
values were evaluated through one-way analysis of variance with a
Dunnett's post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
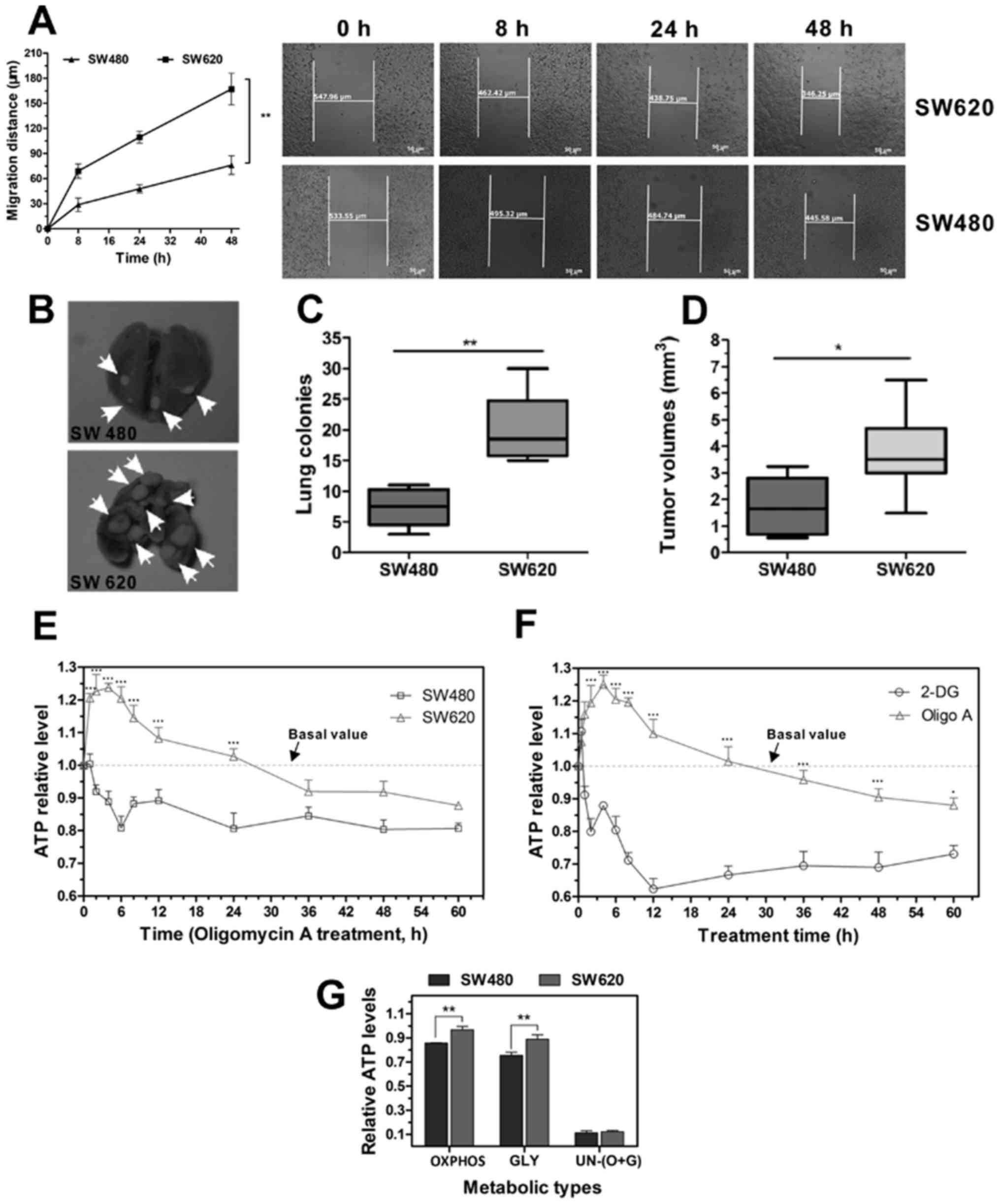
SW620 cells exhibit increased energy
metabolism and metastatic ability compared with SW480 cells
As previously mentioned, the SW480 cell line was
established from a primary tumor, and the SW620 cell line was
derived from a lymph metastatic lesion from the same patient
(22). Results from the
wound-healing assay indicated that SW620 cells exhibited a stronger
migratory ability compared with SW480 cells; SW620 cells migrated
further compared with the SW480 cells over the same period time
(Fig. 1A). In vivo
metastatic ability assay revealed that mice injected with SW620
cells had significantly higher numbers and total volumes of tumor
nodules in the lung compared with mice injected with SW480 cells
(Fig. 1B-D); tumor metastatic
lesions were not observed in other organs.
Metastatic cells that escape from the primary tumor
to distant organs, must possess certain traits that allow them to
survive in a new microenvironment, including its conditions of
nutrient scarcity (5). Cell
viability and survival depend on the efficiency of ATP production
through OXPHOS or glycolysis. To determine the differences in
metabolic performance between SW480 and SW620, cells were treated
with Oligo A, an inhibitor of ATP synthase and the OXPHOS pathway
(26), or with 2-DG, a glucose
molecule that has the 2-hydroxyl group replaced by hydrogen, and is
thus unable to undergo further glycolysis (27), to investigate the effect on
intracellular ATP levels. When OXPHOS was inhibited, the ATP levels
of SW480 cells decreased rapidly and remained stable between 24 and
60 h post-treatment (Fig. 1E); ATP
levels were maintained at ~0.8 of the basal value (basal value was
determined from the same cells cultured in L-15 medium containing
FBS and glucose, without Oligo A or 2-DG; in these conditions cell
metabolism is through both OXPHOS and glycolysis). By contrast, at
4 h following Oligo A treatment, the ATP levels of SW620 cells
increased to 1.2 of the basal value and decreased to the basal
value at 24 h following treatment and remained stable at ~0.9 of
the basal value between 36 and 60 h post-treatment. Therefore, it
was speculated that when treated with Oligo A in the first 24 h,
SW620 cells may be able to initiate emergency cellular metabolism
pathways to compensate for the depleted energy supply.
To evaluate whether SW620 cells upregulated OXPHOS
when glycolysis was suppressed, SW620 cells were treated with 2-DG.
It was determined that the ATP levels were below the basal value at
all time points because of the action of 2-DG (Fig. 1F). A similar ATP suppression effect
was not observed in the Oligo A group. These data suggested that
glycolysis may serve a key role when cells are confronted with
microenvironmental changes.
OXPHOS and glycolysis are the two principal
metabolism processes to maintain cell viability and biosynthesis.
In the present study, the potency of OXPHOS and glycolysis were
compared in the two cell lines. Glycolysis and OXPHOS were
inhibited by 2-DG or Oligo A, respectively. Cells were treated with
2-DG for 24 h; the ATP levels in SW480 cells were 0.856 of the NOR,
whereas SW620 cells exhibited a higher level, at 0.967 of the NOR
(Fig. 1G). Similarly, when cells
were treated with Oligo A and metabolized through glycolysis, the
ATP levels were 0.755 and 0.890 of the NOR in SW480 and SW620
cells, respectively. When both OXPHOS and glycolysis were
suppressed at the same time, ATP levels in the two cell lines
dropped to only 0.10 of the NOR. These results indicated that SW620
cells exhibited stronger OXPHOS and glycolysis metabolism potential
compared with SW480 cells.
In summary, SW620 cells exhibited higher energy
metabolism capability compared with SW480 cells; when the cells
were confronted with an energy supply shortage, SW620 cells
exhibited a stronger capability for OXPHOS and glycolysis.
SW620 cells express high levels of
glycolysis-regulated proteins
To further investigate the association between ATP
levels and glycolysis, SW480 and SW620 cells were exposed to three
different environments: i) OXPHOS, ii) NOR and iii) GLY. The
expression levels of regulatory proteins of glycolysis, including
HK1, HK2, GLUT1 and HIF-1α, were measured by western blot analysis.
In the presence of glycolysis metabolism (NOR and GLY), the
expression levels of HK1, HK2 and GLUT1 proteins in SW620 cells
were all higher compared with the OXPHOS group, whereas the
expression levels in SW480 cells were all decreased compared with
the OXPHOS group (Fig. 2A-F). In
addition, the expression levels of HIF-1α, HK1, HK2 and GLUT1
proteins in NOR and GLY treatments in SW620 cells were all
significantly higher compared with the respective expression levels
in SW480; OXPHOS was used as the control. These results suggested,
compared with SW480 cells, SW620 cells are more likely to
metabolize through glycolysis when glucose is sufficient.
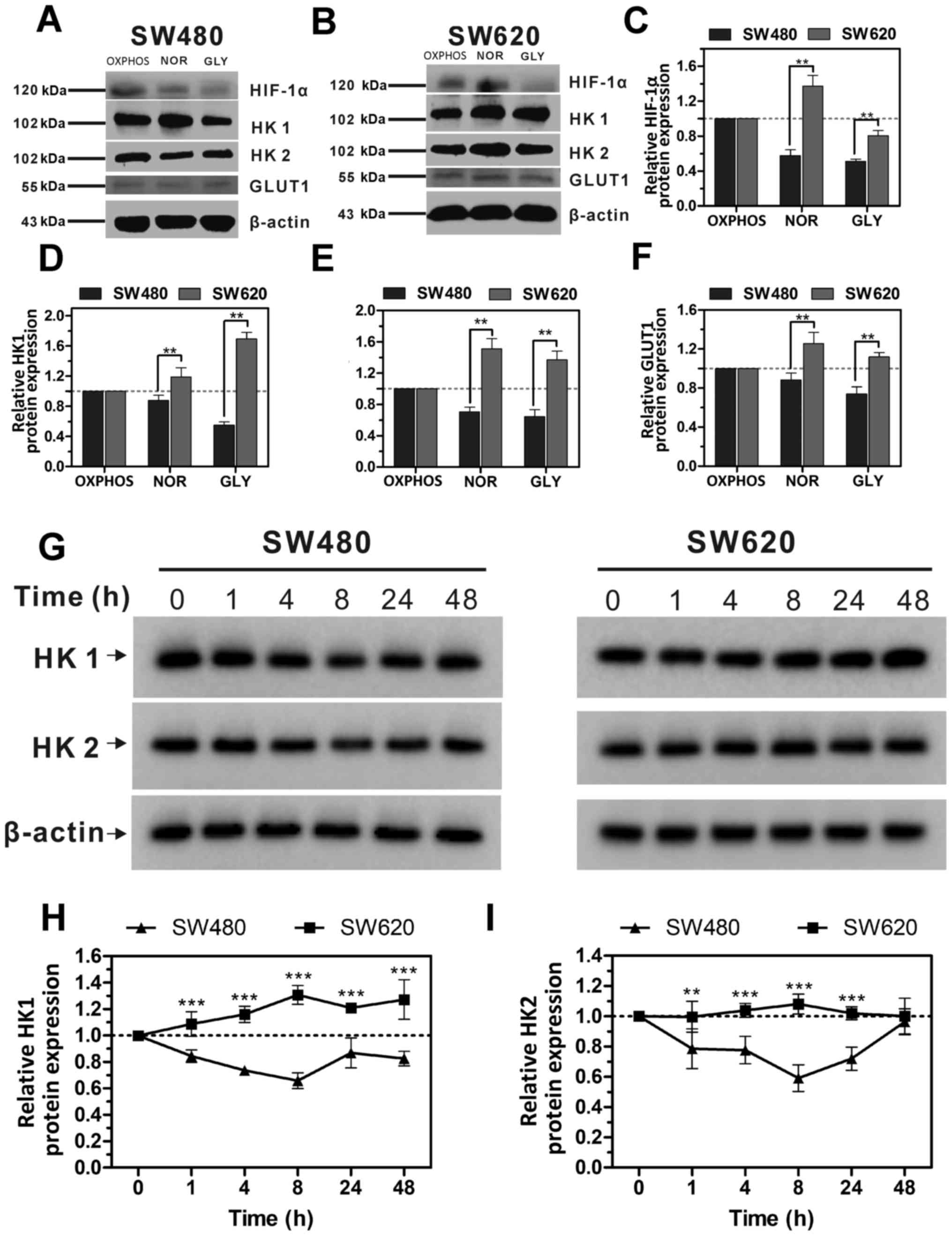 | Figure 2Expression levels of glycolysis
regulation proteins in SW480 and SW620 cells with different
metabolic types. (A and B) Representative western blotting images
of HIF-1α, HK1, HK2 and GLUT1 protein expressions in (A) SW480 and
(B) SW620 cells cultured in different media. (C-F) Protein
expression levels from (A) and (B) were quantified using Image Lab
analysis software; the relative expression levels of HIF-1α, HK1,
HK2 and GLUT1 were calculated as a ratio of OXPHOS; β-actin was
used as the loading control. (G) Western blot analysis of HK1 and
HK2 protein expression levels in SW480 and SW620 cells treated with
Oligomycin A (1 µM) for 0-48 h. (G) Representative western
blots indicating the expression of HK1 and HK2 in SW480 and SW620
cells at different treatment times. (H and I) Protein expression
levels from (G) were quantified using Image Lab analysis software,
and relative expression levels of (H) HK1 and (I) HK2 were
calculated as a ratio of 0 h; β-actin was used as a loading
control. Data are presented as the mean ± standard deviation; n=3;
**P<0.01 and ***P<0.001 vs. SW480.
GLUT1, glucose transporter type 1; HIF-1α, hypoxia-inducible
factor; HK, hexokinase; NOR, normal; GLY, glycolysis; OXPHOS,
oxidative phosphorylation. |
In SW480 and SW620 cells treated with Oligo A for 48
h, the protein expression levels of HK1 and HK2 in SW480 cells
gradually decreased in the first 8 h of treatment and increased
slightly at 24 and 48 h; however, expression remained below the
basal value (0 h post-treatment; Fig.
2G-I). By contrast, the expression of HK1 in SW620 cells
increased gradually in the first 8 h of treatment and subsequently
reduced slightly, remaining above the basal value (the expression
levels at 0 h) up to 48 h post-treatment (Fig. 2G and H). HK2 expression in SW620
did not exhibit any significant changes compared with 0 h (Fig. 2G-I). In conjunction with the data
presented in Fig. 1E, these
results indicated that when OXPHOS was suppressed, the
intracellular ATP levels were positively associated with HK1 and
HK2 expression.
Expression of cell cycle regulatory
proteins is significantly different in different
microenvironments
The study of metabolic regulations of the cell cycle
is a relatively new field that is gaining increasing attention, as
metabolic signals are integrated into, and coupled with, cell cycle
progression (10,28). In the present study, the effects of
different microenvironments on the cell cycle were
investigated.
Tumor-associated cell cycle defects are often
mediated by alterations in CDK activity (29). CDK catalytic activities are often
modulated by interactions with cyclins and CDK inhibitors (28). In the present study, the expression
profiles of cell cycle regulatory proteins were compared in SW480
and SW620 cells following exposure to three different
microenvironments. It was identified that the expression levels of
cell cycle regulatory proteins in these two cell lines were
significantly different (Fig. 3).
When cells metabolized by OXPHOS and glycolysis (NOR), the
expression of Cyclin D3, pcdc2, and CDK4 in SW620 cells was
significantly higher compared with SW480 cells, OXPHOS was used as
the control (Fig. 3). However,
when cells were metabolized by glycolysis (GLY), the expression of
Cyclin D3 and CDK4 in SW620 cells was significantly lower compared
with SW480 cells. Moreover, CDK6 and Cyclin D1 did not express in
SW480 cells but its expression decreased in SW620 cells. CDC2 is a
master regulator of mitosis, as it controls the centrosome cycle as
well as mitotic onset, and can be activated by phosphorylation
(30). Cyclin D1 and Cyclin D3
control the G1-S progression of the cell cycle in complex with CDK4
and CDK6 (28). These data
indicated that SW620 cells may manage their proliferation through
upregulating the expression of cyclins when energy supply is
sufficient. However, in the GLY environment, the expression of
Cyclin D3 and CDK4 in SW620 cells was significantly lower compared
with SW480 cells (Fig. 3C and E,
respectively). These results suggested that SW620 cells may inhibit
the expression of positive cell cycle regulatory proteins to
sustain cell survival when energy supplies are low.
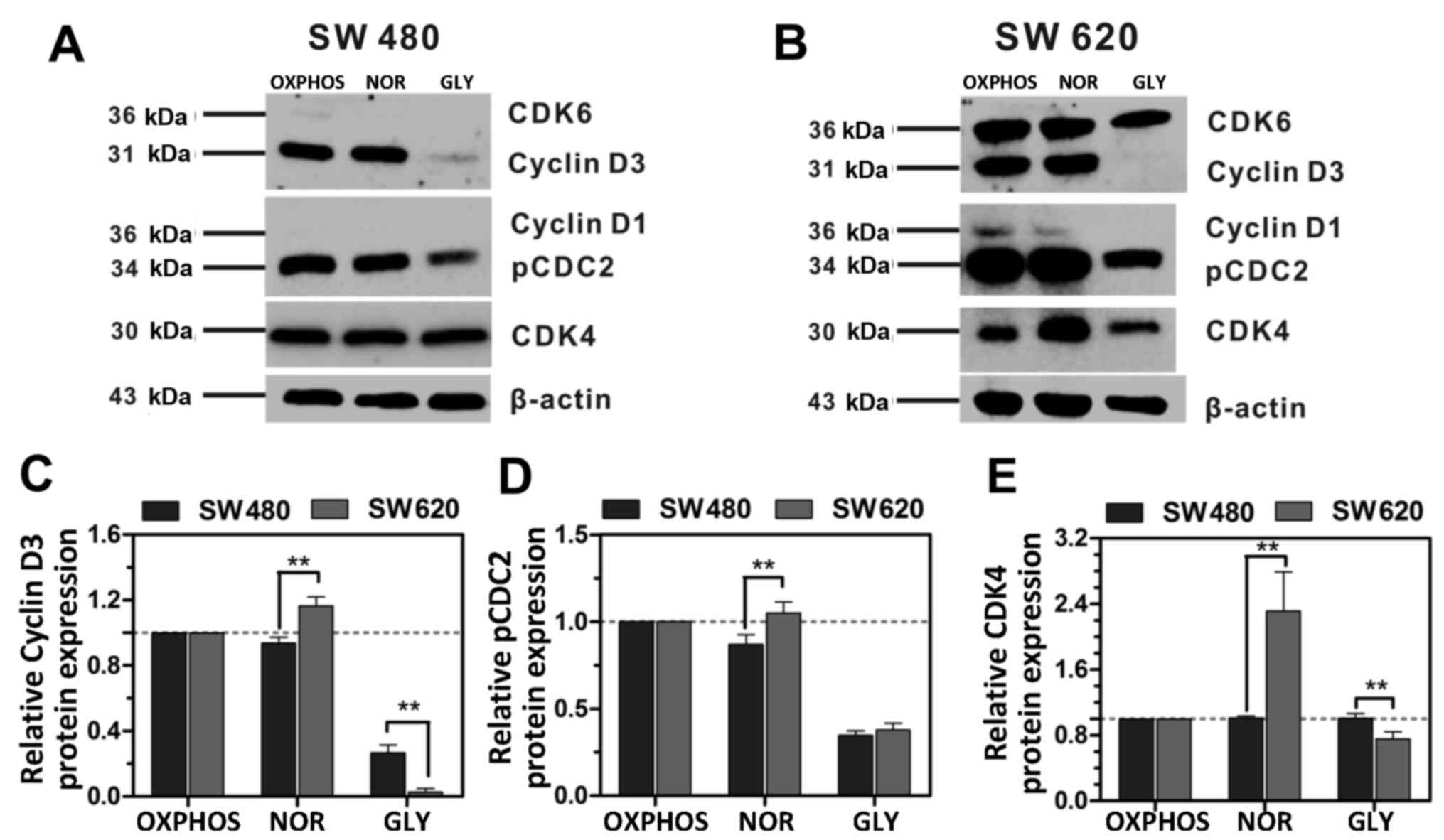 | Figure 3Expression of cell cycle regulation
proteins in SW480 and SW620 cells with different metabolic types.
(A and B) Representative western blotting images of CDK6, Cyclin
D3, Cyclin D1, pCDC2 and CDK4 protein expression in (A) SW480 and
(B) SW620 cells cultured in different media; β-actin was used as a
loading control. (C-E) Protein expression levels from (A) and (B)
were quantified using Image Lab analysis software, and relative
expression levels of (C) Cyclin D3, (D) pCDC2 and (E) CDK4 were
calculated as a ratio of the same protein in OXPHOS. Data are
presented as the mean ± standard deviation; n=3;
**P<0.01. CDC, cell division control protein; CDK,
cyclin-dependent kinase; NOR, normal; GLY, glycolysis; OXPH,
oxidative phosphorylation. |
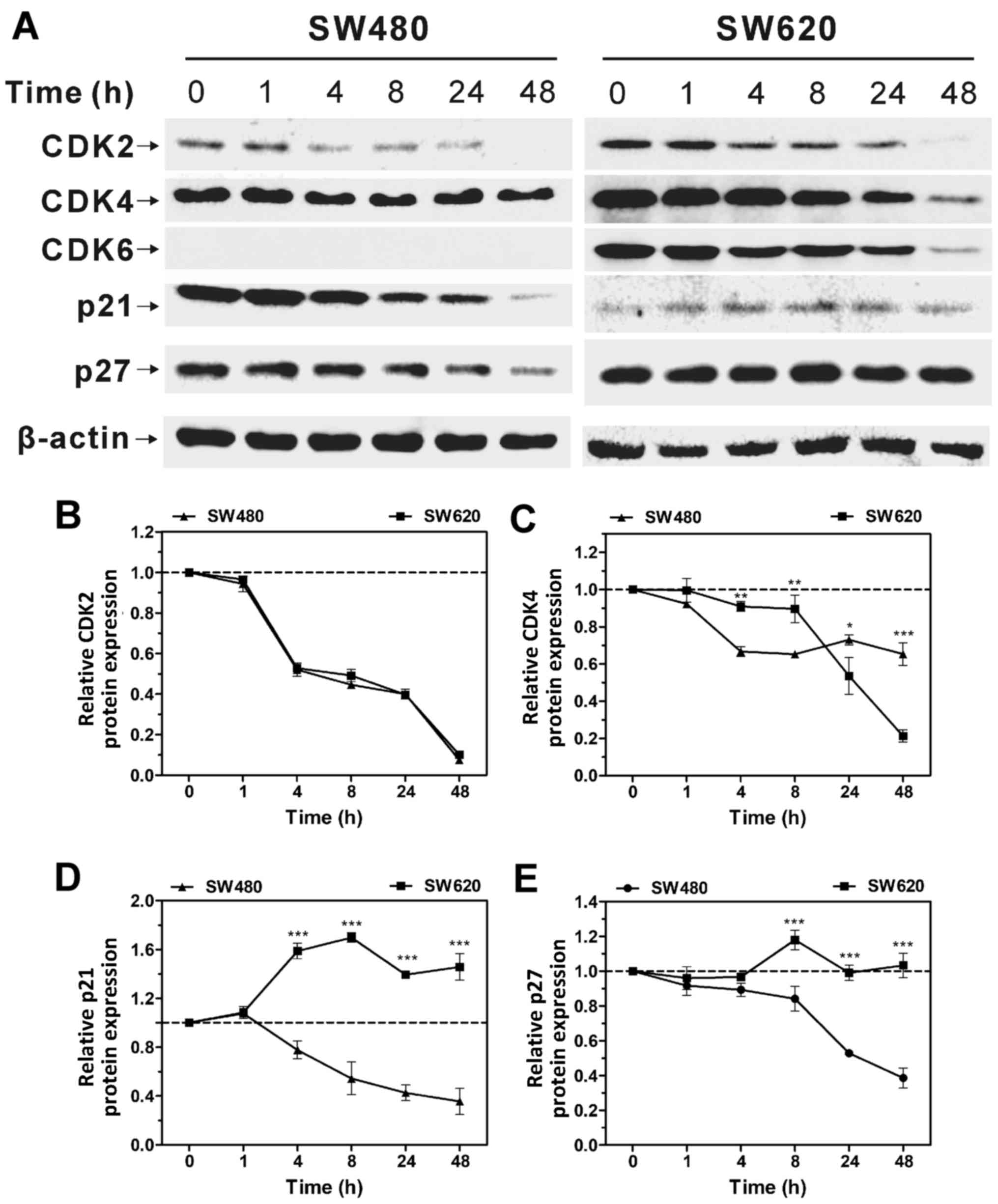
Oligo A was used to inhibit OXPHOS, and the changes
in cell cycle regulation were compared in SW480 and SW620 cells
over a 48-h period. The expression levels of CDK2 and CDK4 in SW480
and SW620 cells decreased over time (Fig. 4), which indicated that the cell
cycle was arrested when the expression of positive cell regulatory
proteins was inhibited. Similarly, the expression of CDK2 and CDK4
exhibited the same trend in SW620 cells when treated with Oligo A.
CDK6 expression was not detected in SW480 cells, whereas its
expression decreased in SW620 cells over time. Furthermore, protein
expression levels of the CDK inhibitors p21 and p27 were
upregulated in SW620 cells as the treatment time increased, whereas
their expression levels in SW480 cells decreased over time
(Fig. 4A, D and E). Cell cycle
upstream regulatory proteins p21 and p27 may inhibit the expression
of CDKs and promote cell cycle arrest (31,32).
These results indicated that cell cycle progression may be closely
associated with intracellular metabolism.
SW620 cells exhibit stronger adaptation
to different micro-environments compared with SW480 cells
The bioenergetics adaptability of SW480 and SW620
cells in different microenvironment was evaluated by apoptosis and
∆Ψm analysis. Cells in the NOR group metabolized through both
OXPHOS and glycolysis; both SW480 and SW620 maintain a considerable
proportion of viable cells (Fig.
5A). In the OXPH group, cells metabolized through OXPHOS and
the apoptotic rates increased. In the UN-(O+G) group, cells did not
metabolized through either OXPHOS or glycolysis; almost all cells
are apoptotic or necrotic. In the GLY group, cells metabolized
through glycolysis, and the proportion of apoptotic cells was
significantly lower in SW620 cells compared with that in SW480
cells. These results indicated that when the cells metabolized by
glycolysis, SW620 cells exhibited a stronger anti-apoptotic ability
compared with SW480 cells.
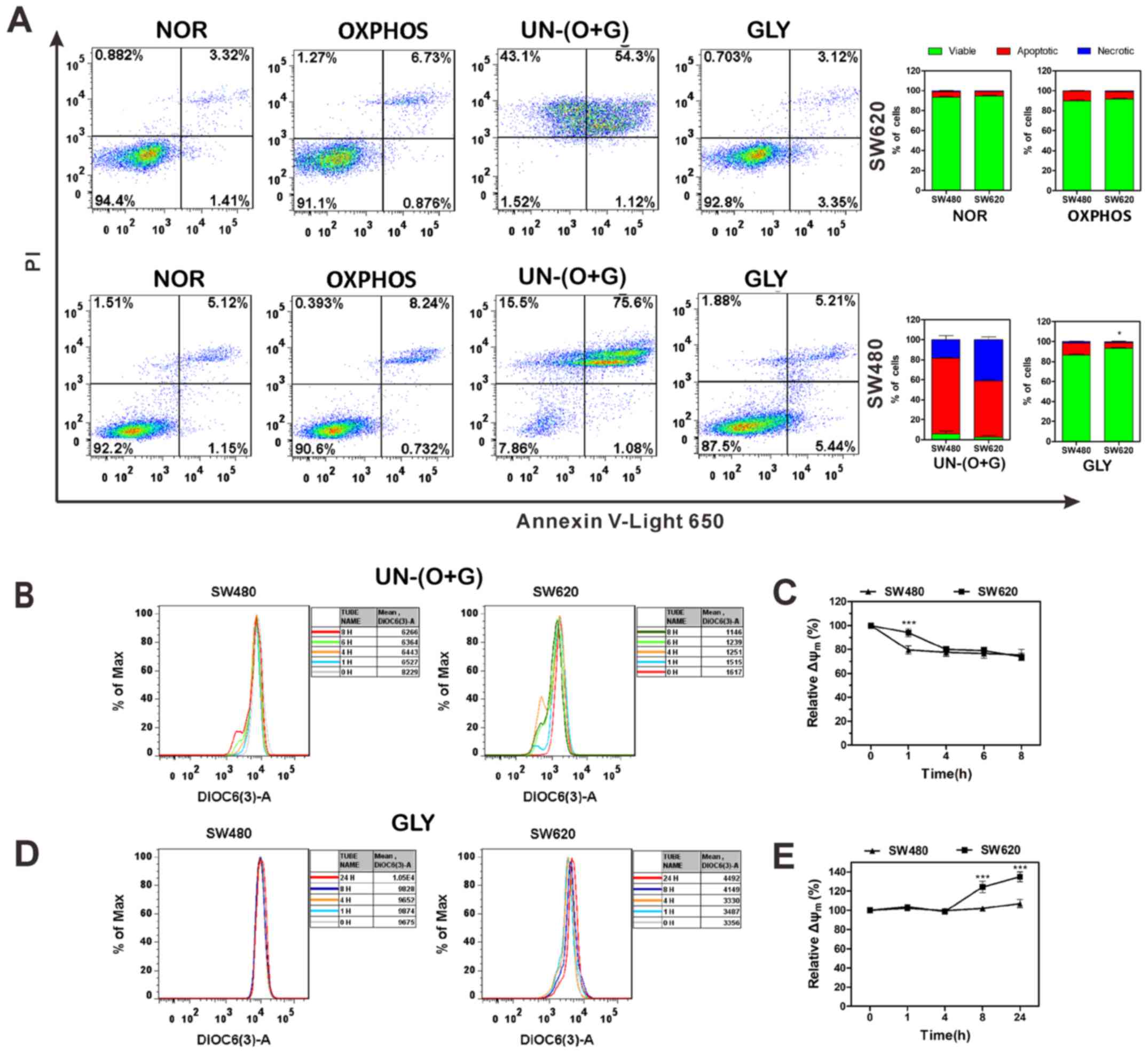 | Figure 5Apoptosis and ∆Ψm analyses of SW480
and SW620 cells in different culture environments. (A) Apoptosis
was examined in SW620 and SW480 cells cultured in differing culture
microenvironments. Following treatment, cells were collected,
stained with PI and Annexin V and analyzed by flow cytometry. (B)
∆Ψm was examined in SW480 and SW620 cells treated with Oligo A
without glucose. Following treatment, cells were collected, stained
with DiOC6(3) and analyzed by flow
cytometry. (C) Relative ΔΨm of SW480 and SW620 from (B). (D) ∆Ψm
was examined in SW480 and SW620 cells treated with Oligo A and
glucose. (E) Relative ∆Ψm of SW480 and SW620 from (D). Data are
expressed as the mean fluorescence intensities. Data are presented
as the mean ± standard deviation; n=3; *P<0.05 and
***P<0.001 vs. SW480. ∆Ψm, mitochondrial membrane
potential; DiOC6(3),
3,3'-dihexyloxacarbocyanine iodide; Oligo A, Oligomycin A; PI,
propidium iodide; NOR, normal; OXPH, oxidative phosphorylation;
GLY, glycolysis; UN-(O+G), no metabolism through OXPHOS or
glycolysis. |
ΔΨm is vital for sustaining the physiological
function of the cellular respiration/electron transport chain to
yield ATP; decreases in ΔΨm will lead to mitochondrial dysfunction
and result in apoptosis (33).
Changes in ΔΨm of SW480 and SW620 cells were evaluated when both
glycolysis and OXPHOS were inhibited [UN-(O+G) group]. The ΔΨm of
SW480 cells exhibited a sudden drop at 1 h and gradually decreased
between 1 and 8 h, whereas the ΔΨm of SW620 cells declined
gradually during the entire experimental period when both OXPHOS
and glycolysis were restrained. In addition, as UN-(O+G) SW480 and
SW620 cells exhibited increased rates of apoptosis and necrosis at
24-h culturing, ΔΨm could not be detected in cells at 24 h
post-treatment (Fig. 5B and C).
When only OXPHOS was suppressed (GLY group), the ΔΨm of SW620 cells
was significantly increased at 8 and 24 h following Oligo A
treatment compared with 0 h (Fig. 5D
and E). The elevation of ΔΨm indicates the alleviation of
mitochondrion impairment, and cells were protected from apoptosis
(34). These data suggested that
SW620 cells exhibit stronger adaptability under unfavorable
conditions (Oligo A treatment) to prevent apoptosis by increasing
ΔΨm.
SW620 cells exhibit stronger mesenchymal
phenotypic properties compared with SW480 cells
SW620 cells are a type of metastatic cell derived
from a lymph node lesion, which may possess stronger mesenchymal
phenotypic properties compared with primary tumor-derived SW480
cells. To investigate the association between metabolism and EMT,
the mRNA transcription levels of glycolysis regulatory proteins
(HK1, HK2 and HIF-1) and EMT regulatory factors (CDH1, FN1, VIM,
CD133, SNAI1, SNAI2, ZEB1, CDH2 and TWIST) were compared in
different metabolic microenvironment. When cells were cultured in
medium without glucose and metabolized through OXPHOS, the mRNA
transcriptional levels of the mesenchymal phenotype-associated
proteins FN1 and VIM, and the key glycolysis enzyme HK1 were lower
in SW620 cells compared with expression levels in SW480 cells; the
other mRNA transcriptional levels were significantly higher in
SW620 cells (Fig. 6A). When cells
metabolized through both OXPHOS and glycolysis (NOR), the
transcriptional levels of FN1 and SNAI2 were lower compared with
transcriptional levels in SW480 cells; for all other genes, the
transcriptional levels were significantly higher in SW620 cells
(Fig. 6B). However, when OXPHOS
was inhibited by treating with glucose and Oligo A, in which cells
metabolized through glycolysis, all the mRNA transcriptional levels
were upregulated in SW620 cells compared with SW480 cells (Fig. 6C). These data suggested that SW620
cells may regulate the expression of HK1 according to alterations
in the culture environment. The HK2 mRNA expression level was
higher in SW620 cells compared with SW480 cells in each type of
culture condition (Fig. 6A-C),
which indicated that HK2 may be a key enzyme that is upregulated in
highly metastatic tumor cells. Furthermore, compared with SW480
cells, SW620 cells exhibited higher mRNA transcriptional levels of
the EMT promoting factors SNAI1, TWIST and ZEB1 (Fig. 6D), which suggested that SW620 cells
exhibited stronger mesenchymal cell properties and metastatic
potential.
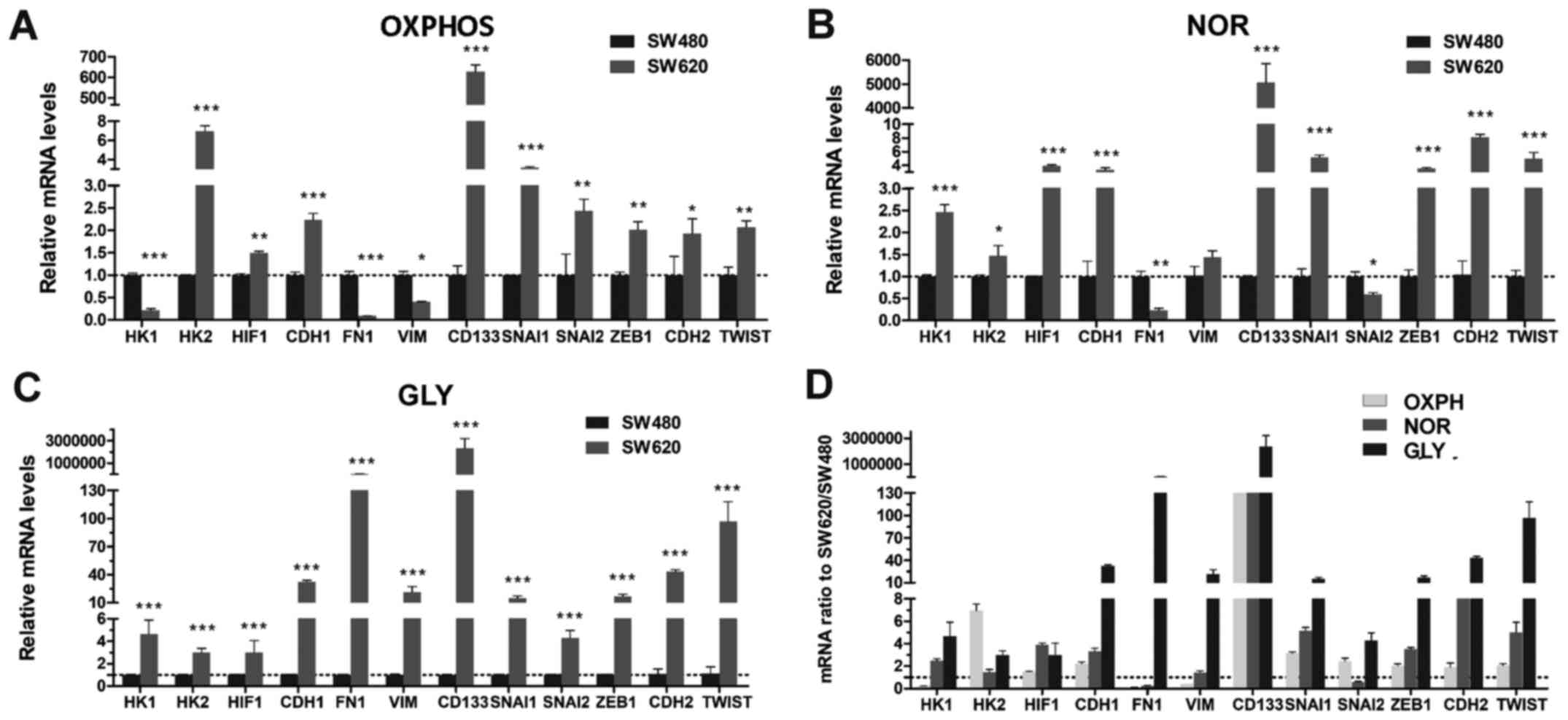 | Figure 6RT-qPCR analysis of SW480 and SW620
cells in different culture microenvironments. (A-C) RT-qPCR
analysis of the relative mRNA expression levels of HK1, HK2,
HIF-1α, CDH1, FN1, VIM, CD133, SNAI1, SNAI2, ZEB1, CDH2 and TWIST1
in SW480 and SW620 cells cultured in different media conditions;
mRNA expressions were normalized to β-actin. (D) SW620/SW480 mRNA
expression ratios in different types of metabolism. Data are
presented as the mean ± standard deviation; n=3;
*P<0.05, **P<0.01 and
***P<0.001 vs. SW480. CDH1, Epithelial cadherin; FN1,
fibronectin; HIF-1α, hypoxia-inducible factor; HK, hexokinase;
Oligo A, Oligomycin A; OXPHOS, oxidative phosphorylation; SNAI,
snail family transcriptional repressor; VIM, vimentin; ZEB,
Zinc-finger E-box-binding homeobox; NOR, normal; GLY, glycolysis;
OXPH, oxidative phosphorylation. |
When OXPHOS was inhibited by Oligo A for 48 h, the
mRNA transcriptional levels of the key glycolysis enzymes HIF-1α,
HK1 and HK2 were significantly upregulated in SW620 cells compared
with SW480 cells (Fig. 7). These
results were consistent with the data presented in Figs. 1E-H and 2G-I, which further indicated that SW620
cells may regulate the expression of HK1 and HK2 to enhance
glycolysis to supplement the energy supply under conditions of
energy shortage.
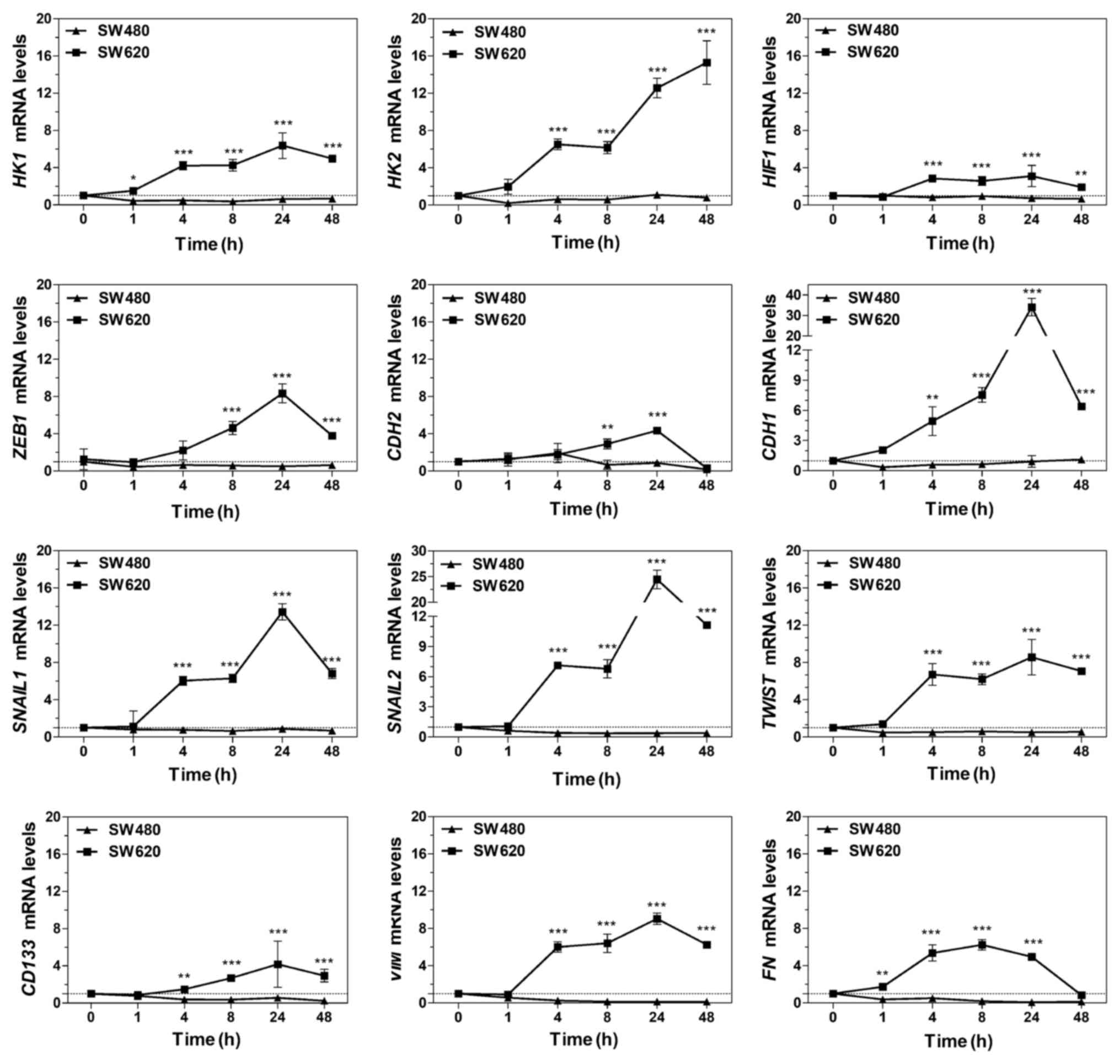 | Figure 7mRNA levels in SW480 and SW620 cells
treated with Oligo A over time. Reverse transcription-quantitative
polymerase chain reaction analysis of mRNA expression levels of
HK1, HK2, HIF-1α, CDH1, CDH2, ZEB1, SNAI1, SNAI2, TWIST1, CD133,
VIM and FN in SW480 and SW620 cells treated with Oligomycin A (1
µM) for 0-48 h. mRNA expression levels were normalized to
β-actin. Data are presented as the mean ± standard deviation; n=3;
**P<0.01; ***P<0.001 vs. SW480. CDH1,
epithelial cadherin; CDH2, Neural-cadherin; FN1, fibronectin;
HIF-1α, hypoxia-inducible factor; HK, hexokinase; SNAI, snail
family transcriptional repressor; VIM, vimentin; ZEB, Zinc-finger
E-box-binding homeobox. |
In addition, in SW620 cells, mRNA expression levels
of the EMT promoting factors SNAI1, SNAI2, ZEB1 and TWIST, the
mesenchymal phenotype-associated proteins FN1 and VIM and other
EMT-related proteins CDH1, CDH2 and CD133 were upregulated over
time (Fig. 7). This may result in
an enhancement of migration and metastasis ability. By contrast,
SW480 cells exhibited the opposite trends.
SW620 cells express CD133 and CD166
In the present study, the expression of
metastasis-associated proteins CD29, CD44, CD47, CD54, VIM, CD49d,
CD49e, CD49f, CD51/61, FN, CD133, CD166, CD324, CD325 and CD326
were compared in SW480 and SW620 cells by flow cytometry. The
expression levels of CD44, CD54, CD133, CD166 and CD324 between
SW480 and SW620 cells were markedly different; the most notable
differences were that CD133 and CD166 were highly expressed in
SW620 cells, but their expressions were not be detected in SW480
cells (Fig. 8). Also, the
expression of CD324 in SW620 is higher than that in SW480.
Conversely, the expression of CD44 and CD54 was lower in SW620
cells compared with expression in SW480 cells.
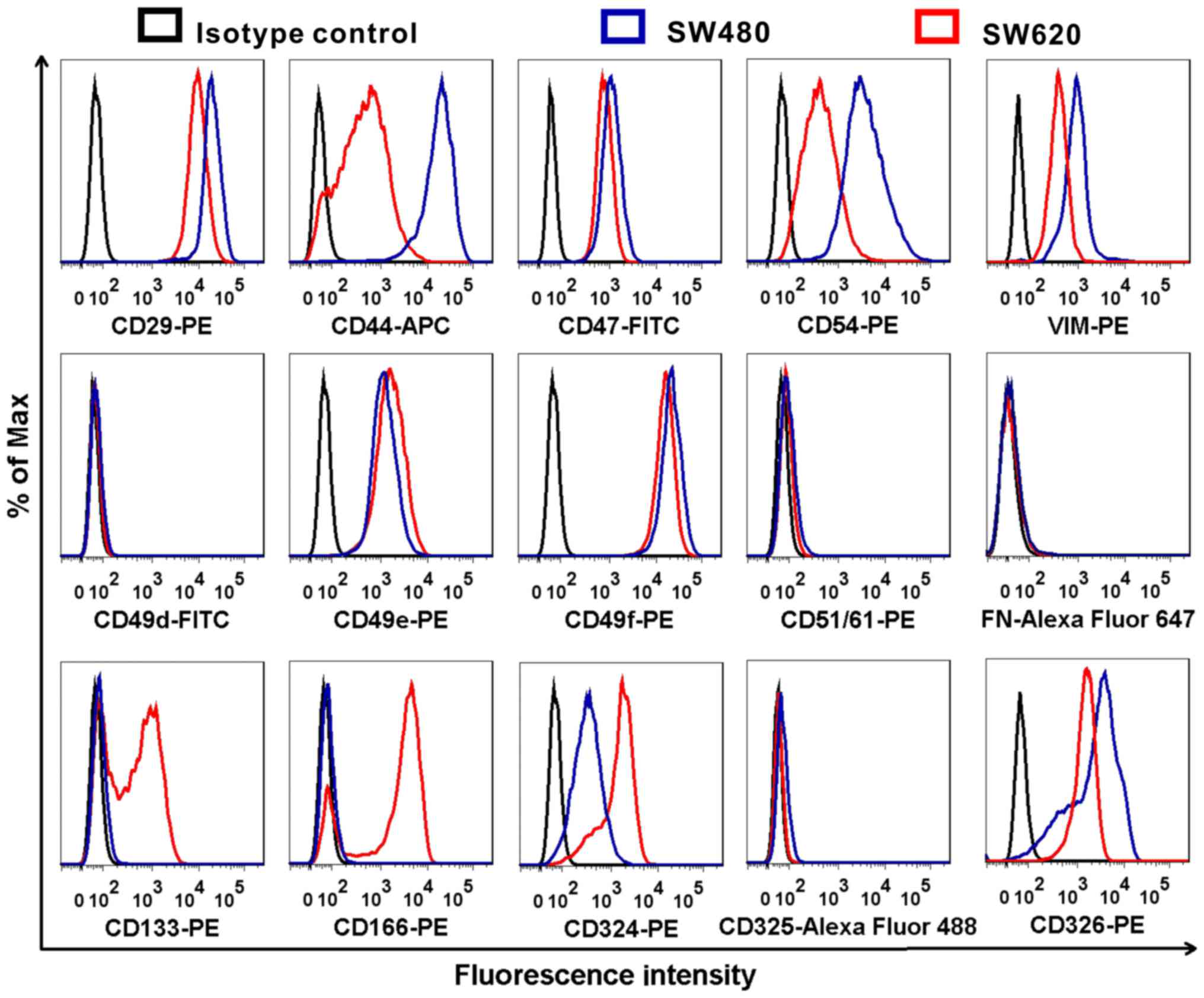 | Figure 8Comparison of different
metastasis-associated protein expression levels between SW480 and
SW620 cells. SW480 and SW620 cells were stained with CD29-PE,
CD44-APC, CD47-FITC, CD54-PE, VIM-PE, CD49d-FITC, CD49e-PE,
CD51/61-PE, FN-Alexa Fluor 647, CD133-PE, CD166-PE, CD324-PE,
CD325-Alexa Fluor 488 and CD326-PE antibodies, and analyzed by flow
cytometry. APC, allophycocyanin; FITC, fluorescein isothiocyanate;
FN, fibronectin; PE, phycoerythrin; VIM, vimentin. |
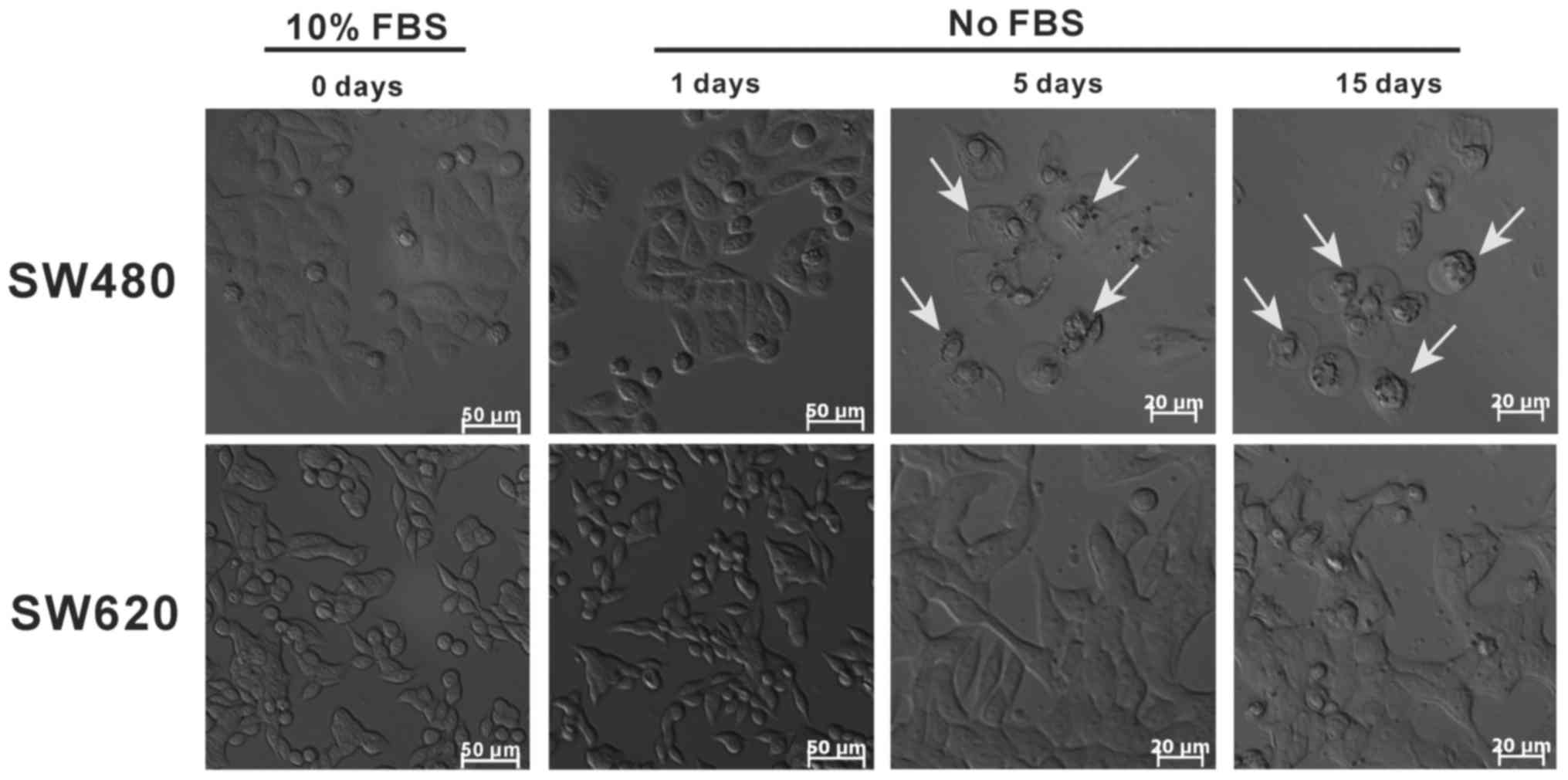
SW620 cells exhibit a stronger tolerance
to FBS-free culture compared with SW480 cells
Serum is commonly used as a cell culture supplement
and provides macromolecules, carrier proteins, trace elements,
attachment and spreading factors, low molecular weight nutrients,
hormones and growth factors; FBS is the most widely used serum and
is often used to maintain cell growth and proliferation (35). In the present study, the cell
growth status of SW480 and SW620 cells in FBS-free medium was
compared. SW480 cells exhibited low adherence, nuclear condensation
and a small degree of apoptosis (white arrows) following 5 days of
starvation culturing, and almost all cells had undergone apoptosis
at day 15 (Fig. 9). By contrast,
SW620 cells maintained high adherence, integrated morphology and a
seemingly high viability following starvation culturing for 5 or 15
days. These results indicated that SW480 cells were sensitive to
FBS and that SW620 cells were strongly tolerant of FBS-free medium.
By comparison with SW480 cells, SW620 cells appear to be better
adapted to survive in nutrient-scarce microenvironments.
Discussion
In the present study, it was observed that
metastatic SW620 cells exhibited stronger bioenergetic adaptation
in unfavorable situations compared with primary tumor-derived SW480
cells, through sustained elevation of glycolysis and regulation of
the cell cycle. The notable glycolytic ability of SW620 cells may
be associated with high expression levels of HK1, HK2, GLUT1 and
HIF-1α. Furthermore, SW620 cells exhibited a stronger mesenchymal
phenotype and stem cell characteristics, which may promote the
metastatic process. These findings suggest that metastatic cancer
cells have better adaptability and survivability in certain
microenvironments owing to the upregulation of glycolysis,
optimization of the cell cycle and metabolic reprogramming. In
addition, a previously study established the PC3-Epi primary
prostate cancer and PC3-EMT metastatic prostate cancer cell models,
and reported similar findings in that glycolysis was the primary
bioenergetic pathway for cell motility and cytoskeletal remodeling
in mesenchymal (metastatic) cancer cells (36).
HK1 and HK2 are two isoforms of hexokinase that
phosphorylate glucose to produce glucose-6-phosphate, the first
step in the majority of glucose metabolism processes (37). Cellular protection and energy
metabolism utilize common signaling pathways, and it has been
suggested that HK2 serves a critical role not only in glycolysis,
but also in cell survival (38,39).
In the present study, it was demonstrated that when OXPHOS was
inhibited by Oligo A in the presence of glucose, SW620 cells were
able to upregulate the expression of glucose metabolic enzyme HK1
and HK2, and increased the ATP levels in a short period of time. By
contrast, in SW480 cells the ATP levels were decreased, and the
expression of HK1 and HK2 was significantly inhibited. It has been
reported that HK2 may induce cancer cell autophagy to cope with the
poor micro-environment when energy supply is insufficient (39,40).
In addition, previous studies have also revealed that HK2 serves a
significant role in AKT-mediated mitochondrial protection against
opening of the mitochondrial permeability transition pores
(41,42), and that HK2 competes with apoptotic
Bcl-2 family proteins to prevent outer mitochondrial membrane
rupture (43,44). These data are consistent with our
present study results; when cells metabolized through OXPHOS or
glycolysis, the proportion of apoptotic cells was lower among the
SW620 cell population, compared with that among the SW480 cells.
Flow cytometric analysis indicated that the ΔΨm of SW620 cells was
upregulated at 8 and 24 h inhibition of OXPHOS by Oligo A,
indicating that mitochondria impairment was alleviated. This may be
due to a difference in ΔΨm between SW620 and SW480 cells. When
stimulated by Oligo A, SW620 cells may have initiated a protective
mechanism. The alteration of mitochondria membrane permeability may
enhance cell survival, by increasing the tolerance to mitochondrial
impairment. In addition, culture in FBS-free medium demonstrated
that SW620 cells exhibit a stronger tolerance to nutrient-poor
microenvironments, which suggested that metastatic SW620 cells
possess stronger adaptability and viability in unfavorable
environments. Based on these findings, it was hypothesized that
SW620 cells have improved microenvironment adaptability and
survivability through the upregulation of HK2 expression.
Metabolic reprogramming is considered to be a
hallmark of cancer (8). Oncogene
activation and loss of tumor suppressors was reported to promote
metabolic reprogramming in cancers, which results in increased
nutrient uptake for energetic and biosynthetic pathways (6). HIF-1α serves a role in metabolic
reprogramming by activating the transcription of key genes encoding
metabolic enzymes (45,46), including L-lactate dehydrogenase A
chain (47),
3-phosphoinositide-dependent protein kinase 1 (48,49)
and BCL2 (50). In addition,
limited nutrients within solid tumors may induce metastatic cancer
cells to exhibit metabolic flexibility to sustain growth and
survival (6). In the present
study, HIF-1α expression in SW620 cells was higher in the presence
of glycolysis metabolism (normal and glycolysis environments)
compared with SW480 cells, when OXPHOS was used as the control.
SW620 cells exhibited stronger mesenchymal cell properties and
metabolic flexibility to sustain survival through mediation of the
cell cycle and EMT regulatory factors. It was observed that the ATP
levels of SW480 cells cultured in Oligo A declined quickly within
24 h, whereas the ATP levels of SW620 cells at 24 h were higher
compared with at 0 h. Therefore, it was speculated that, during the
first 24 h of OXPHOS inhibition, SW620 cells may initiate
glycolysis immediately to generate ATP and to enhance invasion and
metastasis to enable cells to escape the hostile environment.
Complex networks regulate metastasis, including
tumor-stroma interactions at the primary site, cancer cell
dissemination and the microenvironment at the metastatic sites
(5). To establish metastasis,
cancer cells need to survive in the circulation and possess
adherence and invasion properties, which are coupled to certain
proteins, including CD29, CD44, CD133 and CD166 (51-53).
Differences in cell surface protein expressions between SW620 and
SW480 cells may serve important roles in the metastatic process.
For example, CD133 is a five-transmembrane domain molecule that
marks stem-like cells of various tissues and cancer types, and
CD133 expression in colon cancer strongly correlates with patient
survival (54). CD166 is a type of
adhesion molecule and the expression of CD166 introduces a more
general switch in developmental program, which connects the
regulation of cell growth and cell migration (55-57).
The present study results demonstrated that CD133 and CD166 were
highly expressed in SW620 cells, but not be detected in SW480
cells, which indicated increased metastasis, migration and
invasion, and suggested that CD133 and CD166 may be the potential
targets in cancer therapy.
In conclusion, data from the present study
demonstrated that metastatic cancer cells exhibited stronger
metabolic flexibility and microenvironmental adaptability through
metabolic reprogramming. A complex networks of cell surface
proteins, the glycolysis-associated proteins HK1, HK2, GLUT1 and
HIF-1α, cell cycle regulators and mitochondria regulate the
adaptation of cancer cells in the metastatic process. However, the
methods of the present study had certain limitations; the
regulatory mechanisms of glycolysis-associated proteins for
metastatic cancer cells bioenergetic adaptation and metabolic
reprogramming will be studied through gene knockdown and
overexpression, and preclinical animal experiments, in future
studies. Although the detailed mechanisms underlying the observed
cell behaviors require further investigation, the present study may
provide the groundwork for a novel approach to cancer metastasis
therapy.
Funding
This research was supported by The Ministry of
Science and Technology of China (grant no. 2015CB931804), The
National Natural Science Foundation of China (grant nos. NSFC
81773063, U1505225, 81703555, and 81702988), The China Postdoctoral
Science Foundation (grant no. 2017M620268) and The Strait
Postdoctoral Exchange Grant Program of Fujian Province (grant no.
02510515).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' Contributions
YL and LJ conceived and designed the experiments.
YL, YC, SL and HL performed the flow cytometry experiments. YL, DZ,
SLian, YC and JX performed cells culture experiments. YL, YC, SL,
SL and HL performed western blotting experiments. YL, YC, SL, RX
and JC performed reverse transcription-quantitative polymerase
chain reaction experiments. YL, SL and YC performed adenosine
5'-triphosphate measurement experiments. YL, SL, YY and YC acquired
and analyzed the experimental data. JX and XX reviewed and provide
essential comments for the final version to be published. YL and YC
wrote the manuscript, and LJ revised it critically for important
intellectual content. All authors reviewed the manuscript.
Ethical approval and consent to
participate
All animals used in this study were handled in
accordance with the Guide for the Care and Use of Laboratory
Animals (National Research Council, 1996). The Institutional Animal
Care and Use Committee of Fuzhou University (Fuzhou, China)
reviewed and approved all animal use procedures based on the above
regulations.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would like to thank The Ministry of
Science and Technology of China, The National Natural Science
Foundation of China, The China Postdoctoral Science Foundation and
The Strait Postdoctoral Exchange Grant Program of Fujian
Province.
References
|
1
|
Sethi N and Kang Y: Unravelling the
complexity of metastasis -molecular understanding and targeted
therapies. Nat Rev Cancer. 11:735–748. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chambers AF, Groom AC and MacDonald IC:
Dissemination and growth of cancer cells in metastatic sites. Nat
Rev Cancer. 2:563–572. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lu Y, Liang H, Yu T, Xie J, Chen S, Dong
H, Sinko PJ, Lian S, Xu J, Wang J, et al: Isolation and
characterization of living circulating tumor cells in patients by
immunomagnetic negative enrichment coupled with flow cytometry.
Cancer. 121:3036–3045. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Labelle M and Hynes RO: The initial hours
of metastasis: The importance of cooperative host-tumor cell
interactions during hematogenous dissemination. Cancer Discov.
2:1091–1099. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Joyce JA and Pollard JW:
Microenvironmental regulation of metastasis. Nat Rev Cancer.
9:239–252. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Boroughs LK and DeBerardinis RJ: Metabolic
pathways promoting cancer cell survival and growth. Nat Cell Biol.
17:351–359. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Loo JM, Scherl A, Nguyen A, Man FY,
Weinberg E, Zeng Z, Saltz L, Paty PB and Tavazoie SF: Extracellular
metabolic energetics can promote cancer progression. Cell.
160:393–406. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vander Heiden MG, Cantley LC and Thompson
CB: Understanding the Warburg effect: The metabolic requirements of
cell proliferation. Science. 324:1029–1033. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Phan L, Chou PC, Velazquez-Torres G,
Samudio I, Parreno K, Huang Y, Tseng C, Vu T, Gully C, Su CH, et
al: The cell cycle regulator 14-3-3σ opposes and reverses cancer
metabolic reprogramming. Nat Commun. 6:75302015. View Article : Google Scholar
|
|
11
|
Phan LM, Yeung SC and Lee MH: Cancer
metabolic reprogramming: Importance, main features, and potentials
for precise targeted anti-cancer therapies. Cancer Biol Med.
11:1–19. 2014.PubMed/NCBI
|
|
12
|
Zheng J: Energy metabolism of cancer:
Glycolysis versus oxidative phosphorylation (Review). Oncol Lett.
4:1151–1157. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Buchakjian MR and Kornbluth S: The engine
driving the ship: Metabolic steering of cell proliferation and
death. Nat Rev Mol Cell Biol. 11:715–727. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gatenby RA and Gillies RJ: Why do cancers
have high aerobic glycolysis? Nat Rev Cancer. 4:891–899. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Alderton GK: Metastasis: Metabolic
reprogramming in disseminated cells. Nat Rev Cancer. 14:7032014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Postovit LM, Adams MA, Lash GE, Heaton JP
and Graham CH: Oxygen-mediated regulation of tumor cell
invasiveness. Involvement of a nitric oxide signaling pathway. J
Biol Chem. 277:35730–35737. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Singh A and Settleman J: EMT, cancer stem
cells and drug resistance: An emerging axis of evil in the war on
cancer. Oncogene. 29:4741–4751. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Asnaghi L, Gezgin G, Tripathy A, Handa JT,
Merbs SL, van der Velden PA, Jager MJ, Harbour JW and Eberhart CG:
EMT-associated factors promote invasive properties of uveal
melanoma cells. Mol Vis. 21:919–929. 2015.PubMed/NCBI
|
|
19
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Thiery JP and Sleeman JP: Complex networks
orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell
Biol. 7:131–142. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guan RJ, Ford HL, Fu Y, Li Y, Shaw LM and
Pardee AB: Drg-1 as a differentiation-related, putative metastatic
suppressor gene in human colon cancer. Cancer Res. 60:749–755.
2000.PubMed/NCBI
|
|
23
|
Lu Y, Yu T, Liang H, Wang J, Xie J, Shao
J, Gao Y, Yu S, Chen S, Wang L, et al: Nitric oxide inhibits
hetero-adhesion of cancer cells to endothelial cells: Restraining
circulating tumor cells from initiating metastatic cascade. Sci
Rep. 4:43442014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lian S, Lu Y, Cheng Y, Yu T, Xie X, Liang
H, Ye Y and Jia L: S-nitrosocaptopril interrupts adhesion of cancer
cells to vascular endothelium by suppressing cell adhesion
molecules via inhibition of the NF-KB and JAK/STAT signal pathways
in endothelial cells. Eur J Pharmacol. 791:62–71. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
26
|
Kroemer G and Pouyssegur J: Tumor cell
metabolism: Cancer's Achilles' heel. Cancer Cell. 13:472–482. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wick AN, Drury DR, Nakada HI and Wolfe JB:
Localization of the primary metabolic block produced by
2-deoxyglucose. J Biol Chem. 224:963–969. 1957.PubMed/NCBI
|
|
28
|
Lim S and Kaldis P: Cdks, cyclins and
CKIs: Roles beyond cell cycle regulation. Development.
140:3079–3093. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Malumbres M and Barbacid M: Cell cycle,
CDKs and cancer: A changing paradigm. Nat Rev Cancer. 9:153–166.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Elledge SJ: Cell cycle checkpoints:
Preventing an identity crisis. Science. 274:1664–1672. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shin I, Yakes FM, Rojo F, Shin NY, Bakin
AV, Baselga J and Arteaga CL: PKB/Akt mediates cell-cycle
progression by phosphorylation of p27(Kip1) at threonine 157 and
modulation of its cellular localization. Nat Med. 8:1145–1152.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sheaff RJ, Singer JD, Swanger J,
Smitherman M, Roberts JM and Clurman BE: Proteasomal turnover of
p21Cip1 does not require p21Cip1 ubiquitination. Mol Cell.
5:403–410. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gottlieb E, Armour SM, Harris MH and
Thompson CB: Mitochondrial membrane potential regulates matrix
configuration and cytochrome c release during apoptosis. Cell Death
Differ. 10:709–717. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Beltrán B, Mathur A, Duchen MR,
Erusalimsky JD and Moncada S: The effect of nitric oxide on cell
respiration: A key to understanding its role in cell survival or
death. Proc Natl Acad Sci USA. 97:14602–14607. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gstraunthaler G: Alternatives to the use
of fetal bovine serum: Serum-free cell culture. ALTEX. 20:275–281.
2003.PubMed/NCBI
|
|
36
|
Shiraishi T, Verdone JE, Huang J, Kahlert
UD, Hernandez JR, Torga G, Zarif JC, Epstein T, Gatenby R,
McCartney A, et al: Glycolysis is the primary bioenergetic pathway
for cell motility and cytoskeletal remodeling in human prostate and
breast cancer cells. Oncotarget. 6:130–143. 2015. View Article : Google Scholar :
|
|
37
|
Cárdenas ML, Cornish-Bowden A and Ureta T:
Evolution and regulatory role of the hexokinases. Biochim Biophys
Acta. 1401:242–264. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pastorino JG and Hoek JB: Hexokinase II:
The integration of energy metabolism and control of apoptosis. Curr
Med Chem. 10:1535–1551. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Roberts DJ, Tan-Sah VP, Ding EY, Smith JM
and Miyamoto S: Hexokinase-II positively regulates glucose
starvation-induced autophagy through TORC1 inhibition. Mol Cell.
53:521–533. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tan VP and Miyamoto S: HK2/hexokinase-II
integrates glycolysis and autophagy to confer cellular protection.
Autophagy. 11:963–964. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Miyamoto S, Murphy AN and Brown JH: Akt
mediates mitochondrial protection in cardiomyocytes through
phosphorylation of mitochondrial hexokinase-II. Cell Death Differ.
15:521–529. 2008. View Article : Google Scholar
|
|
42
|
Sun L, Shukair S, Naik TJ, Moazed F and
Ardehali H: Glucose phosphorylation and mitochondrial binding are
required for the protective effects of hexokinases I and II. Mol
Cell Biol. 28:1007–1017. 2008. View Article : Google Scholar :
|
|
43
|
Robey RB and Hay N: Mitochondrial
hexokinases, novel mediators of the antiapoptotic effects of growth
factors and Akt. Oncogene. 25:4683–4696. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Pastorino JG, Shulga N and Hoek JB:
Mitochondrial binding of hexokinase II inhibits Bax-induced
cytochrome c release and apoptosis. J Biol Chem. 277:7610–7618.
2002. View Article : Google Scholar
|
|
45
|
Semenza GL: Tumor metabolism: Cancer cells
give and take lactate. J Clin Invest. 118:3835–3837.
2008.PubMed/NCBI
|
|
46
|
Faubert B, Vincent EE, Griss T, Samborska
B, Izreig S, Svensson RU, Mamer OA, Avizonis D, Shackelford DB,
Shaw RJ, et al: Loss of the tumor suppressor LKB1 promotes
metabolic reprogramming of cancer cells via HIF-1α. Proc Natl Acad
Sci USA. 111:2554–2559. 2014. View Article : Google Scholar
|
|
47
|
Semenza GL, Jiang BH, Leung SW, Passantino
R, Concordet JP, Maire P and Giallongo A: Hypoxia response elements
in the aldolase A, enolase 1, and lactate dehydrogenase A gene
promoters contain essential binding sites for hypoxia-inducible
factor 1. J Biol Chem. 271:32529–32537. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kim JW, Tchernyshyov I, Semenza GL and
Dang CV: HIF-1-mediated expression of pyruvate dehydrogenase
kinase: A metabolic switch required for cellular adaptation to
hypoxia. Cell Metab. 3:177–185. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Papandreou I, Cairns RA, Fontana L, Lim AL
and Denko NC: HIF-1 mediates adaptation to hypoxia by actively
downregulating mitochondrial oxygen consumption. Cell Metab.
3:187–197. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang H, Bosch-Marce M, Shimoda LA, Tan
YS, Baek JH, Wesley JB, Gonzalez FJ and Semenza GL: Mitochondrial
autophagy is an HIF-1-dependent adaptive metabolic response to
hypoxia. J Biol Chem. 283:10892–10903. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Dalerba P, Dylla SJ, Park IK, Liu R, Wang
X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, et al:
Phenotypic characterization of human colorectal cancer stem cells.
Proc Natl Acad Sci USA. 104:10158–10163. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Clevers H: The cancer stem cell: Premises,
promises and challenges. Nat Med. 17:313–319. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Du L, Wang H, He L, Zhang J, Ni B, Wang X,
Jin H, Cahuzac N, Mehrpour M, Lu Y, et al: CD44 is of functional
importance for colorectal cancer stem cells. Clin Cancer Res.
14:6751–6760. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Horst D, Kriegl L, Engel J, Kirchner T and
Jung A: CD133 expression is an independent prognostic marker for
low survival in colorectal cancer. Br J Cancer. 99:1285–1289. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Lunter PC, van Kilsdonk JW, van Beek H,
Cornelissen IM, Bergers M, Willems PH, van Muijen GN and Swart GW:
Activated leukocyte cell adhesion molecule (ALCAM/CD166/MEMD), a
novel actor in invasive growth, controls matrix metalloproteinase
activity. Cancer Res. 65:8801–8808. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Levin TG, Powell AE, Davies PS, Silk AD,
Dismuke AD, Anderson EC, Swain JR and Wong MH: Characterization of
the intestinal cancer stem cell marker CD166 in the human and mouse
gastrointestinal tract. Gastroenterology. 139:2072–2082.e2075.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Hansen AG, Arnold SA, Jiang M, Palmer TD,
Ketova T, Merkel A, Pickup M, Samaras S, Shyr Y, Moses HL, et al:
ALCAM/ CD166 is a TGF-β-responsive marker and functional regulator
of prostate cancer metastasis to bone. Cancer Res. 74:1404–1415.
2014. View Article : Google Scholar : PubMed/NCBI
|