Introduction
Epithelial ovarian cancer (EOC) is the second most
common form of gynecological cancer and is currently the first
leading cause of gynecological cancer-related mortality (1). The 5-year survival rate of patients
with advanced disease is merely 30% (2). Despite the intra-tumor heterogeneity,
patients with EOC are usually treated with standard therapy,
including cytoreductive surgery and platinum-based chemotherapy
(3,4). However, conventional treatment
usually causes chemoresistance, resulting in tumor recurrence and
even death (3). Therefore, it is
imperative to identify novel molecular mechanisms and potential
therapeutic targets in order to better control the severity of
EOC.
Circular RNAs (circRNAs) are regarded as widespread
endogenous non-coding RNAs (5,6).
They were discovered >20 years ago, but were mostly
misinterpreted as mis-splicing errors due to low abundance
(7). Only in 2012/2013, their
substantial presence in eukaryotic cells was demonstrated using
high-throughput sequencing data (8). circRNAs are characterized by
covalently closed loop structures without 5′ caps or 3′
polyadenylated tails (9). The
formation of these structures involves back-splicing events by the
spliceosome at the expense of canonical mRNA isoforms (10,11).
Such circularization calls for the covalent binding of upstream 5′
acceptors to downstream 3′ donors (12). Although the concrete mechanisms of
back-splicing events remain unclear, flanking introns containing
ALU repeats are believed to be essential for the process (13). It is becoming increasingly evident
that, instead of being regarded as splicing errors, circRNAs are
rather the products of regulated back-splicing events (14). Accordingly, a body of evidence has
indicated that circRNAs are of huge regulatory potency (15). Previous studies have found that a
number of circRNAs play crucial roles in human
epithelial-mesenchymal transition and porcine embryonic brain
development (16,17). Recently, an increasingly number of
studies have demonstrated that circRNAs can regulate gene
expression by acting as microRNA sponges, such as CDR1as (18).
Of note, a growing body of evidence has indicated
that circRNAs are closely associated with the pathogenesis of a
number of human diseases, particularly cancer (19). To date, abnormal expression levels
of circRNAs have been revealed in various types of cancer,
including breast, liver, gastric, colorectal and bladder cancer
(20-24). However, the characterization of
circRNAs in ovarian cancer remains largely unknown. To date, and at
least to the best of our knowledge, only two articles have been
published in the literature regarding the association between
circRNAs and ovarian cancer (12,25).
In the study by Ahmed et al (25), differential expression profiles of
circRNAs were defined by performing paired-end RNA sequencing of
primary sites, peritoneal and lymph node metastases from 3 patients
with stage IIIC ovarian cancer. In the study by Bachmayr-Heyda
et al (12), the expression
of circular and linear RNAs and proliferation were analyzed in
matched normal colon mucosa and tumor tissues. They then validated
the correlation of global circular RNA abundance (the circRNA
index) and the proliferation in ovarian cancer cells compared to
cultured normal ovarian epithelial cells. However, neither of these
two articles investigated the differential expression profiles of
circRNAs in EOC specimens compared to normal ovarian tissues. In
this study, we aimed to examine the pathogenesis of EOC and to
identify potential therapeutic targets and circRNA biomarkers by
comprehensively profiling circRNA expression in EOC specimens.
Materials and methods
Human samples
Four EOC specimens and four normal ovarian tissues
for circRNA-sequencing analysis were obtained from the Department
of Obstetrics and Gynecology in Peking Union Medical College
Hospital (PUMCH) between 2013 and 2017. A total of 54 EOC specimens
and 54 normal ovarian tissues, including those used for
circRNA-sequencing analysis were collected for circRNA validation
by reverse transcription-quantitative polymerase chain reaction
(RT-qPCR). Cancer specimens were collected from patients who had
histologically proven to suffer from EOC and had received wedge
biopsy of ovaries or cytoreductive surgery. Normal ovarian tissues
were collected from patients who underwent adnexectomy due to
myoma. Cancer tissues were collected from primary sites, and the
normal ovarian tissues were collected from the surface epithelium.
Fresh tissues were collected during surgery, frozen in liquid
nitrogen within 5 min following resection, and stored at −80°C
until use. All patients were consecutive and did not receive any
pre-operative chemotherapy, radiotherapy or target therapy. All the
EOC specimens and normal ovarian tissues were confirmed by
experienced pathologists. Clinicopathological data, including age
at diagnosis, International Federation of Gynecology and Obstetrics
(FIGO) stage, residual tumor diameter, lymph node metastasis (LNM),
CA125 level, and ascites are presented in Table I. The study protocol conformed to
the ethical guidelines of the 1975 Declaration of Helsinki, and was
approved by the Ethics Committee of PUMCH. All experiments were
performed in accordance with relevant guidelines. Written informed
consents were obtained from all patients for research purpose.
 | Table IThe association between differentially
expressed circRNAs and various clinicopathological features of
epithelial ovarian cancer. |
Table I
The association between differentially
expressed circRNAs and various clinicopathological features of
epithelial ovarian cancer.
| Variables | N | circBNC2 | circEXOC6B | circFAM13B | circN4BP2L2 | circRHOBTB3 | circCELSR1 |
|---|
| means ± SD | P-value | means ± SD | P-value | means ± SD | P-value | means ± SD | P-value | means ± SD | P-value | means ± SD | P-value |
|---|
| Age, years | | | 0.027a | | 0.018a | | 0.032a | | 0.047a | | 0.023a | | 0.448 |
| <50 | 23 | 9.37±2.01 | | 11.10±3.48 | | 12.28±0.96 | | 5.97±1.28 | | 6.95±1.81 | | 11.35±1.63 | |
| ≥50 | 31 | 10.83±2.50 | | 13.29±2.37 | | 13.62±2.15 | | 7.01±1.56 | | 8.45±2.25 | | 10.56±1.66 | |
| FIGO stage | | | 0.045a | | 0.145 | | 0.044a | | 0.046a | | 0.288 | | 0.043a |
| FIGO I-II | 14 | 10.98±2.04 | | 12.86±2.10 | | 13.94±1.99 | | 7.07±1.24 | | 7.84±1.50 | | 10.44±1.65 | |
| FIGO III-IV | 40 | 9.74±2.82 | | 11.56±2.98 | | 12.52±1.13 | | 6.06±1.50 | | 7.01±2.06 | | 11.67±1.82 | |
| Residual tumor
diameter (cm) | | | 0.598 | | 0.849 | | 0.701 | | 0.900 | | 0.873 | | 0.035a |
| <1 | 43 | 10.12±2.69 | | 13.11±2.15 | | 13.65±1.94 | | 6.86±1.53 | | 8.01±2.18 | | 10.69±1.92 | |
| ≥1 | 11 | 9.54±1.55 | | 12.90±1.96 | | 13.24±1.03 | | 6.72±0.96 | | 7.74±1.44 | | 12.13±1.48 | |
| Lymph node
metastasis | | | 0.005a | | 0.032a | | 0.034a | | 0.003a | | 0.009a | | 0.172 |
| No | 35 | 10.90±2.72 | | 13.32±2.24 | | 13.71±2.12 | | 7.04±1.70 | | 8.27±2.54 | | 10.70±1.41 | |
| Yes | 19 | 8.80±2.64 | | 11.75±2.02 | | 12.46±1.34 | | 5.69±2.35 | | 6.72±2.98 | | 11.51±1.76 | |
| CA125 level
(U/ml) | | | 0.081 | | 0.288 | | 0.438 | | 0.885 | | 0.826 | | 0.425 |
| <600 | 31 | 10.82±2.73 | | 12.95±2.46 | | 13.22±1.95 | | 6.43±1.68 | | 8.08±2.37 | | 10.68±1.35 | |
| ≥600 | 23 | 9.30±1.79 | | 11.70±3.51 | | 12.90±1.79 | | 6.17±1.53 | | 7.41±2.03 | | 11.12±1.74 | |
| Ascites (ml) | | | 0.034a | | 0.211 | | 0.610 | | 0.245 | | 0.434 | | 0.021a |
| <100 | 13 | 11.54±3.29 | | 13.73±2.99 | | 13.32±2.75 | | 7.08±2.11 | | 8.45±3.48 | | 9.671±1.86 | |
| ≥100 | 41 | 9.59±2.30 | | 12.12±2.91 | | 12.65±1.26 | | 6.17±1.48 | | 7.44±2.10 | | 11.21±1.67 | |
RNA-seq analysis
Total RNA was extracted from the tissues using
TRIzol reagent (Life Technologies/Thermo Fisher Scientific,
Waltham, MA, USA) according to the manufacturer’s instructions.
Briefly, ribosomal RNA was removed from total RNA using the
Epicentre Ribo-Zero rRNA Removal kit (Illumina, San Diego, CA,
USA). The ribosome-depleted RNA was then treated with RNase R
(Epicentre) and fragmented to approximately 200 bp. Subsequently,
the purified RNA was subjected to first-strand cDNA synthesis
according to the instructions provided with the NEBNext Ultra
Directional RNA Library Prep kit for Illumina (NEB, Beverly, MA,
USA). The cDNA fragments were repaired using an End-It DNA End
Repair kit and then modified by the Klenow fragment to add an A to
the 3′ end of the DNA fragments, and were finally ligated to
adapters. Purified first-strand cDNA was then subjected to 13-15
cycles of PCR amplification. The PCR protocol included an initial
denaturation step (98°C for 30 sec) and 15 cycles of denaturation
(98°C for 10 sec, 65°C for 75 sec) and annealing (65°C for 5
min).The library products were evaluated using the Agilent 2200
TapeStation and Qubit 2.0 (Life Technologies, Carlsbad, CA, USA)
and then diluted to 10 pM for cluster generation in situ on
the HiSeq3000 pair-end flow cell followed by sequencing (2×150 bp)
on HiSeq3000 (Illumina).
Identification and quantification of
circRNAs
Firstly, we used TopHat2 (26) to map the RNA-seq FASTQ reads to the
human reference genome (GRCh37/hg19) obtained from the UCSC genome
database (http://genome.ucsc.edu/). Subsquently,
as previously described, the unmapped reads were used to identify
circRNAs (16). In brief, all the
unmapped reads were processed to 20-nucleotide anchors from both
ends of the sequencing read. Anchors aligning in the head-to-tail
orientation indicated a back-spliced junction. Subsequently, the
anchor alignment was extended, so that the complete read alignment
and the breakpoint were flanked by a GT/AG splice site. The circRNA
abundance was measured using the total number of reads that spanned
back-spliced junctions. RefSeq was used to annotate the genomic
regions that mapped to inferred circRNAs. Gene coordinates were
obtained from the RefGene tables in the UCSC Genome Browser
(downloaded on 03/28/2014). A custom script was used to determine
the host genes of circRNAs. The longest transcript fragment whose
boundaries (5′ or 3′ end) exactly matched both ends of each
circRNAs in the same strand was searched. The corresponding gene of
this transcript fragment was then defined as the host gene of this
circRNA.
RNA preparation and RT-qPCR
Total RNA was extracted from 30-50 mg of the EOC
specimens and normal ovarian tissues using TRIzol reagent (Life
Technologies/Thermo Fisher Scientific) according to the
manufacturer’s instructions. RT-qPCR was performed using M-MLV
Reverse Transcriptase (Promega, Madison, WI, USA), PowerUp™ SYBR™
Green Master Mix (Thermo Fisher Scientific) according to the
manufacturer’s instructions. GAPDH was used as an internal
reference gene. The RT-qPCR protocol included an initial
denaturation step (95°C for 20 sec) and 40 cycles of denaturation
(95°C for 3 sec) and annealing (60°C for 30 sec). The relative
expression levels were calculated using 2−ΔΔCq method
(27) as previously described
(28). The primer sequences were
as follows: circBNC2 (forward, 5′-GCAGTTCGGAACCAGAACGAC-3′ and
reverse, 5′-ATG CTGGCCAGTCTTGCTCAC-3′), circEXOC6B (forward, 5′-TGC
CTCAAGTAAGCCACTATC-3′ and reverse, 5′-TTCCTT TCCCTCATGTTGTAG-3′),
circFAM13B (forward, 5′-GTGAA AGTAACAGAGACTGTTC-3′ and reverse,
5′-ACTTACTCT GTAGAGGCTGG-3′), circN4BP2L2 (forward, 5′-CATGGT
GTGTCTCGAAAGAAG-3′ and reverse, 5′-CTGTACCCATC TTGATGGTGA-3′),
circRHOBTB3 (forward, 5′-GCTTTTCA GTGGGAAGAATTG-3′ and reverse,
5′-ACATGGCTTACAGC GCAGAGA-3′), circCELSR1 (forward,
5′-CGATTCGACACCAT CCATGAAG-3′ and reverse, 5′-GTCCACCTGCGTCTCAT
TGC-3′) and GAPDH (forward, 5′-AACGTGTCAGTGGTGGAC CTG-3′ and
reverse, 5′-GAGACCACCTGGTGCTCAGTG-3′).
Bioinformatics analysis
Gene Ontology (GO) analysis was performed to explore
the functional roles of target genes in terms of biological
processes, cellular components and molecular functions. Biological
pathways defined by Kyoto Encyclopedia of Genes and Genomes (KEGG),
Biocarta and Reactome (http://www.genome.jp/kegg/) were identified by
Database for Annotation, Visualization and Integrated Discovery
(DAVID; http://www.david.abcc.ncifcrf.gov/).
Statistical analysis
In the circRNA-seq analysis, circRNAs were
identified as differentially expressed between the EOC specimens
and normal ovarian tissues if the expression change was ≥2-fold
with a P-value of <0.05. Data from at least 3 independent
experiments are expressed as the means ± standard deviation. The
Mann-Whitney U test was used to determine the statistical
significance for comparisons between 2 groups, and a P-value less
than α which was adjusted by Bonferroni correction (adjusted α was
0.05/6, i.e., 0.0083) was considered statistically significant.
Overall survival (OS) and progress-free survival (PFS) were
calculated from the date of surgery to the date of death, progress
or last follow-up (April 2018). The average follow-up time was 29.0
months (range, 4 to 52 months). Kaplan-Meier survival analysis was
used for the analysis of the survival rate; The P-value was
calculated by the log-rank test and a P-value <0.05 was
considered to indicate a statistically significant difference.
Statistical analyses were carried out using SPSS statistical
software version 23.0 (SPSS, Chicago, IL, USA) and GraphPad Prism
5.0. All tests were two-sided.
Results
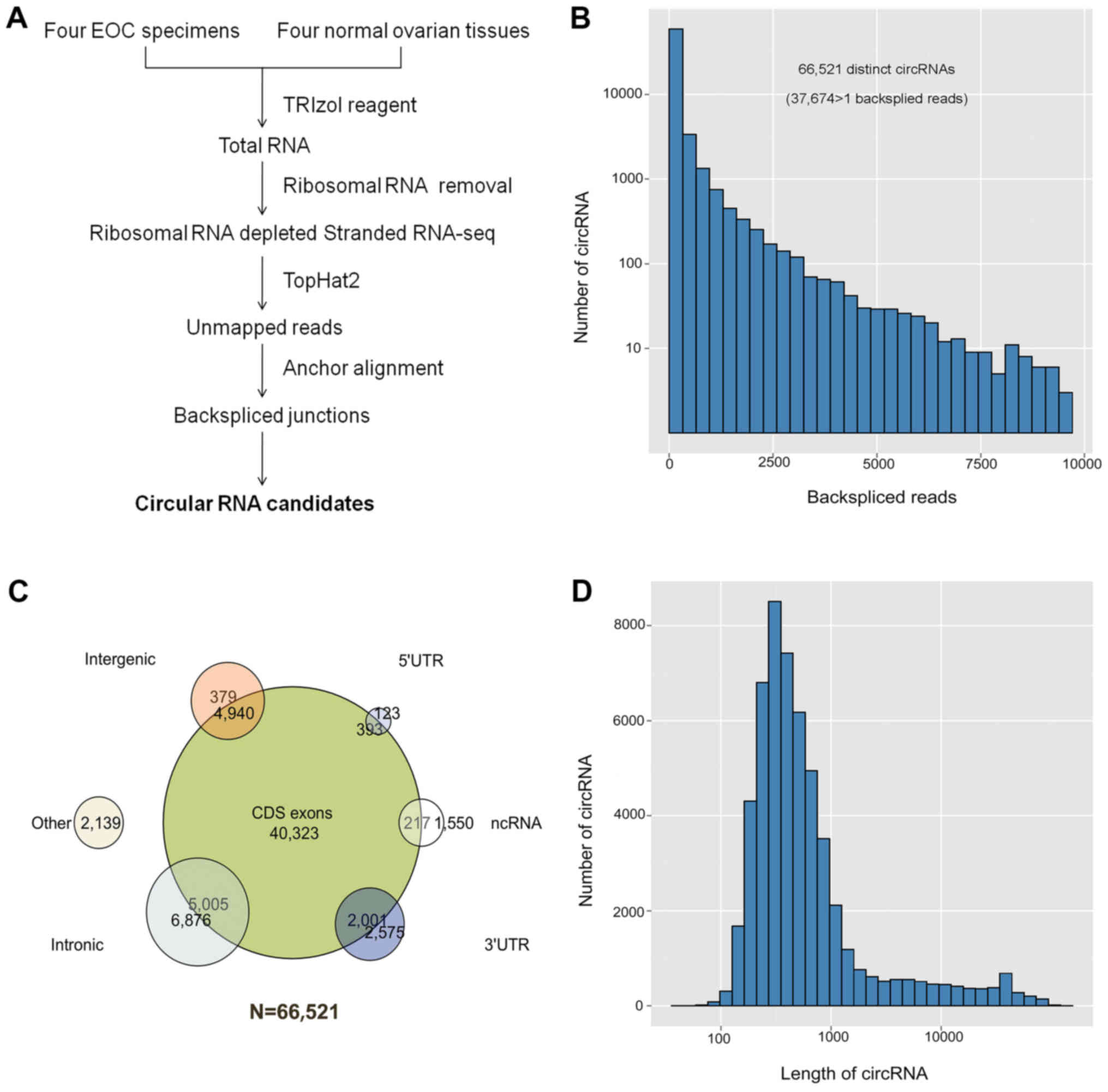
Identification of circRNAs by RNA-seq
analysis in EOC
Firstly, circRNAs were characterized using the
RNA-seq analysis of 4 EOC specimens and 4 normal ovarian tissues
(Fig. 1A). A total of 66,521
distinct circRNA candidates were identified from these samples. Of
these, 37,674 circRNAs contained at least 2 unique back-spliced
reads (Fig. 1B). The circRNA
candidates were annotated using the RefSeq database. More than 50%
of these circRNAs were transcribed from protein-coding exons, and a
small fraction of these were from introns, non-coding RNAs, 5′ UTR,
3′ UTR, and other sources (Fig.
1C). The median length of the exonic circRNAs was ~500
nucleotides (nt) (Fig. 1D).
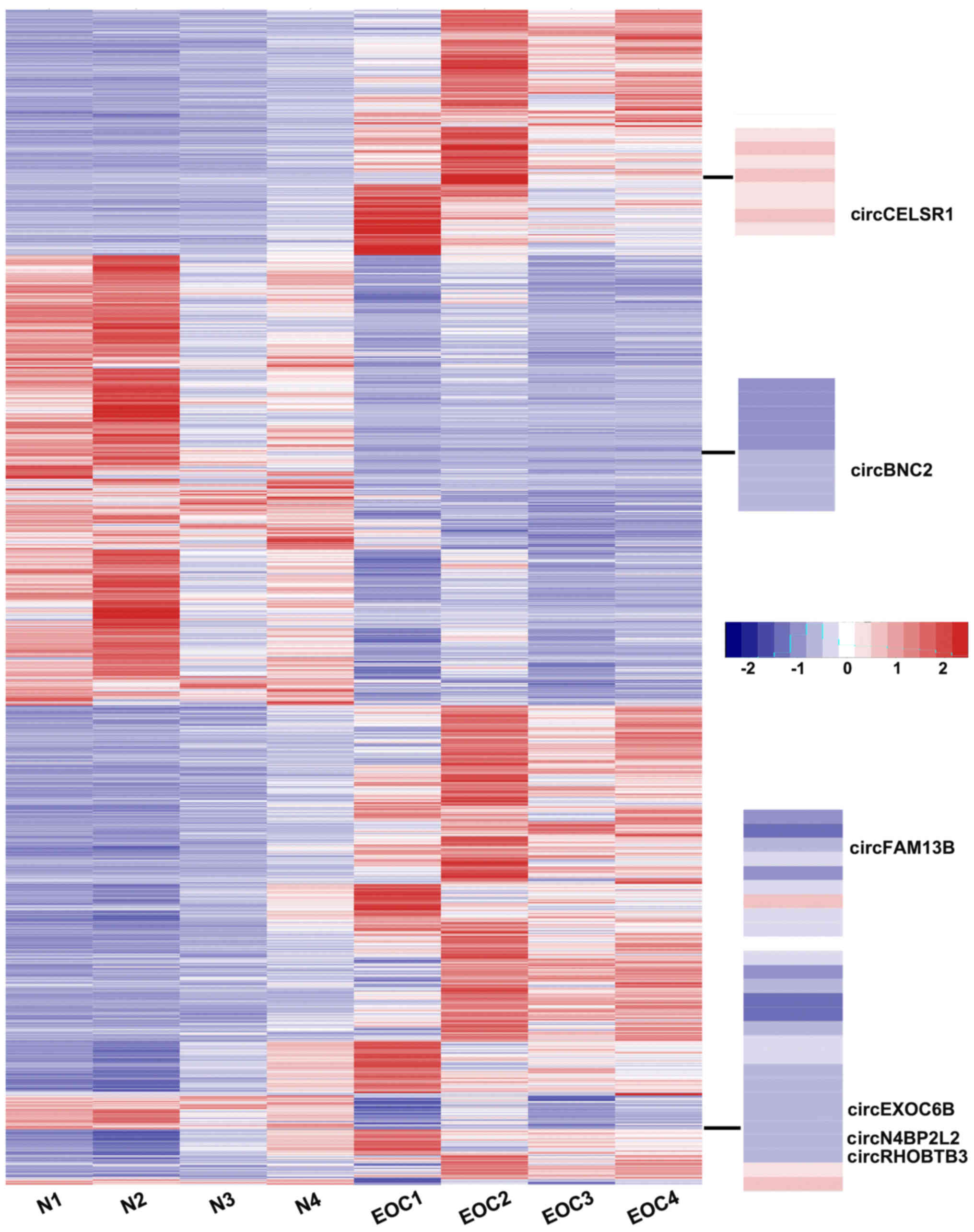
The relative expression analysis of these circRNAs
revealed a series of differentially expressed circRNAs in the EOC
specimens compared with the normal ovarian tissues (Fig. 2). Based on the RNA-seq results,
circRNAs with a signal altered by 2-fold and with value of
P<0.05 were identified as statistically altered. On the whole,
4,388 circRNAs were identified to be differentially expressed in
the EOC specimens compared with the normal ovarian tissues. Of
these, 2,556 circRNAs were upregulated, while 1,832 circRNAs were
downregulated in the EOC specimens (data not shown).
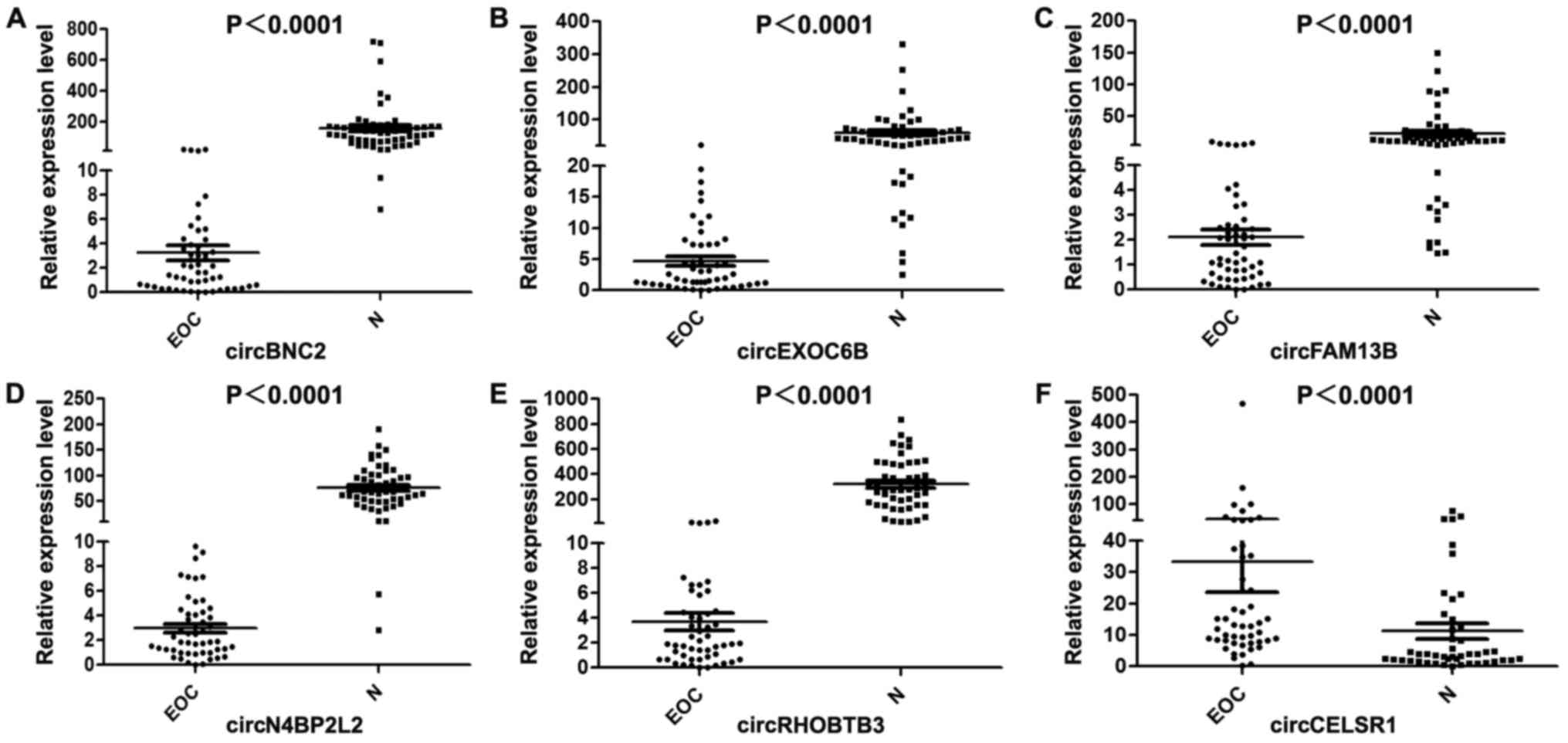
Validation of the differential expression
levels of circRNAs
In order to verify the reliability of the
circRNA-seq results, the 4,388 variable expressed circRNAs were
ranked based on calculating fold change and abundance, and the top
6 of the ranked circRNAs were selected for validation in the 54 EOC
specimens and 54 normal ovarian tissues by RT-qPCR. Finally, 5
downregulated circRNAs and 1 upregulated circRNA were selected
randomly from the 4,388 dysregulated circRNA transcripts. The
results of RT-qPCR revealed that these circRNAs were significantly
differentially expressed between the 2 groups. In particular,
circBNC2 (hsa_circ_0008732) (P<0.0001, Fig. 3A), circEXOC6B (hsa_circ_0009043)
(P<0.0001, Fig. 3B), circFAM13B
(hsa_circ_0001535) (P<0.0001, Fig.
3C), circN4BP2L2 (hsa_circ_0000471) (P<0.0001, Fig. 3D) and circRHOBTB3
(hsa_circ_0007444) (P<0.0001, Fig.
3E) were significantly downregulated in the EOC specimens,
whereas circCELSR1 (hsa_circ_0063809) (P<0.0001, Fig. 3F) was significantly upregulated in
the EOC group (Fig. 4). The
results of qRT-PCR were consistent with the circRNA-seq results,
which further verified the reliability of our circRNA-seq
results.
The association between circRNAs and the
clinicopathological features of EOC
The association between circRNAs and various
clinicopathological features of EOC was evaluated in the 54
patients with EOC. The median expression levels of circRNAs were
used to divide the circRNAs into the high level group and the low
level group. Our data revealed that the low level of circBNC2 was
associated with younger age (P=0.027), an advanced FIGO stage
(P=0.045), LNM (P=0.005) and massive ascites (P=0.034). The low
level of circEXOC6B was associated with younger age (P=0.018) and
LNM (P=0.032). The low level of circFAM13B was associated with
younger age (P=0.032), an advanced FIGO stage (P=0.044) and LNM
(P=0.034). The low level of circN4BP2L2 was associated with younger
age (P=0.047), an advanced FIGO stage (P=0.046) and LNM (P=0.003).
The low level of circRHOBTB3 was associated with younger age
(P=0.023) and LNM (P=0.009). In addition, the high level of
circCELSR1 was associated with an advanced FIGO stage (P=0.043), a
larger residual tumor diameter (P=0.035) and massive ascites
(P=0.021) (Table I).
Prognostic value of circRNAs in EOC
We further investigated the prognostic value of
various clinicopathological features and differentially expressed
circRNAs in patients with EOC. The results from univariate analysis
revealed that, an advanced FIGO stage (P=0.003, log-rank test) and
LNM (P=0.024, log-rank test) were predictive of a worse PFS.
Moreover, a high level of circN4BP2L2 (P=0.025, log-rank test) was
associated with a better PFS. In addition, a high level of
circEXOC6B (P=0.045, log-rank test) was associated with a better OS
(Table II).
 | Table IIThe prognostic values of various
clinicopathological features and differentially expressed circRNAs
in patients with epithelial ovarian cancer. |
Table II
The prognostic values of various
clinicopathological features and differentially expressed circRNAs
in patients with epithelial ovarian cancer.
| Variables | N | OS Rate (%) | P-value | PFS Rate (%) | P-value |
|---|
| All cases
(n=54) | | | | | |
| Age (years) | | | | | |
| <50 | 23 | 69.6 | 0.526 | 39.1 | 0.334 |
| ≥50 | 31 | 80.6 | | 51.6 | |
| FIGO stage | | | | | |
| I-II | 14 | 92.9 | 0.13 | 78.6 | 0.003a |
| III-VI | 40 | 70.0 | | 35.0 | |
| Residual tumor
diameter (cm) | | | | | |
| <1 | 43 | 74.4 | 0.866 | 44.2 | 0.570 |
| ≥1 | 11 | 81.8 | | 54.5 | |
| Lymph node
metastasis | | | | | |
| No | 35 | 80.0 | 0.354 | 60.0 | 0.024a |
| Yes | 19 | 63.2 | | 21.1 | |
| CA125 level
(U/ml) | | | | | |
| <600 | 31 | 74.2 | 0.817 | 48.4 | 0.860 |
| ≥600 | 23 | 78.3 | | 43.5 | |
| Ascites (ml) | | | | | |
| <100 | 13 | 76.9 | 0.661 | 53.8 | 0.350 |
| ≥100 | 41 | 75.6 | | 43.9 | |
| circBNC2 | | | | | |
| High | 27 | 70.4 | 0.814 | 51.9 | 0.809 |
| Low | 27 | 81.5 | | 40.7 | |
| circEXOC6B | | | | | |
| High | 27 | 88.9 | 0.045a | 48.1 | 0.636 |
| Low | 27 | 63.0 | | 44.4 | |
| circFAM13B | | | | | |
| High | 27 | 77.8 | 0.956 | 48.1 | 0.939 |
| Low | 27 | 74.1 | | 44.4 | |
| circN4BP2L2 | | | | | |
| High | 27 | 85.2 | 0.059 | 59.3 | 0.025a |
| Low | 27 | 66.7 | | 33.3 | |
| circRHOBTB3 | | | | | |
| High | 27 | 81.5 | 0.052 | 51.9 | 0.121 |
| Low | 27 | 70.4 | | 40.7 | |
| circCELSR1 | | | | | |
| High | 27 | 74.1 | 0.856 | 44.4 | 0.944 |
| Low | 27 | 77.8 | | 48.1 | |
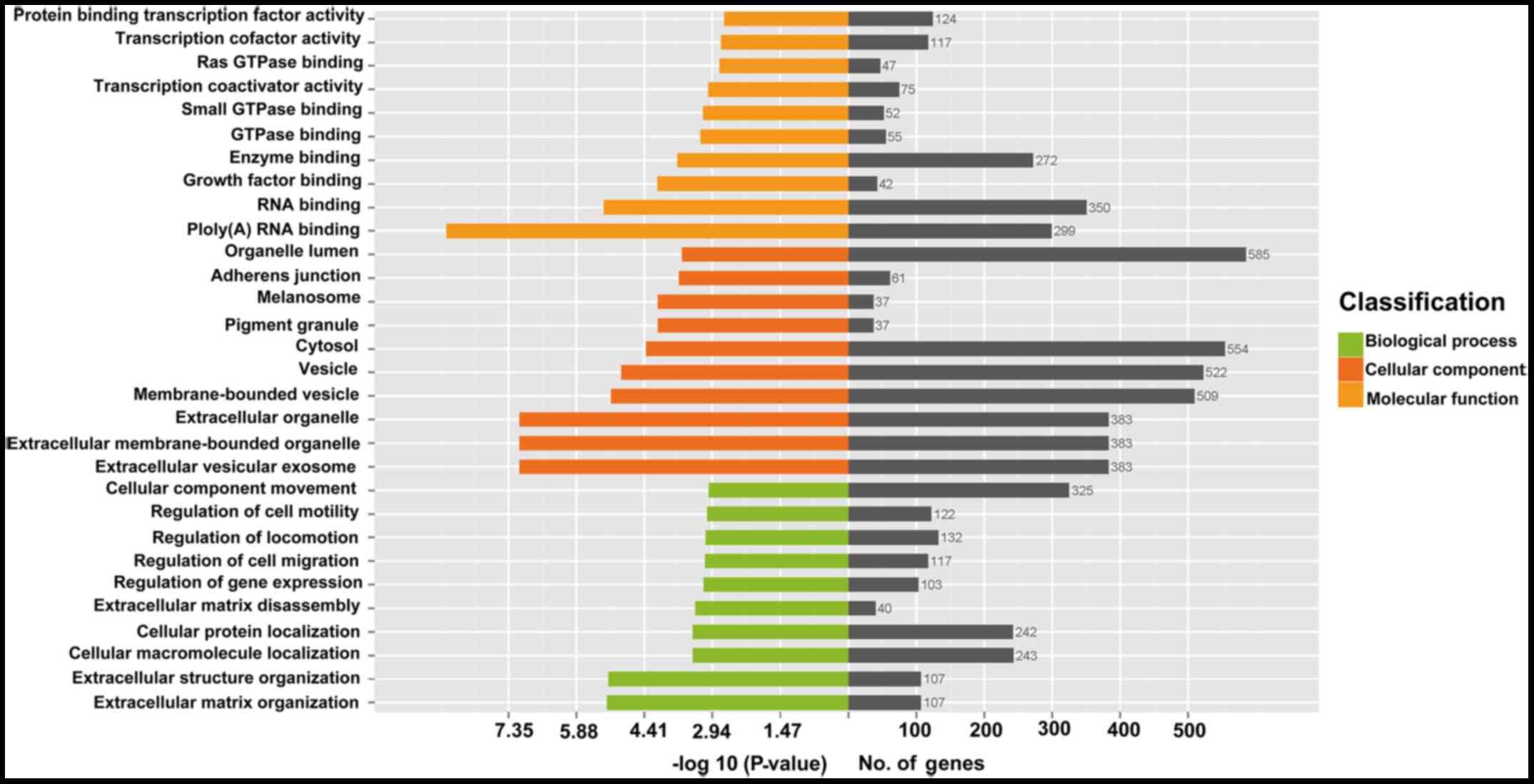
Bioinformatics analyses
The results of GO enrichment analysis of the
differentially expressed circRNAs with identified target genes are
shown in Fig. 5. GO analysis
revealed that numerous target genes were involved in the biological
processes, such as cellular process, regulation of biological
process, metabolic process, etc. These processes were associated
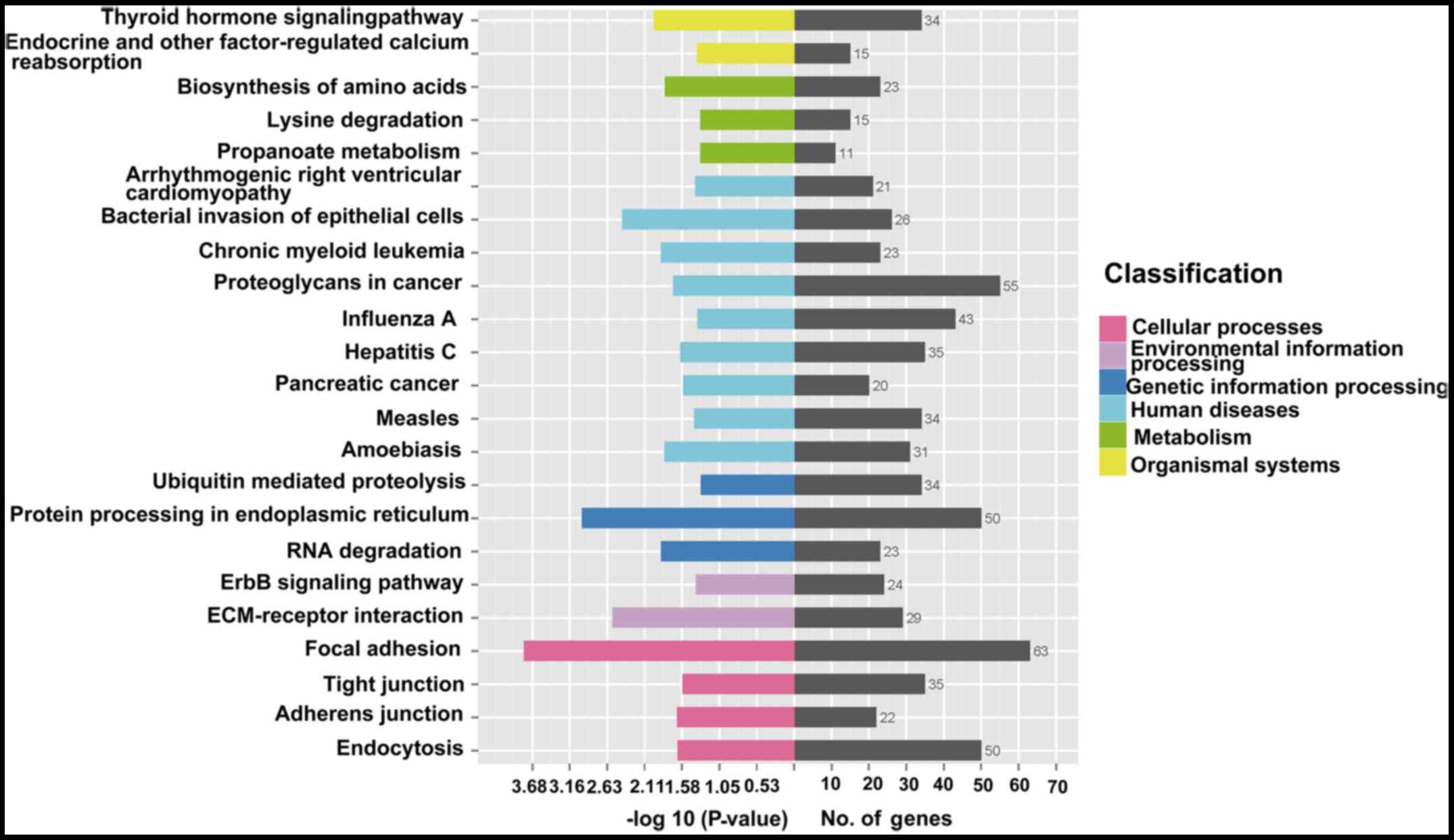
with human tumorigenesis and metastasis. KEGG analysis revealed
that there were 23 pathways related to differentially expressed
circRNAs, including the ErbB signaling pathway, ECM-receptor
interaction and focal adhesion (Fig.
6).
Discussion
In this study, we successfully identified a large
number of circRNAs in EOC specimens. We also found that a
considerable portion of these circRNAs were differentially
expressed in cancer specimens compared with normal ovarian tissues.
Our findings suggest that these differentially expressed circRNAs
may be regulated and may play an important role in the development
of EOC. We further evaluated the prognostic value of circRNAs in
EOC and found that circEXOC6B and circN4BP2L2 may have potential
for use as prognostic biomarkers in EOC. To the best of our
knowledge, we are the first to perform a detailed circRNA-seq
analysis of EOC species and normal ovarian tissues.
The circNRA expression profiles in this study
demonstrated that 2,556 circRNAs were abnormally upregulated and
1,832 circRNAs were downregulated in the EOC specimens compared
with the normal ovarian tissues. In particular, circBNC2,
circEXOC6B, circFAM13B, circN4BP2L2, circRHOBTB3 and circCELSR1
were validated to be significantly dysregulated in the EOC
specimens compared with the normal ovarian tissues by RT-qPCR.
Recently, Bahn et al (26)
successfully identified >400 circRNAs in human cell-free saliva.
It has also been shown that, compared to their corresponding linear
counterparts, circRNAs have a higher expression levels in blood.
While the liner mRNAs are actually absent, the circRNA isoforms are
easily detected, thus enabling circRNAs to embody disease
information which conventional RNA assay fails to determine
(19). Given the abundance of
circRNAs, their tissue-specific expression patterns and their high
stability in vivo, circRNAs may be used as promising
biomarkers for clinical applications in the future. Accordingly,
the results of this study suggest that circBNC2, circEXOC6B,
circFAM13B, circN4BP2L2, circRHOBTB3 and circCELSR1 may be
potential diagnostic biomarkers for EOC. We further tested these 6
circRNAs in our laboratory with tissues and plasma.
By performing outcome analysis, we found that the
expression levels of circEXOC6B and circN4BP2L2 were significantly
associated with OS and PFS, respectively. The OS of patients with
EOC with a low circEXOC6B expression was significantly worse than
that of patients with a high circEXOC6B expression. Moreover,
patients with EOC with a low circN4BP2L2 expression had a
significantly worse PFS than those with a high circN4BP2L2
expression. These results suggested that circEXOC6B and circN4BP2L2
may act as important and useful prognostic biomarkers in patients
with EOC. Our data demonstrated that circEXOC6B and circN4BP2L2
were negatively associated with FIGO stage and/ or LNM. In
addition, we found that FIGO stage and LNM were negatively
associated with the survival of patients with EOC. Thus, circEXOC6B
and circN4BP2L2 may play a profound role in regulating tumor
invasion and metastasis. Prognostic biomarkers can help in
selecting personalized treatment regimens for patients with EOC.
circRNAs are highly stable in cells, and their tissue-specific
character make them more sensitive to reflect the real condition
in vivo (6). Therefore, our
findings verified the promising clinical values of circRNAs in
tumor diagnosis and treatment.
This study has some limitations. Since the sample
size in our study was relatively small and was limited to Han
ethnicity Chinese patients from a single institution, the
specificity and accuracy of the model cannot be estimated
accurately. Therefore, prospectively multicenter studies with a
larger sample size and heterogeneous cohorts of patients should be
conducted in the future to verify our results. Furthermore,
functional experimental analyses of cancer cell lines and
experimental animals should be further conducted to determine
whether these circRNAs are involved in the tumorigenesis and
progression of EOC.
In conclusion, this study identified the circRNA
profile of EOC specimens. We demonstrated that circBNC2,
circEXOC6B, circFAM13B, circN4BP2L2, circRHOBTB3 and circCELSR1
were significantly dysregulated in EOC specimens and may be
potential diagnostic biomarkers for EOC. We also found that
circEXOC6B and circN4BP2L2 may serve as novel prognostic biomarkers
in patients with EOC. However, future experiments with a larger
sample are required to verify the current findings and to elucidate
the regulatory mechanisms of action of circRNAs in the
tumorigenesis of EOC.
Funding
This study was funded by the Chinese Academy of
Medical Sciences Initiative for Innovative Medicine (grant no.
CAMS-2017-I2M-1-002).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors’ contributions
LN, BL, YH and JHL contributed to study conception
and design, data analysis, and manuscript preparation. WZ
contributed to data analysis. MY, SW, DC, JY and KS contributed to
data collection and data analysis. All authors have read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
The use of human tissues was approved by the Ethics
Committee of PUMCH and patient consent was obtained.
Consent for publication
Not applicable.
Competing Interests
The authors declared that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bowtell DD, Böhm S, Ahmed AA, Aspuria PJ,
Bast RC Jr, Beral V, Berek JS, Birrer MJ, Blagden S, Bookman MA, et
al: Rethinking ovarian cancer II: Reducing mortality from
high-grade serous ovarian cancer. Nat Rev Cancer. 15:668–679. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kroeger PT Jr and Drapkin R: Pathogenesis
and heterogeneity of ovarian cancer. Curr Opin Obstet Gynecol.
29:26–34. 2017. View Article : Google Scholar :
|
|
5
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang PL, Bao Y, Yee MC, Barrett SP, Hogan
GJ, Olsen MN, Dinneny JR, Brown PO and Salzman J: Circular RNA is
expressed across the eukaryotic tree of life. PLoS One.
9:e908592014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cocquerelle C, Mascrez B, Hétuin D and
Bailleul B: Mis-splicing yields circular RNA molecules. FASEB J.
7:155–160. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Salzman J, Gawad C, Wang PL, Lacayo N and
Brown PO: Circular RNAs are the predominant transcript isoform from
hundreds of human genes in diverse cell types. PLoS One.
7:e307332012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lasda E and Parker R: Circular RNAs:
Diversity of form and function. RNA. 20:1829–1842. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Starke S, Jost I, Rossbach O, Schneider T,
Schreiner S, Hung LH and Bindereif A: Exon circularization requires
canonical splice signals. Cell Rep. 10:103–111. 2015. View Article : Google Scholar
|
|
11
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bachmayr-Heyda A, Reiner AT, Auer K,
Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW,
Zeillinger R and Pils D: Correlation of circular RNA abundance with
proliferation - exemplified with colorectal and ovarian cancer,
idiopathic lung fibrosis, and normal human tissues. Sci Rep.
5:80572015. View Article : Google Scholar
|
|
13
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar :
|
|
14
|
Chen LL and Yang L: Regulation of circRNA
biogenesis. RNA Biol. 12:381–388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Conn SJ, Pillman KA, Toubia J, Conn VM,
Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA and
Goodall GJ: The RNA binding protein quaking regulates formation of
circRNAs. Cell. 160:1125–1134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Venø MT, Hansen TB, Venø ST, Clausen BH,
Grebing M, Finsen B, Holm IE and Kjems J: Spatio-temporal
regulation of circular RNA expression during porcine embryonic
brain development. Genome Biol. 16:2452015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu L, Gong X, Sun L, Zhou Q, Lu B and Zhu
L: The circular RNA Cdr1as act as an oncogene in hepatocellular
carcinoma through targeting miR-7 expression. PLoS One.
11:e01583472016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen Y, Li C, Tan C and Liu X: Circular
RNAs: A new frontier in the study of human diseases. J Med Genet.
53:359–365. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nair AA, Niu N, Tang X, Thompson KJ, Wang
L, Kocher JP, Subramanian S and Kalari KR: Circular RNAs and their
associations with breast cancer subtypes. Oncotarget.
7:80967–80979. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shang X, Li G, Liu H, Li T, Liu J, Zhao Q
and Wang C: Comprehensive circular RNA profiling reveals that
hsa_circ_0005075, a new circular RNA biomarker, is involved in
hepatocellular carcinoma development. Medicine (Baltimore).
95:e38112016. View Article : Google Scholar
|
|
22
|
Chen J, Li Y, Zheng Q, Bao C, He J, Chen
B, Lyu D, Zheng B, Xu Y, Long Z, et al: Circular RNA profile
identifies circPVT1 as a proliferative factor and prognostic marker
in gastric cancer. Cancer Lett. 388:208–219. 2017. View Article : Google Scholar
|
|
23
|
Zhu M, Xu Y, Chen Y and Yan F: Circular
BANP, an upregulated circular RNA that modulates cell proliferation
in colorectal cancer. Biomed Pharmacother. 88:138–144. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhong Z, Lv M and Chen J: Screening
differential circular RNA expression profiles reveals the
regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in
bladder carcinoma. Sci Rep. 6:309192016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ahmed I, Karedath T, Andrews SS, Al-Azwani
IK, Mohamoud YA, Querleu D, Rafii A and Malek JA: Altered
expression pattern of circular RNAs in primary and metastatic sites
of epithelial ovarian carcinoma. Oncotarget. 7:36366–36381. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bahn JH, Zhang Q, Li F, Chan TM, Lin X,
Kim Y, Wong DT and Xiao X: The landscape of microRNA,
Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem.
61:221–230. 2015. View Article : Google Scholar :
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
28
|
Wang P and Lv L: miR-26a induced the
suppression of tumor growth of cholangiocarcinoma via KRT19
approach. Oncotarget. 7:81367–81376. 2016. View Article : Google Scholar : PubMed/NCBI
|