Introduction
Breast cancer is a major cause of mortality among
women worldwide (1). Despite much
progress being made in therapeutic approaches, including early
detection, surgery, radiation, as well as endocrine and
anti-HER-2 therapies, cancer patients succumb to the disease
due to metastasis, which is incurable (2,3).
Therefore, the better understanding of the biological pathways that
contribute to disease progression and the development of novel
therapeutic modalities is of paramount importance.
MicroRNAs (miRNAs or miRs), are small,
single-stranded, endogenous, non-coding RNA molecules, 20-23
nucleotides in length, which play an essential role in the
regulation of gene expression at the post-transcriptional level. In
several malignant tumors, such as breast, lung, hepatocellular and
pancreatic cancer, significant differences have been found in the
expression profiles of miRNAs compared to normal tissues (4-7).
These miRNAs regulate the expression of tumor suppressor genes
and/or oncogenes. For example, the overexpression of miR-506 has
been shown to inhibit the expression of tumor suppressor gene, IQ
motif containing GTPase activating protein 1 (IQGAP1)
(8); conversely, the lower
expression or absence of miR-9 has been shown to block the
downregulation of the HER-2 oncogene, leading to breast
cancer progression (9).
In our pervious study, we observed that miR-193b was
significantly downregulated in breast cancer cell lines, which
promoted tumor cell proliferation, migration and invasion;
RAB22A was also identified as a potential downstream target
of this miRNA (10). RAB22A is a
small GTPase, belonging to the RAB protein family, RAB5 subfamily
(11), which mainly presents in
early endosomes, Golgi bodies and late endosomes. RAB proteins are
involved in the regulation of vesicular traffic and exosome
formation (12). Studies have
found that the RAB5 subfamily (including RAB5, RAB21, RAB22A and
RAB22B) is mainly involved in the endocytosis, transport and
metabolism of growth factor receptors, and may thus be associated
with cancer progression (13-15).
In different cell lines, RAB22A is known as an endosomal-associated
protein, involved in exocrine vehicle formation (16,17).
At the plasma membrane of MDA-MB-231 cells, RAB22A has been shown
to be a component of microvesicles. In breast cancer cells, the
knockdown of the RAB22A gene has been shown to decrease
hypoxia-induced exocrine vehicle formation and the cell invasive
ability (17).
Exosomes are a type of exocrine vehicle, ranging in
diameter from 30 to 100 nm, originating from the intracellular
body. Exosomes contain membrane-anchored receptors, adhesion
molecules, signaling proteins, active oncogenes and nucleic acids.
Exosomes can alter the biological behavior of these recipient cells
by transferring their cargo to recipient cells and are now
considered to be key players in cell-to-cell communications. For
example, cancer cells release exosomes carrying proteins, lipids
and nucleic acids that promote tumor progression, reprogram tumor
microenvironment and modulate immune response (18).
Our previous study on miR-193b demonstrated its
relevance in cancer development, progression and metastasis
(10). However, the mechanisms
through which miR-193b interacts with its targets, leading to
cancer development/progression, remain largely unknown. In this
study, we thus investigated the role of miR-193b in the regulation
of its target gene, RAB22A, and in the alteration of the
exosomal biosynthesis pathway by altering the biological behavior
of recipient cells. The findings of this study may aid in the
development of novel targeted therapies.
Materials and methods
Human tissues
We collected 23 groups of samples (20 used for
RT-qPCR and 3 for western blot analysis) we with breast cancer
lesions (including tumor and metastatic lymph node) and their
matched adjacent normal breast tissues from patients with both
breast cancer and axillary lymph node metastases at the Clinic of
the Breast Surgery Department of China-Japan Union Hospital
affiliated to Jilin University (Jilin, China). All the clinical
procedures were approved by the Committees of Clinical Ethics of
China-Japan Union Hospital of Jilin University (2017022218, Jilin,
China). Written informed consents were obtained from individual or
guardian participants.
Cell lines and transfection
All cell lines were purchased from the American Type
Culture Collection (ATCC, Manassas, VA, USA), including human
normal mammary epithelial MCF-10A cells were grown in DMEM/F12
Ham’s Mixture supplemented with 5% horse serum (Gemini Bio,
Woodland, CA, USA), EGF 20 ng/ml, insulin 10 µg/ml,
hydrocortisone 0.5 mg/ml and cholera toxin 100 ng/ml (all from
Sigma-Aldrich, St. Louis, MO, USA). The primary human breast cancer
cell lines, MCF-7, MDA-MB-231 and MDA-MB-453, were cultured as
previously described (10). The
SK-BR-3, ZR-75 and T47D cells were grown in RPMI supplemented with
10% fetal bovine serum (FBS; Gibco/Thermo Fisher Scientific,
Waltham, MA, USA); the MDA-MB-468 cells were grown in L-15 medium
with 10% FBS. All cell lines were maintained at 37°C and 5%
CO2.
The synthetic mimic pre-miR-193b (stem-loop
sequence: GUGGUCUCAGAAUCGGGGUUUUGAGGGCGAGAUGA
GUUUAUGUUUUAUCCAACUGGCCCUCAAAGUCCCG CUUUUGGGGUCAU), the
negative-scramble control RNAs (SC), pcDNA3.1-RAB22A ORF plasmid
and blank pcDNA3.1 vectors were purchased from Ambion/Thermo Fisher
Scientific. The MCF-7 or SK-BR-3 cells were seeded in 6-cm dishes
until approximately 70% conf luency, and Lipofectamine 2000
(Invitrogen/Thermo Fisher Scientific) was then added to the diluted
miR and plasmid at a final concentration of 40 nmol/l, followed by
incubation for 20 min at room temperature, and was then added to
the cell medium. The cells were then incubated at 37°C, 5%
CO2 for 6 h. Subsequently, fresh medium was added. The
cells were harvested 24 h later for used in RT-qPCR, western blot
analysis and in vitro experiments.
Short hairpin RNA (shRNA) sequences were designed by
Hanbio Biotechnology Co. Ltd. (HH20170612LLN-LV01, Shanghai, China)
to target human RAB22A. After annealing, double strands of shRNA
were inserted into lentiviral pHBLV-Scramble-Puro vector (Hanbio,
Shanghai, China). This construct, named shRAB22A, and a
negative-scramble control, named SC were used according to the
manufacturer’s instructions. In brief, 2×105 cells were
incubated with 2×107 transducing units (TU) lentivirus
and 8 µg Polybrene (Hanbio) in 1 ml cell medium at 37°C, 5%
CO2 for 4 h. Subsequently, 1 ml fresh medium was added
for 24 h prior to changing back to normal media. Stable cell lines
were generated after 48 h by using puromycin (Hanbio)
selection.
RT-qPCR
Human tissues were obtained and analyzed. The
MCF-10A, MCF-7, MDA-MB-231, MDA-MB-453, SK-BR-3, ZR-75, T47D and
MDA-MB-468 were cultured in indicated medium. For mRNA expression,
total RNA was purified using TRIzol reagent (Invitrogen/Thermo
Fisher Scientific) according to the manufacturer’s instructions.
Following quan-titation using Synergy HT (BioTek, Highland Park,
Illinois, USA), 1 µg of total RNA was reverse transcribed
using SuperScript III Reverse Transcriptase (Invitrogen/Thermo
Fisher Scientific) according to the manufacturer’s instructions.
The cDNA (100 ng) was added to the RT-qPCR mixture containing SYBR
Premix ExTaq II (Takara Bio Inc., Kusatsu, Shiga, Japan) and the
appropriate primers. All RT-qPCR analyses were performed using the
ABI PRISM 7900 Sequence Detection System (Applied Biosystems,
Foster City, CA, USA). The cycling parameters are as follows:
Initial melting at 95°C for 30 sec, followed by 40 cycles of 95°C
for 5 sec, 57°C for 30 sec and 68°C for 30 sec. The relative fold
change in RNA expression was calculated using the 2-ΔΔCq
method (19), GAPDH was
used as an endogenous control for normalization. The expression of
miRNA was conducted by using TaqMan MicroRNA Assay (Applied
Biosystems) specific for hsa-miR-193b and RNU44. Prior to
RT, the RNA extracted using the mir-Vana kit (Ambion/Thermo Fisher
Scientific). Firstly, 5 µl total RNA with 3 µl
specific primers were added to TaqMan MicroRNA Reverse
Transcription kit (Applied Biosystems), incubated at 16°C for 30
min, 42°C for 30 min, 85°C for 5 min and hold in 4°C. Then, the
products were subsequently amplified by incubated at 95°C for 10
min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min,
RNU44 was used as an endogenous control for normalization.
The primer sequences used in this study were as follows:
RAB22A forward (5′-3′), ttgtagttgccattgcagga and reverse
(5′-3′), aggctgtcttcggagtttga; GAPDH forward (5′-3′),
gtctcctctgacttcaacagcg and reverse (5′-3′),
accaccctgttgctgtagccaa.
Cell proliferation and clonogenic
assay
The SK-BR-3 or MCF-7 cells cultured for the
indicated periods of time (0-5 days), and cell viability was
detected using the cell counting kit-8 (CCK-8) according to the
manufacturer’s instructions (Beyotime Institute of Biotechnology,
Shanghai, China) and read using a microplate reader (Synergy HT,
BioTek), at 450 nm.
The clonogenicity of a single cell was detected by
colony assay. Following treatment with 0.25% trypsin, the SK-BR-3
or MCF-7 cells were collected and adjusted to a concentration of
300 cells/Petri dish, and 2 ml of pre-heated culture medium were
then added. The cells were cultured at 37°C with 5% CO2
for 2 weeks. After the colonies were visible to the naked eye, the
cells were washed twice with phosphate-buffered saline (PBS),
fixated with 4% paraformaldehyde for 15 min before Giemsa
(Sigma-Aldrich) staining for 10 min at room temperature and the
number of colonies was counted.
Transwell assays
The SK-BR-3 cells were maintained in RPMI
supplemented with 10% FBS, and 1×105 cells were plated
on BD BioCoat MATRIGEL Invasion Chambers with Matrigel and control
inserts with polyethylene terephthalate membrane (BD Biosciences,
San Jose, CA, USA) to assess the cell invasive and migratory
abilities as previously described (10,20).
Exosome isolation and purification
Exosomes were isolated from the supernatant either
of the SK-BR-3 or MCF-7 cell culture media. In brief,
2×106 cells were plated in a Petri dish with RPMI
supplemented with 10% FBS and 1% penicillin/streptomycin. The
following day, the medium was changed to exosome-free RPMI
supplemented with 10% exosome-depleted FBS. The culture medium was
collected until the cells reached 70-80% confluence and exosomes
were isolated by ultracentrifugation with a rotor (L8-80M; Beckman,
Brea, CA, USA) at the centrifugal force of 110,000 × g for 70 min.
Exosomes were re-washed in PBS at 110,000 × g for 70 min to
eliminate contaminating proteins, then re-suspended in 100
µl PBS and immediately tested or stored at −80°C for further
analysis. TSG101 and HSP70 levels were detected by western blot
analysis as exosome markers.
To dissolve exosome components, exosomes were
treated (37°C, 60 min) with 500 µg/ml proteinase K
(Sigma-Aldrich) dissolved in RNase-free water, followed by heat
inactivation of the protease (60°C, 10 min) and incubation (37°C,
10 min) with 2 µg/ml protease-free RNase A
(Sigma-Aldrich).
Western blot analysis
For western blot analysis, aliquots of total protein
extracts (25 µg) from human tissues, cells or exosomes were
loaded and separated by SDS-PAGE (15%), and samples were
transferred onto PVDF membranes (Millipore, Billerica, MA, USA).
The membranes were then blocked in 5% bovine serum albumin and
incubated at 4°C overnight with specific primary antibodies. The
secondary antibodies (diluted 1:2,000), anti-rabbit or anti-mouse
IgG antibodies (cat. no. 7074 or 7076) from Cell Signaling
Technology (Beverly, MA, USA), were added followed by incubation at
room temperature for 2 h, and the detected signals were visualized
using BeyoECL Plus (Beyotime Institute of Biotechnology, Haimen,
Jiangsu, China), as recommended by the manufacturer. Densitometry
was performed using ImageJ software (Bundled with 64-bit Java for
Windows, version 1.8, Bethesda, MD, USA). The densitometry value
for each sample was normalized against the value for GAPDH or
β-actin to obtain the intensities for RAB22A reported in the
figures. To examine the concentrations of proteins in the lysates,
the following antibodies were used: RAB22A (1:500, cat. no.
16181-1-AP; Proteintech, Rosemont, IL, USA), TSG101 (1:500, cat.
no. sc-7964), HSP70 (1:500, cat. no. sc-32239), Calnexin (1:500,
cat. no. sc-23954), GAPDH (1:1,000, cat. no. sc-47724) (all from
Santa Cruz Biotechnology, Santa Cruz, CA, USA), TFIIB (1:1,000,
cat. no. 4169; Cell Signaling Technology) and β-actin (1:5,000,
cat. no. A5441; Sigma-Aldrich).
Co-culture experiments
A total of 1×106 SK-BR-3 or MCF-7 cells were plated
in a Petri dish in exosome-depleted culture medium and then
supplemented with the indicated exosomes (20 µg/ml), every
day for 2 days. Subsequently, 2×104 cells were seeded
for the indicated days and then subjected to cell density assay.
For the Transwell assay, after priming with the exosomes,
1×105 cells were plated on the chambers to assess the
cell invasive and migratory abilities.
Fluorescence microscopy analysis
The SK-BR-3 cells grown on coverglass slips were
incubated for 1 h with the fluorescent probe DiO (Vybrant DiO
Cell-Labling Solution from Invitrogen/Thermo Fisher Scientific)
labeled exosomes in RPMI at 37°C. The cells were then washed with
PBS to remove unbound labeled exosomes, fixed with 4%
paraformaldehyde for 15 min and stained nuclei with DAPI
(4′,6-diamidino-2-phenylindole, Invitrogen/Thermo Fisher
Scientific) at room temperature for 5 min and subsequently imaged
with a Zeiss LSM 510 Axiovert 200M microscope (Carl Zeiss Inc.,
Oberkochen, Germany) to produce confocal-like images.
Statistical analysis
Experimental cultures were set up in triplicate and
each experiment was repeated at least 3 times. Data are presented
as the means ± standard error unless otherwise indicated. Unpaired
two-tailed student t-tests or analysis of variance (ANOVA) were
used to determine the P-values for differences between samples.
One-way ANOVA, followed by the LSD post hoc test was used to
compare mean differences between 3 or more groups. Two-way ANOVA,
followed by Dunnett’s or Sidak’s multiple comparisons test was used
for multiple comparisons. Means with P-values <0.05 were
considered statistically significant. The ratio of the protein band
intensities relative to that of GAPDH or β-actin, the
quantification of colonies in clonogenic assay and cells in
Transwell assays was carried out for each sample using ImageJ
software (Bethesda, MD, USA).
We used KM Plotter (http://kmplot.com/analysis/) to determine the
prognostic values of RAB22A in breast cancer (21). Gene expression data and survival
information of 3,951 breast cancer patients downloaded from GEO by
KM Plotter. Cancer patients were divided into the high and low
expression groups by the median values of RAB22A mRNA
expression, and the 2 groups were then compared by log-rank test
followed by Cox proportional hazards regression, and a Kaplan-Meier
plot was drawn. The number of cases, median values of mRNA
expression level, HR, 95% CI and log-rank P-value were extracted
from the KM plotter webpage.
Results
Upregulation of RAB22A is associated with
tumor production and lymph node metastasis
To further examine the role of miR-193b and
RAB22A in predicting breast cancer proliferation and
metastasis (10), the expression
levels of these genes were analyzed simultaneously by RT-qPCR using
20 groups of samples with breast cancer lesions and their matched
adjacent normal breast tissues. In this analytical approach, we
identified a signature of RAB22A and miR-193b, exhibiting a
negative association between the tumor and peritumoral tissues
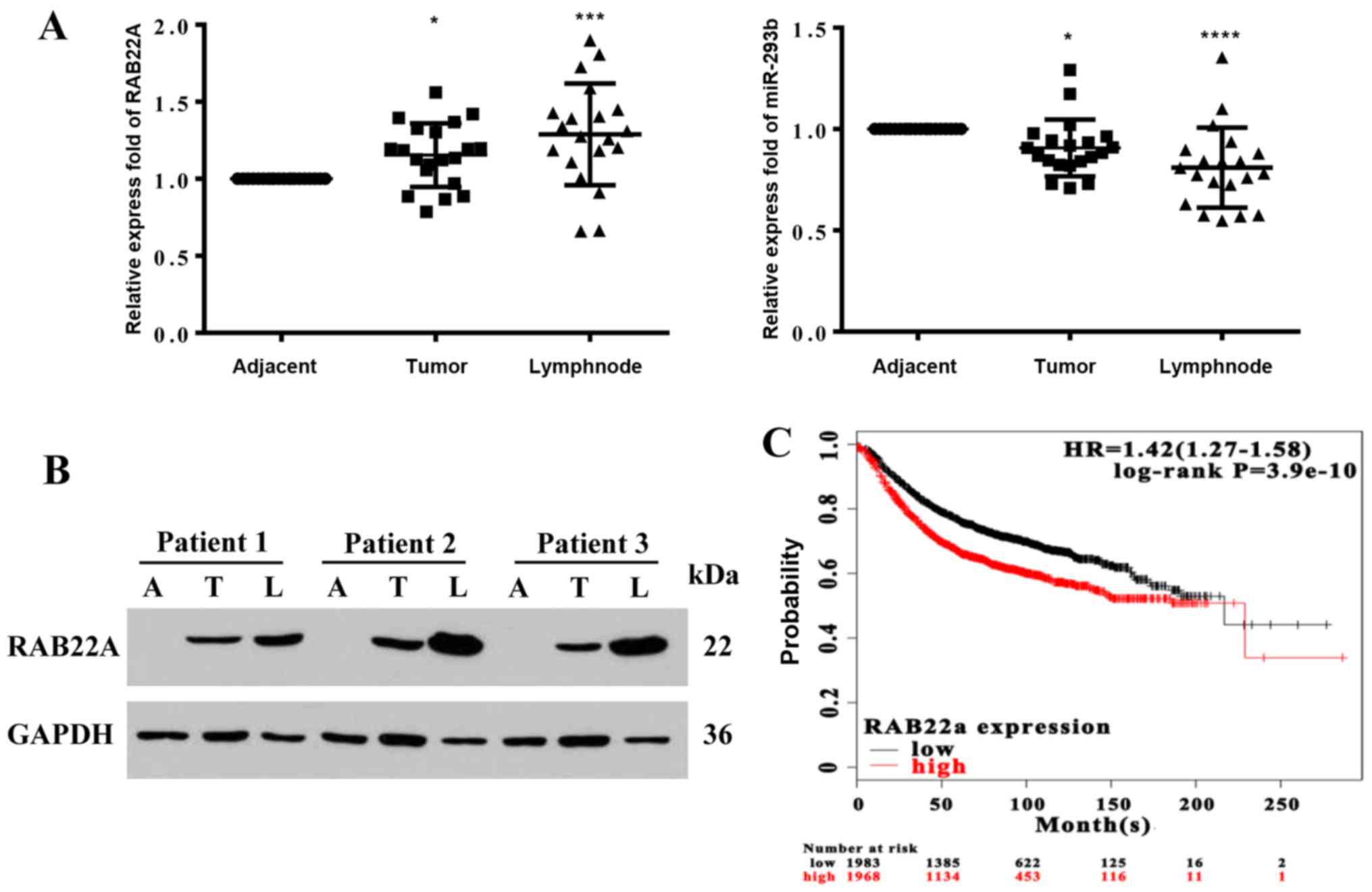
(Fig. 1A). Whereas the gene
expression level of miR-193b was decreased, that of RAB22A
was increased in the cancer lesions when compared with the adjacent
normal tissues. RAB22A protein extraction from a random selection
of 3 groups of samples revealed that its level was increased in the
cancer lesions, particularly in the metastatic tissues with lymph
node metastasis, compared to the corresponding adjacent normal
tissues (Fig. 1B). Gene expression
data and survival information of 3,951 breast cancer patients
downloaded from GEO, patients were divided into high and low
expression group by the median values of RAB22A mRNA
expression. The Kaplan-Meier survival curve was used to confirm the
prognostic power of RAB22A mRNA (Fig. 1C), which revealed that a lower
expression of RAB22A was associated with a longer survival
time of the patients. These results confirm our previous
observation showing a similar negative association between
RAB22A and miR-193b in breast cancer cell lines (10).
RAB22A is the regulatory target gene of
miR-193b
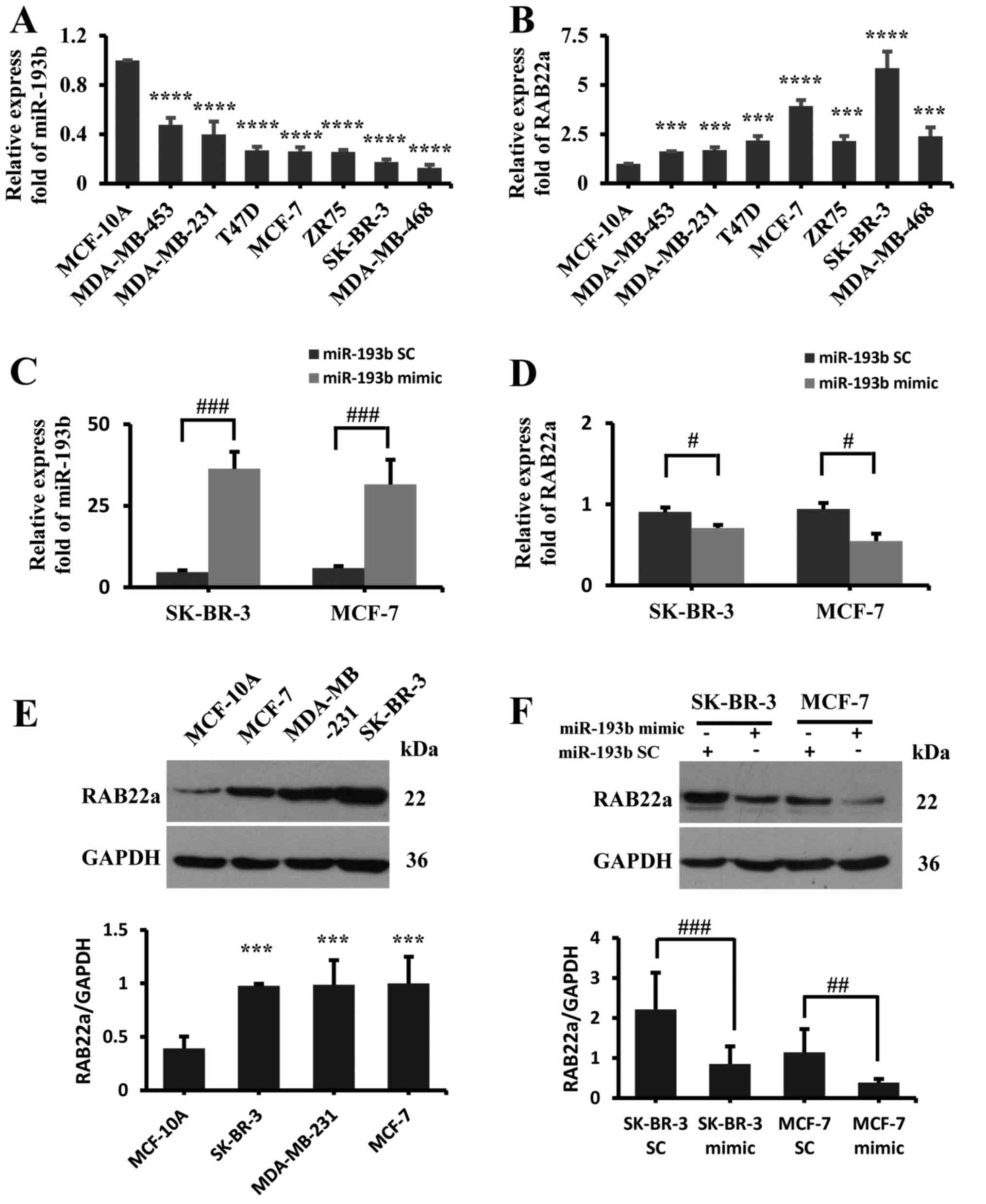
A negative association between miR-193b and
RAB22A expression was also observed in the normal breast
epithelial MCF-10A cells compared to the other human breast cancer
cell lines, such as MDA-MB-453, MDA-MB-231, SK-BR-3, MCF-7,
MDA-MB-468, ZR-75 and T47D (Fig. 2A, B
and E). This result supports our previous observation
suggesting RAB22A as a downstream target of miR-193b in
breast cancer (10). To further
confirm this observation, in this study, miR-193b mimic was
transfected into the MCF-7 and SK-BR-3 cell lines. The results
revealed that miR-193b overexpression (Fig. 2C) markedly downregulated
RAB22A gene expression, as detected by RT-qPCR (Fig. 2D) and western blot analysis
(Fig. 2F).
RAB22A knockdown inhibits the cell
proliferative ability of breast cancer cells
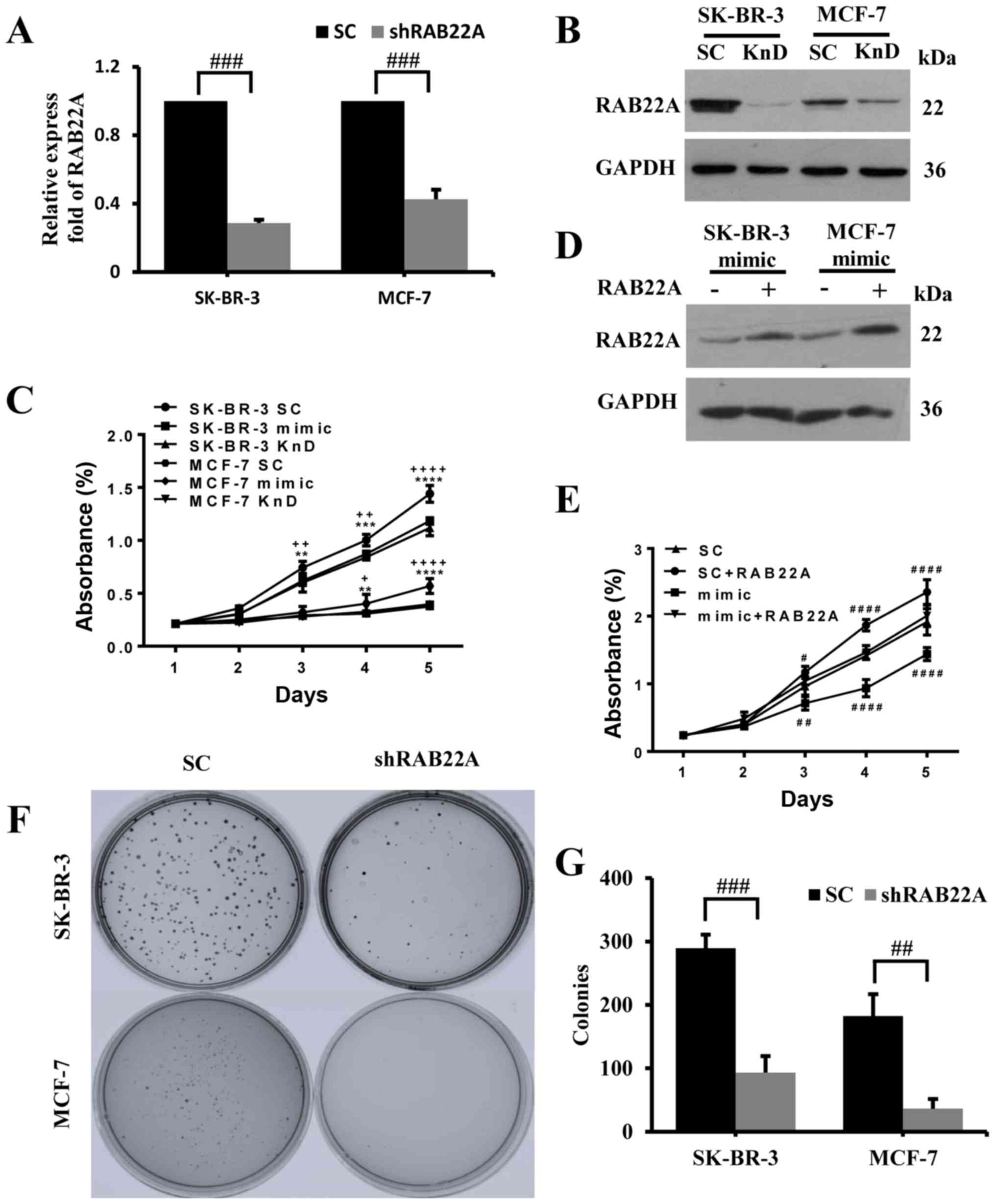
To demonstrate the role of RAB22A in breast
cancer, this gene was knocked down in both the SK-BR-3 and MCF-7
cells. The results of RT-qPCR analysis revealed that the decreased
in RAB22A mRNA expression was close to 70% of the levels
observed in the control cells (Fig.
3A). While the mRNA levels of RAB22A were similar
between the SK-BR-3 and MCF-7 cells, the decrease in the RAB22A
protein level was more evident in the SK-BR-3 than in the MCF-7
cells (Fig. 3B). The knockdown of
RAB22A or the overexpression of miR-193b by transfection
with miR-193b mimic in the MCF-7 and SK-BR-3 cells resulted in a
significant reduction in proliferation compared to the control,
after 5 days in culture (Fig. 3C).
To further confirm whether the tumor-suppressive role of miR-193b
is mediated by RAB22A, a gain-of-function analysis was
performed. Firstly, we transfected the MCF-7 and SK-BR-3 cells with
either negative-scramble control or mimic pre-miR-193b along with
either pcDNA3.1 or pcDNA3.1-RAB22A to detect the protein level of
RAB22A (Fig. 3D). We found that
the overexpression of RAB22A completely abolished the
effects of miR-193b on the proliferation and invasion of the
SK-BR-3 cells (Fig. 3E). To
examine the long-term effects of RAB22A gene knockdown on
the MCF-7 and SK-BR-3 cells, these cells and their corresponding
controls were cultured at low density for 2 weeks. Quantification
of the colonies formed from the single cells revealed that the
knockdown of RAB22A reduced the number of colonies when
compared to the control cells (Fig. 3F
and G).
The RAB22A regulation of exosomes
mediates the migratory and invasive ability of the SK-BR-3
cells
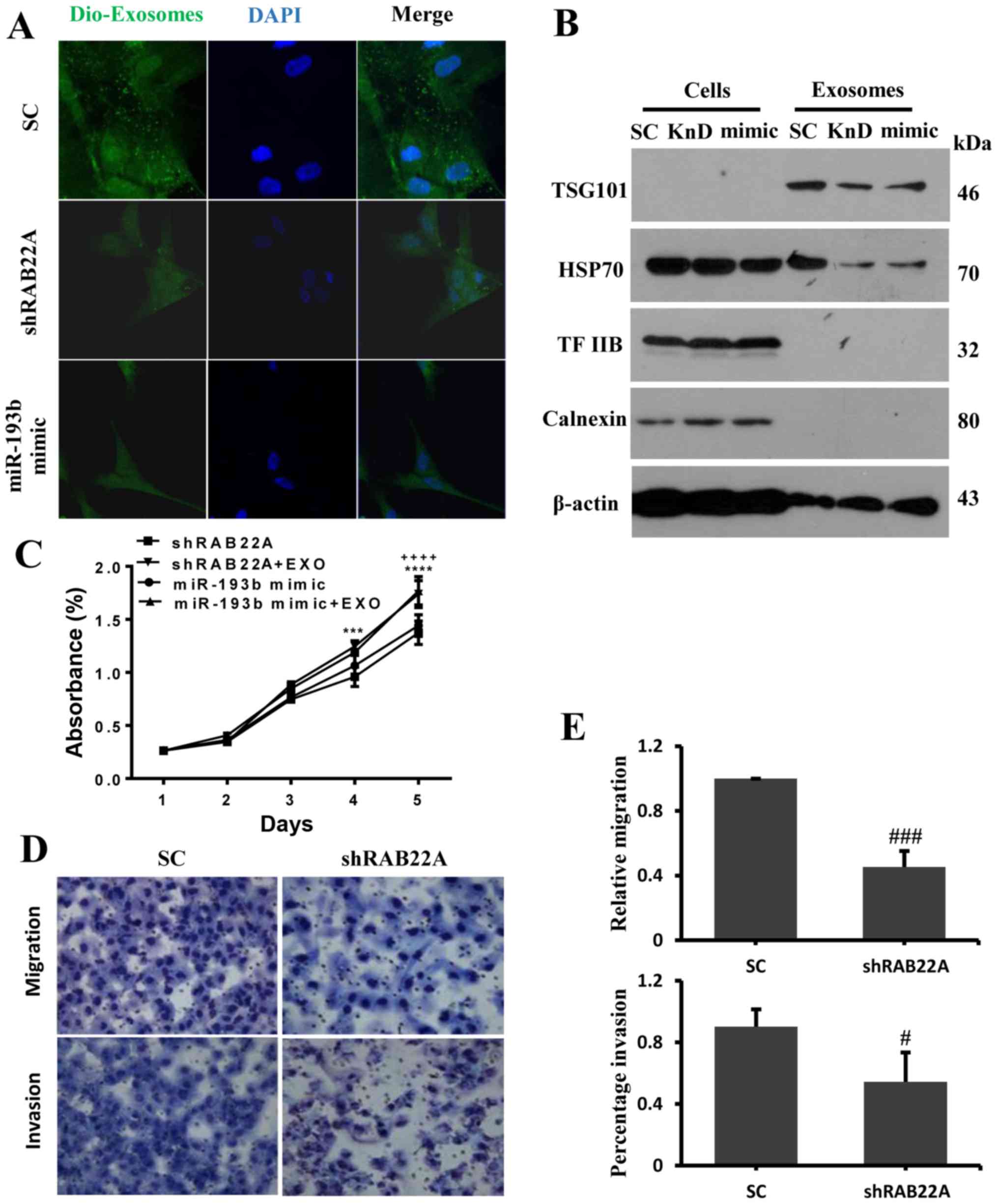
Consistent with the known involvement of
RAB22A in exosome function and trafficking (17), the knockdown of RAB22A or
overexpression of miR-193b by transfection with miR-193b mimic in
the SK-BR-3 cells decreased the level of exosomes in the cancer
cells (Fig. 4A). Western blot
analysis also revealed the reduced expression of the exocrine
marker proteins, namely the multi-vesicular body formation protein
TSG101 and the exosomal cargo protein HSP70 in the exosomes from
the cells in which RAB22A was knocked down and the cells
transfected with the miR-193b mimic; however, this was not found in
the total protein isolated from these cells. However, no changes in
the expression of TFIIB and Calnexin were detected in both the
exosomes and extracts from the total cells (Fig. 4B). To further confirm whether the
RAB22A-related exosomes affect cell growth, exosomes secreted from
the SK-BR-3 cells were added back to the culture medium of the
cells in which RAB22A was knocked down and the cells
transfected with miR-193b mimic; increased cell growth was observed
(Fig. 4C). To examine whether
RAB22A affects cell migration and invasion to another organ,
as observed in patients via lymph node metastasis, Transwell assays
were performed. The results shown in Fig. 4D and E revealed that RAB22A
knockdown significantly inhibited cell migration and invasion.
To further confirm the role of RAB22A in
proliferation and invasion, exosomes were prepared from the
supernatant of the SK-BR-3 and MCF-7 cells in which RAB22A
was knocked down, or transfected with the miR-193b mimic or
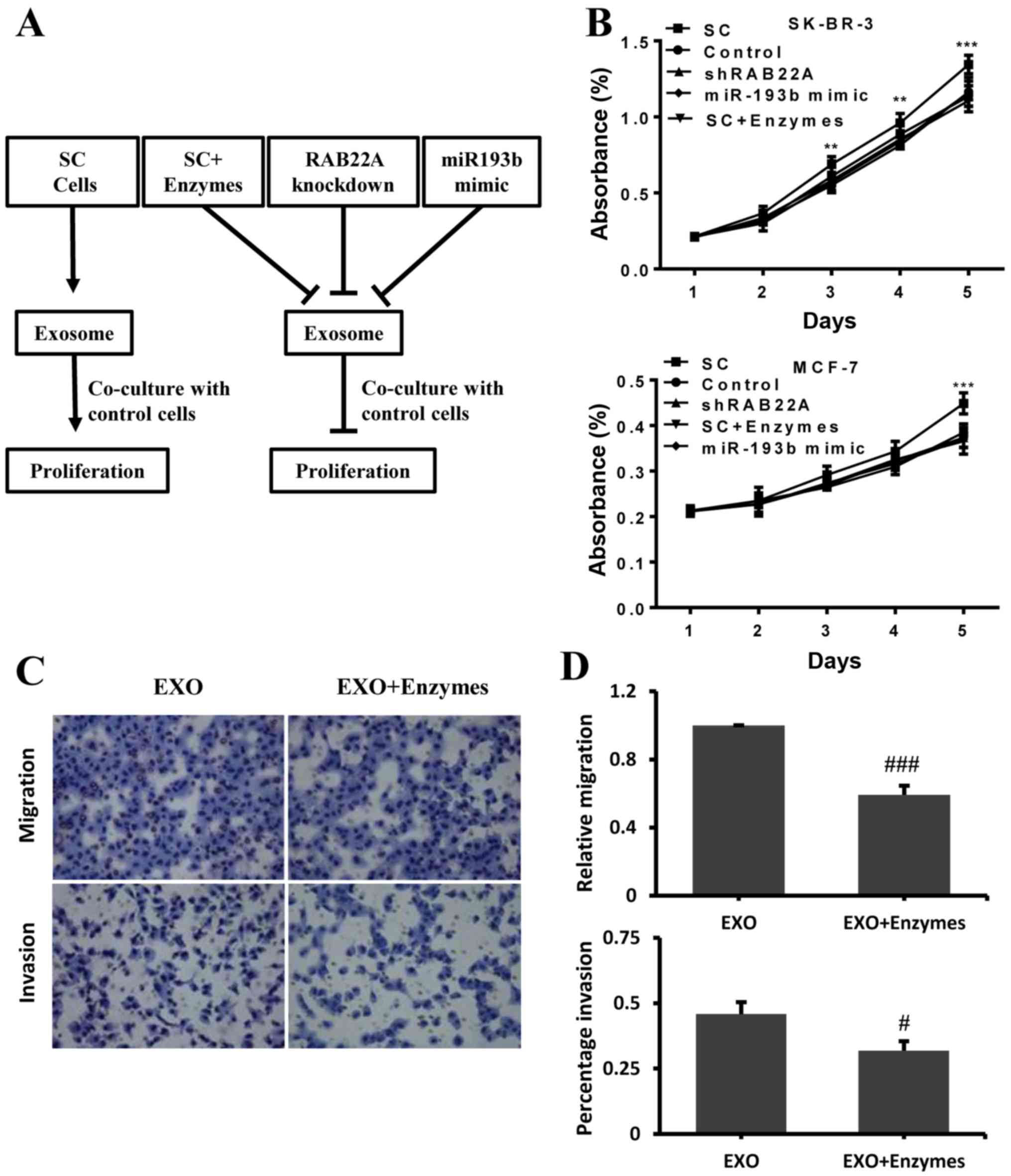
scrambled (SC) control cells, as depicted in Fig. 5A. A total of 2×104 cells
from both the SK-BR-3 and MCF-7 cells in which RAB22A was
knocked down and the controls were plated in a Petri dish in
exosome-depleted culture medium, and their cultures were
supplemented with the isolated exosomes for the indicated days. A
greater cell proliferation was observed following co-culture with
the exosomes generated from the cells transfected with the scramble
control (SC group), but not from the cells in which RAB22A
was knocked down or the cells transfected with the miR-193b mimic.
Treatment of the cells in the SC group with proteinase K/RNase (to
dissolve the exosomes), also reduced the proliferative ability of
these cells to similar levels observed with RAB22A knockdown
and trans-fection with miR-193b mimic (Fig. 5B). To examine whether exosome
production affects cell migration and invasion to other organs,
exosomes extracted from the SK-BR-3 cells were harvested and
treated with or without proteinase K/RNase, then added to the
culture of SK-BR-3 cells in which RAB22A was knocked down in
the Transwell assay. The results shown in Fig. 5C and D revealed that the addition
of exosomes not treated with proteinase K/RNase increased the
migration and invasion of these cells compared to the addition of
exosomes that were treated with the enzyme.
Discussion
In a previous study, we identified RAB22A as a
target of miR-193b by luciferase reporter assay, that their
expression levels were negatively associated in cell lines and
tumors from breast cancer patients, particularly in tissues with
lymph node metastasis, as opposed to the surrounding normal cells.
We found that the knockdown of RAB22A in breast cancer cells
attenuated tumor growth and invasion, a process that was mediated
through exosomal function. These results support the role of
RAB22A in tumor progression and invasion, and suggests that
it may have therapeutic implications for breast cancer.
Recent studies have demonstrated that miRNAs play
important roles in cancer growth and metastasis, including breast
cancer, and are associated with clinical characteristics and
outcomes (10,22). Among these miRNAs, emerging
evidence suggests that miR-193b acts as a tumor suppressor in
various types of cancer, such as breast, pancreatic, esophageal,
ovarian, prostate cancer and Ewing sarcomas, through the
downregulation of proto-oncogenes, RAB22A, CCND1,
KRAS, STMN1, Cyclin D1 and ERBB4
(10,23-28).
This study demonstrated that the reconstitution of miR-193b
expression in a breast cancer cell line resulted in decreased cell
proliferation, clonogenicity, migration and invasion, and RAB22A
was confirmed as a direct target of this miRNA (10); the recovery of RAB22A completely
abolished the tumor suppressive effects of miR-193b. We further
demonstrated the association between the low expression of miR-193b
and a higher production of RAB22A in a sample of patients,
which was associated with a poor prognosis.
Despite the higher expression of RAB22A in
tumor tissue, its role in breast cancer remains unclear. With
increasing malignancy and a poor prognosis, RAB22A is highly
expressed in various types of cancer. For instance, Su et al
demonstrated that the upregulation of RAB22A promoted
melanoma cell growth (29).
Proliferative effects can be observed from gastric and colorectal
cancer cells via the silencing miR-204-5p expression which also
targets RAB22A (30,31).
Wang et al also analyzed the association between clinical
data and RAB22A in >700 cases of breast cancer and found
that a high mRNA expression of RAB22A was associated with a
shortened overall survival time. They also found that RAB22A-coated
extracellular vesicles promoted the infiltration and migration of
breast cancer under hypoxic conditions (17). Thus, RAB22A may be a good
target for therapeutic intervention in breast cancer.
As a ‘new way’ of cell-to-cell communication,
extracellular vesicles within exosomes transmit special signals
(including cell proliferation, migration and infiltration
activities) between cells and tissues (32). Although exosomes can be secreted by
a variety of cell types, the number and content of the exosomes are
closely related to the physiological state of the parental cell
(33,34). A growing number of studies have
indicated that exosomes are related to the occurrence and
progression of tumor cells (35-37),
which can regulate immune response, promote tumor angiogenesis,
invasion, and metastasis, and even act directly on other tumor or
non-tumor cells (38). Studies
have also demonstrated that the level of exosomes released by cells
(particularly tumor cells) under pathological conditions is
significantly increased, and the components of exosomes differ from
those under normal physiological conditions, which not only
emphasizes the parental cell precision adjustment to the contents
of exosomes, but also suggests their important role in tumor
formation and development (39).
For example, under hypoxic conditions, breast cancer cells exhibit
an increased secretion of exosomes by the hypoxia-inducible factor
(HIF)-1 and exosomes secreted by squamous cell carcinoma cells
contain higher levels of angiogenesis-related proteins, such as
RAB22A that play important role in this process (17,40).
However, the role of exosomes the modulation of the immune
response, tumor microenvironment reprogramming and metastasis, to
reprogram recipient cells in breast cancer, remains undefined. In
this study, the amounts of exosomes secreted by the SK-BR-3 cells
were reduced following the knockdown of RAB22A or
transfection with pre-miR-193b mimic, that also caused the
downregulation of the RAB22A gene, resulting in diminished
migratory and infiltrative abilities of the cancer cells. The
re-addition of the exosomes extracted from the supernatant of
SK-BR-3 cells abrogated the tumor suppressor role of miR-193b or
the lack of the RAB22A gene. Moreover, when exosomes
extracted from supernatant of breast cancer cells were co-cultured
with SK-BR-3 and MCF-7 cells in which RAB22A was knocked
down, the abilities of the cells to proliferate, infiltrate and
migrate were significantly enhanced, but this ability was
neutralized by treating the exosomes with protein and RNA degrading
enzymes. These results suggested that exosome components, including
proteins and RNAs, the expression of which is affected by miR-193b
and RAB22A, were likely responsible for tumor growth. The
unknown nature of these components, which play a key role in tumor
growth and metastasis, suggests an interesting area of research
that warrants further investigation. Overall, these results,
suggest a role for RAB22A in exosome secretion and
function.
In conclusion, in this study, we demonstrate that
oncogenic RAB22A, regulated by miR-193b, affects the
exosome-mediated growth, invasion, and metastasis of the recipient
breast cancer cells. RAB22A may thus prove to be a novel
target for breast cancer therapy.
Funding
This study was supported by an NSFC (China) grant
81372456 (to YJL) and by the Foundation for Training Young Talents
in Science and Technology of Jilin Province Health and Family
Planning Commission 2017Q035 (to ZYY).
Availability of data and materials
All data generated or analyzed in this study are
included in this manuscript.
Authors’ contributions
LS initiated the experiment, analyzed data and wrote
the manuscript. MH, NX, DHX and YBD provided support with
experimental techniques. YBD also contributed to manuscript writing
and revision. ZYY and YJL conceived and designed the study and
supervised all the experiments. All authors have read and approved
the final manuscript.
Ethics approval and consent to
participate
All the clinical procedures were approved by the
Committees of Clinical Ethics of China-Japan Union Hospital of
Jilin University (2017022218, Jilin, China). Written informed
consents were obtained from individual or guardian
participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would like to thank Professor David
Spaner (Department of Medicine, University of Toronto, ON, Canada)
for providing helpful and critical comments during the preparation
of this manuscript. The authors would also like to thank Professor
Ling Zhang (Department of Medical Laboratory, The First Hospital of
Jilin University, Jilin, China) for technical editing.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Howlader N, Noone A, Krapcho M, Miller D,
Bishop K and Altekruse S: SEER Cancer Statistics Review, 1975–2015.
National Cancer Institute; Bethesda, MD: 2015
|
|
3
|
Chaffer CL and Weinberg RA: A perspective
on cancer cell metastasis. Science. 331:1559–1564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kanno S, Nosho K, Ishigami K, Yamamoto I,
Koide H, Kurihara H, Mitsuhashi K, Shitani M, Motoya M, Sasaki S,
et al: MicroRNA-196b is an independent prognostic biomarker in
patients with pancreatic cancer. Carcinogenesis. 38:425–431. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lu M, Kong X, Wang H, Huang G, Ye C and He
Z: A novel microRNAs expression signature for hepatocellular
carcinoma diagnosis and prognosis. Oncotarget. 8:8775–8784. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Peng Z, Pan L, Niu Z, Li W, Dang X, Wan L,
Zhang R and Yang S: Identification of microRNAs as potential
biomarkers for lung adenocarcinoma using integrating genomics
analysis. Oncotarget. 8:64143–64156. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ahmadinejad F, Mowla SJ, Honardoost MA,
Arjenaki MG, Moazeni-Bistgani M, Kheiri S and Teimori H: Lower
expression of miR-218 in human breast cancer is associated with
lymph node metastases, higher grades, and poorer prognosis. Tumour
Biol. 39:10104283176983622017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sun G, Liu Y, Wang K and Xu Z: miR-506
regulates breast cancer cell metastasis by targeting IQGAP1. Int J
Oncol. 47:1963–1970. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun G, Sun L, Liu Y, Xing H and Wang K:
Her-2 expression regulated by downregulation of miR-9 and which
affects chemotherapeutic effect in breast cancer. Cancer Gene Ther.
24:194–202. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang Z, He M, Wang K, Sun G, Tang L and Xu
Z: Tumor suppressive microRNA-193b promotes breast cancer
progression via targeting DNAJC13 and RAB22A. Int J Clin Exp
Pathol. 7:7563–7570. 2014.
|
|
11
|
Delprato A and Lambright DG: Structural
basis for Rab GTPase activation by VPS9 domain exchange factors.
Nat Struct Mol Biol. 14:406–412. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Antonyak MA, Wilson KF and Cerione RAR:
R(h)oads to microvesicles. Small GTPases. 3:219–224. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mesa R, Magadán J, Barbieri A, López C,
Stahl PD and Mayorga LS: Overexpression of Rab22a hampers the
transport between endosomes and the Golgi apparatus. Exp Cell Res.
304:339–353. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Weigert R, Yeung AC, Li J and Donaldson
JG: Rab22a regulates the recycling of membrane proteins
internalized independently of clathrin. Mol Biol Cell.
15:3758–3770. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kauppi M, Simonsen A, Bremnes B, Vieira A,
Callaghan J, Stenmark H and Olkkonen VM: The small GTPase Rab22
interacts with EEA1 and controls endosomal membrane trafficking. J
Cell Sci. 115:899–911. 2002.PubMed/NCBI
|
|
16
|
Johnson DL, Wayt J, Wilson JM and
Donaldson JG: Arf6 and Rab22 mediate T cell conjugate formation by
regulating clathrin-independent endosomal membrane trafficking. J
Cell Sci. 130:2405–2415. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang T, Gilkes DM, Takano N, Xiang L, Luo
W, Bishop CJ, Chaturvedi P, Green JJ and Semenza GL:
Hypoxia-inducible factors and RAB22A mediate formation of
microvesicles that stimulate breast cancer invasion and metastasis.
Proc Natl Acad Sci USA. 111:E3234–E3242. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ruivo CF, Adem B, Silva M and Melo SA: The
Biology of Cancer Exosomes: Insights and New Perspectives. Cancer
Res. 77:6480–6488. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−ΔΔC(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
20
|
Justus CR, Leffler N, Ruiz-Echevarria M
and Yang LV: In vitro cell migration and invasion assays. J Vis
Exp. Jun 1–2014.Epub ahead of print. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Györffy B, Lanczky A, Eklund AC, Denkert
C, Budczies J, Li Q and Szallasi Z: An online survival analysis
tool to rapidly assess the effect of 22,277 genes on breast cancer
prognosis using microarray data of 1,809 patients. Breast Cancer
Res Treat. 123:725–731. 2010. View Article : Google Scholar
|
|
22
|
Li X, Zhang Y, Shi Y, Dong G, Liang J, Han
Y, Wang X, Zhao Q, Ding J, Wu K, et al: MicroRNA-107, an oncogene
microRNA that regulates tumour invasion and metastasis by targeting
DICER1 in gastric cancer. J Cell Mol Med. 15:1887–1895. 2011.
View Article : Google Scholar
|
|
23
|
Li J, Kong F, Wu K, Song K, He J and Sun
W: miR-193b directly targets STMN1 and uPA genes and suppresses
tumor growth and metastasis in pancreatic cancer. Mol Med Rep.
10:2613–2620. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nyhan MJ, O’Donovan TR, Boersma AW, Wiemer
EA and McKenna SL: MiR-193b promotes autophagy and non-apoptotic
cell death in oesophageal cancer cells. BMC Cancer. 16:1012016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nakano H, Yamada Y, Miyazawa T and Yoshida
T: Gain-of-function microRNA screens identify miR-193a regulating
proliferation and apoptosis in epithelial ovarian cancer cells. Int
J Oncol. 42:1875–1882. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jin X, Sun Y, Yang H, Li J, Yu S, Chang X,
Lu Z and Chen J: Deregulation of the MiR-193b-KRAS Axis Contributes
to Impaired Cell Growth in Pancreatic Cancer. PLoS One.
10:e01255152015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kaukoniemi KM, Rauhala HE, Scaravilli M,
Latonen L, Annala M, Vessella RL, Nykter M, Tammela TL and
Visakorpi T: Epigenetically altered miR-193b targets cyclin D1 in
prostate cancer. Cancer Med. 4:1417–1425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Moore C, Parrish JK and Jedlicka P:
MiR-193b, downregulated in Ewing Sarcoma, targets the ErbB4
oncogene to inhibit anchorage-independent growth. PLoS One.
12:e01780282017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Su F, Chen Y, Zhu S, Li F, Zhao S, Wu L,
Chen X and Su J: RAB22A overexpression promotes the tumor growth of
melanoma. Oncotarget. 7:71744–71753. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang B, Yin Y, Hu Y, Zhang J, Bian Z,
Song M, Hua D and Huang Z: MicroRNA-204-5p inhibits gastric cancer
cell proliferation by downregulating USP47 and RAB22A. Med Oncol.
32:3312015. View Article : Google Scholar
|
|
31
|
Yin Y, Zhang B, Wang W, Fei B, Quan C,
Zhang J, Song M, Bian Z, Wang Q, Ni S, et al: miR-204-5p inhibits
proliferation and invasion and enhances chemotherapeutic
sensitivity of colorectal cancer cells by downregulating RAB22A.
Clin Cancer Res. 20:6187–6199. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Maia J, Caja S, Strano Moraes MC, Couto N
and Costa-Silva B: Exosome-based cell-cell communication in the
tumor microen-vironment. Front Cell Dev Biol. 6:182018. View Article : Google Scholar
|
|
33
|
Yáñez-Mó M, Siljander PR, Andreu Z, Zavec
AB, Borràs FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J,
et al: Biological properties of extracellular vesicles and their
physiological functions. J Extracell Vesicles. 4:270662015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kishore R and Khan M: More than tiny
sacks: Stem cell exosomes as cell-free modality for cardiac repair.
Circ Res. 118:330–343. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yu DD, Wu Y, Shen HY, Lv MM, Chen WX,
Zhang XH, Zhong SL, Tang JH and Zhao JH: Exosomes in development,
metastasis and drug resistance of breast cancer. Cancer Sci.
106:959–964. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lowry MC, Gallagher WM and O’Driscoll L:
The Role of Exosomes in Breast Cancer. Clin Chem. 61:1457–1465.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Rivoltini L, Chiodoni C, Squarcina P,
Tortoreto M, Villa A, Vergani B, Bürdek M, Botti L, Arioli I, Cova
A, et al: TNF-related apoptosis-inducing ligand (TRAIL)-armed
exosomes deliver proapoptotic signals to tumor site. Clin Cancer
Res. 22:3499–3512. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Boelens MC, Wu TJ, Nabet BY, Xu B, Qiu Y,
Yoon T, Azzam DJ, Twyman-Saint Victor C, Wiemann BZ, Ishwaran H, et
al: Exosome transfer from stromal to breast cancer cells regulates
therapy resistance pathways. Cell. 159:499–513. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Riches A, Campbell E, Borger E and Powis
S: Regulation of exosome release from mammary epithelial and breast
cancer cells - a new regulatory pathway. Eur J Cancer.
50:1025–1034. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Languino LR, Singh A, Prisco M, Inman GJ,
Luginbuhl A, Curry JM and South AP: Exosome-mediated transfer from
the tumor microenvironment increases TGFβ signaling in squamous
cell carcinoma. Am J Transl Res. 8:2432–2437. 2016.
|