Introduction
Colorectal cancer (CRC) is one of the most malignant
diseases worldwide, with high morbidity and mortality rates. In
recent years, early diagnostic techniques and novel therapeutic
methods have been used to improve the general outcome of patients
with CRC at stages I to IIIB; however, the prognosis remains poor
for patients with metastases, with a 5-year survival rate of
<10% (1). Approximately 50-60%
of patients diagnosed with CRC will develop metastases (2,3).
Gaining an understanding of the underlying mechanisms responsible
for metastasis and CRC progression is critical in order to improve
patient prognosis.
Helicase-like transcription factor (HLTF), which is
a member of the SWItch/sucrose non-fermenting (SWI/SNF) family, has
been reported to be associated with the prognosis of a number of
types of cancer. However, the role of HLTF as a tumor suppressor or
promoter remains controversial (4). Previous studies have reported that
HLTF functions as a tumor suppressor in DNA repair, genome
stability maintenance and gene transcription (4-7). A
recent study reported that a high HLTF expression protected cells
against apoptosis by impairing the effects of lysosomal autophagy
inhibitors (8). The majority of
studies on human CRC have focused on the hypermethylation of the
HLTF promoter, which is a biomarker of a poor prognosis (9). However, the functional significance
and underlying mechanisms of tumor suppression remain unclear.
The transforming growth factor (TGF)-β signaling
pathway plays a paradoxical role in carcinogens, promoting
progression in late CRC (10,11)
and inhibiting cell proliferation and apoptosis in early CRC
(12,13). Furthermore, it has been reported
that TGF-β signaling plays a critical role in suppressing tumor
metastasis (14,15). Whether TGF-β signaling acts as an
inhibitor or stimulator depends on the function of its core
members, including SMAD4. The loss of SMAD4 enhances tumorigenicity
and promotes metastasis (16,17).
In this study, to investigate the function of HLTF
in CRC cells, we first detected the expression of HLTF in CRC and
then investigated the effects of HLTF on the motility of CRC cells.
Furthermore, we identified a novel mechanism through which HLTF
affects the migration and invasion of CRC cells, by regulating
TGF-β/SMAD signaling. These findings suggest that HLTF may be a
useful prognostic biomarker and a potential target for the
diagnosis and treatment of CRC.
Materials and methods
Clinical specimens and follow-up
A total of 86 paraffin-embedded specimens of primary
CRC were obtained by surgical resection at the Department of
Pathology, Xiangya Hospital (Changsha, China) between February and
December, 2011. The patient clinical data are presented in Table I. Patients were not pre-treated
with radiotherapy or chemotherapy prior to surgery. All patients
were followed-up until October 30, 2017, and their complete
clinical data were collected. Overall survival (OS) is defined as
time to death, irrespective of cause, and censored is defined as
loss to follow-up. All procedures were performed according to the
ethical guidelines of Xiangya Hospital. Informed consent was
obtained from all participating patients. For the use of human
samples, the protocol for this study was approved by the Xiangya
Hospital Ethics Committee (Changsha, China). The 7th edition of the
AJCC TNM staging system for CRC was used to classify tumor
stage.
 | Table IAssociation between HLTF expression
and the clinicopathological characteristics of 86 patients with
CRC. |
Table I
Association between HLTF expression
and the clinicopathological characteristics of 86 patients with
CRC.
| Clinicopathological
characteristics | HLTF expression
| P-value |
|---|
| n | Low | High |
|---|
| Age (years) | | | | |
| <65 | 28 | 12 | 16 | 0.0927a |
| ≥65 | 58 | 36 | 22 | |
| Sex | | | | |
| Male | 45 | 22 | 23 | 0.1755a |
| Female | 41 | 26 | 15 | |
| Tumor
differentiation | | | | |
| Well-moderate | 47 | 16 | 31 | <0.0001a |
| Poor | 39 | 32 | 7 | |
| TNM stage | | | | |
| I-II | 38 | 11 | 27 | <0.0001a |
| III-IV | 48 | 37 | 11 | |
| Depth of
invasion | | | | |
| T1-T2 | 18 | 1 | 17 | <0.0001b |
| T3-T4 | 68 | 47 | 21 | |
| Lymph node
metastasis | | | | |
| N0 | 41 | 13 | 28 | <0.0001a |
| N1-N2 | 45 | 35 | 10 | |
| Distant
metastasis | | | | |
| M0 | 65 | 31 | 34 | 0.0076a |
| M1 | 21 | 17 | 4 | |
Immunohistochemical staining
Sections (5-μm-thick) were prepared from the
formalin-fixed and paraffin-embedded tissues. Immunohistochemical
staining was performed as follows: After dewaxing and rehydrating
the tissue slides, antigen retrieval was performed using boiling
citrate buffer (pH 6.0) in a pressure cooker for 2.5 min. The
slides were subsequently incubated with 3%
H2O2 for 15 min at room temperature.
Following incubation with rabbit anti-HLTF polyclonal antibody
(1:100; ab183042; Abcam, Cambridge, UK) overnight (4°C), the slides
were washed 3 times with PBS to remove excess primary antibodies.
The tissues were subsequently incubated with horseradish
peroxidase-conjugated anti-mouse/anti-rabbit secondary antibody
(cat. no. SPN-9000; undiluted; included in the Biotin-Streptavidin
HRP Detection System, Beijing Zhongshan Golden Bridge
Biotechnology, Beijing, China) for 30 min at 37°C.
3,3′-Diaminobenzidine (DAB) was used to visualize positive immune
reactions and hematoxylin was used as a counterstain. The primary
antibody was omitted for the negative control. Briefly, the
sections were scored using a Fourier scale according to the
percentage of positive cells and staining intensity: (-) or 0,
tissue specimens without staining (0-10%); (+) or 1, tissue
specimens with weak staining (10-25%); (++) or 2, tissue specimens
with moderate staining (25-50%) and (+++) or 3, tissue specimens
with strong staining (>50%). (-) and (+) were defined as low
expression, while (++) and (+++) were defined as high expression or
overexpression (18). Two
board-certified clinical pathologists, who were blinded to the
clinical parameters, independently evaluated the staining results.
Any disagreement between the two evaluators was resolved by
re-evaluation and careful discussion.
Cell culture and treatment
The HCT-8, HCT-116, RKO, SW480 and HT-29 cell lines
were obtained from the Cancer Research Institute, Central South
University (Changsha, China). LoVo cells were purchased from the
Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All
cells were cultured in RPMI-1640 medium containing 10% fetal bovine
serum (FBS) and maintained in an incubator at 37°C in an atmosphere
containing 5% CO2 (both from Gibco/Thermo Fisher
Scientific, Inc., Waltham, MA, USA). The cells were seeded in
6-well plates at a density of 5×104/cells/ml and
cultured for 24 h prior to exposure to TGF-β1 (cat. no.
AF-100-21C-2; Peprotech, Inc., Rocky Hill, NJ, USA) at the
concentration of 0, 0.5, 5 ng/ml. Cells were collected after 48 h
for use in western blot analysis.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
TRIzol reagent (Invitrogen/Thermo Fisher Scientific,
Inc.) was used to extract total RNA from the cells. A Prime Script
RT reagent kit with a gDNA Eraser (Takara Biotechnology Co., Ltd.,
Dalian, China) was used to perform the reverse transcription. The
resulting cDNA was used for qPCR with a CFX96 Real-Time System
(Bio-Rad Laboratories, Inc., Hercules, CA, USA) with SYBR Premix Ex
Taq II (Takara Biotechnology Co., Ltd.). All processes were
performed according to the corresponding manufacturer’s
instructions. The 2-ΔΔCq method (19) was used to calculate the relative
abundance of RNA for each gene compared with GAPDH expression. Each
reaction was performed in triplicate. The primer sequences used are
presented in Table II.
 | Table IISequences of the primers for reverse
transcription-quantitative polymerase chain reaction. |
Table II
Sequences of the primers for reverse
transcription-quantitative polymerase chain reaction.
| Gene | Sequence |
|---|
| HLTF | F:
5′-GCTTAGACGGTTCCATGGCTCAAA-3′ |
| R:
5′-TCCAGGCTGGGTCCATTAAGAACA-3′ |
| SMAD4 | F:
5′-CTCATGTGATCTATGCCCGTC-3′ |
| R:
5′-AGGTGATACAACTCGTTCGTAGT-3′ |
| GAPDH | F:
5′-CCCTCAAGATTGTCAGCAATG-3′ |
| R:
5′-GTCCTCAGTGTAGCCCAGGAT-3′ |
Western blot analysis
The CRC cells were washed and lysed in strong RIPA
buffer supplemented with proteinase and phosphatase inhibitors
(Thermo Fisher Scientific, Inc.). Total protein was extracted and
the concentration was determined by bicinchoninic acid protein
assay. Equal amounts of proteins (40 μg per sample) were
loaded and separated by 8% SDS-PAGE. Following electrophoresis,
proteins were transferred onto a 0.22 μm PVDF membrane (EMD
Millipore, Billerica, MA, USA). Following blocking with 5% non-fat
milk, the membranes were incubated with specific primary antibodies
at 4°C overnight, washed extensively and incubated with secondary
antibodies (1:6,000 dilution; cat. no. 7074S; Cell Signaling
Technology, Inc., Danvers, MA, USA) for 1 h. The protein bands were
visualized and quantified using an ECL Advanced Detection System
(EMD Millipore). GAPDH was used as loading control. The primary
antibodies used were as follows: HLTF (1:1,000; cat. no.
14286-1-AP; ProteinTech Group, Inc., Chicago, IL, USA), SMAD4
(1:1,000, cat. no. 38454T), SMAD2/3 (1:1,000, cat. no. 8685T),
Vimentin (1:1,000, cat. no. 5741T) (all from Cell Signaling
Technology, Inc.), p-SMAD2/3 (1:500; cat. no. wl02305), zinc finger
e-box binding homeobox 1 (ZEB1; 1:500; cat. no. wl01657; both from
Wanleibio, China) and (GAPDH; 1:5,000; cat. no. SAB2701826;
Sigma-Aldrich, Merck KGaA, Darmstadt, Germany).
shRNAs, siRNA and vector
transfection
shRNAs targeting HLTF and a control shRNA were
purchased from GeneChem (Shanghai, China). The shRNA vector
(GV493), also from GeneChem, was hU6-MCS-CBh-gcGFP-IRES-puromycin.
Moreover, the shRNAs obtained targeted the following sequences of
HTLF (NM_139048): shRNA 1# targets AGGT GGAGTTGGTTTGAAT and shRNA
2# targets TATTAGAG AACCGGCCTTA. The HCT-8 and HCT-116 cells were
infected with the purified lentiviruses containing the shRNA
sequence targeting HLTF or the control sequence lentivirus for 24
h. Stably transfected cells were isolated using 2 μg/ml
puromycin (Sigma; Merck KGaA) for 7 days. In addition, human SMAD4
siRNA was purchased from GenePharma (Shanghai, China). The
SMAD4-siRNA sequences were as follows: sense,
5′-GAGAAGTTCTCAAAGTTAA-3′ and anti-sense,
5′-TTAACTTTGAGAACTTCTC-3′. The pGV141-HLTF plasmid (purchased from
GeneChem), is an expression vector, containing HLTF cDNA and the
structure of the vector is CMV-MCS-3FLAG-SV40-Neomycin. When the
cells are transfected with pGV141-HLTF, they then overexpress HLTF.
ViaFect™ Transfection reagent (cat. no. E4981; Promega Corp.,
Madison, WI, USA) was used to transfect the plasmid into RKO cells
according to the manufacturer’s instructions. The siRNA and plasmid
were co-transfected into the cells using Lipofectamine®
2000 reagent (cat. no. 11668027; Invitrogen; Thermo Fisher
Scientific, Inc.).
Wound healing assay
The cells were cultured on 6-well plates with
RPMI-1640 containing 10% FBS. When the cell density reached 70-80%,
the bottom of the plate was scratched with a 100 μl pipette
tip to create a cell-free gap, following which the cells were
incubated for 48 (HCT-116 and RKO) or 72 h (HCT-8) with RPMI-1640
medium containing 1% FBS. An inverted Olympus IX50 microscope
(Olympus Corp., Tokyo, Japan) was used to obtain phase-contrast
images of the wound healing process at different time-points after
scratching. The size of the healed wound was then compared with the
size of the initial wound.
Transwell assay
A total of 5×104 cells suspended in 100
μl serum-free RPMI-1640 medium were seeded into the upper
chamber of a Transwell apparatus (Corning Inc., Corning, NY, USA; 8
μm pore) with 50 μl Matrigel (BD Biosciences,
Franklin Lakes, NJ, USA). A total of 600 μl RMPI-1640 medium
containing 10% FBS was added to the lower chamber. Following
incubation at 37°C for 48 h (HCT-116 and RKO) or 60 h (HCT-8),
cells in the upper chamber were removed with a cotton swab and
cells on the lower surface were fixed in 1% paraformaldehyde
followed by staining with 0.1% crystal violet solution (Beyotime
Institute of Biotechnology, Haimen, China) at room temperature. The
number of invading cells was determined for 5 randomly selected
fields (×200 magnificatoin) under a microscope (Leica inverted
microscope DMi1; Leica, Wetzlar, Germany). Three independent
experiments were performed and the mean was calculated.
Prediction of transcription factor
binding sites
GCBI online software (https://www.gcbi.com.cn/gclib/html/index) was used to
predict the putative binding sites for HLTF on SMAD4 promoter
region.
Statistical analysis
Data were analyzed using GraphPad Prism 5.0 software
(GraphPad Software, Inc., La Jolla, CA, USA). Differences between 2
groups were assessed using a Student’s t-test. For 3 or more
groups, one-way ANOVA test was used followed by Tukey’s post hoc
test for multiple comparisons. The associations between HLTF
expression and the patient clinicopathological characteristics were
determined using the Chi-square and Fisher’s exact tests. Overall
survival (OS) curves were plotted according to the Kaplan-Meier
method, and differences in survival were examined using the
log-rank test. P<0.05 was considered to indicate a statistically
significant difference.
Results
HLTF expression is negatively associated
with the progression of CRC
To verify the expression of HLTF in CRC,
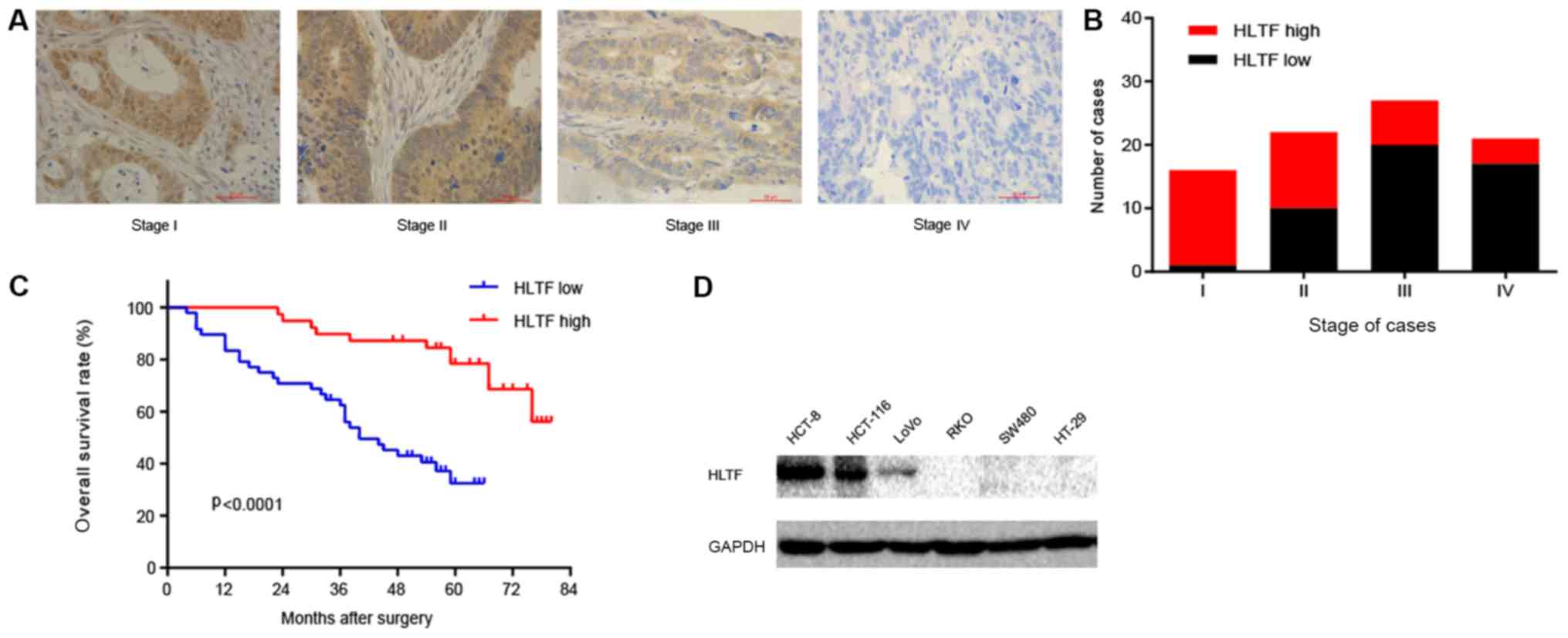
immunochemistry was performed on CRC tissues at various stages. The
results revealed that HLTF expression decreased with the increasing
TNM stage (Fig. 1A and B). This
suggests that HLTF expression is negatively correlated with CRC
progression. To explore the association between HLTF and CRC, we
analyzed the association between HLTF expression and the patient
clinicopathological characteristics. As shown in Table I, the expression of HLTF was
significantly associated with the differentiation status
(P<0.0001), TNM stage (P<0.0001), depth of invasion
(P<0.0001), lymph node metastasis (P<0.0001) and distant
metastasis (P<0.01). However, no significant association was
observed between HLTF expression and age (P=0.0927) or sex
(P=0.1755). To further assess the prognostic value of HLTF in CRC,
we performed a survival analysis. Kaplan-Meier analysis revealed
that patients with CRC with a high HLTF expression had a
significantly longer OS compared with patients with a low HLTF
expression (Fig. 1C, log-rank
test, P<0.0001), suggesting that a low HLTF protein expression
contributes to CRC progression and is associated with a poor
prognosis. Based on these results, we thus concluded that the
expression of HLTF is negatively associated with the progression of
CRC.
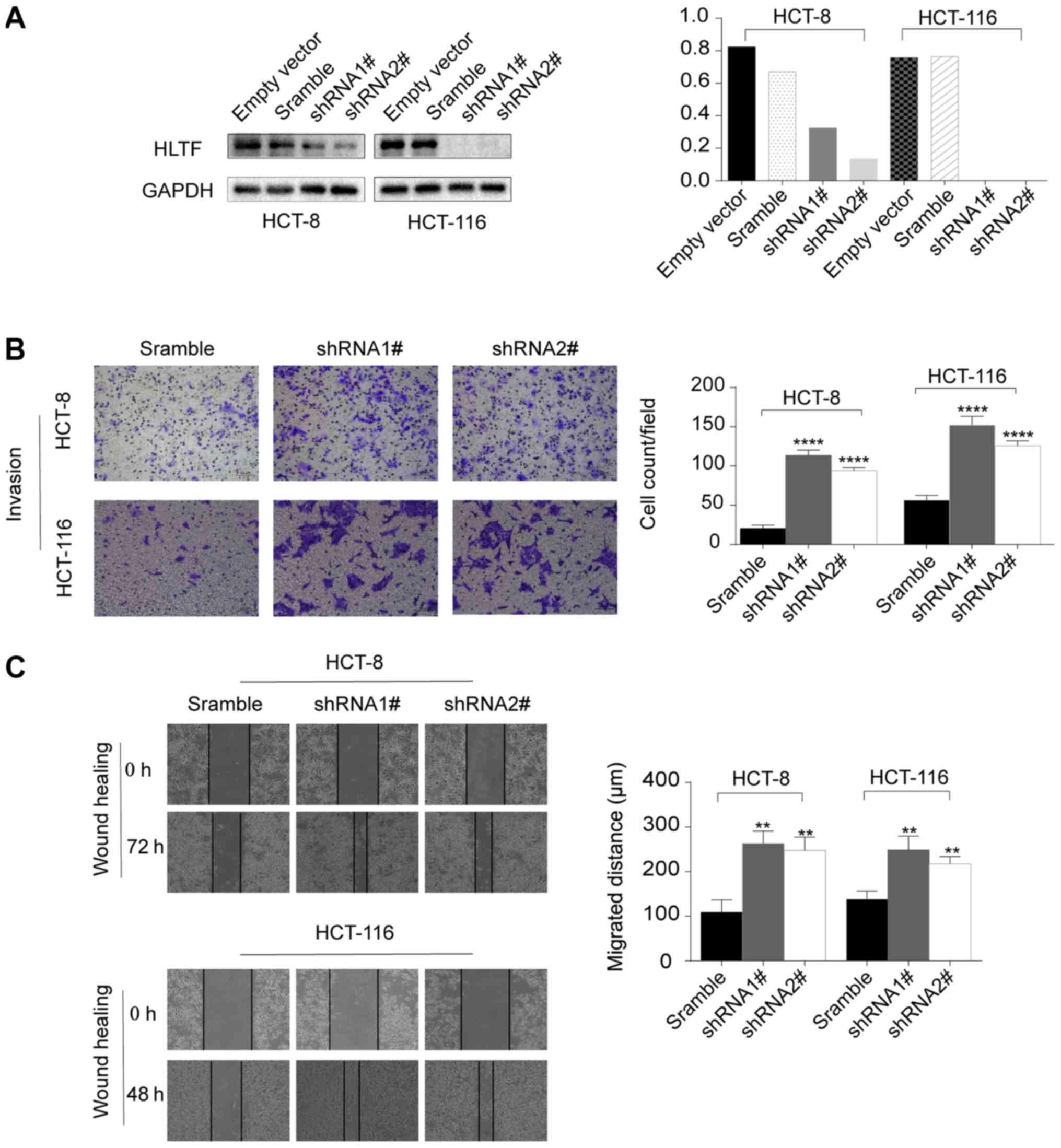
HLTF knockdown in CRC cells promotes cell
migration and invasion
The HCT-8 and HCT-116 cell lines were used to assess
the effects of HLTF on cell migration and invasion due to their
high expression of HLTF (Fig. 1D).
We manipulated the cells by knocking down HLTF (Fig. 2A). The results of wound healing
(P<0.01; Fig. 2C) and Transwell
assays (P<0.0001; Fig. 2B)
demonstrated that HLTF knockdown significantly enhanced the
migration and invasion of CRC cells in vitro.
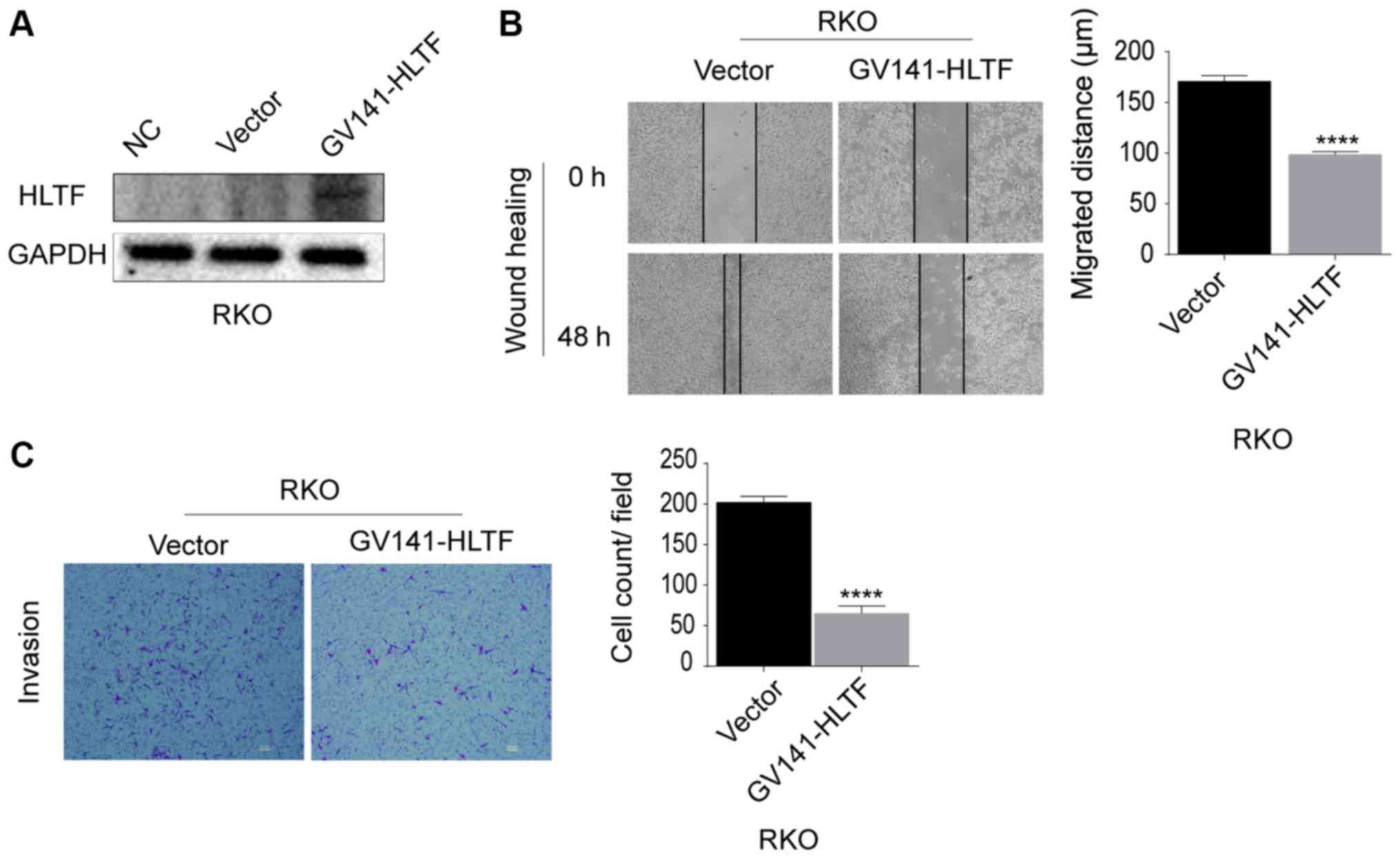
HLTF overexpression in RKO cells
suppresses cell migration and invasion
HLTF was overexpressed in RKO cells to assess
whether the migration and invasion abilities would decrease.
Evidence of HLTF overexpression in RKO cells is shown in Fig. 3A. HLTF upregulation significantly
suppressed the migration (P<0.0001; Fig. 3B) and invasion (P<0.0001;
Fig. 3C) of the CRC cells. These
results confirmed that HLTF overexpression suppressed the migration
and invasion of CRC cells in vitro.
HLTF affects the TGF-β/SMAD pathway in
CRC cells
In order to elucidate the mechanisms through which
HLTF suppresses the migration and invasion of CRC cells, we
investigated metastasis-related pathways. The results of western
blot analysis revealed that the expression of HLTF, SMAD4 and
p-SMAD2/3, but not that of total SMAD2/3 were increased when the
HCT-8, HCT-116 and RKO cells were stimulated with TGF-β1 (Fig. 4A), suggesting that HLTF is
regulated by the TGF-β signaling pathway. We also investigated the
mechanism through which HLTF affects the TGF-β signaling pathway.
As shown in Fig. 4B, we observed
that HLTF knockdown decreased the expression of SMAD4 and
p-SMAD2/3, but not that of total SMAD2/3 in both the HCT-8 and
HCT-116 cells. The results of RT-qPCR confirmed that the mRNA
expression of SMAD4 was also decreased (Fig. 4D). In addition, when using GCBI
online software, we predicted that the SMAD4 gene promoter
presented putative binding sites for HLTF (51056636 - 51056653,
aaaaAAAAAgagggttc; 51027033 - 51027050, aggcACTAAgaagctga)
(Fig. 4C). Furthermore, in the
HCT-8 and HCT-116 cells, HLTF knockdown upregulated the expression
of Vimentin and ZEB1, which are downstream targets of TGF-β
signaling (Fig. 4E). The opposite
result was observed in the RKO cells overexpressing HLTF (Fig. 4B, D and E). These results suggest
that HLTF is associated with the TGF-β/SMAD pathway, and that a
high HLTF expression suppresses the migration and invasion of CRC
cells by targeting TGF-β/SMAD signaling.
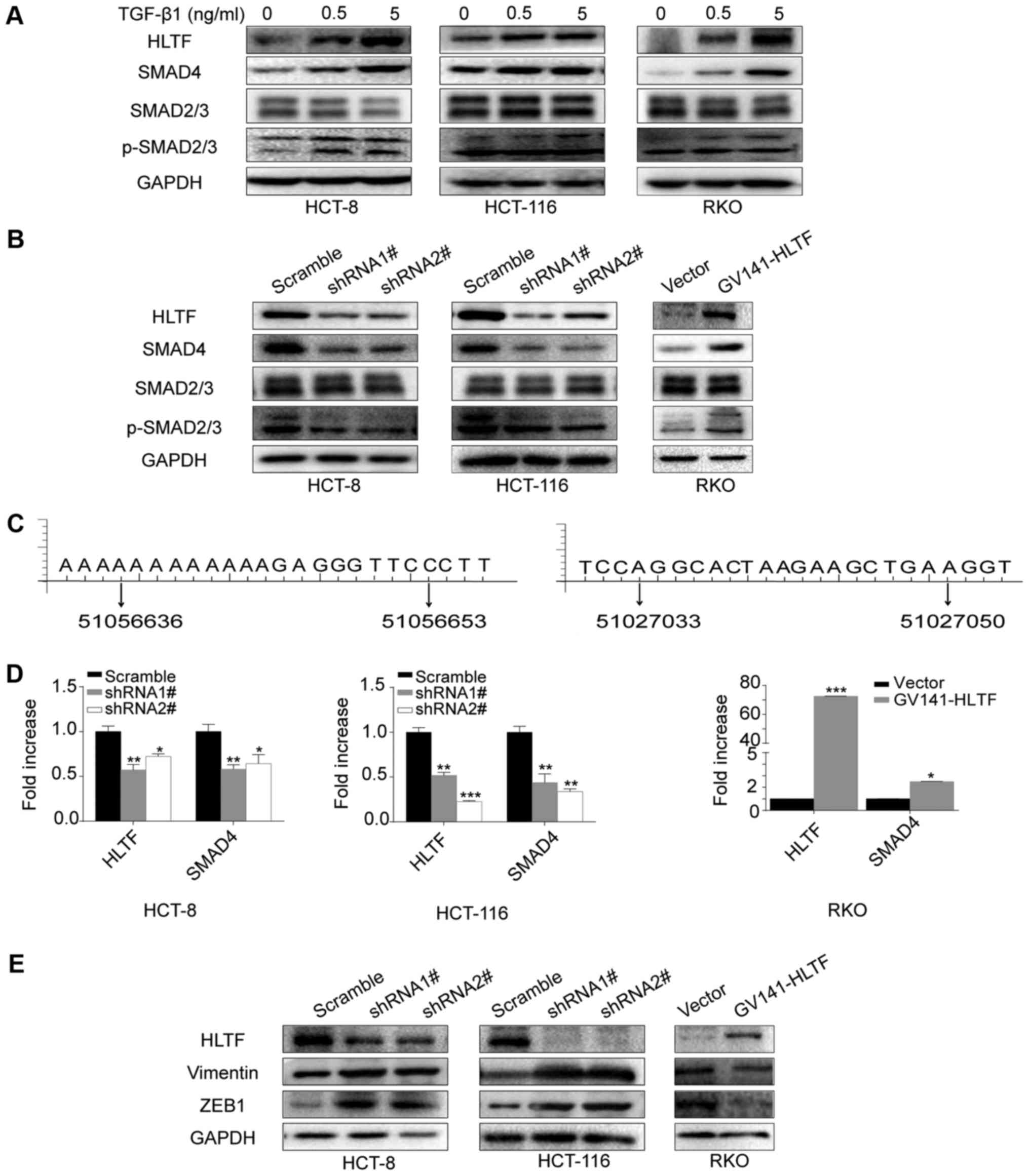 | Figure 4HLTF expression is associated with
TGF-β/SMAD signaling. (A) HLTF protein level increased in CRC cells
following stimulation with 0, 0.5 or 5 ng/ml TGF-β1. (B) HLTF
knockdown in HCT-8 and HCT-116 cells reduced SMAD4 and p-SMAD2/3
expression, whereas HLTF upregulation in RKO cells increased SMAD4
and p-SMAD2/3 expression. The total SMAD2/3 level was not
significantly affected in the HCT-8, HCT-116 and RKO cells. (C)
Predicted sequences of putative binding sites for HLTF on SMAD4
promoter region by using GCBI online software. Left panel:
aaaaAAAAAgagggttc, the region band from beginning to end is
51056636 - 51056653; right panel: aggcACTAAgaagctga, the region
band from beginning to end is 51027033 - 51027050. (D) Reverse
transcription-quantitative polymerase chain reaction conformed that
the SMAD4 mRNA levels were reduced in the HCT-8 and HCT-116 cells
following HLTF knockdown (*P<0.05 and
**P<0.01 vs. scramble), while they were increased in
the HLTF-RKO cells (*P<0.05 vs. vector). (E) HLTF
knockdown promoted the expression of Vimentin and ZEB1 protein,
whereas HLTF overexpression had the opposite effect. HLTF,
helicase-like transcription factor; CRC, colorectal cancer, TGF-β,
transforming growth factor-β. |
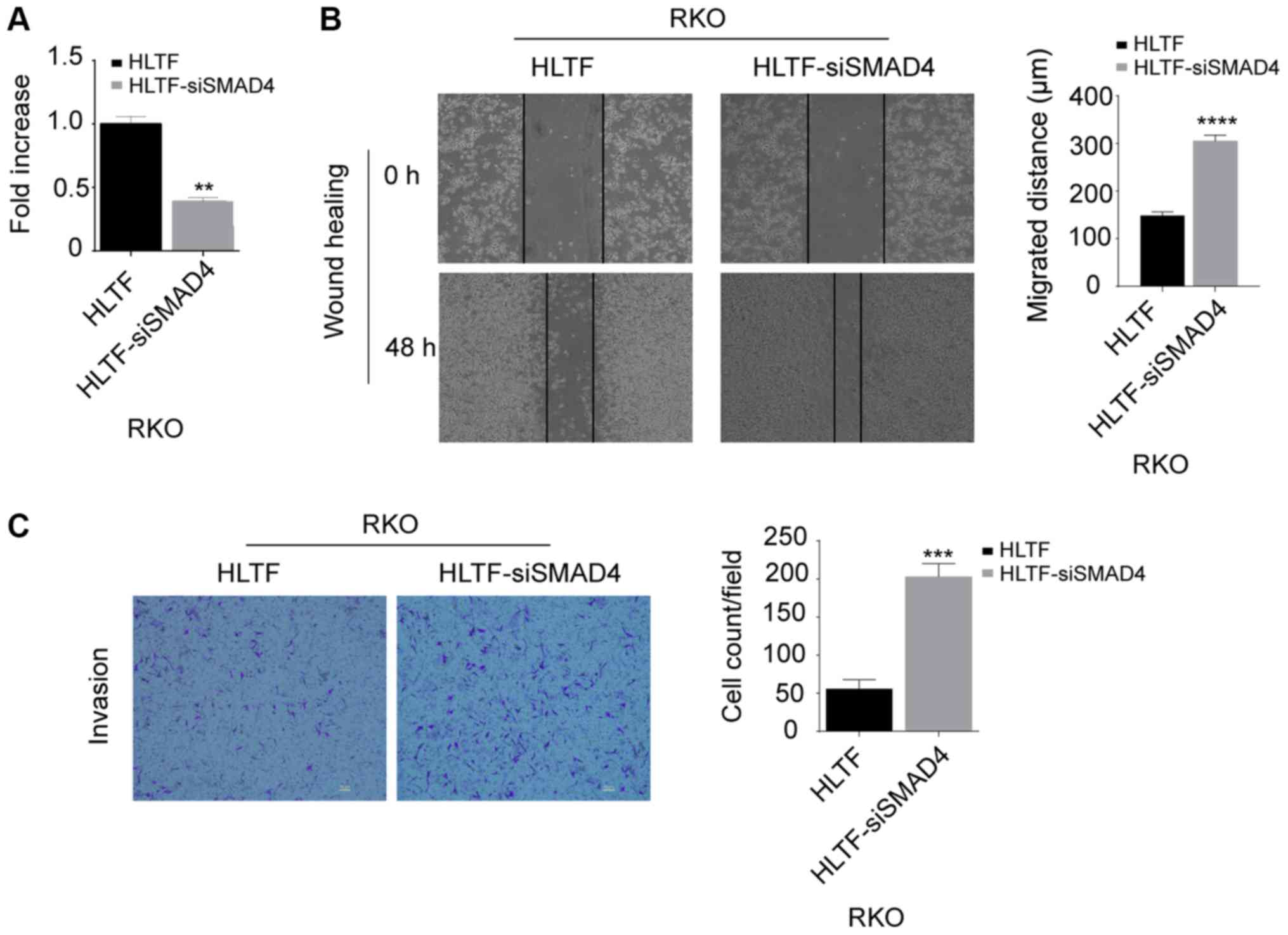
HLTF suppresses tumor aggressiveness
through TGF-β/SMAD signaling
To confirm whether HLTF affects tumor aggressiveness
via the TGF-β/SMAD pathway, SMAD4 was knocked down using siRNA in
HLTF-transduced RKO (HLTF-siSMAD4-RKO) cells. With 3 siRNAs
targeting SMAD4, we finally selected the most effective siRNA
(siRNA1#) for the following experiments (data not shown). As shown
in Fig. 5A, the SMAD4 mRNA level
was significantly decreased in the HLTF-siSMAD4-RKO cells compared
with the HLTF-RKO cells (P<0.01). Furthermore, the results of
wound healing (P<0.0001; Fig.
5B) and Transwell assays (P<0.001; Fig. 5C) revealed that the motility of the
HLTF-siSMAD4-RKO cells was increased compared with that of the
HLTF-RKO cells. These results further support the conclusion that
HLTF affects CRC cells via the TGF-β/SMAD signaling pathway.
Discussion
The role of HLTF in cancer progression is unclear;
previous studies have stated that it functions as an oncogene or
tumor suppressor. In the kidneys, hypopharynx and cervical cancer,
HLTF serves as an oncogene (20-23),
while in the thyroid and digestive carcinoma, HLTF functions as a
tumor suppressor (24,25). Furthermore, the functional loss of
HLTF promotes intestinal carcinogenesis (26). In CRC, promoter hypermethylation is
frequently observed (27-29). It is significantly associated with
tumor stage, metastatic disease (30,31)
and is regarded as a biomarker of poor prognosis (32,33)
in CRC. However, the potential mechanisms through which HLTF
functions in CRC have not yet been reported. This study revealed an
association between HLTF expression and metastasis in CRC.
Mechanistic studies have revealed that HLTF suppresses the motility
of CRC cells via the TGF-β signaling pathway.
In 2016, Cheng et al demonstrated that HLTF
expression was reduced in adult acute myeloid leukemia patients
with more aggressive disease phenotypes (34). In this study, the results of
immunohistochemistry revealed that HLTF expression was negatively
associated with the differentiation status, tumor invasive depth,
lymph metastasis and distant metastasis in CRC. A poor
differentiation status, lymph node and distant metastasis have been
reported to be negative prognostic factors in CRC (35,36).
These results suggest that CRC tissues with a low HLTF expression
are more aggressive and are associated with severe cancer
progression. Survival analysis revealed that a low HLTF expression
was associated with a poor prognosis. Metastasis is characterized
by a series of pathological events, including deep invasion into
the intestinal wall and the lymphovascular or circulatory systems
(37). We then examined the
motility of CRC cells when HLTF was knocked down or overexpressed.
The results indicated that silencing HLTF markedly enhanced the
migration and invasion of HCT-8 and HCT-116 cells, while HLFT
overexpression suppressed the the migratory and invasive ability of
the RKO cells in vitro. From these results, it can be
concluded that HLTF is a biomarker of CRC aggressiveness.
There is evidence to indicate that HLTF functions as
a tumor suppressor mainly through two mechanisms: Promoter
hypermethylation or alternative mRNA splicing (4). The results of the present study
provide novel insight into the suppressive role of HLTF in CRC.
Firstly, a previous study employing a MetaCoret™ enrichment pathway
analysis suggested that HLTF is closely asscociated with cell
adhesion and TGF-β signaling (38). Our study showed that TGF-β
stimulation induced HLTF overexpression in CRC cells. In addition,
HLTF knockdown in HCT-8 and HCT-116 cells resulted in a decrease in
SMAD4 and p-SMAD2/3 expression and a reduction in the expression of
their downstream targets, Vimentin and ZEB1. The opposite was
observed in HLTF-RKO cells.
SMAD4, which is a CRC suppressor, is an important
factor in TGF-β signaling. SMAD4 mutation or loss can strongly
affect TGF-β/SMAD signaling and promote CRC cell metastasis
(17,39-42).
There is evidence to indicate that the loss of SMAD4 promotes CRC
cell metastasis (43,44). Furthermore, the expression of SMAD4
is positively associated with survival in colon cancer (45). In addition, we predicted that the
SMAD4 gene promoter presented putative binding sites for HLTF using
GCBI online software. The results of this study indicated that HLTF
depletion reduced the expression of SMAD4 and p-SMAD2/3 in HCT-8
and HCT-116 cells, while their expression was increased in the
HLTF-RKO cells. Furthermore, transfection with siRNA targeting
SMAD4 promoted cell motility. We therefore concluded that HLTF
inhibits the migration and invasion of CRC cells via the
TGF-β-HLTF-SMAD signaling pathway, and we hypothesize that HLTF may
be a direct transcriptional activator of the SMAD4 gene. However,
further investigation is required to elucidate the detailed
mechanisms.
Evidence indicate that both HLTF and TGF-β signaling
play a paradoxical role in cancer progression (4,11).
Whether TGF-β-HLTF-SMAD signaling plays a similar role in CRC is
unclear. Further experiments in vivo and the analysis of
clinical samples are required for a more in depth
investigation.
In conclusion, the results of the present study
suggest that HLTF functions as a tumor suppressor gene in CRC. HLTF
downregulation promotes the migration and invasion of CRC cells
in vitro. Furthermore, HLTF expression is negatively
associated with OS in patients with CRC. HLTF may suppress the
migration and invasion of CRC cells by targeting TGF-β/SMAD
signaling. Taken together, these results suggest that HLTF may be a
potential prognostic biomarker for CRC.
Funding
This study was supported by the Natural Science
Foundation of China (nos. 81372428, and 81702956).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article or are available from the
corresponding author on reasonable request.
Authors’ contributions
HZ and FT conceived and designed the study. LL, HL,
YaZ, JH and QL performed the experiments. LL drafted the
manuscript. MZ and JW revised the manuscript. DY collected the
clinical data. MZ, JW, YuZ and HPP participated in data analysis
and interpretation of the results. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Informed consent was obtained from all participating
patients. For the use of human samples, the protocol for this study
was approved by the Xiangya Hospital Ethics Committee (Changsha,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Van Cutsem E, Nordlinger B, Adam R, Köhne
CH, Pozzo C, Poston G, Ychou M and Rougier P; European Colorectal
Metastases Treatment Group: Towards a pan-European consensus on the
treatment of patients with colorectal liver metastases. Eur J
Cancer. 42:2212–2221. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee WS, Yun SH, Chun HK, Lee WY, Yun HR,
Kim J, Kim K and Shim YM: Pulmonary resection for metastases from
colorectal cancer: Prognostic factors and survival. Int J
Colorectal Dis. 22:699–704. 2007. View Article : Google Scholar
|
|
4
|
Dhont L, Mascaux C and Belayew A: The
helicase-like transcription factor (HLTF) in cancer: Loss of
function or oncomorphic conversion of a tumor suppressor? Cell Mol
Life Sci. 73:129–147. 2016. View Article : Google Scholar
|
|
5
|
Poole LA and Cortez D: Functions of
SMARCAL1, ZRANB3, and HLTF in maintaining genome stability. Crit
Rev Biochem Mol Biol. 52:696–714. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kile AC, Chavez DA, Bacal J, Eldirany S,
Korzhnev DM, Bezsonova I, Eichman BF and Cimprich KA: HLTF’s
ancient HIRAN domain binds 3′ DNA ends to drive replication fork
reversal. Mol Cell. 58:1090–1100. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Taglialatela A, Alvarez S, Leuzzi G,
Sannino V, Ranjha L, Huang JW, Madubata C, Anand R, Levy B, Rabadan
R, et al: Restoration of replication fork stability in BRCA1- and
BRCA2-deficient cells by inactivation of SNF2-family fork
remodelers. Mol Cell. 68:414–430.e8. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Piao S, Ojha R, Rebecca VW, Samanta A, Ma
XH, Mcafee Q, Nicastri MC, Buckley M, Brown E, Winkler JD, et al:
ALDH1A1 and HLTF modulate the activity of lysosomal autophagy
inhibitors in cancer cells. Autophagy. 13:2056–2071. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Debauve G, Capouillez A, Belayew A and
Saussez S: The helicase-like transcription factor and its
implication in cancer progression. Cell Mol Life Sci. 65:591–604.
2008. View Article : Google Scholar
|
|
10
|
Tauriello DVF and Batlle E: Targeting the
microenvironment in advanced colorectal cancer. Trends Cancer.
2:495–504. 2016. View Article : Google Scholar
|
|
11
|
Drabsch Y and ten Dijke P: TGF-β
signalling and its role in cancer progression and metastasis.
Cancer Metastasis Rev. 31:553–568. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Massagué J: TGFβ signalling in context.
Nat Rev Mol Cell Biol. 13:616–630. 2012. View Article : Google Scholar
|
|
13
|
Wakefield LM and Hill CS: Beyond TGFβ:
Roles of other TGFβ superfamily members in cancer. Nat Rev Cancer.
13:328–341. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang L, Liu Z, Tan J, Dong H and Zhang X:
Multispectral imaging reveals hyper active TGF-β signaling in
colorectal cancer. Cancer Biol Ther. 19:105–112. 2018. View Article : Google Scholar
|
|
15
|
Fritzmann J, Morkel M, Besser D, Budczies
J, Kosel F, Brembeck FH, Stein U, Fichtner I, Schlag PM and
Birchmeier W: A colorectal cancer expression profile that includes
transforming growth factor beta inhibitor BAMBI predicts metastatic
potential. - PubMed - NCBI. Gastroenterology. 137:165–175. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Papageorgis P, Cheng K, Ozturk S, Gong Y,
Lambert AW, Abdolmaleky HM, Zhou JR and Thiagalingam S: Smad4
inactivation promotes malignancy and drug resistance of colon
cancer. Cancer Res. 71:998–1008. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang B, Halder SK, Kashikar ND, Cho YJ,
Datta A, Gorden DL and Datta PK: Antimetastatic role of Smad4
signaling in colorectal cancer. Gastroenterology. 138:969–80.e1.
32010. View Article : Google Scholar
|
|
18
|
Rizzardi AE, Johnson AT, Vogel RI,
Pambuccian SE, Henriksen J, Skubitz AP, Metzger GJ and Schmechel
SC: Quantitative comparison of immunohistochemical staining
measured by digital image analysis versus pathologist visual
scoring. Diagn Pathol. 7:422012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
20
|
Debauve G, Nonclercq D, Ribaucour F,
Wiedig M, Gerbaux C, Leo O, Laurent G, Journé F, Belayew A and
Toubeau G: Early expression of the Helicase-Like Transcription
Factor (HLTF/SMARCA3) in an experimental model of estrogen-induced
renal carcinogenesis. Mol Cancer. 5:232006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Capouillez A, Decaestecker C, Filleul O,
Chevalier D, Coppée F, Leroy X, Belayew A and Saussez S:
Helicase-like transcription factor exhibits increased expression
and altered intracellular distribution during tumor progression in
hypopharyngeal and laryngeal squamous cell carcinomas. Virchows
Arch. 453:491–499. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Capouillez A, Debauve G, Decaestecker C,
Filleul O, Chevalier D, Mortuaire G, Coppée F, Leroy X, Belayew A
and Saussez S: The helicase-like transcription factor is a strong
predictor of recurrence in hypopharyngeal but not in laryngeal
squamous cell carcinomas. Histopathology. 55:77–90. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ye C, Sun NX, Ma Y, Zhao Q, Zhang Q, Xu C,
Wang SB, Sun SH, Wang F and Li W: MicroRNA-145 contributes to
enhancing radiosensitivity of cervical cancer cells. FEBS Lett.
589:702–709. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Arcolia V, Paci P, Dhont L, Chantrain G,
Sirtaine N, Decaestecker C, Remmelink M, Belayew A and Saussez S:
Helicase-like transcription factor: A new marker of
well-differentiated thyroid cancers. BMC Cancer. 14:4922014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kim JJ, Chung SW, Kim JH, Kim JW, Oh JS,
Kim S, Song SY, Park J and Kim DH: Promoter methylation of
helicase-like transcription factor is associated with the early
stages of gastric cancer with family history. Ann Oncol.
17:657–662. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sandhu S, Wu X, Nabi Z, Rastegar M, Kung
S, Mai S and Ding H: Loss of HLTF function promotes intestinal
carcinogenesis. Mol Cancer. 11:182012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hibi K, Nakayama H, Kanyama Y, Kodera Y,
Ito K, Akiyama S and Nakao A: Methylation pattern of HLTF gene in
digestive tract cancers. Int J Cancer. 104:433–436. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Borinstein SC, Conerly M, Dzieciatkowski
S, Biswas S, Washington MK, Trobridge P, Henikoff S and Grady WM:
Aberrant DNA methylation occurs in colon neoplasms arising in the
azoxymethane colon cancer model. Mol Carcinog. 49:94–103. 2010.
|
|
29
|
Moinova HR, Chen WD, Shen L, Smiraglia D,
Olechnowicz J, Ravi L, Kasturi L, Myeroff L, Plass C, Parsons R, et
al: HLTF gene silencing in human colon cancer. Proc Natl Acad Sci
USA. 99:4562–4567. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wallner M, Herbst A, Behrens A, Crispin A,
Stieber P, Göke B, Lamerz R and Kolligs FT: Methylation of serum
DNA is an independent prognostic marker in colorectal cancer. Clin
Cancer Res. 12:7347–7352. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Leung WK, To KF, Man EP, Chan MW, Bai AH,
Hui AJ, Chan FK and Sung JJ: Quantitative detection of promoter
hypermethylation in multiple genes in the serum of patients with
colorectal cancer. Am J Gastroenterol. 100:2274–2279. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Herbst A, Rahmig K, Stieber P, Philipp A,
Jung A, Ofner A, Crispin A, Neumann J, Lamerz R and Kolligs FT:
Methylation of NEUROG1 in serum is a sensitive marker for the
detection of early colorectal cancer. Am J Gastroenterol.
106:1110–1118. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Philipp AB, Stieber P, Nagel D, Neumann J,
Spelsberg F, Jung A, Lamerz R, Herbst A and Kolligs FT: Prognostic
role of methylated free circulating DNA in colorectal cancer. Int J
Cancer. 131:2308–2319. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cheng CK, Chan NP, Wan TS, Lam LY, Cheung
CH, Wong TH, Ip RK, Wong RS and Ng MH: Helicase-like transcription
factor is a RUNX1 target whose downregulation promotes genomic
instability and correlates with complex cytogenetic features in
acute myeloid leukemia. Haematologica. 101:448–457. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Caliskan C, Guler N, Karaca C, Makay O,
Firat O and Korkut MA: Negative prognostic factors in colorectal
carcinoma: An analysis of 448 patients. Indian J Surg. 72:243–248.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bosch SL, Teerenstra S, de Wilt JH,
Cunningham C and Nagtegaal ID: Predicting lymph node metastasis in
pT1 colorectal cancer: A systematic review of risk factors
providing rationale for therapy decisions. Endoscopy. 45:827–834.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Leake I: Colorectal cancer. Understanding
the routes of metastasis in colorectal cancer. Nat Rev
Gastroenterol Hepatol. 11:2702014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Helmer RA, Foreman O, Dertien JS, Panchoo
M, Bhakta SM and Chilton BS: Role of helicase-like transcription
factor (hltf) in the G2/m transition and apoptosis in brain. PLoS
One. 8:e667992013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Huang D, Sun W, Zhou Y, Li P, Chen F, Chen
H, Xia D, Xu E, Lai M, Wu Y, et al: Mutations of key driver genes
in colorectal cancer progression and metastasis. Cancer Metastasis
Rev. 37:173–187. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chandrasinghe P, Cereser B, Moorghen M, Al
Bakir I, Tabassum N, Hart A, Stebbing J and Warusavitarne J: Role
of SMAD proteins in colitis-associated cancer: From known to the
unknown. Oncogene. 37:1–7. 2018. View Article : Google Scholar
|
|
41
|
Xu Y and Pasche B: TGF-β signaling
alterations and susceptibility to colorectal cancer. Hum Mol Genet.
16R:R14–R20. 2007. View Article : Google Scholar
|
|
42
|
Coates RF, Gardner JA, Gao Y, Cortright
VM, Mitchell JM, Ashikaga T, Skelly J and Yang MX: Significance of
positive and inhibitory regulators in the TGF-β signaling pathway
in colorectal cancers. Hum Pathol. 66:34–39. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Itatani Y, Kawada K, Fujishita T, Kakizaki
F, Hirai H, Matsumoto T, Iwamoto M, Inamoto S, Hatano E, Hasegawa
S, et al: Loss of SMAD4 from colorectal cancer cells promotes CCL15
expression to recruit CCR1+ myeloid cells and facilitate
liver metastasis. Gastroenterology. 145:1064–1075.e11. 2013.
View Article : Google Scholar
|
|
44
|
Voorneveld PW, Kodach LL, Jacobs RJ, Liv
N, Zonnevylle AC, Hoogenboom JP, Biemond I, Verspaget HW, Hommes
DW, de Rooij K, et al: Loss of SMAD4 alters BMP signaling to
promote colorectal cancer cell metastasis via activation of Rho and
ROCK. Gastroenterology. 147:196–208.e13. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Isaksson-Mettävainio M, Palmqvist R,
Dahlin AM, Van Guelpen B, Rutegård J, Öberg A and Henriksson ML:
High SMAD4 levels appear in microsatellite instability and
hyper-methylated colon cancers, and indicate a better prognosis.
Int J Cancer. 131:779–788. 2012. View Article : Google Scholar
|