Introduction
Liver cancer ranked in the top 10 among estimated
new cases of cancer and associated worldwide in 2018, across 20
world regions, with 841,080 (4.7%) new cases and 781,631 (8.2%)
mortalities (1). Hepatocellular
carcinoma (HCC) is not only the predominant histological type of
liver cancer, but also accounts for the highest proportion of ~80%
of all primary liver cancer incidences (2). In China, HCC is a common type of
tumor and is the second leading cause of cancer mortality (3). Approximately 80-90% of all HCC cases
are a result of liver cirrhosis, while the second highest
percentage is a result of persistent hepatitis B or C virus (HBV)
infection (4). Other risk factors
for HCC include obesity, iron overload, alcohol abuse,
environmental pollutants and aflatoxin contaminations (5,6).
Early-stage HCC can be diagnosed and effectively treated through
curative resection and liver transplantation, but treatments for
advanced HCC are limited and have unsatisfactory outcomes (7,8). HCC
tumor recurrence, drug resistance, and disease relapse after
therapy are critical issues that result in poor prognosis (8,9).
Long noncoding RNAs (lncRNAs), of >200
nucleotides in length, are a subclass of functional noncoding RNAs
that are capable affecting protein expression (10,11).
These lncRNAs share several characteristics of mRNAs: LncRNAs are
5′capped, equipped with a 3′polyadenylate tail, are made up of a
variety of exons and are transcribed by RNA polymerase II (11,12).
Previous studies have indicated that lncRNAs play a pivotal role in
many biological processes, including cell cycle regulation, cardiac
development and X chromosome inactivation (11-14).
In addition, lncRNAs are involved in several diseases (15). Microarray technology has identified
both upregulated and downregulated lncRNAs in a large number of
malignancies, such as breast cancer (16), prostate cancer (17), lung cancer (18) and HCC (19).
LncRNA LINC00668 has been identified to be
associated with tumor progression and prognosis: LINC00668, along
with LINC00710 and LINC00607, are the three most significantly
downregulated lncRNAs in lung adenocarcinoma (20). LINC00668 has been identified as a
potentially carcinogenic lncRNA and its knockdown can inhibit the
proliferation, invasion and migration abilities of laryngeal
squamous cell carcinoma (LSCC) cell lines (21). Induced by E2F transcription factor
1, upregulated LINC00668 can predict poor prognosis of gastric
cancer (GC) and promote cell proliferation by epigenetically
silencing cyclin-dependent protein kinase inhibitors (22). However, the aforementioned studies
did not report the tissues specificities of LINC00668 in tumor
cells. Databases (https://portals.broadinstitute.org/ccle/page?gene=LINC00668)
indicate that tumor cells of these organs expressing LINC00668
highly are meningioma, colorectal, stomach, bile duct and liver. In
addition, the association between LINC00668 and HCC remains
unclear. Therefore, we conducted an analysis to explore the
potential roles of LINC00668 in HCC diagnosis, prognosis and its
molecular mechanism.
Materials and methods
Data source and genome-wide co-expression
correlated genes
Clinical data and the gene expressions of HCC
patients were obtained from The Cancer Genome Atlas (TCGA;
https://cancergenome.nih.gov/). The
treatments of these patients underwent can be accessed at
https://xenabrowser.net/datapages/?host=https%3A%2F%2Ftcga.xenahubs.net&removeHub=https%3A%2F%2Fxena.treehouse.gi.ucsc.edu%3A443.
The co-expression correlation coefficient was used to evaluate the
correlation between LINC00668 and genome-wide genes, using R 3.5.0
(https://www.r-project.org/). LncRNAs do
not encode proteins alone, and their function has been associated
with co-expressed protein coding genes (PCGs) (23,24).
LINC00668 and its top 10 correlated genes, known as PCGs, were
employed for further analysis based on the median levels of
expression which served as the cut-off value; they were further
divided as low and high expression PCGs.
Expression of LINC00668 and genes in
tumor and non-tumor tissue
The expression of LINC00668 and its top 10 PCGs in
tumor and non-tumor tissues were obtained from the Metabolic gEne
RApid Visualizer (http://merav.wi.mit.edu/) (25). Scatter plots were then created in
TCGA database using these data and were visualized using GraphPad
7.0 (GraphPad Software, Inc.).
Diagnostic, prognostic and joint-effect
analysis of LINC00668 and its PCGs
The diagnostic value of LINC00668 and its top 10
PCGs were visualized in GraphPad 7.0, using receiver operating
characteristic (ROC) curves. An area under curve (AUC) value of
<0.7 was considered significant for HCC diagnosis. Then,
joint-effect analysis was performed between significant genes and
LINC0068.
Thereafter, their prognostic value for overall
survival (OS) were analyzed using SPSS 16.0 (SPSS, Inc.) and the
results were presented using Kaplan-Meier plots visualized using
GraphPad 7.0. Joint-effect analysis with LINC00668 was performed on
genes that were of prognostic significance for HCC.
Gene set enrichment analysis (GSEA)
GSEA (http://software.broadinstitute.org/gsea/index.jsp.),
which includes Gene Ontology (GO): Biological process (BP),
cellular component (CC), molecular function (MF) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) metabolic pathway
analyses, was performed to explore the potential molecular
mechanisms of LINC00668 and genes that are responsible for the
development and progression of HCC. Then, the KEGG set (c2.cp.kegg.
v6.1.symbols.gmt) and GO sets (c5.bp.v6.1.symbols.gmt, c5.cc.
v6.1.symbols.gmt, c5.mf.v6.1.symbols.gmt) obtained were used for
analysis.
Nomogram, co-expression matrix, gene-gene
interaction (GGI) and GO interaction network
Prognosis-related genes, LINC00668 and clinical
factors were included in the nomogram. The nomogram was constructed
and used for 1 year, 3 year, and 5 year OS prediction. Afterwards,
the co-expression matrix between top 10 genes and LINC00668 was
constructed using R 3.5.0 software. The interaction network between
the genes and LINC00668 was presented using the geneMANIA plugin of
Cytoscape software (26,27). Moreover, GO terms were visualized
using the BinGO plugin of Cytoscape software (28).
Pharmacological targets and drug
selection
Genome-wide differentially expressed genes (DEGs),
including upregulated and downregulated genes, as well as heatmaps
and volcano plots were obtained using edgeR (29). The results with a fold change of
>2 and P≤0.05 were used for further analysis. Then, target drugs
were selected from the Connectivity Map (https://portals.broadinstitute.org/cmap/). The
chemical composition of these drugs were acquired from PubChem
Compound (https://www.ncbi.nlm.nih.gov/pccompound/). GO terms
were visualized based on the DEGs using BinGO. Then, enrichment
analysis was performed based on the DEGs using the Database for
Annotation, Visualization and Integrated Discovery v6.8 (DAVID;
https://david.ncifcrf.gov/) (30,31).
Statistical analysis
Survival analyses was performed using SPSS 16.0
software. Median survival time, log-rank P-values, 95% confidence
intervals (CI) and hazard ratios (HR) were calculated using the
Kaplan-Meier method and Cox proportional hazards regression models.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Co-expression of correlated genome-wide
genes and clinicopathological characteristics of HCC patients
Co-expression of correlated genome-wide genes with
LINC00668 were calculated and are shown in Table SI. The top 10 PCGs of LINC00668
were pyrimidineregic receptor P2Y4 (P2RY4), signal peptidase
complex subunit 2 (SPCS2), family with sequence similarity
86 member C1 (FAM86C1), tudor domain containing 5
(TDRD5), ferritin light chain (FTL), stratifin
(SFN), nucleolar complex associated 2 homolog
(NOC2L), peroxiredoxin 1 (PRDX1), cancer/testis
antigen 2 (CTAG2) and leucine zipper and CTNNBIP1 domain
containing (LZIC) (Table
I). Then, LINC00668 and the top 10 PCGs were further explored
for their diagnostic, prognostic significance, along with their
molecular mechanisms in HCC. A total of 370 HCC patients were
enrolled in the analysis. HBV status, tumor stage and radical
resection status were found to be associated with OS (log-rank
P<0.0001, P<0.0001, P 0.007, respectively; Table II).
 | Table ITop 10 PCGs associated with
LINC00668. |
Table I
Top 10 PCGs associated with
LINC00668.
| LncRNA | PCG | Coefficient | P-value | 95% CI |
|---|
| LINC00668 | P2RY4 | 0.46 | 1.30E-20 | 0.37-0.54 |
| LINC00668 | SPCS2 | 0.4 | 6.54E-16 | 0.31-0.49 |
| LINC00668 | FAM86C1 | 0.39 | 1.20E-14 | 0.30-0.47 |
| LINC00668 | TDRD5 | 0.38 | 1.98E-14 | 0.29-0.47 |
| LINC00668 | FTL | 0.38 | 3.16E-14 | 0.29-0.46 |
| LINC00668 | SFN | 0.37 | 9.85E-14 | 0.28-0.46 |
| LINC00668 | NOC2L | 0.37 | 3.84E-13 | 0.27-0.45 |
| LINC00668 | PRDX1 | 0.36 | 1.01E-12 | 0.27-0.45 |
| LINC00668 | CTAG2 | 0.36 | 1.46E-12 | 0.26-0.44 |
| LINC00668 | LZIC | 0.36 | 1.54E-12 | 0.26-0.44 |
 | Table IIDemographic characteristics of
patients with hepatocellular carcinoma in The Cancer Genome Atlas
database. |
Table II
Demographic characteristics of
patients with hepatocellular carcinoma in The Cancer Genome Atlas
database.
| Variables | Patients
(n=370) | Overall survival
|
|---|
| No. of event | MST (days) | HR (95% CI) | P-value |
|---|
| Gender | | | | | 0.262 |
| Female | 121 | 51 | 1,490 | Ref. | |
| Male | 249 | 79 | 2,486 | 0.817
(0.573-1.164) | |
| Age (years) | | | | | 0.217 |
| ≤60 | 177 | 55 | 2,532 | Ref. | |
| >60 | 193 | 75 | 1,622 | 1.246
(0.879-1.766) | |
| Child-pugha | | | | | 0.184 |
| A | 216 | 59 | 2,542 | Ref. | |
| B + C | 22 | 9 | 1,005 | 1.614
(0.796-3.270) | |
| HBV
infectionb | | | | |
<0.001 |
| No | 247 | 104 | 1,210 | Ref. | |
| Yes | 104 | 20 | NA | 0.357
(0.221-0.578) | |
| HCV
infectionc | | | | | 0.730 |
| No | 295 | 105 | 1,791 | Ref. | |
| Yes | 56 | 19 | 1,229 | 1.090
(0.667-1.782) | |
| Histologic
graded | | | | | 0.750 |
| G1 | 55 | 18 | 2,116 | Ref. | |
| G2 | 177 | 60 | 1,685 | 1.181
(0.697-2.000) | 0.537 |
| G3 | 121 | 43 | 1,622 | 1.233
(0.711-2.140) | 0.456 |
| G4 | 12 | 5 | NA | 1.693
(0.626-4.584) | 0.300 |
| Tumor stagee | | | | |
<0.001 |
| I | 171 | 42 | 2,532 | Ref. | |
| II | 85 | 26 | 1,852 | 1.427
(0.874-2.330) | 0.155 |
| III + IV | 90 | 48 | 770 | 2.764
(1.823-4.190) |
<0.001 |
| Ishak fibrosis
scoref | | | | | 0.874 |
| 0 | 74 | 30 | 2,131 | Ref. | |
| 1,2 | 31 | 9 | 1,372 | 0.917
(0.429-1.962) | 0.823 |
| 3,4 | 28 | 6 | NA | 0.682
(0.281-1.654) | 0.397 |
| 5 | 9 | 2 | 1,386 | 0.750
(0.177-3.167) | 0.695 |
| 6 | 69 | 17 | NA | 0.766
(0.418-1.403) | 0.388 |
| AFP (ng/ml)g | | | | | 0.832 |
| ≤400 | 213 | 62 | 2,456 | Ref. | |
| >400 | 64 | 22 | 2,486 | 1.055
(0.645-1.724) | |
| Radical
resectionh | | | | | 0.007 |
| R0 | 323 | 110 | 1,875 | Ref. | |
| R1 + R2 + RX | 40 | 17 | 837 | 2.030
(1.213-3.395) | |
| Vascular
invasioni | | | | | 0.155 |
| No | 206 | 60 | 2,131 | Ref. | |
| Yes | 108 | 36 | 2,486 | 1.351
(0.892-2.047) | |
| Alcohol
historyj | | | | | 0.896 |
| No | 234 | | | Ref. | |
| Yes | 117 | | | 1.026
(0.703-1.496) | |
Expression of LINC00668 and PCGs in tumor
and non-tumor tissues
LINC00668, P2RY4 and CTAG2 exhibited
high expression in non-tumor tissues, whereas other PCGs showed low
expression levels in non-tumor tissues (Fig. 1). TCGA indicated that the
expression of LINC00668, FAM86C1, SFN, NOC2L,
PRDX1 and CTAG2 were significantly different between
tumor and non-tumor tissues (P<0.05; Fig. 2). Moreover, all the PCGs that were
significantly differentially expressed were upregulated in tumor
tissues.
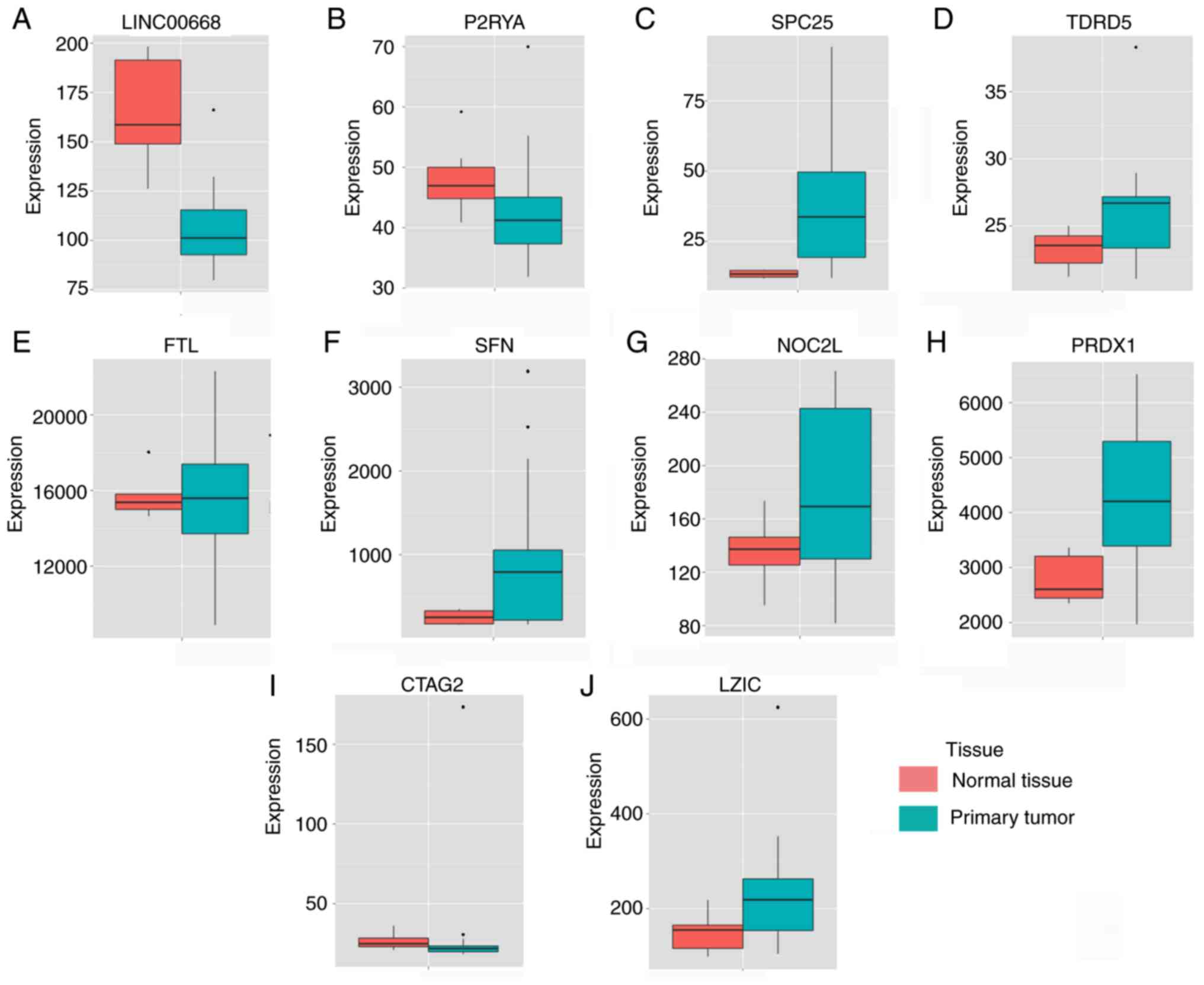 | Figure 1Expressions of LINC00668 and its
co-expression correlated protein-coding genes. (A-J) Expressions
of, LINC00668, P2RY4, SPC25 (SPCS2),
TDRD5, FTL, SFN, NOC2L, PRDX1,
CATG2 and LZIC. CTAG2, cancer/testis antigen 2; FTL,
ferritin light chain; LZIC, leucine zipper and CTNNBIP1 domain
containing; NOC2L, nucleolar complex associated 2 homolog; P2RY4,
pyrimidineregic receptor P2Y4; PRDX1, peroxiredoxin 1; SFN,
stratifin; SPCS2, signal peptidase complex subunit 2; TDRD5, tudor
domain containing 5. |
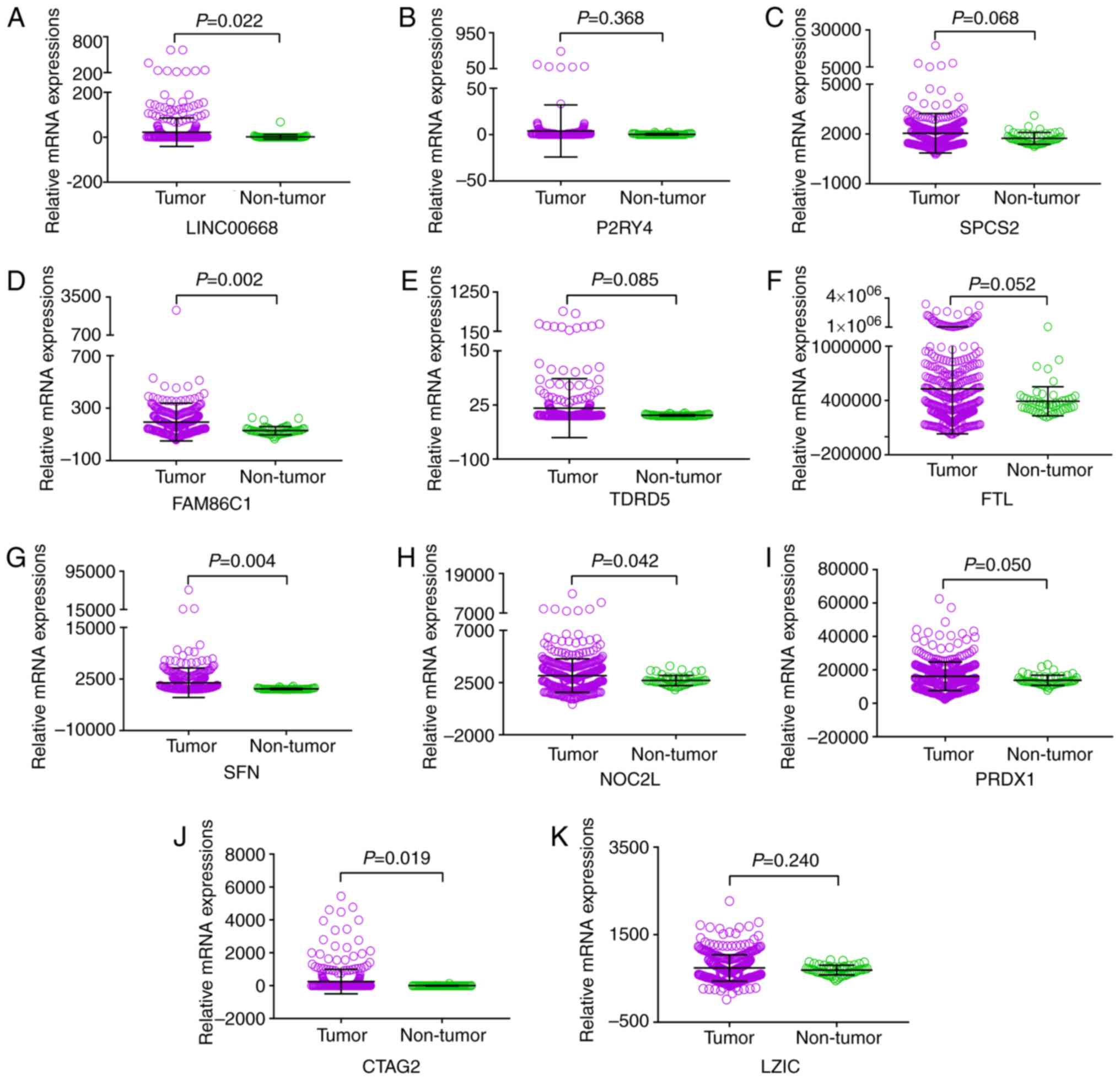 | Figure 2Scatter plots of LINC00668 and its
co-expression correlated protein-coding genes in tumor and
non-tumor tissues. (A-K) Scatter plots of, LINC00668, P2RY4,
SPCS2, FAM86C1, TDRD5, FTL, SFN,
NOC2L, PRDX1, CATG2 and LZIC. CTAG2,
cancer/testis antigen 2; FTL, ferritin light chain; LZIC, leucine
zipper and CTNNBIP1 domain containing; NOC2L, nucleolar complex
associated 2 homolog; P2RY4, pyrimidineregic receptor P2Y4; PRDX1,
peroxiredoxin 1; SFN, stratifin; SPCS2, signal peptidase complex
subunit 2; FAM86C1, family with sequence similarity 86 member C1;
TDRD5, tudor domain containing 5. |
Diagnostic, prognostic and joint-effect
Analysis of LINC00668 and PCGs
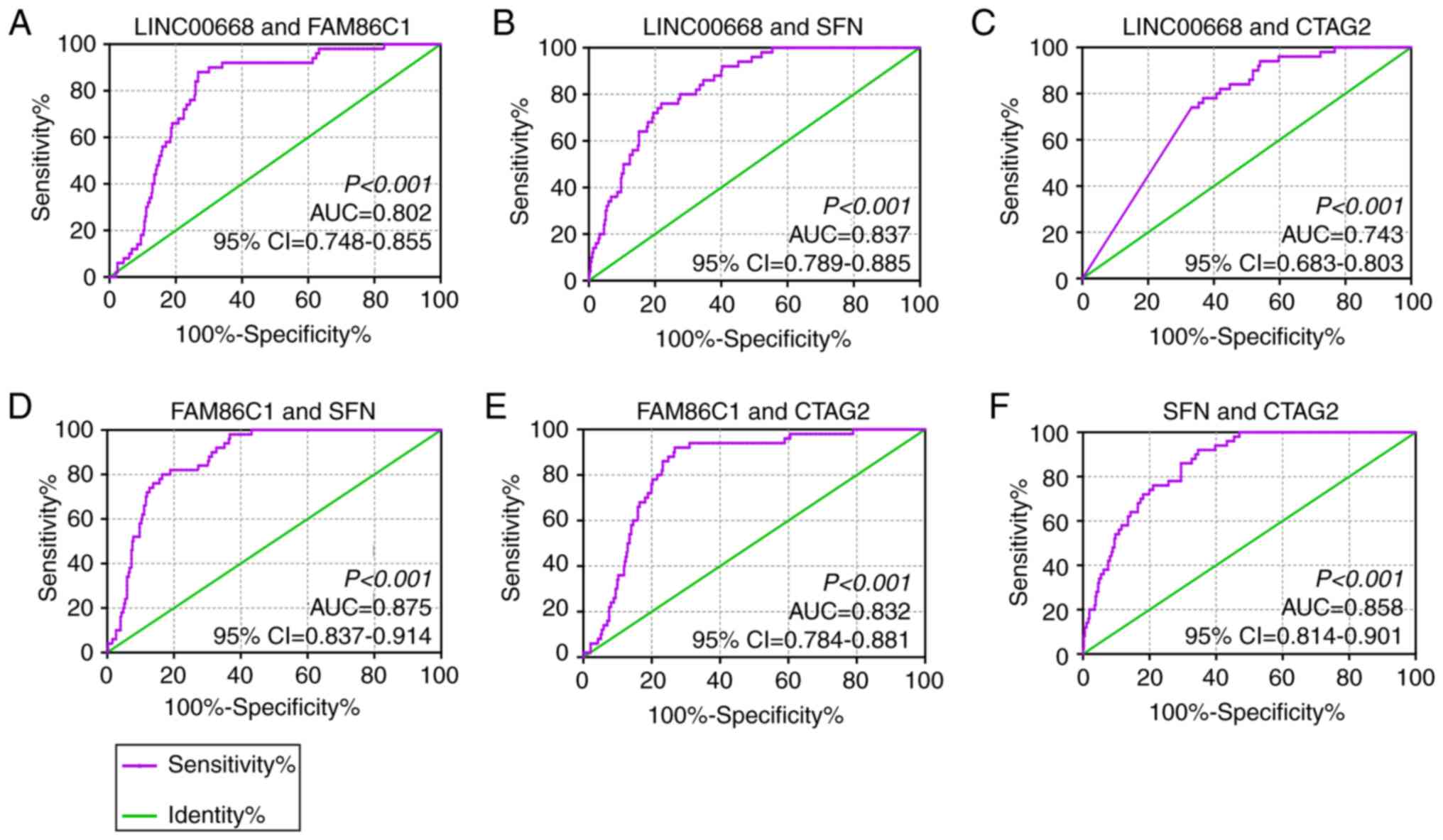
In the diagnostic analysis, FAM86C1, CTAG2 and SFN
were found to be significant for the diagnosis of HCC (Fig. 3D, J and G, AUC=0.766, 0.725 and
0.820; P<0.0001, respectively), while LINC00668, P2RY4, and
SPCS2 were found to be of weak diagnostic significance (Fig. 3A-C, AUC= 0.666, 0.640, and 0.614;
P<0.001, P= 0.001, P= 0.009, respectively). Other PCGs, TDRD5,
FTL, NOC2L, PRDX1 and LZIC, did not show any significance for the
diagnosis of HCC (Fig. 3E-F, H-I,
K, all AUCs<0.600). Then, joint-effect analysis was
performed on LINC00668 and the significant PCGs (Fig. 4). Joint-effect analysis
demonstrated that all of these have a larger AUC value than each
alone.
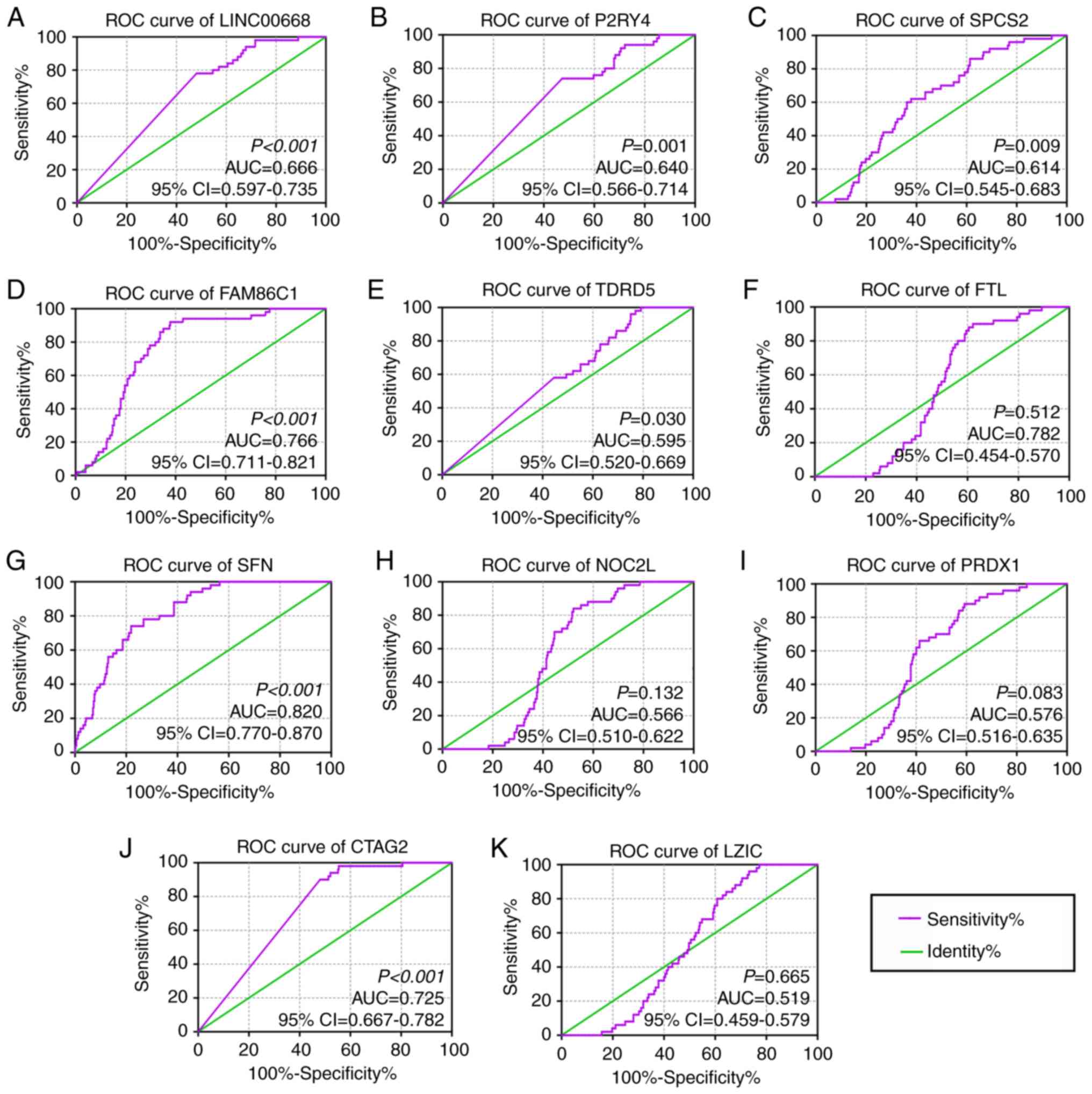 | Figure 3Diagnostic receiver operator curves
of LINC00668 and its co-expression correlated protein-coding genes.
(A-K) Diagnostic ROC curves of, in order, LINC00668, P2RY4,
SPCS2, FAM86C1, TDRD5, FTL, SFN,
NOC2L, PRDX1, CATG2 and LZIC. CTAG2,
cancer/testis antigen 2; FTL, ferritin light chain; LZIC, leucine
zipper and CTNNBIP1 domain containing; NOC2L, nucleolar complex
associated 2 homolog; P2RY4, pyrimidineregic receptor P2Y4; PRDX1,
peroxiredoxin 1; SFN, stratifin; SPCS2, signal peptidase complex
subunit 2; FAM86C1, family with sequence similarity 86 member C1;
TDRD5, tudor domain containing 5; 95% CI, 95% confidence interval;
AUC, area under the curve; ROCs, receiver operator
characteristic. |
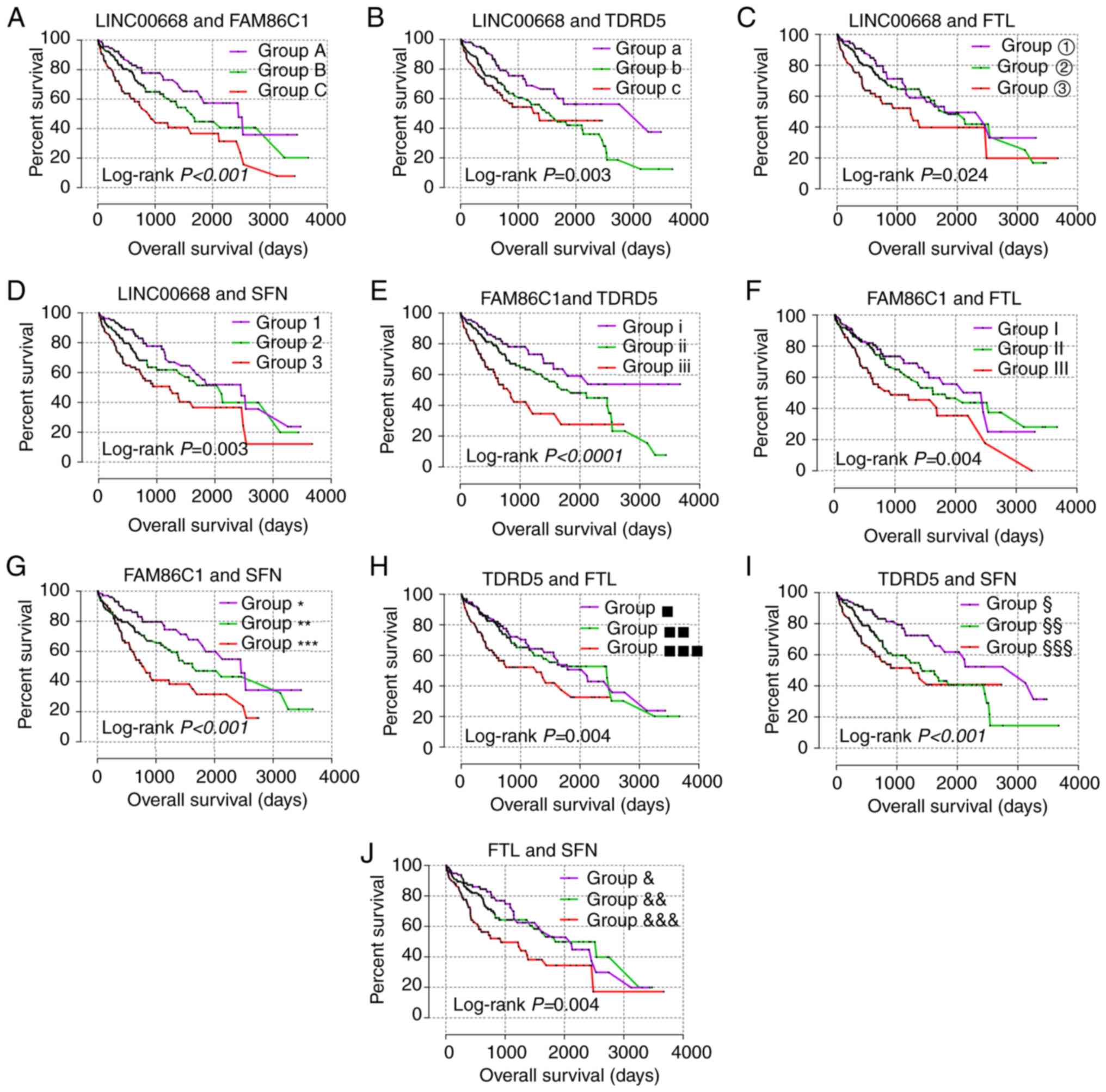
For the prognostic analysis, LINC00668, FAM86C1,
TDRD5, FTL and SFN exhibited prognostic
significance in the multivariate analysis (Table III, adjusted P=0.029, 0.003,
0.012, 0.042 and 0.005, respectively), while LINC00668, FAM86C1,
TDRD5, and SFN exhibited prognostic significance in the
univariate analysis (Table III,
Fig. 5, P=0.025, 0.001, 0.007,
0.003, respectively). Then, joint-effect analysis was performed on
LINC00668 and the significant PCGs (Table IV, Fig. 6). The groups with low expression in
both analyses exhibited the most significance for prognosis; and
groups with high expression in both analyses presented as the
poorest indicators of prognosis; while groups with both low and
high expressions are set in the middle.
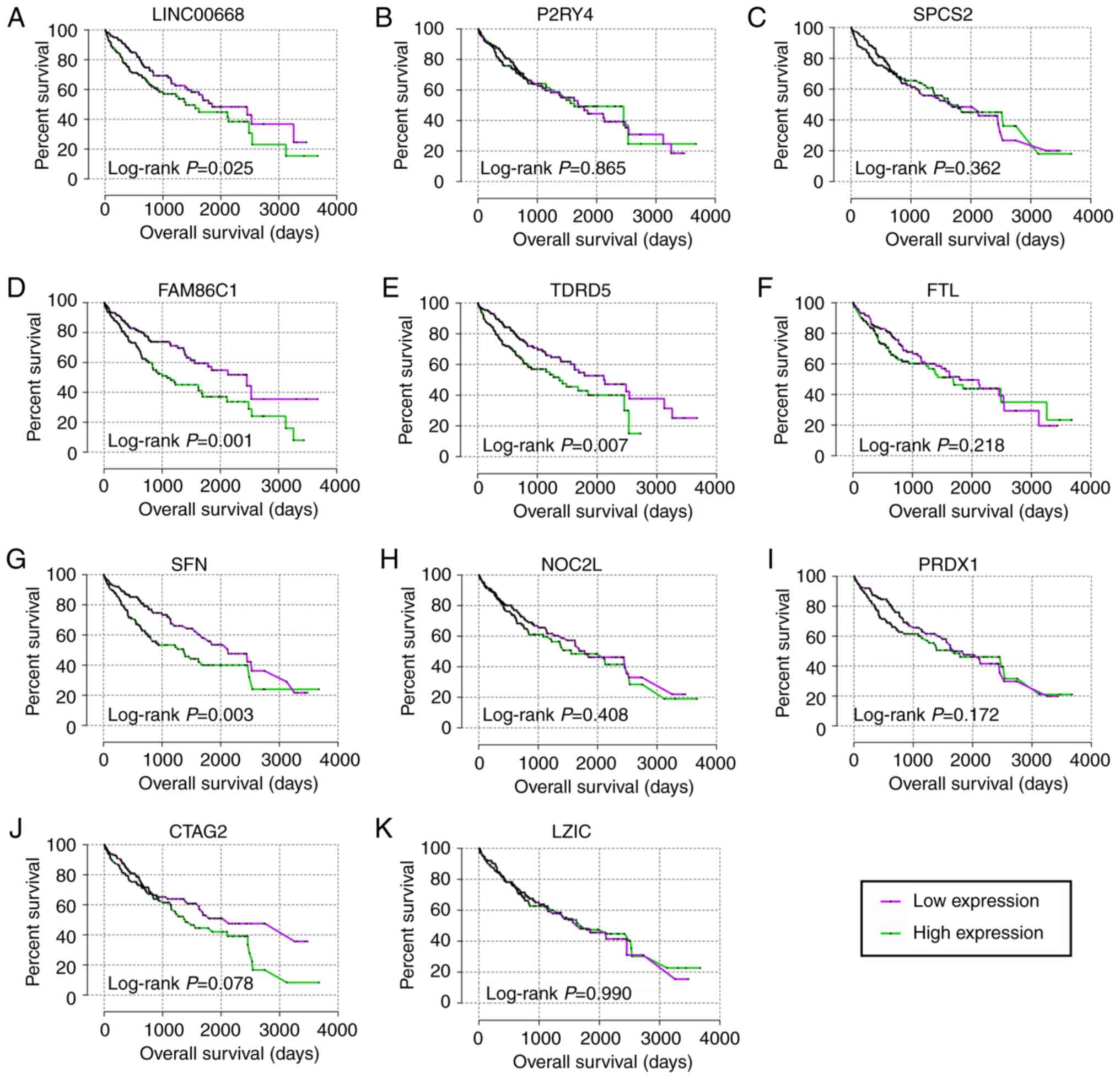 | Figure 5Kaplan-Meier plots of LINC00668 and
its co-expression correlated protein-coding genes. (A-K)
Kaplan-Meier plots of, LINC00668, P2RY4, SPCS2,
FAM86C1, TDRD5, FTL, SFN, NOC2L,
PRDX1, CATG2 and LZIC. CTAG2, cancer/testis
antigen 2; FTL, ferritin light chain; LZIC, leucine zipper and
CTNNBIP1 domain containing; NOC2L, nucleolar complex associated 2
homolog; P2RY4, pyrimidineregic receptor P2Y4; PRDX1, peroxiredoxin
1; SFN, stratifin; SPCS2, signal peptidase complex subunit 2;
FAM86C1, family with sequence similarity 86 member C1; TDRD5, tudor
domain containing 5. |
 | Table IIIPrognostic analysis of
LINC00668 and genes for overall survival in The Cancer
Genome Atlas database. |
Table III
Prognostic analysis of
LINC00668 and genes for overall survival in The Cancer
Genome Atlas database.
| Variables | Patients
(n=370) | Overall survival
|
|---|
| No. of event | MST (days) | HR (95% CI) | Crude P-value | HR (95% CI) | Adjusted
P-valuea |
|---|
| LINC00668 | | | | | 0.025 | | 0.029 |
| Low
expression | 185 | 58 | 1,852 | Ref. | | Ref. | |
| High
expression | 185 | 72 | 1,397 | 1.486
(1.051-2.102) | | 1.540
(1.044-2.270) | |
| P2RY4 | | | | | 0.865 | | 0.646 |
| Low
expression | 185 | 65 | 1,694 | Ref. | | Ref. | |
| High
expression | 185 | 65 | 1,624 | 1.031
(0.729-1.457) | | 0.914
(0.622-1.343) | |
| SPCS2 | | | | | 0.362 | | 0.884 |
| Low
expression | 185 | 70 | 1,694 | Ref. | | Ref. | |
| High
expression | 185 | 60 | 1,685 | 0.851
(0.602-1.203) | | 0.971
(0.659-1.433) | |
| FAM86C1 | | | | | 0.001 | | 0.003 |
| Low
expression | 185 | 54 | 2,456 | Ref. | | Ref. | |
| High
expression | 185 | 76 | 1,088 | 1.796
(1.266-2.550) | | 1.853
(1.241-2.768) | |
| TDRD5 | | | | | 0.007 | | 0.012 |
| Low
expression | 185 | 59 | 2,116 | Ref. | | Ref. | |
| High
expression | 185 | 71 | 1,372 | 1.624
(1.142-2.308) | | 1.680
(1.123-2.514) | |
| FTL | | | | | 0.218 | | 0.042 |
| Low
expression | 185 | 64 | 1,791 | Ref. | | Ref. | |
| High
expression | 185 | 66 | 1,685 | 1.242
(0.880-1.754) | | 1.499
(1.015-2.214) | |
| SFN | | | | | 0.003 | | 0.005 |
| Low
expression | 185 | 54 | 2,131 | Ref. | | Ref. | |
| High
expression | 185 | 76 | 1,372 | 1.706
(1.201-2.421) | | 1.777
(1.194-2.646) | |
| NOC2L | | | | | 0.408 | | 0.996 |
| Low
expression | 185 | 64 | 1,791 | Ref. | | Ref. | |
| High
expression | 185 | 66 | 1,560 | 1.157
(0.819-1.633) | | 0.999
(0.677-1.473) | |
| PRDX1 | | | | | 0.172 | | 0.160 |
| Low
expression | 185 | 62 | 1,685 | Ref. | | Ref. | |
| High
expression | 185 | 68 | 1,694 | 1.272
(0.901-1.795) | | 1.318
(0.897-1.936) | |
| CTAG2 | | | | | 0.078 | | 0.283 |
| Low
expression | 185 | 59 | 2,131 | Ref. | | Ref. | |
| High
expression | 185 | 71 | 1,397 | 1.366
(0.966-1.931) | | 1.235
(0.840-1.816) | |
| LZIC | | | | | 0.990 | | 0.898 |
| Low
expression | 185 | 64 | 1685 | Ref. | | Ref. | |
| High
expression | 185 | 66 | 1694 | 0.998
(0.706-1.410) | | 0.975
(0.662-1.435) | |
 | Table IVJoint-effect analysis of
LINC00668 and genes for overall survival. |
Table IV
Joint-effect analysis of
LINC00668 and genes for overall survival.
| Group | LINC00668
expression | FAM86C1 | TDRD5 | FTL | SFN | Overall survival
|
|---|
| Events/total | MST (days) | Adjusted HR (95%
CI) | Adjusted
P-valuea |
|---|
| A | Low | Low | | | | 26/97 | 2456 | Ref. |
<0.001 |
| B | Low | High | | | | 60/176 | 1624 | 1.604
(0.946-2.721) | 0.080 |
| High | Low | | | | | | | |
| C | High | High | | | | 44/97 | 899 | 2.861
(1.618-5.058) |
<0.001 |
| a | Low | | Low | | | 27/110 | 3258 | Ref. | 0.004 |
| b | Low | | High | | | 63/150 | 1560 | 2.190
(1.314-3.649) | 0.003 |
| High | | Low | | | | | | |
| c | High | | High | | | 40/110 | 1372 | 2.380
(1.350-4.196) | 0.003 |
| ① | Low | | | Low | | 29/91 | 1791 | Ref. | 0.005 |
| ② | Low | | | High | | 64/188 | 1852 | 1.297
(0.785-2.142) | 0.310 |
| High | | | Low | | | | | |
| ③ | High | | | High | | 37/91 | 1229 | 2.350
(1.356-4.073) | 0.002 |
| 1 | Low | | | | Low | 31/107 | 2456 | Ref. | 0.006 |
| 2 | Low | | | | High | 50/156 | 2116 | 1.512
(0.908-2.518) | 0.112 |
| High | | | | Low | | | | |
| 3 | High | | | | High | 49/107 | 1229 | 2.284
(1.370-3.806) | 0.002 |
| i | | Low | Low | | | 24/94 | NA | Ref. |
<0.001 |
| ii | | Low | High | | | 65/182 | 1852 | 1.691
(0.996-2.873) | 0.052 |
| | High | Low | | | | | | |
| iii | | High | High | | | 41/94 | 837 | 3.415
(1.882-6.195) |
<0.0001 |
| I | | Low | | Low | | 33/107 | 2456 | Ref. | 0.003 |
| II | | Low | | High | | 52/156 | 1624 | 1.351
(0.823-2.216) | 0.234 |
| | High | | Low | | | | | |
| III | | High | | High | | 45/107 | 931 | 2.321
(1.399-3.851) | 0.001 |
| * | | Low | | | Low | 26/105 | 2456 | Ref. |
<0.001 |
| ** | | Low | | | High | 56/160 | 1624 | 1.814
(1.068-3.079) | 0.027 |
| | High | | | Low | | | | |
| *** | | High | | | High | 48/105 | 837 | 2.856
(1.662-4.910) |
<0.001 |
| ▪ | | | Low | Low | | 35/97 | 2116 | Ref. |
<0.001 |
| ▪▪ | | | Low | High | | 53/176 | 2456 | 1.127
(0.684-1.857) | 0.640 |
| | | High | Low | | | | | |
| ▪▪▪ | | | High | High | | 42/97 | 1271 | 2.613
(1.508-4.525) |
<0.001 |
| § | | | Low | | Low | 27/109 | 3125 | Ref. | 0.002 |
| §§ | | | Low | | High | 59/152 | 1423 | 2.093
(1.253-3.495) | 0.005 |
| | | High | | Low | | | | |
| §§§ | | | High | | High | 44/109 | 1271 | 2.683
(1.534-4.693) |
<0.001 |
| & | | | | Low | Low | 32/100 | 2116 | Ref. |
<0.001 |
| && | | | | Low | High | 54/170 | 1852 | 1.031
(0.627-1.694) | 0.904 |
| | | | High | Low | | | | |
|
&&& | | | | High | High | 44/100 | 931 | 2.445
(1.461-4.092) |
<0.001 |
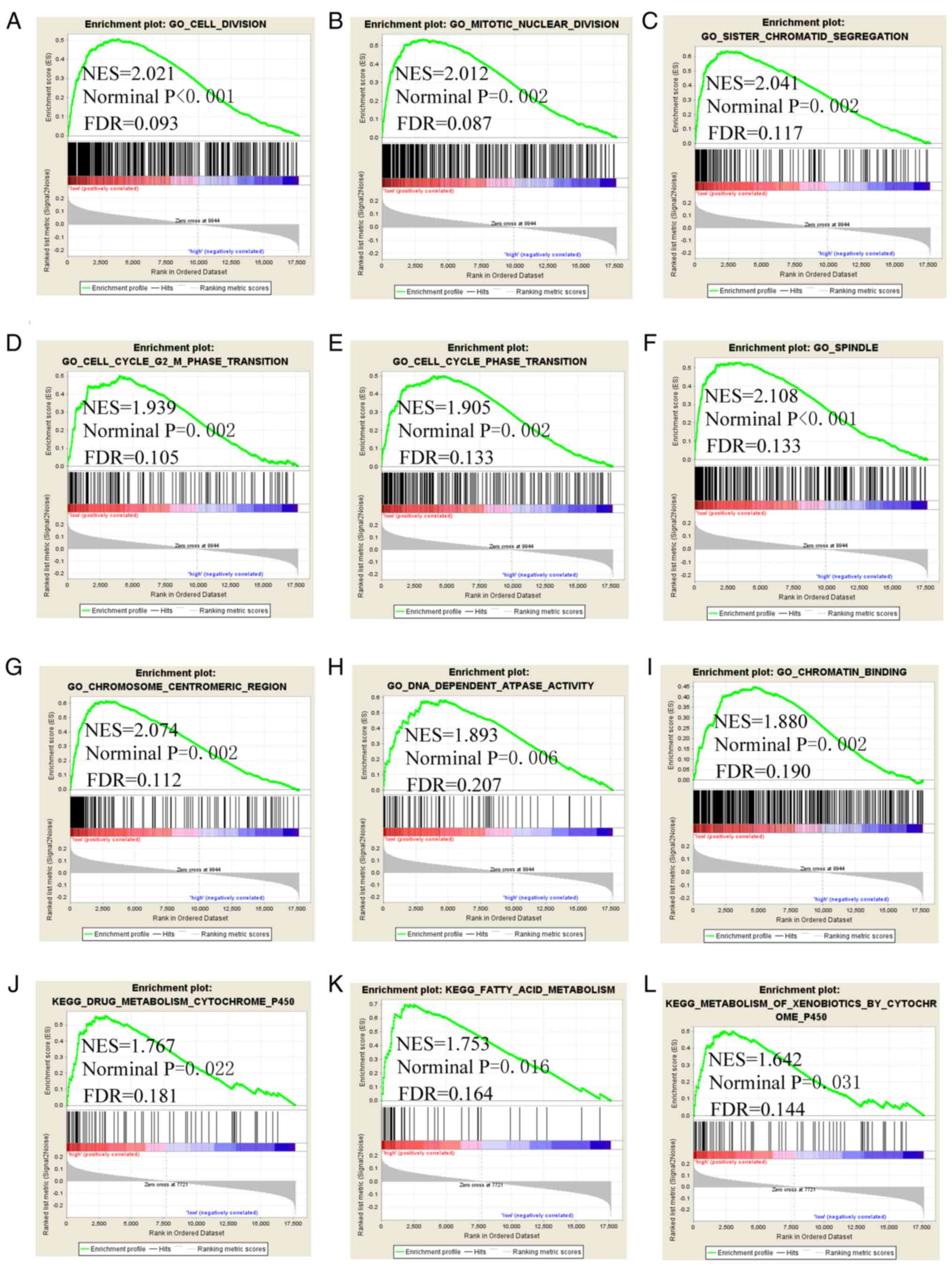
GSEA
GSEA was conducted to explore the genome-wide
potential molecular mechanisms of LINC00668 and its PCGs. The GSEA
of LINC00668 indicated that it is involved in 'cell division',
'mitotic nuclear division', 'sister chromatid segregation', cell
cycle phase transition, 'cell cycle G2 M phase transition',
'spindle', 'chromosome centromeric region', 'DNA dependent ATPase
activity', 'chromatin binding', 'drug metabolism cytochrome P450',
and 'fatty acid metabolism' (Fig.
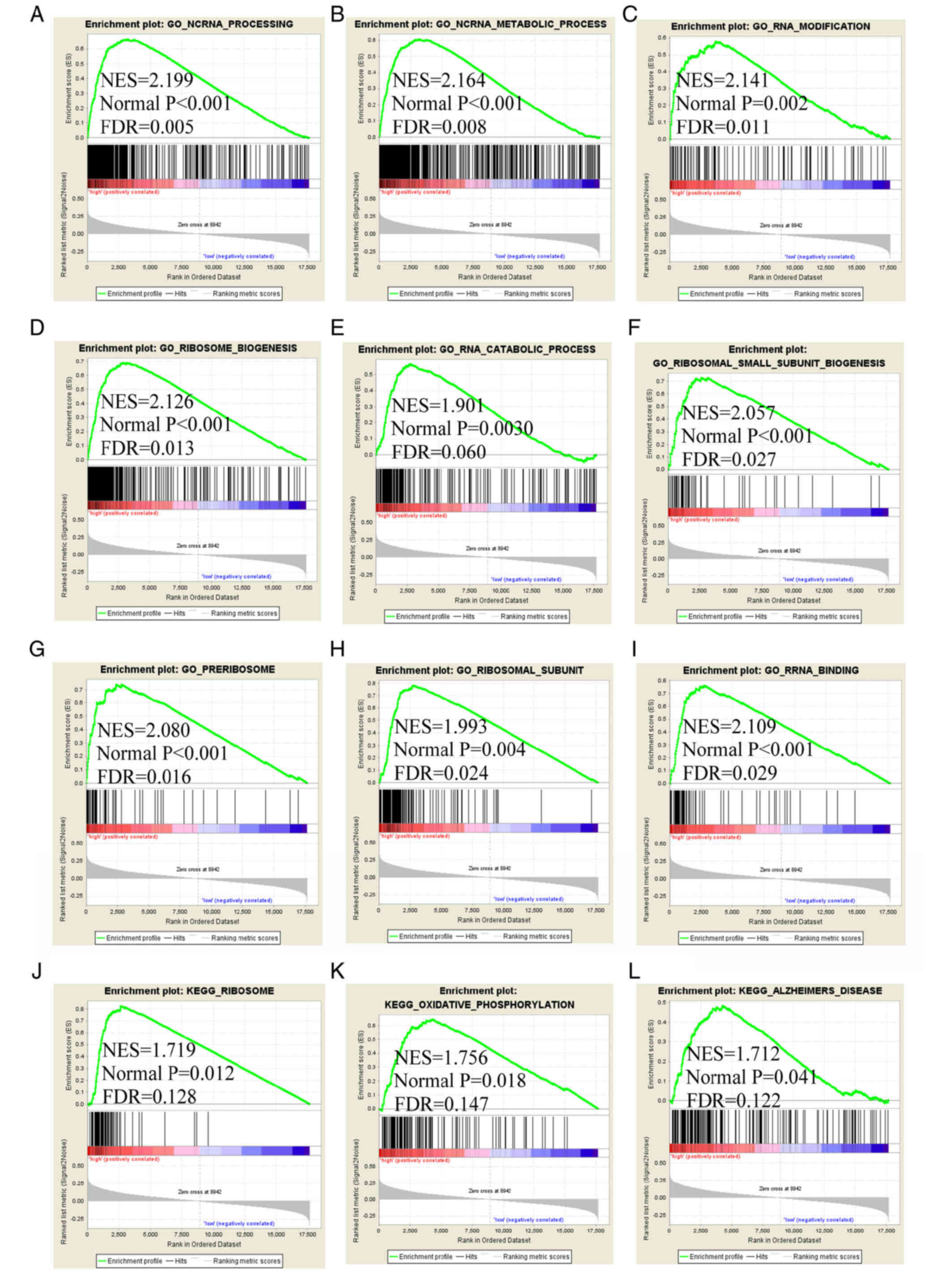
7). The GSEA of FAM86C1 indicated that it is involved in
'ncRNA processing', 'RNA modification', 'RNA catabolic process',
'ncRNA metabolic process', 'ribosome biogenesis', 'ribosomal small
subunit biogenesis', 'preribosome', 'ribosomal subunit', 'RRNA
binding', 'ribosome', 'oxidative phosphorylation' and 'Alzheimer's
disease' (Fig. 8). The GSEA of
FTL indicated that it is involved in 'ncRNA metabolic
process', 'mitochondrial translation', 'amide biosynthetic
process', 'ncRNA processing', 'RRNA metabolic process', 'cytosolic
part', 'structural of constituent of ribosome', 'RRNA binding',
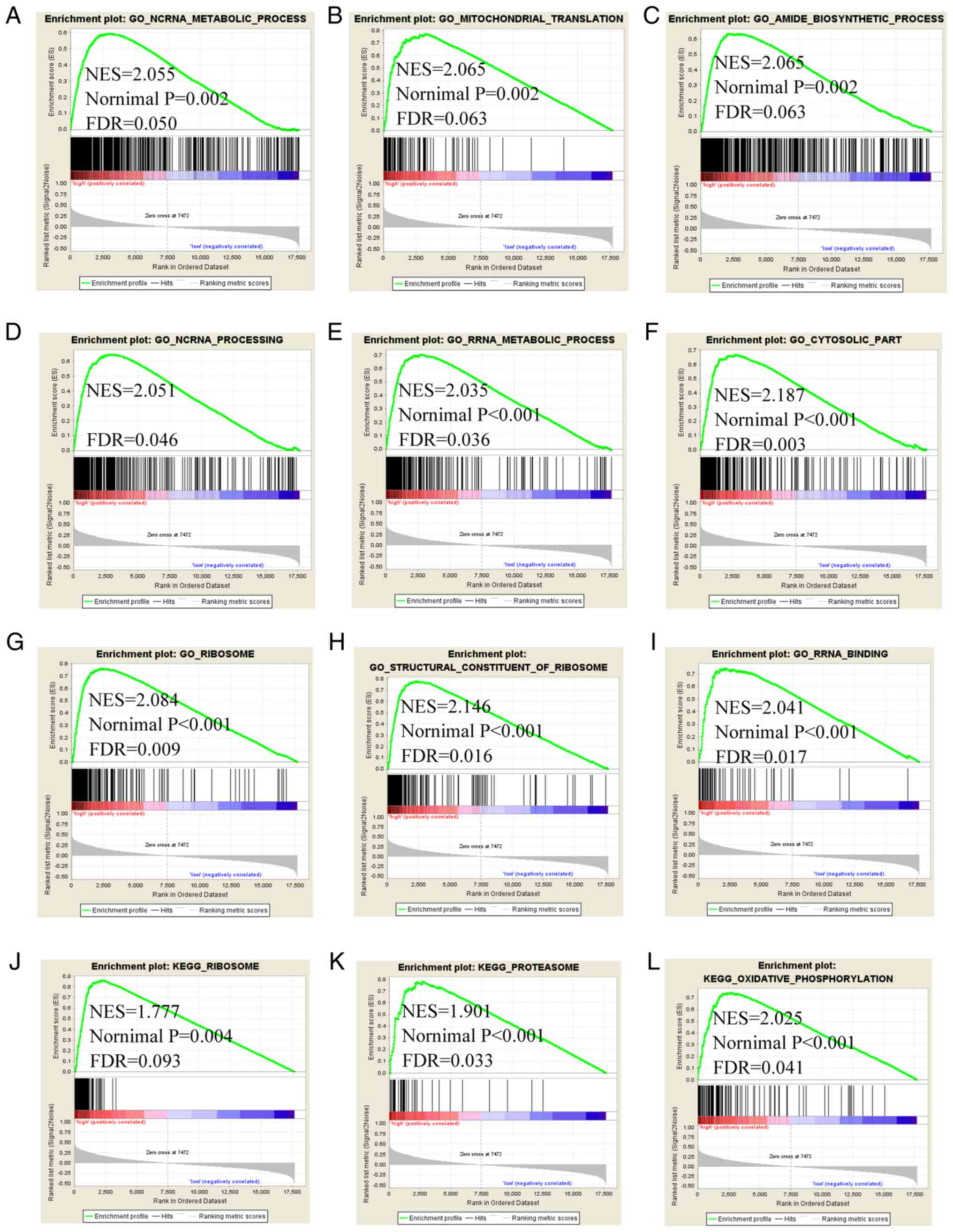
'ribosome', 'proteasome' and 'oxidative phosphorylation' (Fig. 9). The GSEA of SFN and
TDRD5 indicated that they are involved in regulation of cell
cycle, cell cycle phase transition, 'DNA repair', 'regulation of
nuclear division', 'cellular respiration', 'mitochondrial
translation', 'oxidative phosphorylation', 'respiratory chain',
'PPAR signaling pathway', 'Alzheimer's disease', 'fatty acid
metabolism', as well as 'complement and coagulation cascades'
(Figs. S1 and 2).
Nomogram, co-expression matrix, GGI and
GO network
A nomogram was constructed using tumor stage,
radical resection, HBV infection, LINC00668, FAM86C1, TDRD5, FTL,
and SFN (Fig. 10A). Low
expression of LINC00668, FAM86C1, TDRD5, FTL, and SFN had fewer
points, while radical resection, without HBV infection, and a tumor
stage of III and IV accounted for fewer points as well.
Additionally, fewer points suggest better OS. The co-expression
matrix among LINC00668 and the PCGs (Fig. 10B) was also constructed. Most of
them were positively correlated and showed statistical
significance. GGI showed the co-expression relationships among
these PCGs (Fig. 10C). In
addition, CC and MF were visualized, and complex BPs were found
using 10 PCGs (Fig. S3). The
intracellular ferritin complex, signal peptidase complex and
protein kinase C inhibitor activity were enriched in the
network.
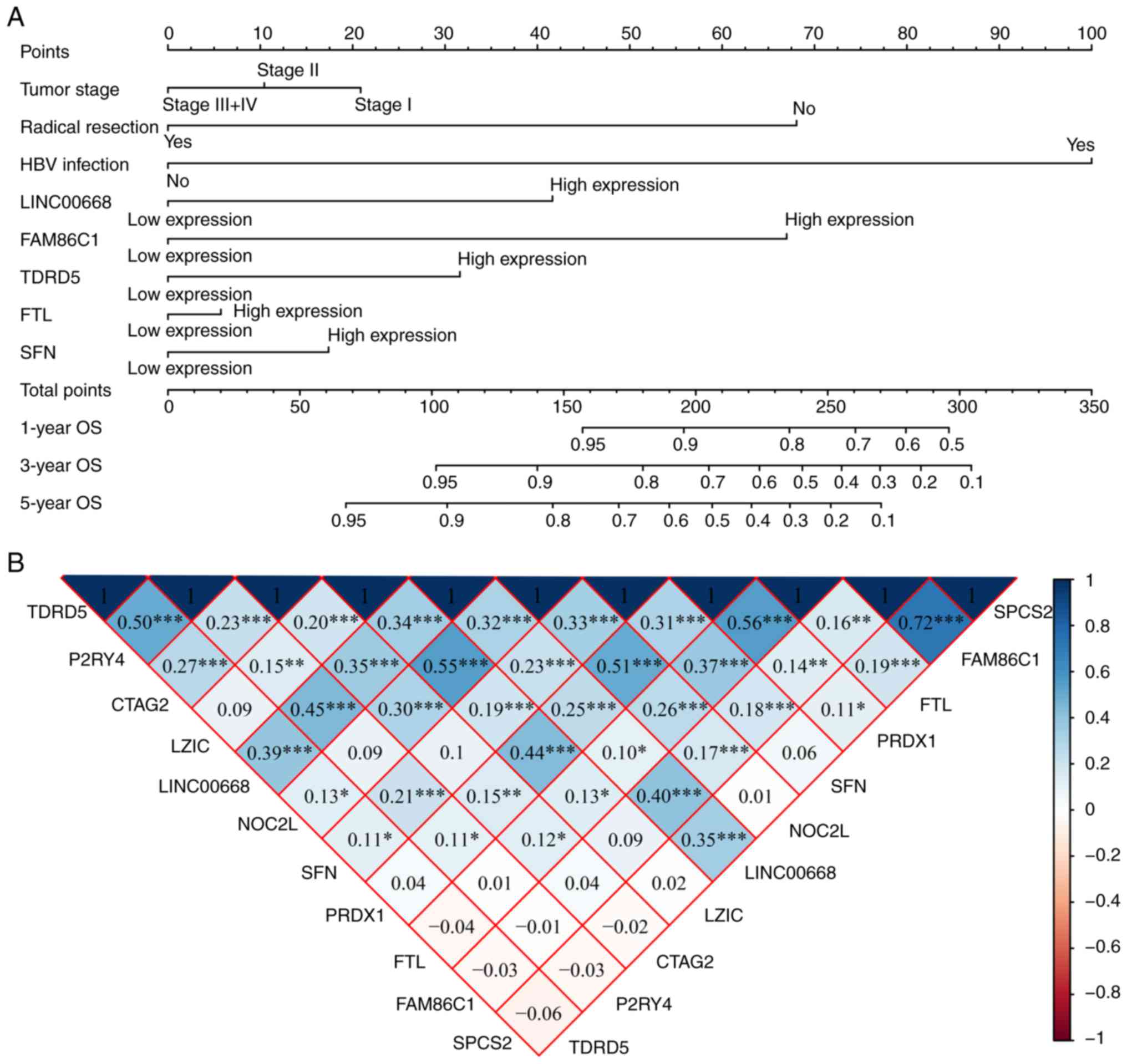 | Figure 10Nomogram, co-expression matrix and
gene-gene interaction network of LINC00668 and protein-coding
genes. (A) Nomogram constructed using LINC00668, FAM86C1, TDRD5,
FTL, SFN, tumor stage, radical resection and HBV infection
status; (B) Co-expression matrix of LINC00668 and its
protein-coding genes; blue and red indicate positive and negative
correlation, respectively. *, **, and
*** denote P≤0.05, 0.01, and 0.001, respectively. (C)
Co-expression network of gene-gene interactions of LINC00668 and
its protein-coding genes. HBV, hepatitis B virus; FTL, ferritin
light chain; FAM86C1, family with sequence similarity 86 member C1;
SFN, stratifin; TDRD5, tudor domain containing 5. |
Pharmacological targets and drugs
The DEGs were acquired using edgeR. Pharmacological
targets and drugs were acquired from the Connectivity Map that was
constructed using the DEGs. Negatively associated drugs are
potential pharmacological targets toward LINC00668 (Tables V and SII). Heatmaps and volcano plots of these
DEGs are presented in Fig. S4,
while the chemical composition and 2D structure of these seven
potential target drugs are presented in Fig. S5. Enrichment analysis of the DEGs
was performed using DAVID. The results included 'cell division',
'mitotic nuclear division', 'sister chromatid cohesion', 'cell
cycle' and 'spliceosome enrichment'. Detailed GO terms and KEGG
pathways are presented in Tables SIII
and SIV, respectively. The GO terms visualized by BinGO are
shown in Fig. S6.
 | Table VPharmacological target and drug. |
Table V
Pharmacological target and drug.
| Drug | PubChem CID | Mean | Enrichment | P-value |
|---|
|
Indolylheptylamine | 35874 | −0.82 | −0.974 | 0.00139 |
| Mimosine | 3862 | −0.47 | −0.900 | 0.00188 |
| Disopyramide | 3114 | −0.375 | −0.794 | 0.00358 |
| Lidocaine | 3676 | −0.441 | −0.720 | 0.00374 |
| NU-1025 | 135398517 | −0.585 | −0.947 | 0.00622 |
| Bumetanide | 2471 | −0.39 | −0.692 | 0.01930 |
|
DQNLAOWBTJPFKL-PKZXCIMASA-N | 5279552 | −0.436 | −0.900 | 0.02014 |
Discussion
In the present study, we explored lncRNA LINC00668
and its associated PCGs for their potential implications in HCC. We
found that LINC00668, FAM86C1, CTAG2 and SFN
are of significance for the diagnosis of HCC. Joint-effect analysis
of these genes revealed that their diagnostic significance was
better when combined than alone. Then, prognostic analysis
indicated that LINC00668, FAM86C1, TDRD5, FTL and
SFN are of prognostic significance in HCC. Furthermore,
joint-effect analysis of these genes indicated that their
diagnostic significance was better when combined than alone. In
order to find their potential molecular mechanisms, GSEA found that
LINC00668 and its PCGs have various functions in 'ncRNA
processing', 'DNA repair', 'cell division', 'mitotic nuclear
division', 'cell cycle phase transition', 'oxidative
phosphorylation', 'drug metabolism cytochrome P450', and 'PPAR
signaling pathway'. A nomogram was constructed using clinical
factors, and LINC00668 and its PCGs were used to predict 1, 3 and 5
year HCC OS. Afterwards, pharmacological target drugs were
identified and seven drugs: Indolylheptylamine, mimosine,
disopyramide, lidocaine, NU-1025, bumetanide and
DQNLAOWBTJPFKL-PKZXCIMASA-N, which may serve as potential targets
with respect to LINC00668 for HCC treatment, were identified.
The discovery of many lncRNAs has notably improved
our understanding of the biological behavior of many complicated
diseases, including tumors. Several studies have demonstrated
abnormal expression of lncRNAs in tumors, which may pinpoint to the
spectrum of cancer progression and predict patient prognosis
(32,33). LncRNAs and microRNAs are major
constituents of the ncRNA family, and it has been revealed that
microRNAs serve a pivotal role in HCC progression (34). LncRNAs function as critical
regulators of many biological behaviors via modulating chromatin
organization, as well as regulation at the transcriptional and
post-transcriptional levels (35,36).
In addition, several studies have indicated that lncRNAs function
as critical factors of tumorigenesis, and that their dysregulation
induces tumor initiation, tumor growth and metastasis (37,38).
Particularly in tumor cells, lncRNAs can affect the proliferation,
growth, cycle progression, apoptosis and migration of transformed
cancer cells (39,40). For instance, functioning as a
molecular decoy for microRNA-221-3P, lncRNA GAPLINC modulates
CD44-dependent cell invasion and is associated with poor prognosis
of gastric cancer (41). LncRNA
FAL1 has been identified as an oncogenic lncRNA, and is associated
with BMI1 and suppresses p21 expression in tumors (42). Activated by TGF-β, lncRNA-ATB binds
to interleukin (IL)-11 mRNA, and the autocrine induction of IL-11
and triggering of the STAT3 signaling pathway promotes the
invasion-metastasis cascade in HCC cell lines (43). LncRNAs HULC (44) and LINC00974 (45) have been reported to be involved in
HCC development and progression.
LncRNA LINC00668 (NR_034100.1) is a 1,751 bp lncRNA,
which is located on chromosome 18p11.31 (46). Our study found that LINC00668 is
upregulated in HCC tumor tissues and was associated with poor
prognosis, which indicates that LINC00668 functions as an oncogene
in HCC. Moreover, our present findings found that LINC00668
expression can affect cell division, cell cycle, mitotic nuclear
division, sister chromosome segregation and drug metabolism
cytochrome P450. Therefore, we speculate that LINC00668 may
function by influencing tumor progression and development. Zhao
et al (21) found that
LINC00668 expression is associated with age, T stage, clinical
stage, cervical lymph node metastasis, and pathological
differentiation degrees. Experiments in vitro indicated that
LINC00668 plays an important role by promoting cell proliferation,
migration, and the invasion ability of TU177 and TU212 cell lines
(21). LINC00668 was determined to
function as an oncogene, is upregulated in tumor tissue and may
serve as a potential biomarker for the targeted treatment of LSCC
(21). In brief, we determined
that LINC00668 plays a consistent role as an oncogene in tumors,
and that its expression is upregulated in LSCC and HCC tumor
tissues. Zhang (46) also
indicated that LINC00668 is upregulated in oral squamous cell
carcinoma (OSCC) tissues and cell lines, and induces poor
prognosis. By competitively sponging microRNA-297, LINC00668
upregulates target gene vascular endothelial growth factor A of
microRNA-297 and facilitates the proliferation of OSCC cells, which
demonstrates that LINC00668 plays a role in the competitive
endogenous RNA network (46). It
was speculate that LINC00668 may serve its pivotal role via the
initiation and progression of OSCC (46). These studies also indicated
important roles of LINC00668 in OSCC progression and prognosis. In
total, our findings are consistent with that of Zhang (46), in which LINC00668 is upregulated in
tumor tissues and is an indicator of poor prognosis that may play
important roles in tumor progression. Furthermore, its related top
10 PCGs were explored to investigate their significance in the
diagnosis and prognosis of HCC. We found these genes to have
distinct diagnostic and prognostic values in HCC. Of note,
potential molecular mechanisms of these genes were explored as well
as LINC00668, including FAM86C1, FTL, SFN and
TDRD5. These potential processes included oxidative
phosphorylation, preribosome, ribosome, NCRNA processing, fatty
acid metabolism, complement and coagulation cascades. Subsequently,
we visualized specific biological processes they were involved
in.
In addition, LINC00668 has been found to be
upregulated in GC tissues and functions as an independent prognosis
indicator for OS (22). LINC00668
plays a role in cell cycle by epigenetically silencing cyclin
dependent kinase inhibitors by binding to polycomb repressive
complex 2, regulating cell growth (22). LINC00668 was also found to be a
predictor of poor prognosis of GC, which is consistent with our
present findings (22). LINC00668
has been reported to be downregulated in lung adenocarcinoma but
was determined to no be associated with patient prognosis or a
biomarker for lung adenocarcinoma (20).
LncRNAs function through their co-expressed PCGs,
and accordingly LINC00668 exerts its role via its top 10 PCGs. Our
study indicates that FAM86C1, TDRD5, FTL and
SFN have prognostic value for HCC, while FAM86C1,
SFN, and CATG2 have diagnostic value for HCC.
Joint-effect analysis of LINC00668 and FAM86C1, SFN and
CATG2 was found to have better diagnostic value than any one
of these genes alone. These results indicated their potential
application in HCC. However, the significance of FAM86C1 in
diseases requires further investigation. TDRD5 has been
found to bind to piwi-interacting (pi)RNA precursors and
selectively enhances pachytene piRNA processing in mice; it has
been speculated that it is involved in piRNA biogenesis (47). Therefore, the potential values of
the aforementioned genes need further investigation in other
cancers. In addition, the diagnostic significance of α-fetoprotein
in this dataset was also evaluated. AFP had AUC=0.613, P=0.010
(data not shown), which did not meet the criteria of candidate
diagnostic biomarkers. Therefore, we concluded that some PCGs were
potential diagnostic biomarkers for HCC.
FTL, an iron utilization gene, has been
reported to be associated with OS and its low expression is linked
to the poor prognosis of HCC (48). Our present results of GO analysis
found that FTL was enriched in ferric iron binding
(GO:0008199). However, our results also indicated that the high
expression of FTL was associated with poor prognosis, which
in inconsistent with the results of Shang et al (48). Specifically, Shang et al
(48) identified that the low
expression of FTL leads to poor prognosis, on the basis of
univariate analysis, whereas our results were based on a
multivariate analysis. Liu et al (49) found that FTL was a DEG and
is upregulated in HCC, which is consistent with our the results of
this study. Wang et al (50) reported that tumor-associated
antigens combined with FTL, AHSG and KRT23 had
high sensitivity and specificity, and these antigens can act as
candidate biomarkers for HCC diagnosis. Given the inconsistency in
the prognostic and diagnostic values of FTL, further
investigation should be conducted to determine its role in HCC. Of
note, its potential significance in diseases, especially in
malignancies, should also be evaluated further. Moreover, we
constructed a nomogram to predict possible risk for 1, 3- and
5-year OS. LINC00668, and prognosis-related genes, including
FAM86C1, FTL, SFN and TDRD5, and
clinical factors, including tumor stage, radical resection, HBV
infection status, were employed in the nomogram for survival
prediction at hepatectomy. According to the above findings, we
concluded that this nomogram provided notable results for survival
prediction in HCC. We also identified seven potential target drugs:
Indolylheptylamine, mimosine, disopyramide, lidocaine, NU-1025,
bumetanide and DQNLAOWBTJPFKL-PKZXCIMASA-N of LINC00668 in HCC via
the Connectivity Map. The Connectivity Map database can provide a
unique method of drug development through the comparison of
potential chemical compounds that can be used to treat diseases,
including tumors, and it has been accepted by several researchers
(51,52). Xiao et al (53) utilized expression profile chip data
and a Connectivity Map to explore the molecular mechanisms of
Hirschsprung's disease and candidate target drugs. They found
certain chemical compounds that may helpful for minimizing the
damage induced by the progression of Hirschsprung's disease
(53). We further visualized
specific structures of these potential target drugs for their
candidate clinical application. Further investigations concerning
these potential target drugs may facilitate the development of
novel strategies for the treatment of HCC.
Additionally, genetic variants concerning TP53 and
catenin β-1 (CTNNB1) mutations have been linked to HCC, including
diagnostic significance. Our study found that TP53 mutations did
not indicate diagnostic significance (AUC:0.648, data not shown),
which is less than the cutoff of 0.700. However, CTNNB1 mutations
suggested diagnostic significance (AUC:0.702, data not shown),
which is slightly higher than the cutoff value. In addition, genes
exhibiting diagnostic significance, including FAM86C1, CTAG2 and
SFN, had higher AUCs than CTNNB1 (AUC=0.766, 0.725 and 0.820,
respectively). These results suggested that FAM86C1, CTAG2 and SFN
may have greater diagnostic value for HCC than CTNNB1 and TP53;
although further investigation is required.
There are certain limitations to the present study
that need to be noted. Firstly, our findings need to be validated
in a larger population. Secondly, a multi-center and validation
cohort are warranted in order to explore clinical significance. In
addition, functional trials regarding LINC00668 and its related
PCGs are warranted to verify their function in HCC.
Our present study identified that lncRNA LINC00668
is differentially expressed and upregulated in HCC tissue. It
functions as an oncogene and its high expression leads to poor
prognosis for HCC. Its co-expressed correlated PCGs have been
determined for diagnostic, value including FAM86C1,
CTAG2 and SFN, and prognostic value, including
FAM86C1, TDRD5, FTL and SFN for HCC.
Investigation into the molecular mechanism indicated that LINC00668
affects cell division, cell cycle, mitotic nuclear division, sister
chromosome segregation and drug metabolism cytochrome P450. We
speculate that it serves important roles in the progression and
development of HCC. Analysis of pharmacological targets revealed 7
candidate target drugs: Indolylheptylamine, mimosine, disopyramide,
lidocaine, NU-1025, bumetanide and DQNLAOWBTJPFKL-PKZXCIMASA-N.
Although these drugs need further validation, this study provides
novel insight into potential treatment strategies for HCC.
Additionally, further functional trials and validation with a
larger cohort are warranted to verify the clinical value of these
findings.
Supplementary Data
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
HBV
|
hepatitis B virus
|
|
LSCC
|
laryngeal squamous cell carcinoma
|
|
GC
|
gastric cancer
|
|
OSCC
|
oral squamous cell carcinoma
|
|
DEG
|
differentially expressed gene
|
|
PCG
|
protein-coding gene
|
|
AUC
|
area under curve
|
|
TCGA
|
The Cancer Genome Atlas
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
GSEA
|
gene set enrichment analysis
|
|
BP
|
biological process
|
|
CC
|
cellular component
|
|
MF
|
molecular function
|
|
GO
|
gene ontology
|
|
OS
|
overall survival
|
|
ROC
|
receiver operating characteristic
|
|
GGI
|
gene-gene interaction
|
|
DAVID
|
database for annotation,
visualization and integrated discovery
|
Acknowledgments
Not applicable.
Funding
This work was supported in part by the National
Nature Science Foundation of China (grant nos. 81560535, 81072321,
30760243, 30460143, 30560133 and 81802874), Natural Science
Foundation of Guangxi Province of China (grant no. 2017JJB140189y),
Key laboratory of High-Incidence-Tumor Prevention & Treatment
(Guangxi Medical University), Ministry of Education (GKE2018-01),
2009 Program for New Century Excellent Talents in University
(NCET), Guangxi Nature Sciences Foundation (grant no. GuiKeGong
1104003A-7), and Guangxi Health Ministry Medicine Grant
(Key-Scientific Research-Grant, grant no. Z201018). The present
study is also partly supported by Scientific Research Fund of the
Health and Family Planning Commission of Guangxi Zhuang Autonomous
Region (grant no. Z2016318), The Basic Ability Improvement Project
for Middle-aged and Young Teachers in Colleges and Universities in
Guangxi (grant no. 2018KY0110), 2018 Innovation Project of Guangxi
Graduate Education (grant no. YCBZ2018036). As well as, the present
study is also partly supported by Research Institute of Innovative
Think-tank in Guangxi Medical University (The gene-environment
interaction in hepatocarcinogenesis in Guangxi HCCs and its
translational applications in the HCC prevention). We also
acknowledge the supported by the National Key Clinical Specialty
Programs (General Surgery & Oncology) and the Key Laboratory of
Early Prevention & Treatment for Regional High-Incidence-Tumor
(Guangxi Medical University), Ministry of Education, China.
Availability of data and materials
The datasets analyzed during the current study are
available from the corresponding author on reasonable request.
Author contribution
XW and TP designed this manuscript; XZ, JL, ZL, LZ,
YG, JH, LY, QW, CY, XL, TY, CH, GZ, XY and TP conducted the study
and analyzed the data. XW wrote the manuscript, and TP guided the
writing.
Ethics approval and consent to
participate
This article does not contain any studies with
human participants or animals performed by any of the authors.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that there are no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Testino G, Leone S, Patussi V, Scafato E
and Borro P: Hepatocellular carcinoma: Diagnosis and proposal of
treatment. Minerva Med. 107:413–426. 2016.PubMed/NCBI
|
|
3
|
Li S, Huang Y, Huang Y, Fu Y, Tang D, Kang
R, Zhou R and Fan XG: The long non-coding RNA TP73-AS1 modulates
HCC cell proliferation through miR-200a-dependent HMGB1/RAGE
regulation. J Exp Clin Cancer Res. 36:512017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gomaa AI, Khan SA, Toledano MB, Waked I
and Taylor-Robinson SD: Hepatocellular carcinoma: Epidemiology,
risk factors and pathogenesis. World J Gastroenterol. 14:4300–4308.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Varnholt H, Drebber U, Schulze F,
Wedemeyer I, Schirmacher P, Dienes HP and Odenthal M: MicroRNA gene
expression profile of hepatitis C virus-associated hepatocellular
carcinoma. Hepatology. 47:1223–1232. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Farazi PA and DePinho RA: Hepatocellular
carcinoma pathogenesis: From genes to environment. Nat Rev Cancer.
6:674–687. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Llovet JM: Liver cancer: Time to evolve
trial design after everolimus failure. Nat Rev Clin Oncol.
11:506–507. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tanaka S and Arii S: Molecular targeted
therapy for hepatocellular carcinoma in the current and potential
next strategies. J Gastroenterol. 46:289–296. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
St Laurent G, Wahlestedt C and Kapranov P:
The Landscape of long noncoding RNA classification. Trends Genet.
31:239–251. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Devaux Y, Zangrando J, Schroen B, Creemers
EE, Pedrazzini T, Chang CP, Dorn GW II, Thum T and Heymans S;
Cardiolinc network: Long noncoding RNAs in cardiac development and
ageing. Nat Rev Cardiol. 12:415–425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ma L, Bajic VB and Zhang Z: On the
classification of long non-coding RNAs. RNA Biol. 10:925–933. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Penny GD, Kay GF, Sheardown SA, Rastan S
and Brockdorff N: Requirement for Xist in X chromosome
inactivation. Nature. 379:131–137. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mourtada-Maarabouni M, Hedge VL, Kirkham
L, Farzaneh F and Williams GT: Growth arrest in human T-cells is
controlled by the non-coding RNA growth-arrest-specific transcript
5 (GAS5). J Cell Sci. 121:939–946. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huo X, Han S, Wu G, Latchoumanin O, Zhou
G, Hebbard L, George J and Qiao L: Dysregulated long noncoding RNAs
(lncRNAs) in hepatocellular carcinoma: Implications for
tumori-genesis, disease progression, and liver cancer stem cells.
Mol Cancer. 16:1652017. View Article : Google Scholar
|
|
16
|
Pickard MR and Williams GT: Regulation of
apoptosis by long non-coding RNA GAS5 in breast cancer cells:
Implications for chemotherapy. Breast Cancer Res Treat.
145:359–370. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pickard MR, Mourtada-Maarabouni M and
Williams GT: Long non-coding RNA GAS5 regulates apoptosis in
prostate cancer cell lines. Biochim Biophys Acta. 1832:1613–1623.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xiaoguang Z, Meirong L, Jingjing Z,
Ruishen Z, Qing Z and Xiaofeng T: Long noncoding RNA CPS1-IT1
suppresses cell proliferation and metastasis in human lung cancer.
Oncol Res. 25:373–380. 2017. View Article : Google Scholar
|
|
19
|
Zhang M, Wang W, Li T, Yu X, Zhu Y, Ding
F, Li D and Yang T: Long noncoding RNA SNHG1 predicts a poor
prognosis and promotes hepatocellular carcinoma tumorigenesis.
Biomed Pharmacother. 80:73–79. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhao B, Xu H, Ai X, Adalat Y, Tong Y,
Zhang J and Yang S: Expression profiles of long noncoding RNAs in
lung adenocarcinoma. Onco Targets Ther. 11:5383–5390. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao L, Cao H, Chi W, Meng W, Cui W, Guo W
and Wang B: Expression profile analysis identifies the long
non-coding RNA landscape and the potential carcinogenic functions
of LINC00668 in laryngeal squamous cell carcinoma. Gene. 687:47–55.
2019. View Article : Google Scholar
|
|
22
|
Zhang E, Yin D, Han L, He X, Si X, Chen W,
Xia R, Xu T, Gu D, De W, et al: E2F1-induced upregulation of long
noncoding RNA LINC00668 predicts a poor prognosis of gastric cancer
and promotes cell proliferation through epigenetically silencing of
CKIs. Oncotarget. 7:23212–23226. 2016.PubMed/NCBI
|
|
23
|
Paci P, Colombo T and Farina L:
Computational analysis identifies a sponge interaction network
between long non-coding RNAs and messenger RNAs in human breast
cancer. BMC Syst Biol. 8:832014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liao Q, Liu C, Yuan X, Kang S, Miao R,
Xiao H, Zhao G, Luo H, Bu D, Zhao H, et al: Large-scale prediction
of long non-coding RNA functions in a coding-non-coding gene
co-expression network. Nucleic Acids Res. 39:3864–3878. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shaul YD, Yuan B, Thiru P, Nutter-Upham A,
McCallum S, Lanzkron C, Bell GW and Sabatini DM: MERAV: A tool for
comparing gene expression across human tissues and cell types.
Nucleic Acids Res. 44:D560–D566. 2016. View Article : Google Scholar :
|
|
26
|
Montojo J, Zuberi K, Rodriguez H, Kazi F,
Wright G, Donaldson SL, Morris Q and Bader GD: GeneMANIA Cytoscape
plugin: Fast gene function predictions on the desktop.
Bioinformatics. 26:2927–2928. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Maere S, Heymans K and Kuiper M: BiNGO: A
Cytoscape plugin to assess overrepresentation of gene ontology
categories in biological networks. Bioinformatics. 21:3448–3449.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar
|
|
30
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar
|
|
31
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Maass PG, Luft FC and Bähring S: Long
non-coding RNA in health and disease. J Mol Med (Berl). 92:337–346.
2014. View Article : Google Scholar
|
|
33
|
Yang G, Lu X and Yuan L: LncRNA: A link
between RNA and cancer. Biochim Biophys Acta. 1839:1097–1109. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wong CM, Kai AK, Tsang FH and Ng IO:
Regulation of hepato-carcinogenesis by microRNAs. Front Biosci
(Elite Ed). 5:49–60. 2013. View
Article : Google Scholar
|
|
35
|
Ulitsky I and Bartel DP: lincRNAs:
Genomics, evolution, and mechanisms. Cell. 154:26–46. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nagano T and Fraser P: No-nonsense
functions for long noncoding RNAs. Cell. 145:178–181. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tsai MC, Spitale RC and Chang HY: Long
intergenic noncoding RNAs: New links in cancer progression. Cancer
Res. 71:3–7. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang H, Chen Z, Wang X, Huang Z, He Z and
Chen Y: Long non-coding RNA: A new player in cancer. J Hematol
Oncol. 6:372013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li J, Zhuang C, Liu Y, Chen M, Chen Y,
Chen Z, He A, Lin J, Zhan Y, Liu L, et al: Synthetic
tetracycline-controllable shRNA targeting long non-coding RNA
HOXD-AS1 inhibits the progression of bladder cancer. J Exp Clin
Cancer Res. 35:992016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hu Y, Wang J, Qian J, Kong X, Tang J, Wang
Y, Chen H, Hong J, Zou W, Chen Y, et al: Long noncoding RNA GAPLINC
regulates CD44-dependent cell invasiveness and associates with poor
prognosis of gastric cancer. Cancer Res. 74:6890–6902. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hu X, Feng Y, Zhang D, Zhao SD, Hu Z,
Greshock J, Zhang Y, Yang L, Zhong X, Wang LP, et al: A functional
genomic approach identifies FAL1 as an oncogenic long noncoding RNA
that associates with BMI1 and represses p21 expression in cancer.
Cancer Cell. 26:344–357. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ,
Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al: A long noncoding RNA
activated by TGF-β promotes the invasion-metastasis cascade in
hepatocellular carcinoma. Cancer Cell. 25:666–681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cui M, Xiao Z, Wang Y, Zheng M, Song T,
Cai X, Sun B, Ye L and Zhang X: Long noncoding RNA HULC modulates
abnormal lipid metabolism in hepatoma cells through an
miR-9-mediated RXRA signaling pathway. Cancer Res. 75:846–857.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tang J, Zhuo H, Zhang X, Jiang R, Ji J,
Deng L, Qian X, Zhang F and Sun B: A novel biomarker Linc00974
interacting with KRT19 promotes proliferation and metastasis in
hepatocellular carcinoma. Cell Death Dis. 5:e15492014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang CZ: Long intergenic non-coding RNA
668 regulates VEGFA signaling through inhibition of miR-297 in oral
squamous cell carcinoma. Biochem Biophys Res Commun. 489:404–412.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ding D, Liu J, Midic U, Wu Y, Dong K,
Melnick A, Latham KE and Chen C: TDRD5 binds piRNA precursors and
selectively enhances pachytene piRNA processing in mice. Nat
Commun. 9:1272018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Shen Y, Li X, Zhao B, Xue Y, Wang S, Chen
X, Yang J, Lv H and Shang P: Iron metabolism gene expression and
prognostic features of hepatocellular carcinoma. J Cell Biochem.
119:9178–9204. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Liu Y, Zhu X, Zhu J, Liao S, Tang Q, Liu
K, Guan X, Zhang J and Feng Z: Identification of differential
expression of genes in hepatocellular carcinoma by suppression
subtractive hybridization combined cDNA microarray. Oncol Rep.
18:943–951. 2007.PubMed/NCBI
|
|
50
|
Wang K, Xu X, Nie Y, Dai L, Wang P and
Zhang J: Identification of tumor-associated antigens by using SEREX
in hepatocellular carcinoma. Cancer Lett. 281:144–150. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Brum AM, van de Peppel J, Nguyen L, Aliev
A, Schreuders-Koedam M, Gajadien T, van der Leije CS, van Kerkwijk
A, Eijken M, van Leeuwen JPTM and van der Eerden BCJ: Using the
Connectivity Map to discover compounds influencing human osteoblast
differentiation. J Cell Physiol. 233:4895–4906. 2018. View Article : Google Scholar
|
|
52
|
Busby J, Murray L, Mills K, Zhang SD,
Liberante F and Cardwell CR: A combined connectivity mapping and
pharmacoepidemiology approach to identify existing medications with
breast cancer causing or preventing properties. Pharmacoepidemiol
Drug Saf. 27:78–86. 2018. View Article : Google Scholar
|
|
53
|
Xiao SJ, Zhu XC, Deng H, Zhou WP, Yang WY,
Yuan LK, Zhang JY, Tian S, Xu L, Zhang L and Xia HM: Gene
expression profiling coupled with Connectivity Map database mining
reveals potential therapeutic drugs for Hirschsprung disease. J
Pediatr Surg. 53:1716–1721. 2018. View Article : Google Scholar : PubMed/NCBI
|