Introduction
Loco-regional recurrence subsequent to rectal cancer
resection is difficult to treat and is associated with severe
debilitating symptoms. The prognosis after a local recurrence is
poor, with a median survival period of 12–18 months (1). Pre-operative chemoradiotherapy (CRT)
has been widely used as the major treatment modality to improve
local control of the disease, as well as to preserve anal sphincter
(2). However, response to CRT
differs in tumors.
microRNAs (miRNA) are a family of small non-coding
RNA molecules that downregulate the expression of their
protein-coding gene targets (3).
Despite the relatively small number of miRNAs, computational and
experimental studies have shown that a number of human
protein-coding genes are collectively regulated by miRNAs (4–6).
Thus, miRNAs are considered to be master regulators of several
important biological processes, such as cell growth, apoptosis,
viral infection and cancer development (3,7–10).
miRNA expression profiles have been shown to be promising
biomarkers for the classification or outcome prediction of a wide
array of human cancers (11,12).
While global miRNA expression patterns of several embryologic,
physiologic and oncogenic processes have been thoroughly studied,
no reports are available on the role of miRNAs in pre-operative CRT
in rectal cancer. This is the first study using miRNA microarray
for predicting response to CRT in rectal cancer. To predict
response to CRT, the miRNA expression patterns of rectal cancer
using miRNA microarray prior to pre-operative CRT were examined. In
addition to miRNA microarray analysis, a candidate miRNA was
examined using real-time reverse transcription-polymerase chain
reaction (RT-PCR).
The present study aimed to define the expression
patterns of miRNA for the prediction of response to CRT and to
establish tailor-made therapies in rectal cancer.
Materials and methods
Patients and tissue samples
For miRNA expression profiling, rectal cancer
samples were obtained from 43 patients, who were willing to receive
pre-operative CRT between September 2007 and December 2009, at the
Tokushima University Hospital. The 43 independent rectal tumor
samples included 22 for training and 21 for testing the outcome
prediction model, respectively. The patient characteristics and
response to CRT are summarized in Table I. The study was approved by the
ethics committee of the Tokushima University Hospital, while every
patient provided written informed consent for their samples to be
used. Biopsy specimens from rectal cancer were prospectively
collected during colonoscopic examinations prior to pre-operative
CRT. Parallel tumor specimens were formalin-fixed and
paraffin-embedded for histologic examination, while other specimens
were used for RNA extraction. Samples were used for RNA extraction
when paralleled specimens contained at least 70% tumor cells.
Samples were snap-frozen immediately in liquid nitrogen and stored
at −80°C until RNA extraction.
 | Table IPatient characteristics and response
to CRT. |
Table I
Patient characteristics and response
to CRT.
| Characteristics | Training set
(n=22) | Testing set
(n=21) |
|---|
| Male/female | 17:5 | 14:7 |
| Age (years) | 72 (41–82) | 60 (44–79) |
| Tumor size (cm) | 3.5 (2.5–9.0) | 4.0 (3.0–10.0) |
| Tumor distance from
the anal verge (cm) | 3.0 (0–8.0) | 4.0 (0–7.0) |
| Grade of
differentiation | | |
|
Well/moderately | 22 | 20 |
| Poorly | 0 | 1 |
| Tumor stage | | |
| T3 | 20 | 19 |
| T4 | 2 | 2 |
| Nodal stage (N) | | |
| 0 | 8 | 11 |
| 1 | 7 | 5 |
| 2 | 7 | 5 |
| Pathological response
(grade) | | |
| 0 | 0 | 0 |
| 1 | 7 | 9 |
| 2 | 13 | 11 |
| 3 | 2 | 1 |
| RECIST | | |
| CR | 0 | 0 |
| PR | 14 | 13 |
| SD | 8 | 8 |
| PD | 0 | 0 |
| Downstaging | | |
| Yes | 13 | 11 |
| No | 9 | 10 |
The patients received CRT at a total dose of 4,000
cGy of pelvic irradiation, five times a week, with a daily fraction
of 200 cGy utilizing a four-field technique. Radiation was
delivered concomitantly with S-1, a novel oral fluoropyrimidine
inhibitory for dihydropyrimidine dehydrogenase with a potent
radiosensitizing property. The S-1 was administered on radiation
days. Surgical treatment was performed 6–8 weeks after the
completion of pre-operative CRT.
miRNA microarrays
Total RNA was isolated using the miRNeasy mini kit
(Qiagen, Hilden, Germany). One hundred nanograms of each RNA sample
were hybridized to Agilent Human miRNA Microarray v2.0, containing
821 miRNAs (G4470B; Agilent Technologies, Inc., Santa Clara, CA,
USA). miRNA labeling, hybridization and washing were carried out
according to the manufacturer's instructions. Images of hybridized
microarrays were obtained with a DNA micro-array scanner (G2565BA;
Agilent Technologies), while features were extracted using the AFE
image analysis tool version A.9.5.3.1.
Real-time RT-PCR
Complementary DNA (cDNA) was synthesized from total
RNA to quantify the expression levels of mature miR-223 using
gene-specific primers, according to the TaqMan miRNA Assay protocol
(Applied Biosystems, Carlsbad, CA, USA) in a 15 μl reaction
volume with 10 ng of RNA template. RT was performed using the
following program: 30 min at 16°C, 30 min at 42°C, 5 min at 85°C
and then held at 4°C. Reverse transcription products were diluted
20-fold, while 2 μl were used in a total reaction volume of
20 μl for relative quantification by RT-PCR using an Applied
Biosystems 7500 Sequence Detection system. The thermal cycling
program used for quantification was as follows: 50°C for 2 min and
95°C for 10 min, followed by 50 cycles of 95°C for 15 sec and 60°C
for 1 min. Normalization was performed using the small nuclear RNA
U6 (RNU6B; Applied Biosystems).
Data analysis
To identify genes that were differentially expressed
in the two groups, the data sets were assigned to either responders
or non-responders. Response to CRT was evaluated by three
parameters [histopathological examination, Response Evaluation
Criteria in Solid Tumors (RECIST) and downstaging], while miRNA
microarray was analyzed in the three parameters. The
histopathological examination of surgically resected specimens was
based on a semi-quantitative classification system, as described in
detail previously (13). Tumors
were classified as responder when assigned to regression grade 2 or
3, and non-responder when assigned to grade 0 or 1. Tumors in
RECIST were classified as responder when assigned to complete
response (CR) or partial response (PR), and non-responder when
stable disease (SD) or progressive disease (PD). Tumors in
downstaging were classified as responder when assigned to ‘Yes’,
and non-responder when ‘No’.
The samples of 43 patients were divided into a
training set (22 samples) and a testing set (21 samples). In the
miRNA microarray analysis to evaluate miRNA expression only
training samples were used. The expression patterns were compared
and the fold change value was determined to identify gene markers
that best discriminated between responders and non-responders.
Two-dimensional hierarchical clustering was applied to the
log-transformed data with average-linkage clustering, with a
standard correlation serving as the similarity metric for the
discriminating genes identified as differentially expressed in
responders and non-responders. Using real-time RT-PCR in a training
set, a candidate miRNA detected by miRNA microarray analysis with
regard to the histopathological examination of surgical specimens
was evaluated. At the end. A candidate miRNA was then evaluated in
a testing set using real-time RT-PCR with regard to the
histopathological examination of surgical specimens.
Statistical analysis
Quantitative data were provided as the median
(range). The statistical analysis was performed using statistical
software (JMP 8.0.1., SAS Campus Drive, Cary, NC, USA). Comparisons
of clinicopathological and PCR data were performed using the
Fisher's exact, Chi-square and Mann-Whitney U tests, as
appropriate. The expression patterns in the miRNA microarray were
compared using unpaired t-tests (with Welch's correction for
unequal variances). Receiver-operating characteristics (ROC) curves
were established to evaluate the diagnostic value of candidate
miRNA for differentiating between responders and non-responders.
The statistical tests performed were two-sided and declared at the
5% significance level.
Results
miRNA expression patterns using
microarrays in responders and non-responders
miRNA expression profiling was established using
miRNA array in the training samples. No statistically significant
difference was detected between the training and the testing sets
with regards to clinicopathological factors, such as gender, age,
histopathologicalal classification, pre-operative tumor stage and
response to CRT. The patient characteristics and response to CRT
are summarized in Table I. Of the
22 training samples, 15 were classified as responders and 7 as
non-responders, according to the histopathologicalal examination of
the surgical specimens. Initially, with regard to the
histopathologicalal examination of surgically resected specimens, 2
genes differentially expressed at significant levels (P<0.05) in
responders and non-responders (Table
II) were identified. The 2 genes (miR-223 and miR-142-3p)
showed a higher expression in responders compared to
non-responders. Of the 22 training samples, 14 were classified as
responders and 8 as non-responders, based on the RECIST. Nine genes
were differentially expressed at significant levels in responders
and non-responders, with regard to RECIST (Table III). One gene (miR-223) showed a
higher, while 8 genes a lower expression in responders compared to
non-responders. The list of under-expressed genes included miR-20b,
miR-92a, let-7a*, miR-20a, miR-17*, miR-106a, miR-17 and miR-20a*.
Additionally, of the 22 training samples, 13 were classified as
responders and 9 as non-responders, based on downstaging. Three
genes were differentially expressed at significant levels in
responders and non-responders, with regard to downstaging (Table IV). The 3 genes (miR-223, miR-630
and miR-126*) showed a higher expression in responders compared to
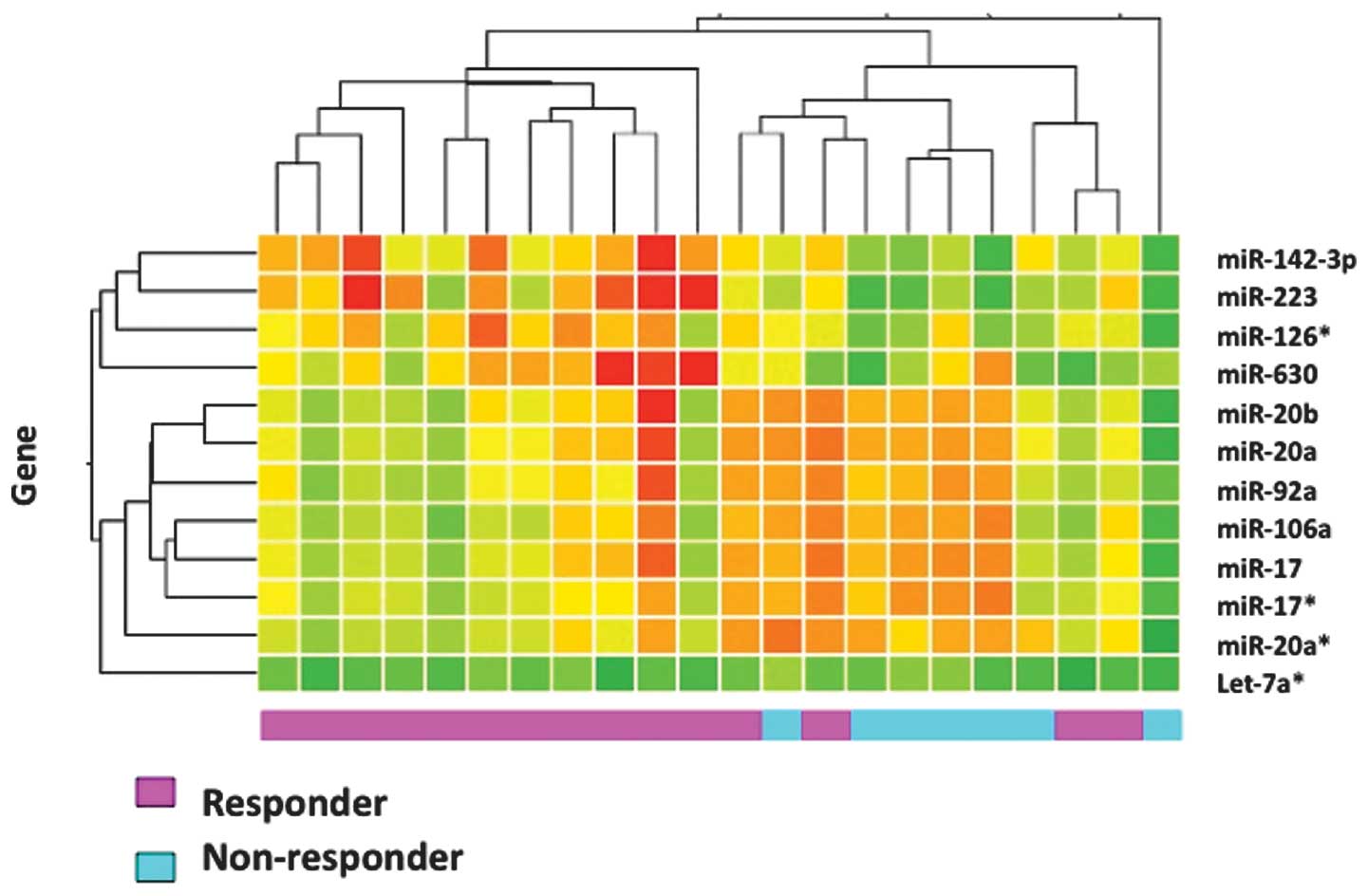
non-responders. Results of a hierarchical cluster analysis of the
12 genes detected by three parameters are shown in Fig. 1. Responders and non-responders were
clustered into two distinct groups, except for four responder
cases.
 | Table IIFrequent differentially expressed
miRNAs in responders and non-responders with regard to
histopathologicalal examination of surgically-resected
specimens. |
Table II
Frequent differentially expressed
miRNAs in responders and non-responders with regard to
histopathologicalal examination of surgically-resected
specimens.
| microRNA | Fold change | P-value |
|---|
| Overexpressed | | |
| miR-223 | 3.13 | 0.026 |
| miR-142-3p | 2.12 | 0.026 |
 | Table IIIFrequent differentially expressed
miRNAs in responders and non-responders with regard to RECIST. |
Table III
Frequent differentially expressed
miRNAs in responders and non-responders with regard to RECIST.
| microRNA | Fold change | P-value |
|---|
| Overexpressed | | |
| miR-223 | 3.13 | 0.034 |
| Underexpressed | | |
| miR-20b | 0.61 | 0.048 |
| miR-92a | 0.61 | 0.024 |
| let-7a* | 0.59 | 0.048 |
| miR-20a | 0.58 | 0.041 |
| miR-17* | 0.55 | 0.012 |
| miR-106a | 0.55 | 0.024 |
| miR-17 | 0.54 | 0.024 |
| miR-20a* | 0.41 | 0.041 |
 | Table IVFrequent differentially expressed
miRNAs in responders and non-responders with regard to
downstaging. |
Table IV
Frequent differentially expressed
miRNAs in responders and non-responders with regard to
downstaging.
| microRNA | Fold change | P-value |
|---|
| Overexpressed | | |
| miR-223 | 3.36 | 0.006 |
| miR-630 | 2.79 | 0.042 |
| miR-126* | 1.87 | 0.049 |
Evaluation of miRNA microarray data by
real-time RT-PCR
miR-223, a candidate gene, showing a higher
expression in responders compared to non-responders in the three
parameters (histopathological examination, RECIST and downstaging)
was chosen for the evaluation of miRNA microarray data using
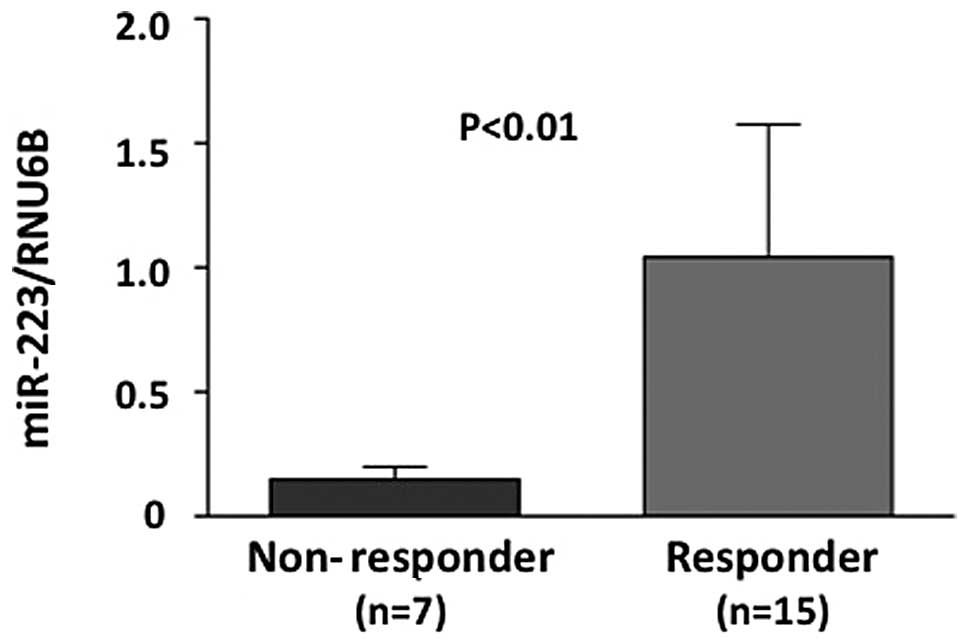
real-time RT-PCR. Initially, the miR-223 level was evaluated using
real-time RT-PCR in a training set. The miR-223 level was
significantly higher in responders compared to non-responders, with
regard to the histopathological examination of surgical specimens
(P<0.01) (Fig. 2). The miR-223
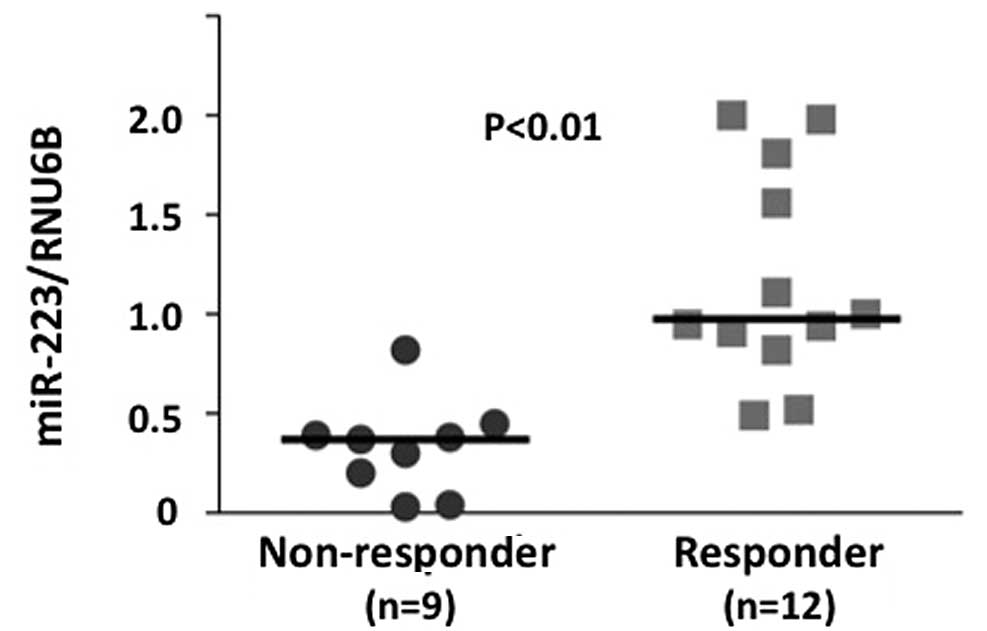
level in a testing set (21 independent rectal cancer samples) was
evaluated. Of the 21 testing samples, 9 were classified as
responders and 12 as non-responders, based on the histopathological
examination of surgical specimens. The miR-223 level was higher in
responders compared to non-responders (P<0.01) (Fig. 3). Additionally, ROC curve analyses
showed that miR-223 might differentiate between responders and
non-responders in 22 training samples with an area under the curve
(AUC) of 0.768 [95% confidence internal (CI), 0.661–0.865]. At the
cut-off value of 0.4 for miR-223, the sensitivity and the
specificity in the 21 testing samples were 100 and 78.0%,
respectively (Fig. 3).
Discussion
Although miRNA expression patterns have been applied
when predicting the outcome of multiple cancers, few studies have
been reported as yet on the application of miRNAs when predicting
the response to CRT using pre-operative biopsy tissue samples in
rectal cancer. The expression patterns of miRNAs for the prediction
of response to CRT by miRNA microarray were defined, while a
candidate miRNA (miR-223) was evaluated by real-time RT-PCR.
In order to determine whether miR-223 showed a high
or low expression when predicting the response to CRT in rectal
cancer, in their study Aslam et al(14) summarized the differential
expression of miRNAs in colorectal cancer compared to healthy
flanking tissue identified by various studies. The results of those
studies suggested that the variability of individual miRNAs with
tumor type and stage probably rendered the combination of miRNAs a
reliable method for detecting cancer status. In this study, the
miR-223 detected as a candidate gene for the prediction of response
to CRT is reported as one of the overexpressed miRNAs in colorectal
cancer compared to healthy tissue (15,16).
The potential value of miRNAs as prognostic and
predictive biomarkers in colon cancer is demonstrated by Schetter
et al(17), who compared
miRNA expression patterns in stage II colonic adenocarcinoma and in
adjacent healthy tissue. A high tumor:healthy expression ratio of
miR-20a, miR-21, miR-106a, miR-181b and miR-203 was associated with
poor survival. Underexpressed miR-20a in a tumor may be associated
with good survival and responders to CRT.
Regarding the correlation between miRNAs and
radiation therapy, higher rectal tumor expression levels of
miR-125b and miR-137 are reported to be associated with a poorer
response to CRT (18). In lung
cancer and healthy lung cell lines, levels of 81 out of 440 miRNAs
demonstrated statistically significant differences subsequent to
irradiation with 23 miRNAs downregulated after treatment. The
members of the let-7 family (except let-7g) decreased markedly by
2–8 h subsequent to irradiation in both cancer and healthy lung
epithelium. Furthermore, let-60/RAS and genes in the DNA damage
response pathway were identified as mechanisms potentially affected
by let-7 (19). In addition to
these results, radiotherapy of two different prostate cancer cell
lines resulted in a differentiated expression of a high number of
miRNAs. The expression of 48 miRNAs altered significantly
subsequent to the application of radiotherapy, with 22 of them
presenting a >3-fold change in levels. Of those 22 miRNAs
hsa-miR-521, hsamiR-196a and hsa-miR-133b were demonstrated to be
decreased, while hsa-miR-34c was shown to be increased in the two
cell lines subsequent to radiation (20). Although inconsistent with the
results of the present study, the data on the let-7 family are
likely to be affected by the differences in cell culture
experiments and clinical research.
Different miRNAs have been found to predict
sensitivity to anticancer treatment. miR-30c, miR-130a and miR-335
have been reported to be downregulated in various chemoresistant
cell lines (21), while the
restoration of miR-34 in p53-deficient human gastric cancer cells
induced chemosensitisation (22).
Irrelevant to the results of this study, regarding the S-1 used for
CRT in the present study, Nakajima et al(23) reported that let-7g and miR-181b
were strongly associated with response to 5-fluorouracil-based
antimetabolite S-1.
miR-223 has been reported to function as an
important modulator of cellular differentiation (24,25).
In bone cells, the enhanced expression of pre-miR-223 completely
abrogated osteoclast differentiation (25). However, few reports are available
on the dysregulation of miR-223 in epithelial cancers. Although
miR-223 transcription in granulopoiesis has been reported to be
regulated by an evolutionarily conserved system driven by the
myeloid transcription factors, PU.1 and CAAT enhancer-binding
protein (26), In their study,
Wong et al(27) identified
stathmin 1 (STMN1) as a putative target of miR-223 in
hepatocellular carcinoma (HCC). STMN1 is a key
microtubule-regulatory protein that controls the microtubule
dynamics, cell proliferation and S-phase of the cell cycle
(28,29). High-levels of STMN1
expression have been reported in leukemia, breast, ovarian and
prostate cancers, in which its increased expression has been
associated with increased histologic grading, shorter patient
survival period and increased resistance to medication (30–33).
By these mechanism, STMN1 as a putative target of miR-223,
may be associated with responders to CRT.
Additionally, miR-223 has recently been found to be
myeloid-specific, negatively regulating the progenitor
proliferation, granulocyte differentiation and activation in mice
(34). Guo et al(35) also reported that the
CD133+ ovarian cancer stem cells in the OVACAR3 cell
line were correlated with miR-223. miR-223, as a candidate miRNA to
regulate stemness gene, may be associated with responders to
CRT.
The present study defined the expression patterns of
miRNAs for the prediction of response to CRT by miRNA microarray
and evaluated miR-223 by real-time RT-PCR using pre-operative
biopsy tissue samples in rectal cancer. Albeit the small samples,
the lack of analysis of target mRNA, of miR-223 and functional
analysis of miR-223, the expression patterns of miRNAs for the
prediction of response to CRT by miRNA microarray were evaluated
using several miRNAs (821 gene). Furthermore, miR-223 was evaluated
by real-time RT-PCR as well as in independent samples. These
results suggest that a candidate common miR-223 shown by three
parameters (histopathological examination, RECIST and existence of
downstaging) may be a new biomarker for the prediction of response
to CRT in rectal cancer.
In conclusion, the candidate miR-223 is a potential
new biomarker for the prediction of response to CRT and is likely
to be useful for establishing tailor-made therapies for rectal
cancer.
Acknowledgements
The present study was partly financed
by the Research Support Foundation of the University of Tokushima
and Taiho Pharmaceutical, Co., Ltd. (research funding provided to
Professor Mitsuo Shimada), and the Grants-in-Aid for Scientific
Research of the Japan Society for the Promotion of Science. A part
of this study was presented at the Gastrointestinal Cancer
Symposium, San Francisco, CA, USA (January 15–17, 2009).
References
|
1.
|
Holm T, Cedermark B and Rutqvist LE: Local
recurrence of rectal adenocarcinoma after ‘curative’ surgery with
and without pre-operative radiotherapy. Br J Surg. 81:452–455.
1994.
|
|
2.
|
Sauer R, Becker H, Hohenberger W, Rödel C,
Wittekind C, Fietkau R, Martus P, Tschmelitsch J, Hager E, Hess CF,
Karstens JH, Liersch T, Schmidberger H and Raab R; German Rectal
Cancer Study Group: Pre-operative versus postoperative
chemoradiotherapy for rectal cancer. N Engl J Med. 351:1731–1740.
2004.
|
|
3.
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004.
|
|
4.
|
Lim LP, Lau NC, Garrett-Engele P, Grimson
A, Schelter JM, Castle J, Bartel DP, Linsley PS and Johnson JM:
Microarray analysis shows that some microRNAs downregulate large
numbers of target mRNAs. Nature. 433:769–773. 2005.
|
|
5.
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005.
|
|
6.
|
Miranda KC, Huynh T, Tay Y, Ang YS, Tam
WL, Thomson AM, Lim B and Rigoutsos I: A pattern-based method for
the identification of microRNA binding sites and their
corresponding heteroduplexes. Cell. 126:1203–1217. 2006.
|
|
7.
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006.
|
|
8.
|
Kent OA and Mendell JT: A small piece in
the cancer puzzle: microRNAs as tumor suppressors and oncogenes.
Oncogene. 25:6188–6196. 2006.
|
|
9.
|
Johnson CD, Esquela-Kerscher A, Stefani G,
Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J,
Shingara J, Chin L, Brown D and Slack FJ: The let-7 micro-RNA
represses cell proliferation pathways in human cells. Cancer Res.
67:7713–7722. 2007.
|
|
10.
|
Miska EA: How microRNAs control cell
division, differentiation and death. Curr Opin Genet Dev.
15:563–568. 2005.
|
|
11.
|
Jay C, Nemunaitis J, Chen P, Fulgham P and
Tong AW: miRNA profiling for diagnosis and prognosis of human
cancer. DNA Cell Biol. 26:293–300. 2007.
|
|
12.
|
Yu SL, Chen HY, Yang PC and Chen JJ:
Unique microRNA signature and clinical outcome of cancers. DNA Cell
Biol. 26:283–292. 2007.
|
|
13.
|
Japanese Society for Cancer of the Colon
and Rectum: Response Assessment of Nonsurgical Treatment for
Colorectal Carcinoma. Japanese classification of colorectal
carcinoma. 1st English edition. Kanehara & Co. Ltd.; Tokyo: pp.
77–82. 1997
|
|
14.
|
Aslam MI, Taylor K, Pringle JH and Jameson
JS: MicroRNAs are novel biomarkers of colorectal cancer. Br J Surg.
96:702–710. 2009.
|
|
15.
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M,
Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M,
Harris CC and Croce CM: A microRNA expression signature of human
solid tumors defines cancer gene targets. Proc Natl Acad Sci USA.
103:2257–2261. 2006.
|
|
16.
|
Monzo M, Navarro A, Bandres E, Artells R,
Moreno I, Gel B, Ibeas R, Moreno J, Martinez F, Diaz T, Martinez A,
Balagué O and Garcia-Foncillas J: Overlapping expression of
microRNAs in human embryonic colon and colorectal cancer. Cell Res.
18:823–833. 2008.
|
|
17.
|
Schetter AJ, Leung SY, Sohn JJ, Zanetti
KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu
CG, Calin GA, Croce CM and Harris CC: MicroRNA expression profiles
associated with prognosis and therapeutic outcome in colon
adenocarcinoma. JAMA. 299:425–436. 2008.
|
|
18.
|
Svoboda M, Izakovicova Holla L, Sefr R,
Vrtkova I, Kocakova I, Tichy B and Dvorak J: Micro-RNAs miR125b and
miR137 are frequently upregulated in response to capecitabine
chemoradiotherapy of rectal cancer. Int J Oncol. 33:541–547.
2008.
|
|
19.
|
Weidhaas JB, Babar I, Nallur SM, Trang P,
Roush S, Boehm M, Gillespie E and Slack FJ: MicroRNAs as potential
agents to alter resistance to cytotoxic anticancer therapy. Cancer
Res. 67:11111–11116. 2007.
|
|
20.
|
Josson S, Sung SY, Lao K, Chung LW and
Johnstone PA: Radiation modulation of microRNA in prostate cancer
cell lines. Prostate. 68:1599–1606. 2008.
|
|
21.
|
Sorrentino A, Liu CG, Addario A, Peschle
C, Scambia G and Ferlini C: Role of microRNAs in drug-resistant
ovarian cancer cells. Gynecol Oncol. 111:478–486. 2008.
|
|
22.
|
Salter KH, Acharya CR, Walters KS, Redman
R, Anguiano A, Garman KS, Anders CK, Mukherjee S, Dressman HK,
Barry WT, Marcom KP, Olson J, Nevins JR and Potti A: An integrated
approach to the prediction of chemotherapeutic response in patients
with breast cancer. PLoS One. 3:e19082008.
|
|
23.
|
Nakajima G, Hayashi K, Xi Y, Kudo K,
Uchida K, Takasaki K, Yamamoto M and Ju J: Non-coding microRNAs
hsa-let-7g and hsa-miR-181b are associated with chemoresponse to
S-1 in colon cancer. Cancer Genom Proteom. 3:317–324. 2006.
|
|
24.
|
Fazi F, Rosa A, Fatica A, Gelmetti V, De
Marchis ML, Nervi C and Bozzoni I: A minicircuitry comprised of
microRNA-223 and transcription factors NFI-A and C/EBPalpha
regulates human granulopoiesis. Cell. 23:819–831. 2005.
|
|
25.
|
Sugatani T and Hruska KA: MicroRNA-223 is
a key factor in osteoclast differentiation. J Cell Biochem.
101:996–999. 2007.
|
|
26.
|
Fukao T, Fukuda Y, Kiga K, Sharif J, Hino
K, Enomoto Y, Kawamura A, Nakamura K, Takeuchi T and Tanabe M: An
evolutionarily conserved mechanism for microRNA-223 expression
revealed by microRNA gene profiling. Cell. 129:617–631. 2007.
|
|
27.
|
Wong QW, Lung RW, Law PT, Lai PB, Chan KY,
To KF and Wong N: MicroRNA-223 is commonly repressed in
hepatocellular carcinoma and potentiates expression of
Stathmin1. Gastroenterology. 135:257–269. 2008.
|
|
28.
|
Melhem RF, Strahler JR, Hailat N, Zhu XX
and Hanash SM: Involvement of OP18 in cell proliferation. Biochem
Biophys Res Commun. 179:1649–1655. 1991.
|
|
29.
|
Rubin CL and Atweh GF: The role of
stathmin in the regulation of the cell cycle. J Cell Biochem.
93:242–250. 2004.
|
|
30.
|
Brattsand G, Roos G, Marklund U, Ueda H,
Landberg G, Nånberg E, Sideras P and Gullberg M: Quantitative
analysis of the expression and regulation of an
activation-regulated phosphoprotein (oncoprotein 18) in healthy and
neoplastic cells. Leukemia. 7:569–579. 1993.
|
|
31.
|
Ghosh R, Gu G, Tillman E, Yuan J, Wang Y,
Fazli L, Rennie PS and Kasper S: Increased expression and
differential phosphorylation of stathmin may promote prostate
cancer progression. Prostate. 67:1038–1052. 2007.
|
|
32.
|
Alli E, Yang JM, Ford JM and Hait WN:
Reversal of stathmin-mediated resistance to paclitaxel and
vinblastine in human breast carcinoma cells. Mol Pharmacol.
71:1233–1240. 2007.
|
|
33.
|
Saal LH, Johansson P, Holm K,
Gruvberger-Saal SK, She QB, Maurer M, Koujak S, Ferrando AA,
Malmström P, Memeo L, Isola J, Bendahl PO, Rosen N, Hibshoosh H,
Ringnér M, Borg A and Parsons R: Poor prognosis in carcinoma is
associated with a gene expression signature of aberrant PTEN tumor
suppressor pathway activity. Proc Natl Acad Sci USA. 104:7564–7569.
2007.
|
|
34.
|
Johnnidis JB, Harris MH, Wheeler RT,
Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD and
Camargo FD: Regulation of progenitor cell proliferation and
granulocyte function by microRNA-223. Nature. 451:1125–1129.
2008.
|
|
35.
|
Guo R, Wu Q, Liu F and Wang Y: Description
of the CD133+ subpopulation of the human ovarian cancer
cell line OVCAR3. Oncol Rep. 25:141–146. 2011.
|