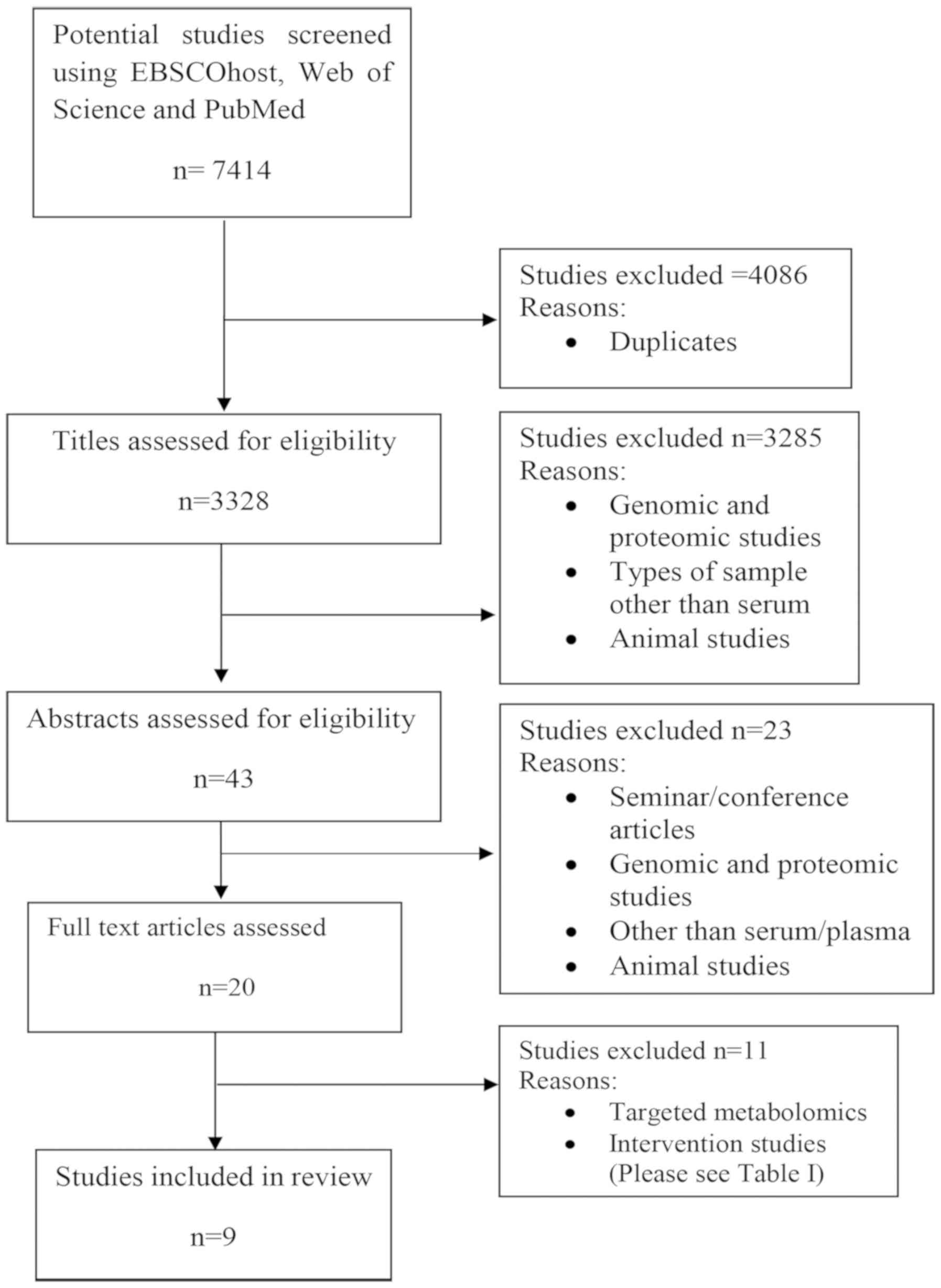
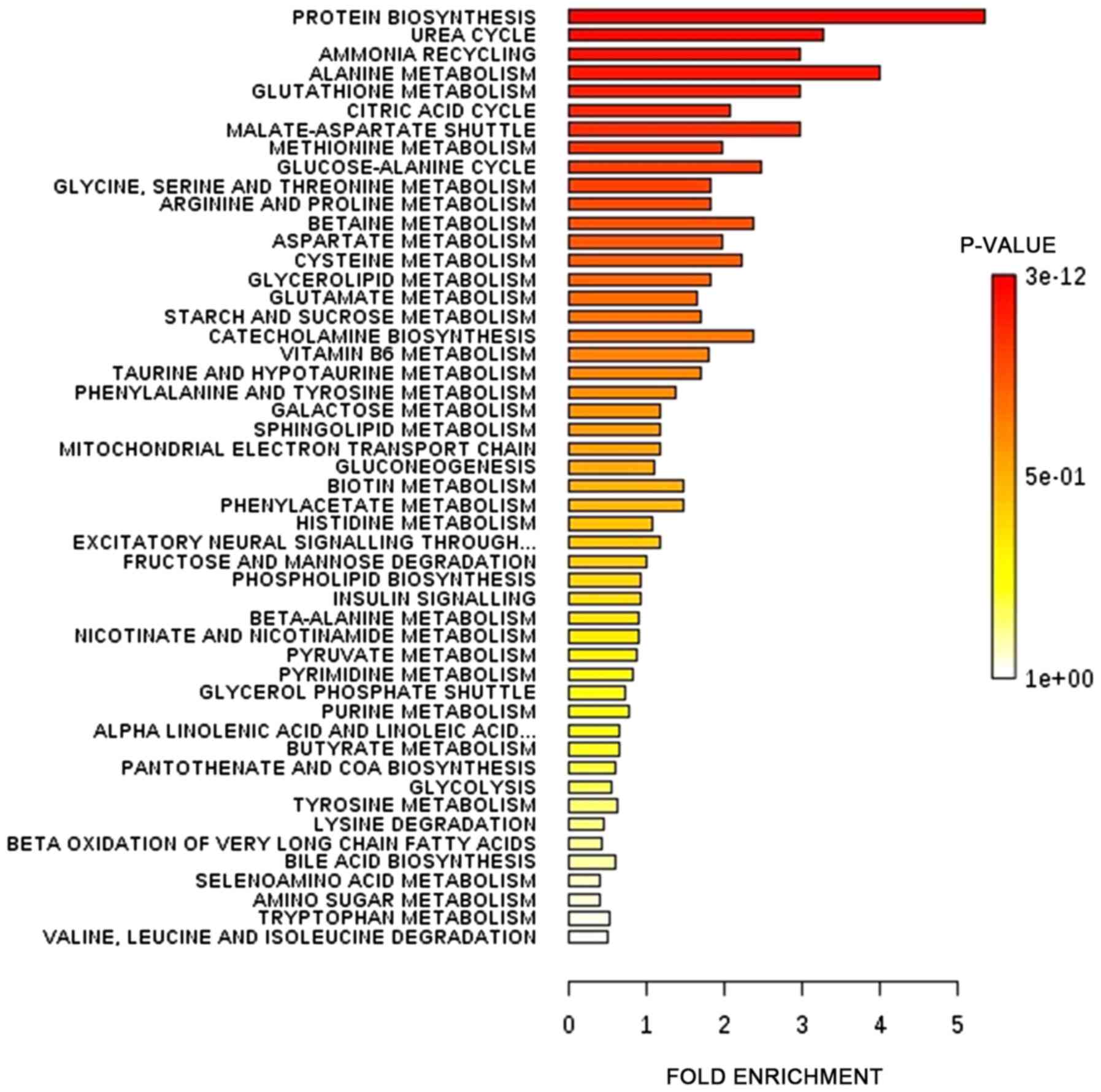
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhu J, Djukovic D, Deng L, Gu H, Himmati
F, Abu Zaid M, Chiorean EG and Raftery D: Targeted serum metabolite
profiling and sequential metabolite ratio analysis for colorectal
cancer progression monitoring. Anal Bioanal Chem. 407:7857–7863.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
American Cancer Society: Cancer facts and
figures 2013. https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2013/cancer-facts-and-figures-2013.pdfAccessed
Month day, year.
|
|
4
|
Wang D, Li W, Zou Q, Yin L, Du Y, Gu J and
Suo J: Serum metabolomic profiling of human gastric cancer and its
relationship with the prognosis. Oncotarget. 8:110000–110015.
2017.PubMed/NCBI
|
|
5
|
Chinnaiyan P, Kensicki E, Bloom G, Prabhu
A, Sarcar B, Kahali S, Eschrich S, Qu X, Forsyth P and Gillies R:
The metabolomic signature of malignant glioma reflects accelerated
anabolic metabolism. Cancer Res. 72:5878–5888. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hadi NI, Jamal Q, Iqbal A, Shaikh F,
Somroo S and Musharraf SG: Serum metabolomic profiles for breast
cancer diagnosis, grading and staging by gas chromatography-mass
spectrometry. Sci Rep. 7:17152017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kumar N, Shahjaman M, Mollah MNH, Islam
SMS and Hoque MA: Serum and plasma metabolomic biomarkers for lung
cancer. Bioinformation. 13:202–208. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Whiting P, Rutjes AW, Reitsma JB, Bossuyt
PM and Kleijnen J: The development of QUADAS: A tool for the
quality assessment of studies of diagnostic accuracy included in
systematic reviews. BMC Med Res Methodol. 3:252003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ma Y, Liu W, Peng J, Huang L, Zhang P,
Zhao X, Cheng Y and Qin H: A pilot study of gas chromatograph/mass
spectrometry-based serum metabolic profiling of colorectal cancer
after operation. Mol Biol Rep. 37:1403–1411. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bertini I, Cacciatore S, Jensen BV, Schou
JV, Johansen JS, Kruhøffer M, Luchinat C, Nielsen DL and Turano P:
Metabolomic NMR fingerprinting to identify and predict survival of
patients with metastatic colorectal cancer. Cancer Res. 72:356–364.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhu J, Djukovic D, Deng L, Gu H, Himmati
F, Chiorean EG and Raftery D: Colorectal cancer detection using
targeted serum metabolic profiling. J Proteome Res. 13:4120–4130.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guertin KA, Loftfield E, Boca SM, Sampson
JN, Moore SC, Xiao Q, Huang WY, Xiong X, Freedman ND, Cross AJ and
Sinha R: Serum biomarkers of habitual coffee consumption may
provide insight into the mechanism underlying the association
between coffee consumption and colorectal cancer. Am J Clin Nutr.
101:1000–1011. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dowling P, Hughes DJ, Larkin AM, Meiller
J, Henry M, Meleady P, Lynch V, Pardini B, Naccarati A, Levy M, et
al: Elevated levels of 14-3-3 proteins, serotonin, gamma enolase
and pyruvate kinase identified in clinical samples from patients
diagnosed with colorectal cancer. Clin Chim Acta. 441:133–141.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Crotti S, Agnoletto E, Cancemi G, Di Marco
V, Traldi P, Pucciarelli S, Nitti D and Agostini M: Altered plasma
levels of decanoic acid in colorectal cancer as a new diagnostic
biomarker. Anal Bioanal Chem. 408:6321–6328. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Farshidfar F, Weljie AM, Kopciuk K, Buie
WD, Maclean A, Dixon E, Sutherland FR, Molckovsky A, Vogel HJ and
Bathe OF: Serum metabolomic profile as a means to distinguish stage
of colorectal cancer. Genome Med. 4:422012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Farshidfar F, Weljie AM, Kopciuk KA,
Hilsden R, McGregor SE, Buie WD, MacLean A, Vogel HJ and Bathe OF:
A validated metabolomic signature for colorectal cancer:
Exploration of the clinical value of metabolomics. Br J Cancer.
115:848–857. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vahabi F, Sadeghi S, Arjmand M, Mirkhani
F, Hosseini E, Mehrabanfar M, Hajhosseini R, Iravani A, Bayat P and
Zamani Z: Staging of colorectal cancer using serum metabolomics
with 1HNMR Spectroscopy. Iran J Basic Med Sci.
20:835–840. 2017.PubMed/NCBI
|
|
18
|
Shu X, Xiang YB, Rothman N, Yu D, Li HL,
Yang G, Cai H, Ma X, Lan Q, Gao YT, et al: Prospective study of
blood metabolites associated with colorectal cancer risk. Int J
Cancer. 143:527–534. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Qiu Y, Cai G, Su M, Chen T, Zheng X, Xu Y,
Ni Y, Zhao A, Xu LX, Cai S and Jia W: Serum metabolite profiling of
human colorectal cancer using GC-TOFMS and UPLC-QTOFMS. J Proteome
Res. 8:4844–4850. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nishiumi S, Kobayashi T, Ikeda A, Yoshie
T, Kibi M, Izumi Y, Okuno T, Hayashi N, Kawano S, Takenawa T, et
al: A novel serum metabolomics-based diagnostic approach for
colorectal cancer. PLoS One. 7:e404592012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tan B, Qiu Y, Zou X, Chen T, Xie G, Cheng
Y, Dong T, Zhao L, Feng B, Hu X, et al: Metabonomics identifies
serum metabolite markers of colorectal cancer. J Proteome Res.
12:3000–3009. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cross A, Moore SC, Boca S, Huang WY, Xiong
X, Stolzenberg-Solomon R, Sinha R and Sampson JN: A prospective
study of serum metabolites and colorectal cancer risk. Cancer.
120:3049–3057. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zamani Z, Arjmand M, Vahabi F, Eshaq
Hosseini SM, Fazeli SM, Iravani A, Bayat P, Oghalayee A,
Mehrabanfar M, Haj Hosseini R, et al: A metabolic study on colon
cancer using (1)h nuclear magnetic resonance spectroscopy. Biochem
Res Int. 2014:3487122014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Deng L, Gu H, Zhu J, Nagana Gowda GA,
Djukovic D, Chiorean EG and Raftery D: Combining NMR and LC/MS
using backward variable elimination: Metabolomics analysis of
colorectal cancer, polyps, and healthy controls. Anal Chem.
88:7975–7983. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Uchiyama K, Yagi N, Mizushima K,
Higashimura Y, Hirai Y, Okayama T, Yoshida N, Katada K, Kamada K,
Handa O, et al: Serum metabolomics analysis for early detection of
colorectal cancer. J Gastroenterol. 52:677–694. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Long Y, Sanchez-Espiridion B, Lin M, White
L, Mishra L, Raju GS, Kopetz S, Eng C, Hildebrandt MAT, Chang DW,
et al: Global and targeted serum metabolic profiling of colorectal
cancer progression. Cancer. 123:4066–4074. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nishiumi S, Kobayashi T, Kawana S, Unno Y,
Sakai T, Okamoto K, Yamada Y, Sudo K, Yamaji T, Saito Y, et al:
Investigations in the possibility of early detection of colorectal
cancer by gas chromatography/triple-quadrupole mass spectrometry.
Oncotarget. 8:17115–17126. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim SJ, Kim SH, Kim JH, Hwang S and Yoo
HJ: Understanding metabolomics in biomedical research. Endocrinol
Metab (Seoul). 31:7–16. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dunn WB, Broadhurst D, Begley P, Zelena E,
Francis-McIntyre S, Anderson N, Brown M, Knowles JD, Halsall A,
Haselden JN, et al: Procedures for large-scale metabolic profiling
of serum and plasma using gas chromatography and liquid
chromatography coupled to mass spectrometry. Nat Protoc.
6:1060–1083. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Serkova NJ, Standiford TJ and Stringer KA:
The emerging field of quantitative blood metabolomics for biomarker
discovery in critical illnesses. Am J Respir Crit Care Med.
184:647–655. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kastenmüller G, Raffler J, Gieger C and
Suhre K: Genetics of human metabolism: An update. Hum Mol Genet.
24:R93–R101. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ahn CS and Metallo CM: Mitochondria as
biosynthetic factories for cancer proliferation. Cancer Metab.
3:12015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hay N: Reprogramming glucose metabolism in
cancer: Can it be exploited for cancer therapy? Nat Rev Cancer.
16:635–649. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jain RK, Munn LL and Fukumura D:
Dissecting tumour pathophysiology using intravital microscopy. Nat
Rev Cancer. 2:266–276. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Semenza GL: Hypoxia-inducible factors in
physiology and medicine. Cell. 148:399–408. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kaelin WG Jr and Ratcliffe PJ: Oxygen
sensing by metazoans: The central role of the HIF hydroxylase
pathway. Mol Cell. 30:393–402. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ananieva E: Targeting amino acid
metabolism in cancer growth and anti-tumor immune response. World J
Biol Chem. 6:281–289. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Prendergast GC, Smith C, Thomas S,
Mandik-Nayak L, Laury-Kleintop L, Metz R and Muller AJ: Indoleamine
2,3-dioxygenase pathways of pathogenic inflammation and immune
escape in cancer. Cancer Immunol Immunother. 63:721–735. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Peters JC: Tryptophan nutrition and
metabolism: An overview. Adv Exp Med Biol. 294:345–358. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Grohmann U and Bronte V: Control of immune
response by amino acid metabolism. Immunol Rev. 236:243–264. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mezrich JD, Fechner JH, Zhang X, Johnson
BP, Burlingham WJ and Bradfield CA: An interaction between
kynurenine and the aryl hydrocarbon receptor can generate
regulatory T cells. J Immunol. 185:3190–3198. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hitosugi T, Fan J, Chung TW, Lythgoe K,
Wang X, Xie J, Ge Q, Gu TL, Polakiewicz RD, Roesel JL, et al:
Tyrosine phosphorylation of mitochondrial pyruvate dehydrogenase
kinase 1 is important for cancer metabolism. Mol Cell. 44:864–877.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pegg AE: Functions of polyamines in
mammals. J Biol Chem. 291:14904–14912. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hoshino Y, Terashima S, Teranishi Y,
Terashima M, Kogure M, Saitoh T, Osuka F, Kashimura S, Saze Z and
Gotoh M: Ornithine decarboxylase activity as a prognostic marker
for colorectal cancer. Fukushima J Med Sci. 53:1–9. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Santos CR and Schulze A: Lipid metabolism
in cancer. FEBS J. 279:2610–2623. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Menendez JA and Lupu R: Fatty acid
synthase and the lipogenic phenotype in cancer pathogenesis. Nat
Rev Cancer. 7:763–777. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Benlloch M, Ortega A, Ferrer P, Segarra R,
Obrador E, Asensi M, Carretero J and Estrela JM: Acceleration of
glutathione efflux and inhibition of gamma-glutamyltranspeptidase
sensitize metastatic B16 melanoma cells to endothelium-induced
cytotoxicity. J Biol Chem. 280:6950–6959. 2005. View Article : Google Scholar : PubMed/NCBI
|