Introduction
Breast cancer is the most frequently diagnosed
cancer in women worldwide (1,2). It has an increasing mortality and
morbidity rate in women <45 years. Every year in China, ~1.6
million women are diagnosed and ~1.2 million people succumb to
breast cancer. Breast cancer results from the accumulation of
abnormal genetic and epigenetic changes in tumor-suppressor genes
and proto-oncogenes (3). Genes, such
as р53, АТМ and human epidermal growth factor receptor 2 (HER2),
are involved in different types of tumors. Breast cancer
susceptibility gene 1 (BRCA1) is another specific gene,
which was identified as a genetic cause of hereditary breast
cancer.
BRCA1 is located on chromosome 17q12-21
(2). It is an important
tumor-suppressor gene associated with human breast cancer (4). The BRCA1 protein plays an important role
in DNA repair of double-strand breaks (5), transcriptional regulation,
ubiquitinylation, as well as other functions (6,7). The
hypermethylation of the BRCA1 promoter has been considered
as an inactivating mechanism of BRCA1 expression (8). This low expression or non-expression of
BRCA1 may not be adequate for repairing DNA damage that
further promotes the accumulation of mutations in cell growth and
division. Certain results suggest that BRCA1 promoter
hypermethylation is associated with poor clinical outcomes. In the
present study, the source data used by the study of Wu et al
(9) was adjusted and augmented to
investigate the association between BRCA1 methylation and
the outcome of breast cancer.
The therapeutic targets for breast cancer are the
receptors. The progesterone receptor (PR) is a nuclear receptor
located inside cells. PR is encoded by chromosome 11q22 in humans.
Estrogen receptors (ER) are receptors that are activated by the
hormone estrogen (10). HER2 is a
member of the epidermal growth factor receptor (EGFR/ERBB) family.
In recent years these proteins have been used as therapy targets in
<30% of breast cancer patients (11). Triple-negative breast cancer is
defined as the absence of ER, PR and HER2 (12). The present treatment for
triple-negative breast cancer is a type of chemotherapy and often
has a poor outcome. Therefore, it is essential to find new and
alternative therapeutic strategies. In the present meta-analysis,
the correlations between therapy target-related negative-receptors
and BRCA1 promoter methylation were also studied. To ensure
the quality of analysis, the Begg's test, χ2-based Q
test, Egger's test, sensitivity analysis and publication bias
analysis were used.
Materials and methods
Literature search
Two investigators independently conducted a
literature search using PubMed, Embase and Google scholar (last
search updated on September 14, 2014). The keywords used included:
BRCA1, breast carcinoma, breast cancer, methylation,
prognosis and survival. In addition, the PubMed additional
function: Related citations; and the references of the selected
studies were scrutinized to identify additional studies.
Eligibility criteria
Studies were included in the meta-analysis only if
they had met the following criteria: i) Evaluated prognostic risk
of patients with BRCA1 methylation; ii) provided overall
survival (OS) or disease-free survival (DFS); iii) hazard ratios
(HR) or odds ratios (OR) with its 95% confidence intervals (CIs);
iv) published in English; and v) data from human subjects. In
addition, studies were excluded if: i) Data was from reviews or
animal studies; and ii) studies had the same population resources
or overlapping datasets.
Data extraction
Following the exclusions, 9 studies met all the
criteria. Two investigators independently extracted the following
data from each study: First author's last name, year of
publication, population, number of study subjects, effects on
clinical outcomes (OS and DFS), and the number of methylated and
unmethylated patients with a different status of ER, PR, HER2 and
triple-negative receptors. OS is a term that denotes the chances of
remaining alive for a group of individuals suffering from a type of
cancer. At a basic level, the OS is representative of cure rates.
DFS was defined as the chances of staying free of disease following
a particular treatment for a group of individuals suffering from a
type of cancer. It is an indication of how effective a particular
treatment is.
Statistical analysis
Random effects and subgroup meta-analysis were
performed according to the DerSimonian Laird method (13), due to the existence of heterogeneity
between studies. The data were divided into two groups by
population: European and Asian. The HRs were used to estimate the
pooled effect of BRCA1 methylation on the prognosis of
patients with breast cancer, and the ORs were pooled to estimate
the strength of the association between BRCA1
hypermethylation and the risk of three negative statuses of
receptors (ER, PR and HER2). When HRs (95% CIs) were shown only in
the figure of survival curves, the authors were contacted for the
exact value or the investigators estimated them according to the
methods provided by Tierney et al (14). The Cochran Q (significant cut-off
point: P=0.10) and I2 (I2>50%, strong
heterogeneity) statistics (15,16) were
used to assess heterogeneity between studies. The Galbraith plot
(17) was used to detect the
potential sources of heterogeneity from the meta-analysis.
Publication bias was assessed by funnel plot and the test of Egger
et al (18). Sensitivity
analyses were performed by the trim-and-fill method (19). All the analyses and graphs were
obtained using STATA 11.0 software (StataCorp LP, College Station,
TX, USA).
Results
Characteristics of studies
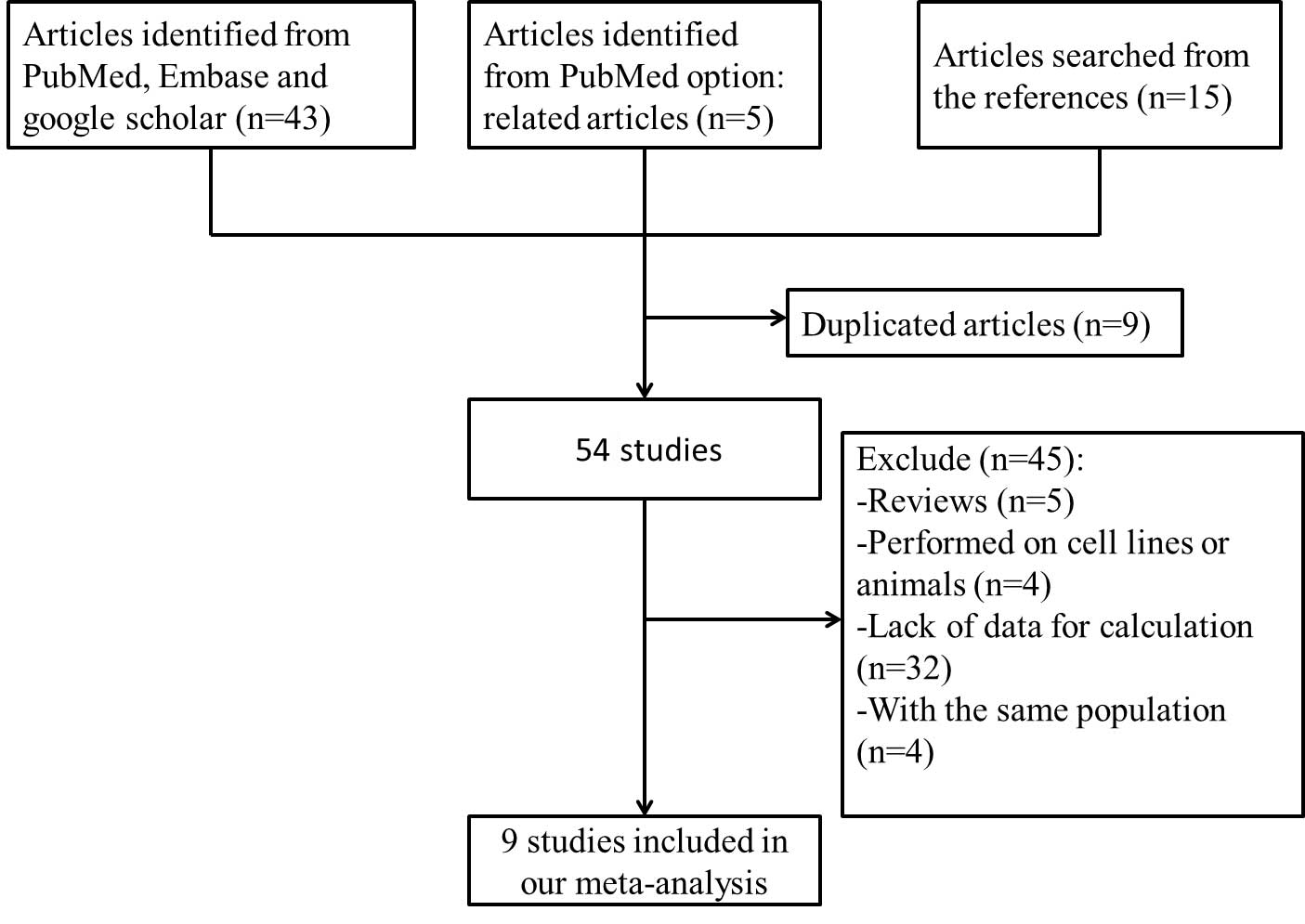
Fig. 1 summarizes the
process of identifying eligible studies. Following the screening by
two investigators independently, according to the inclusion
criteria there were 9 studies with 3,131 study subjects entered
into the meta-analysis (20–28). The characteristics of these studies
are listed in Tables I and II. There were 4 studies from Europe and 5
studies from Asia.
 | Table I.Characteristics of the included
studies. |
Table I.
Characteristics of the included
studies.
|
|
|
|
| OS | DFS |
|---|
|
|
|
|
|
|
|
|---|
| First author,
year | Population | Patients, n | Primer | Univariate analysis
HR (95% CI) | Multivariate
analysis HR (95% CI) | Univariate analysis
HR (95% CI) | Multivariate
analysis HR (95% CI) | (Refs.) |
|---|
| Sharma, 2014 | European |
39 | Esteller | 3.37
(0.23–50.02) | 6.2
(2–19.4) | N/A | 3.5
(1.3–9.8) | (20) |
| Ignatov, 2013 | European |
65 | Baldwin |
N/A |
N/A | 0.325
(0.16–0.662) | 0.224
(0.092–0.546) | (21) |
| Krasteva, 2012 | European |
135 | Baldwin | 0.47
(0.14–1.54) | 0.91
(0.24–3.41) | N/A |
N/A | (22) |
| Xu, 2009 | European |
851 | Others | 1.72
(1.06–2.79) | 1.67
(0.99–2.81) | N/A |
N/A | (23) |
| Xu, 2013 | Asian | 1,163 | Esteller | 1.29
(0.96–1.73) |
N/A | 1.34
(1.06–1.71) |
N/A | (24) |
| Hsu, 2013 | Asian |
139 | Esteller |
N/A | 16.38
(1.37–195.45) | N/A | 12.19
(2.29–64.75) | (25) |
| Sharma, 2009 | Asian |
101 | Esteller | 5.06
(1.58–16.22) | 2.12
(0.47–9.63) | 3.88
(2.05–7.34) | 2.03
(0.96–4.29) | (26) |
| Chen, 2009 | Asian |
536 | Esteller | 1.56
(1.02–2.37) | 1.27
(0.81–1.99) | 1.45
(1.01–2.09) | 1.23
(0.84–1.8) | (27) |
| Jing, 2008 | Asian |
102 | Esteller | 6.4
(2.0–20.5) |
N/A | N/A |
N/A | (28) |
 | Table II.Distribution of the BRCA1
methylation status with different hormone and epidermal growth
factor receptors. |
Table II.
Distribution of the BRCA1
methylation status with different hormone and epidermal growth
factor receptors.
|
|
| BRCA1
methylation, n (total) | BRCA1
non-methylation, n (total) |
|---|
|
|
|
|
|
|---|
| First author,
year | Population | ER negative | PR negative | HER2 negative |
Triple-negative | ER negative | PR negative | HER2 negative |
Triple-negative | (Refs.) |
|---|
| Ignatov, 2013 | European | NA | NA | NA | 43 (86) | NA | NA | NA | 22 (46) | (21) |
| Krasteva, 2012 | European | 11 (23) | 11 (23) | 14 (17) | NA | 48
(112) | 11 (23) | 37 (53) | NA | (22) |
| Xu, 2009 | European | 89
(372) | 135 (372) | NA | NA | 127 (320) | 135 (372) | NA | NA | (23) |
| Xu, 2013 | Asian | 109 (285) | 149 (282) | 220 (279) | 64
(282) | 295 (830) | 149 (282) | 654 (810) | 142 (817) | (24) |
| Hsu, 2013 | Asian | 30 (77) | 36 (77) | 55 (77) | 16 (77) | 21 (61) | 36 (78) | 91
(138) | 5
(61) | (25) |
| Sharma, 2009 | Asian | 22 (27) | 21 (27) | 18 (27) | 15 (27) | 35 (74) | 21 (27) | 60 (74) | 25 (74) | (26) |
| Chen, 2009 | Asian | 55
(138) | 80
(137) | 98
(135) | 31
(136) | 127 (383) | 80
(137) | 298 (378) | 64
(382) | (27) |
| Jing, 2008 | Asian | 21 (33) | 26 (33) | NA | NA | 127 (168) | 26 (33) | NA | NA | (28) |
As shown in Table II,
there were 8 studies that met the inclusion criteria and were
included in the present meta-analysis. The studies involved 337
BRCA1 promoter hypermethylations with an ER-negative status,
458 with a PR-negative status, 405 with an HER2-negative status,
169 with triple-negative receptors, and 780, 458, 1,140 and 258
controls without BRCA1 promoter methylation but with a
negative status, correspondingly.
When the same investigators reported the results
obtained from the same cohort of patients in several studies, only
the largest series was included in the analysis. A cohort of
patients was excluded due to duplicate studies.
Due to insufficient data, HRs on OS could be
extracted from 7 studies for univariate analysis and 6 studies for
multivariate analysis. According to DFS analysis, there were 4
studies with available data for univariate analysis and 5 studies
with available data for multivariate analysis.
Association of BRCA1 methylation with
OS and DFS of patients with breast cancer
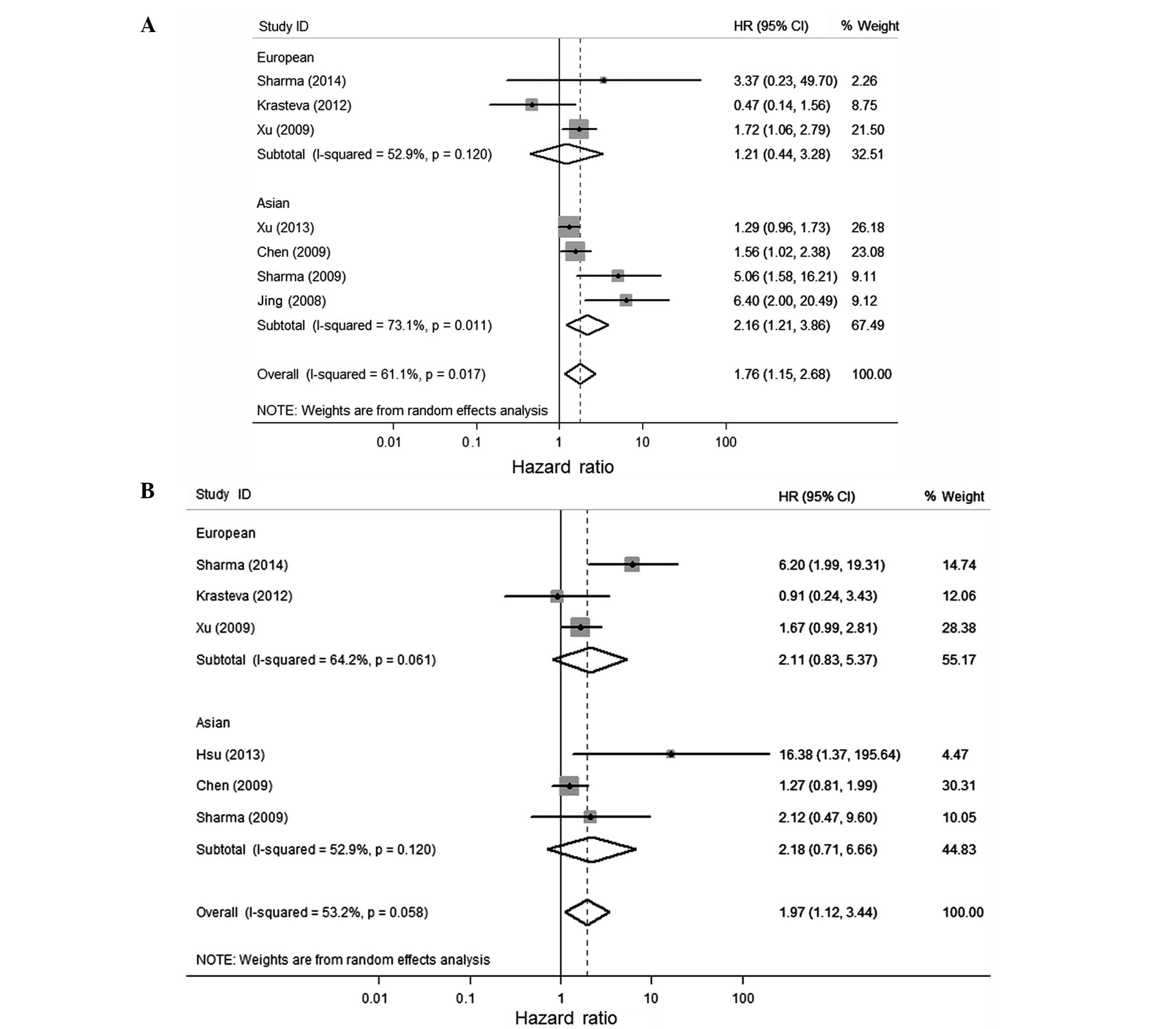
Considering the significant heterogeneity among
studies (P=0.017, I2=61.1%), the random-effect model was
used and a subgroup analysis was performed by considering different
ethnicities or the population of the participants to estimate the
combined effect of BRCA1 methylation. BRCA1
methylation was significantly associated with a poor OS and DFS of
breast cancer in the univariate and multivariate analysis (Figs. 2 and 3
and Table III). The combined HR was
1.76 (1.15–2.68) and 1.97 (1.12–3.44) for univariate analysis and
multivariate analysis of OS, respectively. For studies of DFS, the
pooled HR was 1.28 (0.68–2.43) and 1.64 (0.64–4.19), respectively.
The combined HRs (95% CIs) on OS by univariate analysis was 2.16
(1.21–3.86) for the Asian population, which was significantly
higher than 1.21 (0.44–3.28) for the European population. All the
combined HR scores in the present study are different from the
study of Wu et al (9). They
over-estimated the risk of fatality by using the OR instead of HR
value from the Karray-Chouayekh study (29) and using the incorrect value of OS in
the Xu et al study (23).
 | Table III.HRs and 95% CI for the association
between BRCA1 methylation and OS or DFS. |
Table III.
HRs and 95% CI for the association
between BRCA1 methylation and OS or DFS.
| Analysis | Population | Survival | Pooled HR (95% CI),
P-value | Heterogeneity test
P-value | Eggers test
P-value | Beggs test
P-value |
|---|
| Univariate | European | OS | 1.206
(0.444–3.276), 0.713 | 0.120 | 0.797 | 0.602 |
|
|
| DFS | – | – | – | – |
|
| Asian | OS | 2.164
(1.212–3.863), 0.009 | 0.011 | 0.004 | 0.174 |
|
|
| DFS | 1.795
(1.109–2.907) | 0.009 | 0.236 | 0.117 |
|
| Overall | OS | 1.758
(1.154–2.679), 0.009 | 0.017 | <0.001 | 0.393 |
|
|
| DFS | 1.284
(0.679–2.430), 0.442 | <0.001 | 1.000 | 0.885 |
| Multivariate | European | OS | 2.107
(0.827–5.368), 0.118 | 0.061 | 0.830 | 0.602 |
|
|
| DFS | 0.876
(0.059–12.953), 0.923 | <0.001 | – | 0.317 |
|
| Asian | OS | 2.177
(0.711–6.663), 0.173 | 0.120 | 0.256 | 0.117 |
|
|
| DFS | 2.250
(0.911–5.562), 0.079 | 0.021 | 0.074 | 0.117 |
|
| Overall | OS | 1.966
(1.124–3.438), 0.018 | 0.058 | 0.019 | 0.460 |
|
|
| DFS | 1.635
(0.638–4.191), 0.306 | <0.001 | <0.001 | 0.064 |
Primer for identifying BRCA1 promoter
methylation
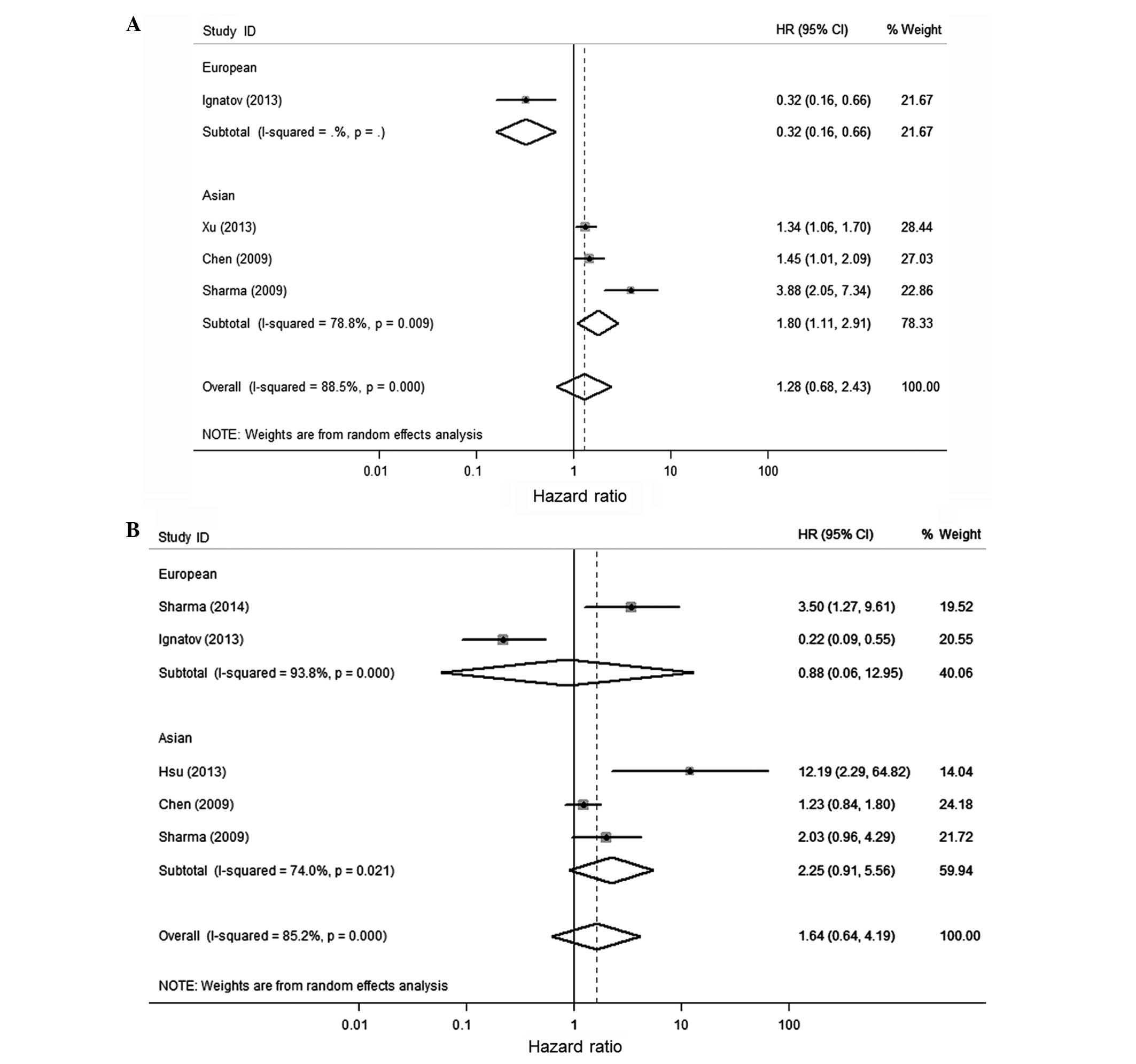
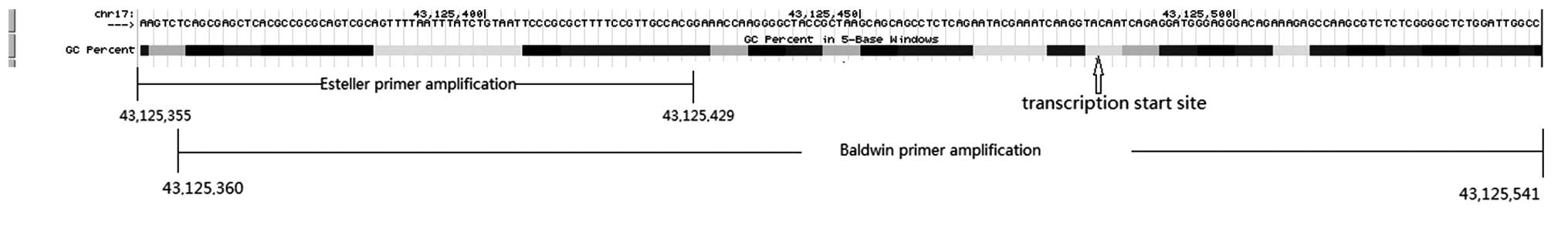
The majority of the studies identified that
BRCA1 promoter methylation is correlated with poor survival,
as shown in the present meta-analysis. However, an opposing opinion
remains as BRCA1 promoter methylation is a protective factor
in 2 studies [Krasteva et al (22) and Ignatov et al (21)]. To explore this difference, we
identified that they used different primers. In the Krasteva et
al (22) and Ignatov et al
(21) studies, which found a better
clinical prognosis in patients with BRCA1 promoter
methylation, they used Baldwin's primer (30) while others used Esteller's primer
(31). The two primer sequences were
blasted to the human genome in the NCBI database to find the
amplified regions. The two primers are for different regions where
there was a different concentration of GC-rich regions. The Baldwin
primer exhibited a larger amplification that contained more GC-rich
regions and included the transcriptional start point (Fig. 4). Therefore, the difference of
BRCA1 promoter methylation in the region of chr17: 43, 125,
429-43, 125, 541 (GRCh38 assembly) may affect the different
prognosis. However, all of these require more testing by
molecular-biological experiments to confirm.
Hormone receptor-negativity
correlation analysis
The number of patients who carried BRCA1
methylation and one of the negative statuses of ER, PR, HER2 or
triple-negative were extracted for the cases, as well as the number
of patients without promoter methylation of BRCA1
correspondingly for the controls. The correlations between
BRCA1 methylation and the negative status of different
breast cancer-related receptors were meta-analyzed separately. The
results of the association between hormone receptors that were
negative, BRCA1 promoter methylation and the heterogeneity
test are shown in Table IV. The
overall results suggested that, particularly for the Asian
populations, all the receptors negativity listed in Table II are associated with the
BRCA1 promoter methylation. Patients with those estrogen,
progesterone and epidermal growth factor-related negative receptors
were more likely to be negative for the BRCA1 protein (OR>1.247,
P<0.014). However, further molecular-biological experiments are
required to determine whether the correlation is spurious, as the
receptor and BRCA1 promoter methylation are associated with
breast cancer.
 | Table IV.Odds ratios and 95% CI for
BRCA1 promoter methylation and breast cancer subtype. |
Table IV.
Odds ratios and 95% CI for
BRCA1 promoter methylation and breast cancer subtype.
|
|
|
| Heterogeneity
test |
|---|
|
|
|
|
|
|---|
| Factors | Pooled OR (95%
CI) |
P-valuea | χ2 | P-value | I2
(%) |
|---|
| ER negative |
|
|
|
|
|
|
Asian | 1.329
(1.084–1.630) | 0.006 | 9.41 | 0.052 | 57.5 |
|
European | 1.040
(0.736–1.471) | 0.824 | 0.14 | 0.704 | 0.0 |
|
Overall | 1.247
(1.045–1.487) | 0.014 | 10.83 | 0.094 | 44.6 |
| PR negative |
|
|
|
|
|
|
Asian | 1.459
(1.195–1.782) | <0.001 | 15.62 | 0.004 | 74.4 |
|
European | 1.010
(0.740–1.379) | 0.948 | 0.28 | 0.599 | 0.0 |
|
Overall | 1.311
(1.108–1.550) | 0.002 | 19.30 | 0.004 | 68.9 |
| HER2 negative |
|
|
|
|
|
|
Asian | 2.834
(2.277–3.528) | <0.001 | 2.98 | 0.394 | 0.0 |
|
European | 2.881
(2.322–3.574) | <0.001 | 3.65 | 0.456 | 0.0 |
|
Triple-negative |
|
|
|
|
|
|
Asian | 1.557
(1.210–2.002) | 0.001 | 2.81 | 0.422 | 0.0 |
|
European | 1.494
(1.178–1.896) | 0.001 | 3.61 | 0.461 | 0.0 |
Sensitivity analysis and publication
bias
The Patsopoulos method (32) was used to test if an individual study
affected the heterogeneity in the OS and DFS analysis. As a result,
if the Jing et al (28) and
Sharma et al (26) studies
were removed the heterogeneity disappeared (I2=17.3%,
P=0.30). Funnel plot and Begg's test were performed to check the
publication bias, and they suggest the absence of publication bias.
The Begg's test for OS and DFS were P=0.187 and P=0.625,
respectively.
Discussion
The present meta-analysis suggested that patients
with BRCA1 promoter methylation had a more significant OS
and DFS disadvantage than those without the methylated status,
similar to the previous meta-analysis by Wu et al (9), which showed that breast cancer patients
with hypermethylation in the promoter of BRCA1 exhibited
poor survival. However, three problems were identified in the Wu
et al (9) study, which were
adjusted in the present study: i) A meta-analysis was performed for
the 5 adjusted HR scores of DFS, but one was an OR (20.7) score
instead of HR, [from the Karray-Chouayekh study (29)]and was much larger than the others; ii)
in the meta-analysis for the 5 adjusted HR scores of OS, the value
observed for all-cause mortality instead of fatality from breast
cancer in the Xu et al (23)
study was used; iii) 3 of the 9 HRs from the Sharma et al
(20,26,33)
studies published in 2009 and 2010 for meta-analysis were from the
same subjects. Following adjustment for these problems and the
addition of a study, the present meta-analysis suggested that the
previous study had over-estimated the risk of fatality from breast
cancer. The pooled HRs in the present study and the Wu et al
(9) study were 1.28 vs. 2.89 and 1.64
vs. 3.92 for univariate and multivariate analysis of DFS,
respectively.
The present meta-analysis additionally assessed the
associations between the promoter methylation of BRCA1 and
breast cancer-related receptors and the results showed that there
are significant correlations between them. This suggested that
potential interactions may exist between the hypermethylation of
the BRCA1 gene and the negative status of ER, PR and HER2
receptors through complex regulation pathways on tumor progression.
However, further studies are also required to explore the
correlations in order to assist in finding a therapeutic target for
breast cancer.
The consistency of two studies that reported the
protective effect for patients with hypermethylation in the
promoter of BRCA1 was found to be due to the different
primers used. Therefore, further studies are required to test
whether the different regions of the promoter methylation of
BRCA1 exhibit different clinical outcomes.
However, certain limitations of the study should be
considered. First, the number of studies contained in the present
meta-analysis is relatively small, particularly in non-African
populations, and the results should be confirmed in large samples.
Second, although the Egger's test did not have statistical
significance, the publication bias may still exist and influence
the results. Asymmetrical appearance of the funnel plot could be
caused by heterogeneity, smaller studies and other factors.
Considering the limitations of the study, the associations among
BRCA1 promoter methylation, prognosis of patients and the
negative status of the breast cancer-related therapeutic target
receptors should be further investigated.
In conclusion, the results revealed that breast
cancer patients with BRCA1 promoter methylation had lower OS
and DFS and had significant correlations with the negative status
of the ER, PR and HER2 receptors. Hypermethylation in different
regions of the BRCA1 promoter may exhibit a different
clinical performance.
Acknowledgements
The present study was financially supported by the
National Science Foundation of China (grant no. 81101547), the
Planned Science and Technology Project of Yunnan Province (grant
nos. 2009CA012 and 2011DH011), and the Fund of State Key Laboratory
of Genetics Resources and Evolution (grant no. GREKF10-07).
References
|
1
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun MJ: Cancer statistics, 2009. CA Cancer J Clin. 59:225–249.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Russo J, Yang X, Hu YF, Bove BA, Huang Y,
Silva ID, Tahin Q, Wu Y, Higgy N, Zekri A, et al: Biological and
molecular basis of human breast cancer. Front Biosci. 3:D944–D960.
1998.PubMed/NCBI
|
|
4
|
Miki Y, Swensen J, ShattuckEidens D,
Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM,
Ding W, et al: A strong candidate for the breast and ovarian cancer
susceptibility gene BRCA1. Science. 266:66–71. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Friedenson B: The BRCA1/2 pathway prevents
hematologic cancers in addition to breast and ovarian cancers. BMC
Cancer. 7:1522007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ralhan R, Kaur J, Kreienberg R and
Wiesmüller L: Links between DNA double strand break repair and
breast cancer: Accumulating evidence from both familial and
nonfamilial cases. Cancer Lett. 248:1–17. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hall JM, Lee MK, Newman B, Morrow JE,
Anderson LA, Huey B and King MC: Linkage of early-onset familial
breast cancer to chromosome 17q21. Science. 250:1684–1689. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Birgisdottir V, Stefansson OA,
Bodvarsdottir SK, Hilmarsdottir H, Jonasson JG and Eyfjord JE:
Epigenetic silencing and deletion of the BRCA1 gene in sporadic
breast cancer. Breast Cancer Res. 8:R382006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu L, Wang F, Xu R, Zhang S, Peng X, Feng
Y, Wang J and Lu C: Promoter methylation of BRCA1 in the prognosis
of breast cancer: A meta-analysis. Breast Cancer Res Treat.
142:619–627. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
DahlmanWright K, Cavailles V, Fuqua SA,
Jordan VC, Katzenellenbogen JA, Korach KS, Maggi A, Muramatsu M,
Parker MG and Gustafsson JA: International Union of Pharmacology.
LXIV. Estrogen receptors. Pharmacol Rev. 58:773–781. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mitri Z, Constantine T and O'Regan R: The
HER2 receptor in breast cancer: Pathophysiology, clinical use, and
new advances in therapy. Chemother Res Pract.
2012:7431932012.PubMed/NCBI
|
|
12
|
Bosch A, Eroles P, Zaragoza R, Viña JR and
Lluch A: Triple-negative breast cancer: Molecular features,
pathogenesis, treatment and current lines of research. Cancer Treat
Rev. 36:206–215. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fleiss JL: The statistical basis of
meta-analysis. Stat Methods Med Res. 2:121–145. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tierney JF, Stewart LA, Ghersi D, Burdett
S and Sydes MR: Practical methods for incorporating summary
time-to-event data into meta-analysis. Trials. 8:162007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
DerSimonian R: Meta-analysis in the design
and monitoring of clinical trials. Stat Med. 15:1237–1252. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Higgins JP, Thompson SG, Deeks JJ and
Altman DG: Measuring inconsistency in meta-analyses. BMJ.
327:557–560. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bax L, Ikeda N, Fukui N, Yaju Y, Tsuruta H
and Moons KG: More than numbers: The power of graphs in
meta-analysis. Am J Epidemiol. 169:249–255. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Egger M, Davey Smith G, Schneider M and
Minder C: Bias in meta-analysis detected by a simple, graphical
test. BMJ. 315:629–634. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lau J, Ioannidis JP and Schmid CH:
Quantitative synthesis in systematic reviews. Ann Intern Med.
127:820–826. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sharma P, Stecklein SR, Kimler BF, Sethi
G, Petroff BK, Phillips TA, Tawfik OW, Godwin AK and Jensen RA: The
prognostic value of BRCA1 promoter methylation in early stage
triple negative breast cancer. J Cancer Ther Res. 3:1–11. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ignatov T, Poehlmann A, Ignatov A,
Schinlauer A, Costa SD, Roessner A, Kalinski T and Bischoff J:
BRCA1 promoter methylation is a marker of better response to
anthracycline-based therapy in sporadic TNBC. Breast Cancer Res
Treat. 141:205–212. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Krasteva ME, Bozhanov SS, Antov GG,
Gospodinova ZI and Angelov SG: Breast cancer patients with
hypermethylation in the promoter of BRCA1 gene exhibit favorable
clinical status. Neoplasma. 59:85–91. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu X, Gammon MD, Zhang Y, Bestor TH,
Zeisel SH, Wetmur JG, Wallenstein S, Bradshaw PT, Garbowski G,
Teitelbaum SL, et al: BRCA1 promoter methylation is associated with
increased mortality among women with breast cancer. Breast Cancer
Res Treat. 115:397–404. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu Y, Diao L, Chen Y, Liu Y, Wang C,
Ouyang T, Li J, Wang T, Fan Z, Fan T, et al: Promoter methylation
of BRCA1 in triple-negative breast cancer predicts sensitivity to
adjuvant chemotherapy. Ann Oncol. 24:1498–1505. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hsu NC, Huang YF, Yokoyama KK, Chu PY,
Chen FM and Hou MF: Methylation of BRCA1 promoter region is
associated with unfavorable prognosis in women with early-stage
breast cancer. PLoS One. 8:e562562013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sharma G, Mirza S, Yang YH, Parshad R,
Hazrah P, Datta Gupta S and Ralhan R: Prognostic relevance of
promoter hypermethylation of multiple genes in breast cancer
patients. Cell Oncol. 31:487–500. 2009.PubMed/NCBI
|
|
27
|
Chen Y, Zhou J, Xu Y, Li Z, Wen X, Yao L,
Xie Y and Deng D: BRCA1 promoter methylation associated with poor
survival in Chinese patients with sporadic breast cancer. Cancer
Sci. 100:1663–1667. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jing F, Jun L, Yong Z, Wang Y, Fei X,
Zhang J and Hu L: Multigene methylation in serum of sporadic
Chinese female breast cancer patients as a prognostic biomarker.
Oncology. 75:60–66. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
KarrayChouayekh S, Trifa F, Khabir A,
Boujelbane N, SellamiBoudawara T, et al: Clinical significance of
epigenetic inactivation of hMLH1 and BRCA1 in Tunisian patients
with invasive breast carcinoma. J Biomed Biotechnol.
2009:3691292009.PubMed/NCBI
|
|
30
|
Baldwin RL, Nemeth E, Tran H, Shvartsman
H, Cass I, et al: BRCA1 promoter region hypermethylation in ovarian
carcinoma: a population-based study. Cancer Res. 60:5329–5333.
2000.PubMed/NCBI
|
|
31
|
Esteller M, Silva JM, Dominguez G, Bonilla
F, MatiasGuiu X, et al: Promoter hypermethylation and BRCA1
inactivation in sporadic breast and ovarian tumors. J Natl Cancer
Inst. 92:564–569. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Patsopoulos NA, Evangelou E and Ioannidis
JP: Sensitivity of between-study heterogeneity in meta-analysis:
proposed metrics and empirical evaluation. Int J Epidemiol.
37:1148–1157. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sharma G, Mirza S, Parshad R, et al:
Clinical significance of promoter hypermethylation of DNA repair
genes in tumor and serum DNA in invasive ductal breast carcinoma
patients. Life Sci. 87:83–91. 2010. View Article : Google Scholar : PubMed/NCBI
|