Introduction
Cancers of unknown primary origin (CUP) pose a
significant challenge for clinicians in terms of identifying the
primary cancer and designing an effective treatment strategy for
the patients (1). CUPs exhibit an
aggressive biological behavior and the prognosis of patients with
CUPs is typically significantly poorer compared with that of
patients with cancers of known origin, with a median survival of
3–4 months after initial diagnosis (2). While CUPs only comprise ~3–5% of new
cancer cases annually, they constitute the fourth most common cause
of cancer-related mortality in Western countries, highlighting the
significance of this diagnostic challenge (3,4). Thus,
there has been a surge in the development of advanced diagnostic
tools for primary tumor identification using molecular testing.
Notably, microRNA expression assays have demonstrated a remarkable
ability to identify the origin of CUPs (5). MicroRNAs are small, non-coding RNAs that
regulate gene expression by RNA degradation or translational
inhibition (6). Aberrant microRNAs
play a critical role in the development and progression of cancer,
making them ideal biomarkers for cancer diagnosis. Multiple studies
have reported high sensitivity and accuracy for microRNA assays in
diagnosing cases with cancers of known and unknown primary,
allowing for their use as a key tool for cancer diagnosis. The case
presented herein is a representative example of the possible
clinical impact of microRNA assays.
Case report
A 54 year-old male patient was presented to his
orthopedist with low back pain in June, 2013. The patient had no
significant past medical history other than mild hypertension and a
history of smoking (35 pack-years). An initial magnetic resonance
imaging (MRI) examination of the lumbar spine revealed two
findings: A mass in the right kidney and a mass in the T9-T10
vertebrae that was impinging on the spinal cord. The patient
underwent surgical resection of the vertebral mass at 4 weeks after
presentation. Initial pathology identified a metastatic low-grade
carcinoma of epithelial origin and the differential diagnosis
included renal carcinoma, neuroendocrine carcinoma, melanoma and
testicular carcinoma. On immunohistochemistry (IHC), the lesion was
strongly positive for cytokeratin AE1/AE3, CAM5.2, epithelial
membrane antigen, E-cadherin, melan-A, synaptophysin and placental
alkaline phosphatase. Additionally, there was mild and/or focal
staining for renal cell carcinoma antigen, CD10, neuron-specific
enolase, S100 and human melanoma black 45. The immunostaining for
cytokeratin (CK)7, CK20, inhibin, thyroid transcription factor 1,
CK5/6, gross cystic disease fluid protein 15, vimentin and CD117,
was negative. A second evaluation also raised the possibility of
carcinoma of prostatic origin. However, the serum prostate-specific
antigen (PSA) levels were within the normal range and IHC for PSA
was negative in the tissue sample. Initial evaluation with computed
tomography (CT) of the brain, chest, abdomen and pelvis and a bone
scan revealed multiple bone lesions, two enlarged mediastinal lymph
nodes, a block of enlarged para-aortic lymph nodes and a mass
located in the upper pole of the right kidney, sized 11×13.5×12.5
cm. The kidney mass exhibited no significant contrast uptake;
following resection, it was found to be a benign cyst.
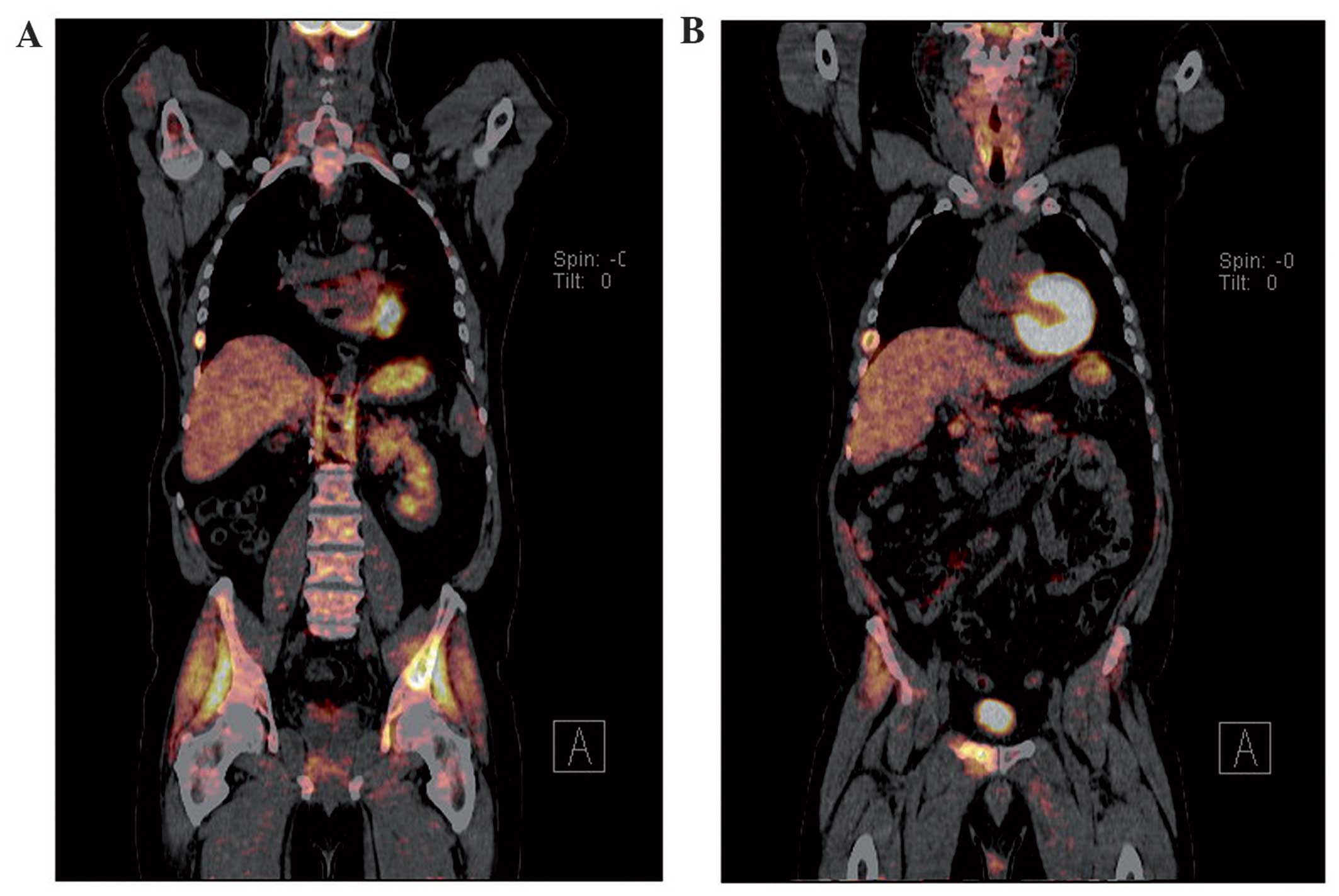
A positron emission tomography (PET)-CT, ordered in
late September, revealed a relatively mild radioactive uptake in
the subcutaneous area of the right shoulder girdle [standardized
uptake value (SUV) = 3.1], and a significantly higher uptake in
several bones (SUVmax = 9.3). Physical examination of the patient
revealed a soft, hairy, pigmented skin lesion on the right anterior
axillary line. An MRI scan of the right shoulder was ordered on the
18th of October, and the mass appeared as a subcutaneous lesion
with homogeneous contrast uptake with an associated group of lymph
nodes. As the primary cancer remained unknown, the microRNA Rosetta
Cancer Origin test (Rosetta Genomics Ltd., Philadelphia, PA, USA)
was ordered 16 weeks after presentation. The test results were
received 18 weeks after presentation and identified the tissue as
breast cancer, with a sensitivity of 90%. The axillary mass was
then resected (20 weeks after presentation) and pathology
identified that mass as a malignant neoplasm similar to the mass
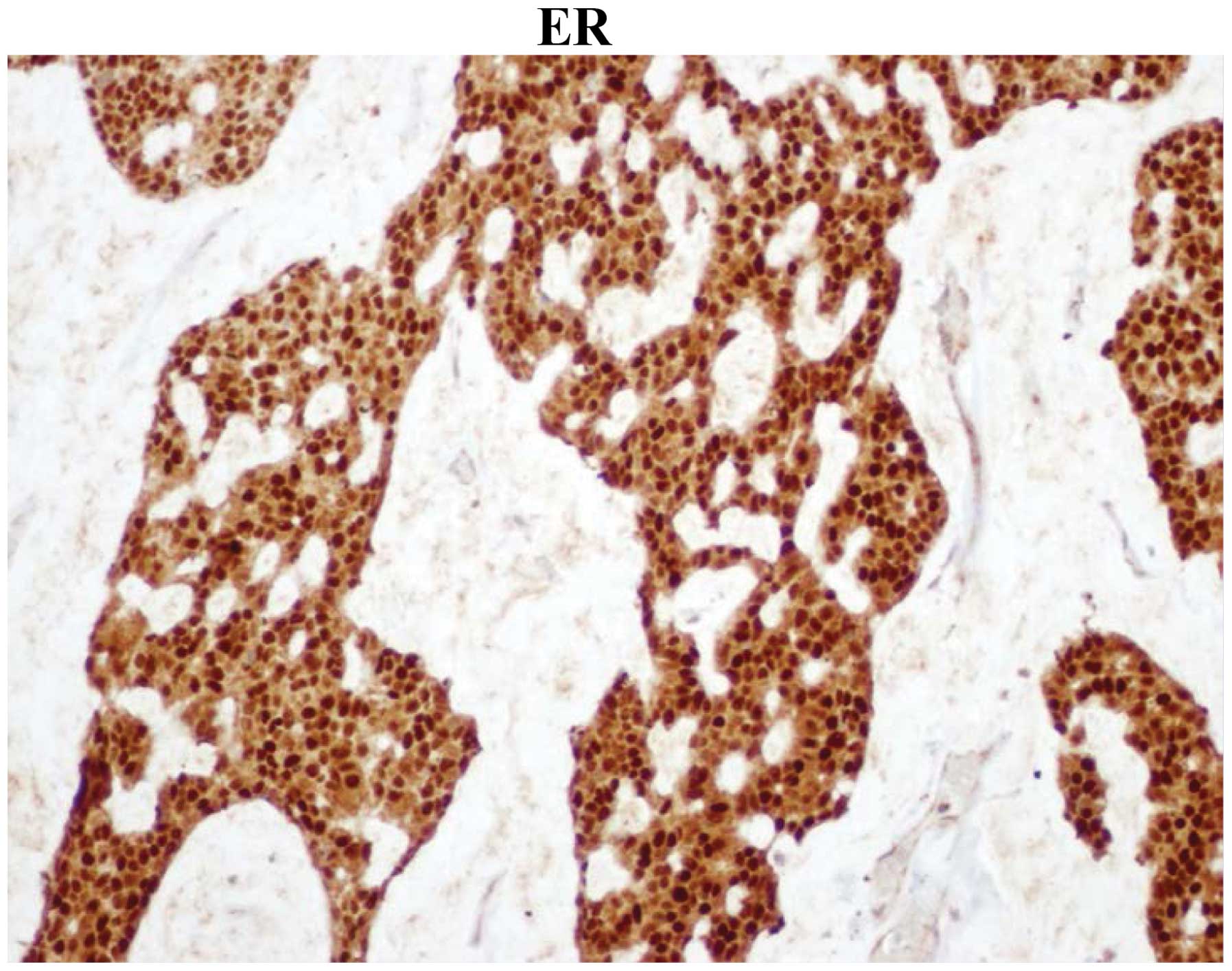
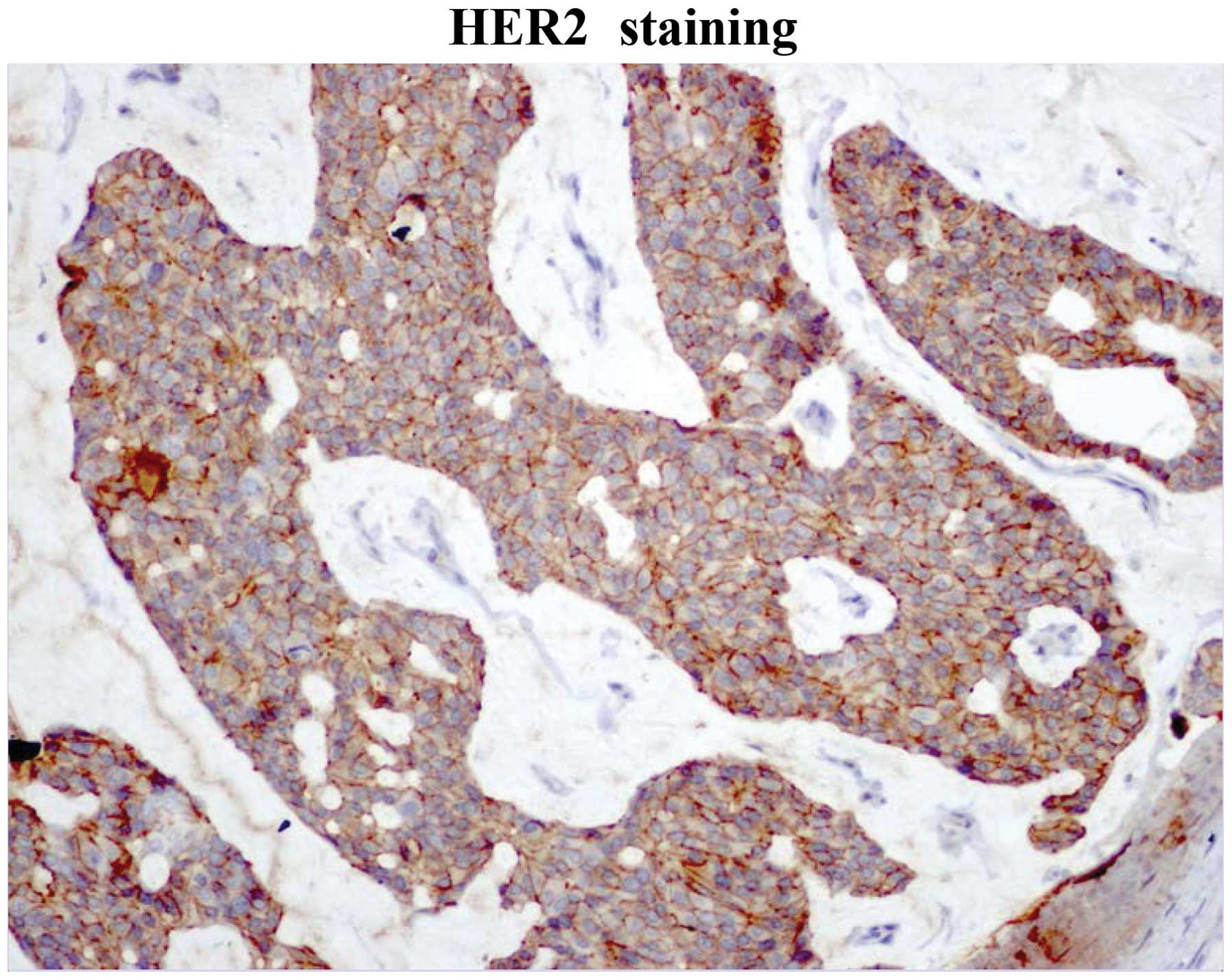
removed from the lumbar area. IHC was 100% positive for estrogen
receptor (ER) and progesterone receptor (PR) and positive for human
epidermal growth factor receptor 2 (HER2) (Figs. 1–3).
The patient was treated for breast cancer with
capecitabine and trastuzumab as the first-line regimen; following
relapse on left iliac and right pubic bone, the regimen was changed
to vinorelbine and trastuzumab. The patient responded well and
proceeded to maintenance trastuzumab and tamoxifen. On December 30,
2014, treatment was switched to letrozole and trastuzumab, due to
an increase in serum marker levels (carcinoembryonic antigen and
carbohydrate antigen 15-3) that was followed by the appearance of a
skin nodule right by the scar of the axillary mass resection. The
nodule was subsequently removed and histology showed local relapse
of the known neoplasm. The patient is completely asymptomatic and
continues on letrozole and trastuzumab with stable bone disease
ever since, while the relevant tumor markers are within normal
limits. The patient currently remains on maintenance letrozole and
trastuzumab, has stable disease (bone lesions alone) and a good
overall clinical status. Last follow-up was on March 16, 2016.
Patient informed consent was obtained for the
publication of the case details.
Discussion
Identifying the primary cancer is crucial in
selecting the optimal therapeutic strategy for the patients. When
clinical and pathological methods fail to identify the primary
tumor, treatment is often delayed and inaccurately targeted,
contributing to the generally very poor prognosis of patients with
CUPs. IHC is the most widely used tool for the diagnosis of several
types of cancer, but fails to identify the tissue of origin in
>30% of the cases (7).
Furthermore, IHC may be biased based on the patient's presentation
and past medical history and the interpretation of the results may
be subjective, further signifying the need for alternative cancer
diagnostic tests. A number of studies have demonstrated that
microRNA profiling may be useful for solving the diagnostic problem
posed by CUPs, with agreement to final diagnosis for microRNA
testing ranging from 84% to as high as 92%, depending on the study
(8–10). MicroRNAs are small, non-coding RNAs of
17–25 nucleotides in length that play an essential regulatory role
in protein translation and expression (6). Since their discovery in 1993, microRNAs
have been implicated in a number of major cell processes and have
been mapped to areas of the genome prone to deletions, mutations
and amplifications (6). Their ability
to act as oncogenes or tumor suppressors, combined with their
unique tissue pattern expression in malignancies, makes microRNAs
ideal cancer biomarkers and provides the basis for their use as a
diagnostic tool (11).
In the present case, microRNA testing was crucial,
as the clinical and pathological findings did not lead to a
diagnosis of breast cancer for a number of reasons. First, the
pathological examination of the vertebral mass did not raise the
suspicion of breast cancer, so this was not included in the
differential diagnosis. Second, while the MRI and PET-CT imaging
found the axillary mass to be suspicious, clinical evidence
regarding the origin of the mass pointed towards the skin rather
than the breast. The clinical evaluation of the skin lesion was
also more consistent with skin cancer, taking into consideration
the appearance and coloration of the mass (Fig. 4). Finally, the patient was male, with
no family history of breast cancer. This is significant, as male
breast cancer (MBC) is a rare entity, constituting <1% of all
breast cancers and <1% of cancers in men (12). MBC is also notably more difficult to
diagnose compared with female breast cancer. Male patients tend to
be older than female patients at diagnosis, and have higher-stage
disease, with more extensive lymph node involvement, contributing
to the high mortality rate of MBC (13,14).
MicroRNA testing proved to be key in identifying the origin of the
primary cancer, when the clinicopathological picture was leading
towards a misdiagnosis. Further examination revealed that the tumor
was ER+, PR+ and HER2+, indicating that the patient had a
particularly good prognosis and could be treated with trastuzumab,
as well as several specific targeted therapies, which would not
have been applied had it not been for the microRNA test.
MicroRNA profiling as a diagnostic test offers
several advantages, the most important being that the assay is
objective and unbiased. Samples may be profiled using quantitative
polymerase chain reaction technology or, more recently, a custom
microRNA array, and classification of tumor types relies on
specific bioinformatics algorithms. The microarray is both powerful
and flexible, it is able to profile thousands of transcripts and
newly identified microRNAs may easily be incorporated into an
updated test design. Furthermore, in terms of the feasibility of
testing, samples of microRNAs may be safely stored for significant
periods of time in formalin-fixed paraffin-embedded tissues and,
when extracted, still exhibit similar profiles to microRNA samples
from fresh tissue (15). Multiple
studies have repeatedly demonstrated high test sensitivity,
specificity and reproducibility, and the turnaround time for
profiles is between 7 and 10 days, rendering this a reliable and
practical diagnostic test. Furthermore, microRNA screening for
early cancer diagnosis shows promising potential with changes in
microRNA profiles implicated in early or pre-tumor development
(6). However, there are challenges to
microRNA testing regarding CUPs. First, the accuracy of testing
cannot be directly validated, as there is no primary cancer
identified to be used as reference; thus, cancers with similar
profiles may be easily confused as being of the same origin
(5,8).
In addition, microRNA assessment may not always agree with the
clinical and pathological diagnosis, with a previous study
reporting a disagreement rate of 16% (8). Furthermore, the assay itself may produce
two different diagnoses, which poses a potential problem for
clinicians if the diagnoses differ regarding the optimal course of
treatment (8). On the other hand, the
test may not be able to predict a result at all if the expression
patterns do not closely correspond to those included in the panel.
It is of great importance to determine the potential benefit that
patients may gain from assigning CUP to a primary tissue of origin.
We would expect that assigning a CUP to a primary would be of great
benefit for such patients; however, evidence thus far does not
strongly support this hypothesis. A recent prospective study by
Hainsworth et al (16), in
which molecular techniques were used to identify the primary,
demonstrated a median survival of 12.5 months for the group of
patients in whom the CUP was assigned a primary tissue of origin,
in contrast to the group of patients who were treated as CUP, in
whom the median survival was 10.8 months. Under the light of the
currently available data, the European Society for Medical Oncology
and National Comprehensive Cancer Network guidelines are quite
conservative regarding the use of molecular tools in the
differential diagnosis of CUP, and they advise careful selection of
the population considered as candidate for such diagnostic tools
(level of evidence, 2B). Finally, while most recent tests may
screen for 42 different primaries, they do not screen for all
cancer origins (8).
MicroRNA profiling may be quite costly, although the
cost-benefit ratio may ultimately be favorable, as several
unnecessary tests may be avoided. For all the abovementioned
reasons, clinicians must consider all data relative to each
individual case, in order to determine the optimal course of
treatment for each patient.
The case presented herein highlights the efficacy of
using microRNA analysis of tumor tissue samples to diagnose CUPs.
As a diagnostic tool, microRNA profiles may be very useful in
identifying the primary tissue of origin and, thus, improving
treatment and outcome for CUP patients.
References
|
1
|
Varadhachary GR and Raber MN: Cancer of
unknown primary site. N Engl J Med. 371:757–765. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Riihimäki M, Thomsen H and Hemminki K,
Sundquist K and Hemminki K: Comparison of survival of patients with
metastases from known versus unknown primaries: Survival in
metastatic cancer. BMC Cancer. 13:362013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pavlidis N and Fizazi K: Cancer of unknown
primary (CUP). Crit Rev Oncol Hematol. 54:243–250. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pavlidis N, Briasoulis E, Hainsworth J and
Greco FA: Diagnostic and therapeutic management of cancer of an
unknown primary. Eur J Cancer. 39:1990–2005. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ferracin M, Pedriali M, Veronese A,
Zagatti B, Gafà R, Magri E, Lunardi M, Munerato G, Querzoli G,
Maestri I, et al: MicroRNA profiling for the identification of
cancers with unknown primary tissue-of-origin. J Pathol. 225:43–53.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 4:143–159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Anderson GG and Weiss LM: Determining
tissue of origin for metastatic cancers: Meta-analysis and
literature review of immunohistochemistry performance. Appl
Immunohistochem Mol Morphol. 18:3–8. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Varadhachary GR, Spector Y, Abruzzese JL,
Rosenwald S, Wang H, Aharonov R, Carlson HR, Cohen D, Karanth S,
Macinskas J, et al: Prospective gene signature study using microRNA
to identify the tissue of origin in patients with carcinoma of
unknown primary. Clin Cancer Res. 17:4063–4070. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pentheroudakis G, Pavlidis N, Fountzilas
G, Krikelis D, Goussia A, Stoyianni A, Sanden M, St Cyr B,
Yerushalmi N, Benjamin H, et al: Novel microRNA-based assay
demonstrates 92% agreement with diagnosis based on
clinicopathologic and management data in a cohort of patients with
carcinoma of unknown primary. Mol Cancer. 12:572013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rosenwald S, Gilad S, Benjamin S, Lebanony
D, Dromi N, Faerman A, Benjamin H, Tamir R, Ezagouri M, Goren E, et
al: Validation of a microRNA-based qRT-PCR test for accurate
identification of tumor tissue origin. Mod Pathol. 23:814–823.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li M, Li J, Ding J, He M and Cheng SY:
MicroRNA and cancer. AAPS J. 12:309–317. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rizzolo P, Silvestri V, Tommasi S, Pinto
R, Danza K, Falchetti M, Gulino M, Frati P and Ottini L: Male
breast cancer: Genetics, epigenetics, and ethical aspects. Ann
Oncol. 24(Suppl 8): viii75–viii82. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schneider S and Sariego J: Male breast
cancer presenting as an axillary mass: A case report and literature
review. South Med J. 102:736–737. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Korde LA, Zujewski JA, Kamin L, Giordano
S, Domchek S, Anderson WF, Bartlett JM, Gelmon K, Nahleh Z, Bergh
J, et al: Multidisciplinary meeting on male breast cancer: Summary
and research recommendations. J Clin Oncol. 28:2114–2122. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rosenfeld N, Aharonov R, Meiri E,
Rosenwald S, Spector Y, Zepeniuk M, Benjamin H, Shabes N, Tabak S,
Levy A, et al: MicroRNAs accurately identify cancer tissue origin.
Nat Biotechnol. 26:462–469. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hainsworth JD, Rubin MS, Spigel DR, Boccia
RV, Raby S, Quinn R and Greco FA: Molecular gene expression
profiling to predict the tissue of origin and direct site-specific
therapy in patients with carcinoma of unknown primary site: A
prospective trial of the Sarah Cannon research institute. J Clin
Oncol. 31:217–223. 2013. View Article : Google Scholar : PubMed/NCBI
|