Introduction
Lipomatous tumors are a highly diverse group of
mesenchymal neoplasms characterized by an overgrowth of adipose
cells or their precursors. Benign lipomas are the most prevalent
type of lipomatous tumor and are the most common soft tissue tumor
(1). Lipomas affect approximately 1%
of the general population, appearing most often in patients aged
40–60 years and generally ranging in size from 1 to 3 cm, although
rare giant lipomas can grow to 20 cm in diameter and weigh up to 5
kg (2,3). These relatively common tumors are
rarely life threatening and in most cases do not require medical
treatment unless the tumor restricts movement or causes pain. By
contrast, malignant liposarcomas are much more rare (approximately
2.5 cases per million individuals), but rank as the second most
common of all soft tissue sarcomas in humans (4). Liposarcomas can be characterized into
multiple, phenotypically diverse subtypes including
well-differentiated, myxoid/round cell liposarcomas, pleiomorphic,
and de-differentiated. These malignant tumors exhibit 5-year
survival rates as low as 39% depending on the particular
histological subtype (5).
Many molecular and clinical similarities exist
between lipomas and low-grade liposarcomas such as the
well-differentiated subtype (6,7). Thus, a
diagnostic dilemma can occur with regard to differentiating these
tumors. When comparing lipomas and low-grade liposarcomas, patients
are misdiagnosed 30–40% of the time following radiological
detection (8–10) and in 7–17% of histological
evaluations (11). Even fluorescent
in situ hybridization (FISH) for MDM2-CDK4 amplification
(considered the gold standard for distinguishing lipomas from
low-grade and de-differentiated liposarcomas) is inaccurate and/or
provides uninterpretable results in 10–15% of cases (12–16).
Diagnostic accuracy is also problematical due to morphological
heterogeneity within particular lipomatous tumors. For instance,
benign pleomorphic lipomas exhibit unusual features such as the
presence of hibernomas or tumor pleomorphism that can lead to
confusion with malignant liposarcomas (17). Many liposarcomas display transitional
features of low- to high-grade lesions or consist of
well-differentiated regions associated with non-lipogenic sarcoma
often resembling malignant fibrous histiocytomas or fibrosarcoma
(17). Further complicating this
issue is a recent study that reveals MDM2 and CDK4
genes are both amplified in up to 5% of large and deep-seated
lipomas, suggesting that the clinical significance of gene
amplifications for lipomatous tumors is unclear and requires
further studies (18).
Accurate, timely, and cost effective diagnosis of
lipomatous tumors is essential for proper patient treatment and
increased long-term survival. In the present study, we utilized a
combination of bioinformatics and immunohistochemistry (IHC)
analysis to accurately and sensitively identify biomarkers that
effectively distinguish benign from low-grade malignant lipomatous
tumors.
Materials and methods
Meta-analysis of lipomatous tumor gene
expression
A whole genome RNA expression dataset [Gene
Expression Omnibus (GEO) reference series GSE6481, deposited by
Nakayama et al (14)] was
analyzed to identify gene expression alterations between benign
lipomas and well-differentiated liposarcomas. This dataset compared
the global gene expression profiles of various soft tissue sarcomas
including lipomas (n=3) and well-differentiated liposarcomas (n=3).
Tab-delimited files from these data were input into Cluster 3.0,
normalized, and clustered using a correlation (uncentered)
similarity metric with a centroid linkage. Data were visualized
using Java Treeview.
Case material
Tissue arrays of formalin-fixed, paraffin-embedded
lipomatous tumor blocks were obtained from Super Bio Chips (Seoul,
Korea) (www.tissue-array.com). These
clinically characterized tumor samples consisted of 2-mm diameter
cores with a section thickness of 4 microns and were composed of
lipomas (n=11) and well-differentiated liposarcomas (n=20). The
cases were blindly reviewed and confirmed by a pathologist.
IHC
Sections were deparaffinized, rehydrated, and
treated for antigen retrieval using Trilogy (cat. no. 920P-10; Cell
Marque, Rocklin, CA, USA). To block non-specific binding, the
sections were incubated in background block solution (cat. no.
927B-05; Cell Marque) at room temperature for 10 min before
application of primary antibody. Antibodies used in this study
included: ATP-binding cassette, sub-family B member 11 (ABCB11)
(cat. no. ab155421), bone morphogenetic protein 8A (BMP8A) (cat.
no. ab60290) and complement component 4 binding protein β (C4BPB)
(cat. no. ab105507) (all from Abcam, Cambridge, MA, USA), class II,
major histocompatibility complex, transactivator (CIITA) (cat. no.
sc48797; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA),
ephrin receptor B2 (EPHB2) (cat. no. ab150652), glutaminase 2
(GLS2) (cat. no. ab113509), homeobox B7 (HOXB7) (cat. no.
ab111018), protein kinase N2 (PKN2) (cat. no. ab32395), RAB6B,
member RAS oncogene family (RAB6B) (cat. no. ab55660),
retinoblastoma-binding protein 5 (RBBP5) (cat. no. ab84511), serine
incorporator 2 (SERINC2) (cat. no. ab134312), G-protein signaling 2
(RGS2) (cat. no. ab36561) γ-synuclein (SNCG) (cat. no. ab55424),
sex determining region Y-box 30 (SOX-30) (cat. no. ab26024), and
thyroid-stimulating hormone receptor (TSHR) (cat. no. ab27974) (all
from Abcam). The sections were stained using the HRP/DAB Detection
IHC kit (cat. no. ab80436; Abcam) and counterstained with
hematoxylin.
Quantification of and statistical
analysis of IHC
Immunopositivity was scored semi-quantitatively for
the percentage of tumor cell staining (0, negative; 1, weak
staining; 2, moderate staining; 3, strong) and intensity (0,
negative; 1, <25% of tumor cells stained; 2, 25–49% of tumor
cells stained; 3, 50–74% of tumor cells stained; 4, >75% of
tumor cells stained). The product of the percentage staining and
intensity values represented the IHC score for each tumor. Mean IHC
values (mean ± SEM) were calculated using the Mann-Whitney rank-sum
test. A logistic regression model was developed for each protein to
test its ability to differentiate between the two tumor types. The
model's discriminatory performance was assessed using area under
the curve (AUC) with a 95% confidence interval. Furthermore, the
threshold for each marker and combined model in differentiating
lipoma from well-differentiated liposarcoma was determined using
receiver operating characteristics analysis. The cut-off value was
chosen where sensitivity and specificity were found to be
similar.
Results
Nakayama et al (19) previously published data comparing the
relative global gene expression profiles of various soft tissue
sarcomas. These data are freely and publically available in the GEO
(no. GSE6481, www.ncbi.nlm.nih.gov/geo/). Included in this genomic
dataset are transcriptional profiles from benign lipomas and
well-differentiated liposarcomas. The data from this analysis can
serve as a guide for further validation experiments using larger
patient datasets to identify biomarkers that can differentiate
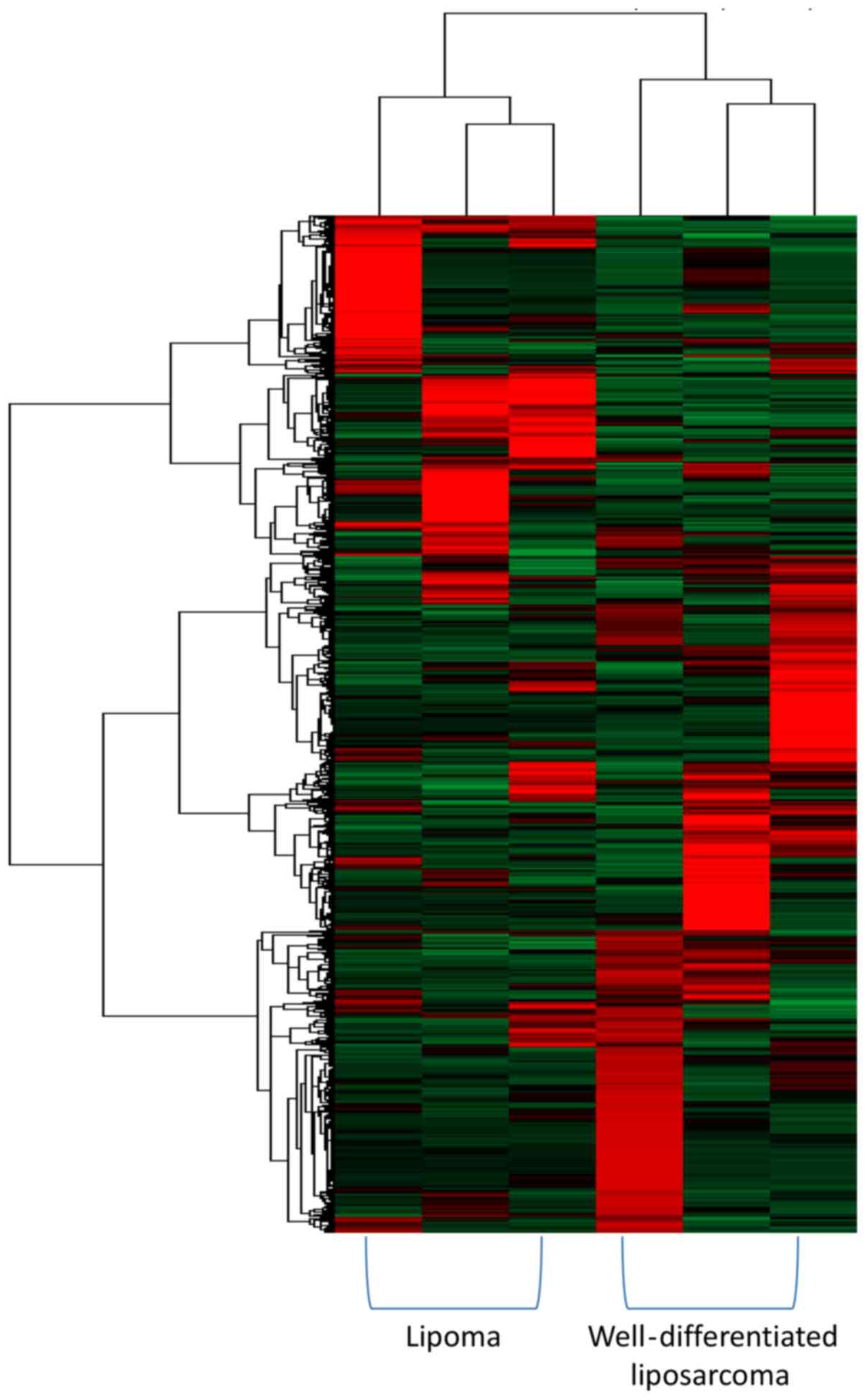
lipomas from well-differentiated liposarcomas. Unsupervised
hierarchical clustering analysis of the lipoma and
well-differentiated liposarcoma gene expression data from Nakayama
et al (14) showed that a
marked divergence in expression profiles occurred even within each
tumor subtype. However, clear clustering occurred for the tumors
based on their histological type (Fig.
1). This result suggests that despite the current clinical
difficulty in distinguishing these tumor types based on radiology,
histology, or cytogenetics, differences in the gene expression
profiles may be identified and utilized to differentiate lipomas
from well-differentiated liposarcomas.
We selected 15 genes from this meta-analysis that
were differentially regulated in well-differentiated liposarcomas
compared to lipomas for further investigation. The genes included
ABCB11, BMP8A, C4BPB, CIITA,
EPHB2, GLS2, HOXB7, PKN2, RAB6B,
RBBP5, RGS2, SERINC2, SNCG,
SOX30 and TSHR. We validated the expression of the 15
genes at the protein level using IHC on an independent panel of
clinically defined lipomatous tumors. This tumor panel consisted of
11 lipoma tumors and 19 well-differentiated liposarcomas. The
clinico-pathological characteristics of this patient dataset are
reported in Table I. The
distribution of age, sex, and location were almost similar between
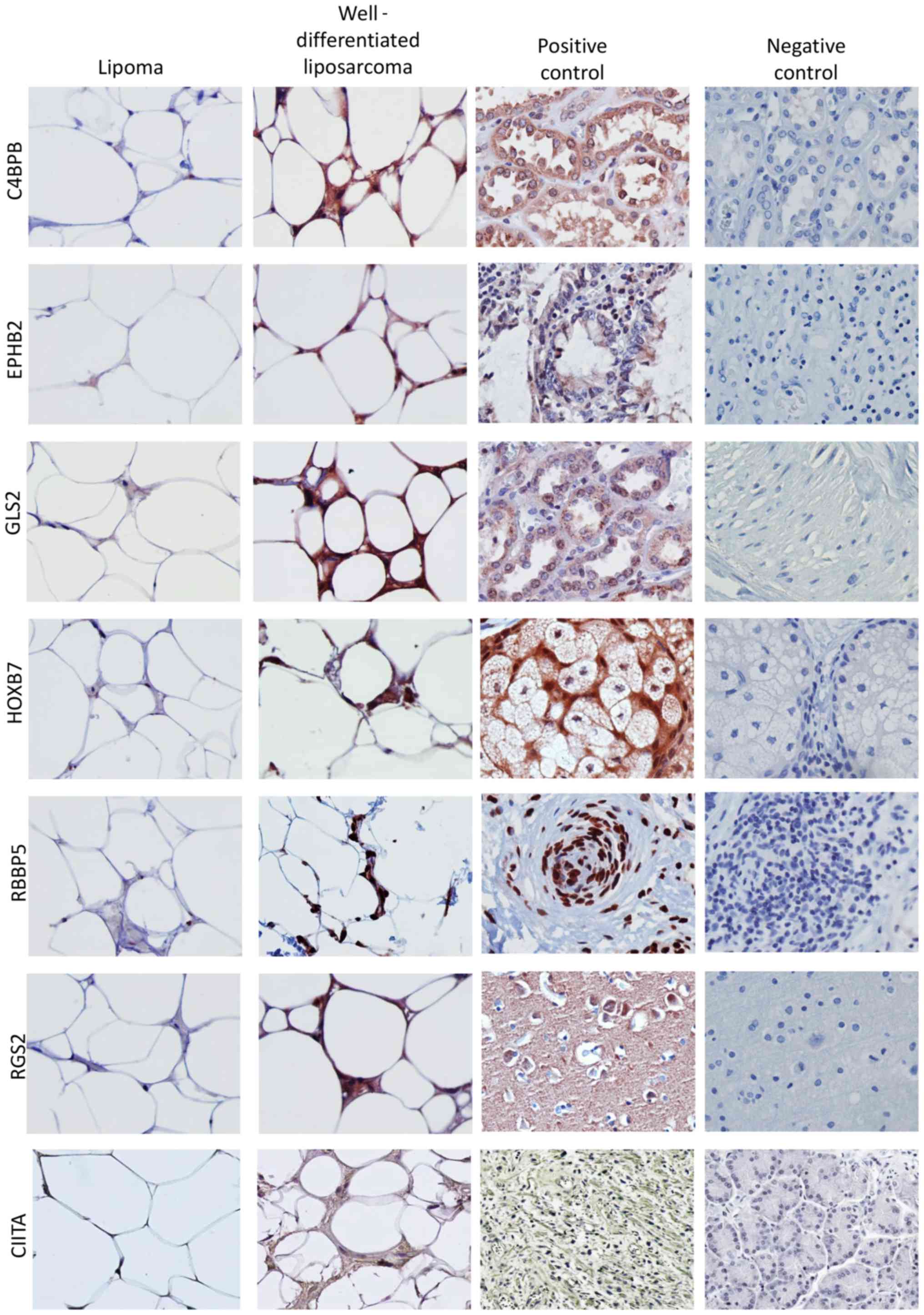
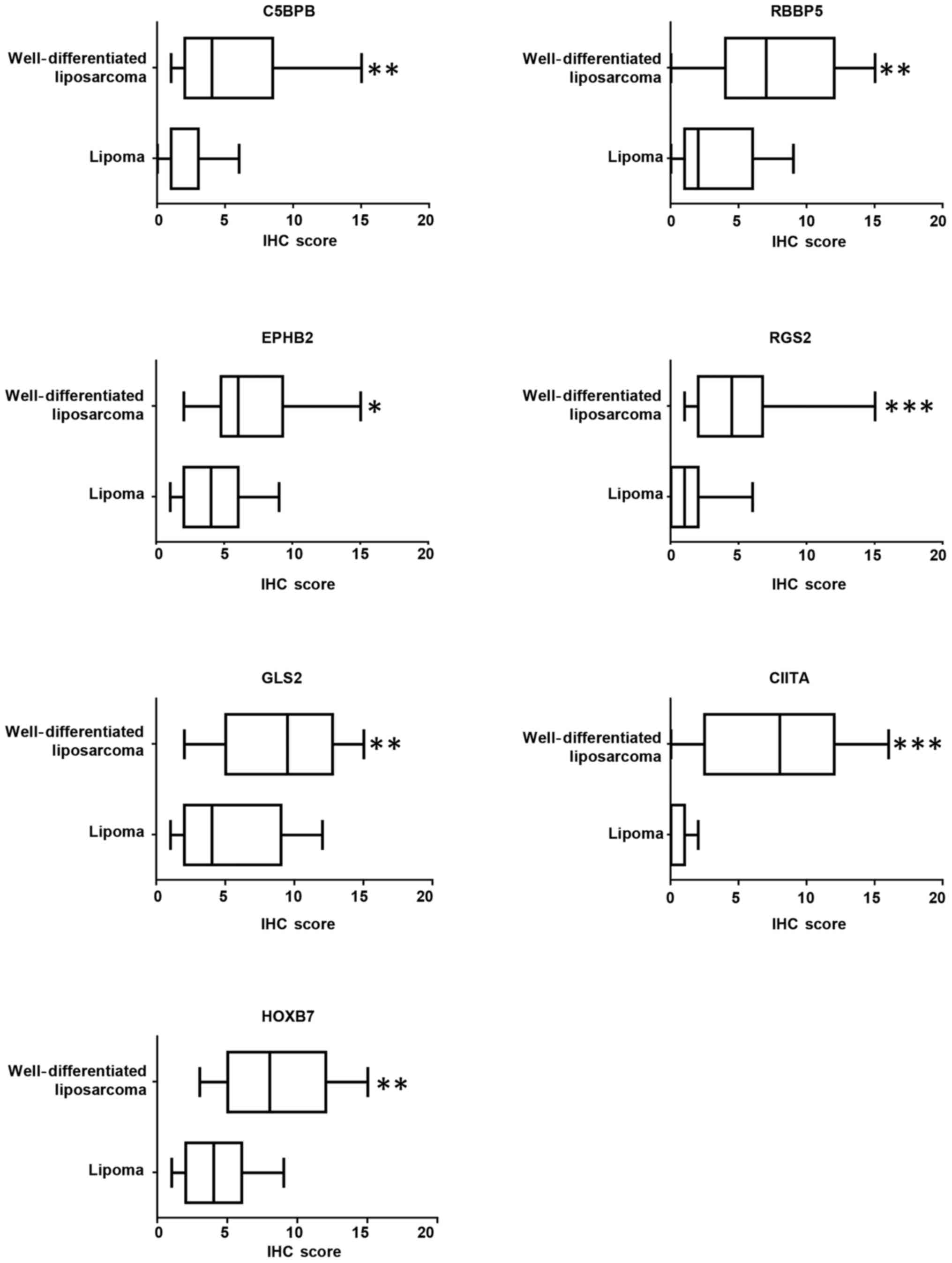
the groups. Semi-quantitative analysis of the staining across the
tumor panel demonstrated that C4BPB, CIITA, EPHB2, HOXB7, GLS2,
RBBP5, and RGS2 protein levels were increased in
well-differentiated liposarcomas compared to lipomas (Figs. 2 and 3). Despite changes in gene expression
observed in our meta-analysis, the IHC analysis did not detect
differences in the protein expression of ABCB11, BMP8A, PKN2,
RAB6B, SERINC2, SNCG, SOX30, or TSHR between lipomas and
well-differentiated liposarcomas (data not shown).
 | Table I.Patient characteristics. |
Table I.
Patient characteristics.
| Variables | Overall | Lipoma | WD liposarcoma |
|---|
| No. of patient
samples | 30 | 11 | 19 |
| Age, years (mean ±
SD) | 51±13 | 50±10 | 51±15 |
| Age (median
years) | 51 (60) | 53 (30) | 49 (60) |
| Sex (F/M) | 21/9 | 8/3 | 13/6 |
| Tumor location (no.
of tumors) |
|
|
|
|
Axilla | 4 | 1 | 3 |
| Back | 2 | 2 | 0 |
|
Breast | 2 | 2 | 0 |
|
Buttock | 1 | 0 | 1 |
| Head and
neck | 4 | 3 | 1 |
| Legs | 7 | 1 | 6 |
|
Mediastinum | 1 | 0 | 1 |
|
Mesentery | 1 | 0 | 1 |
|
Retroperitoneum | 5 | 0 | 5 |
|
Shoulder | 3 | 2 | 1 |
To confirm the statistical significance of our
findings, logistical regression models we utilized to assess the
individual discriminatory value of each protein to distinguish
between lipoma and well-differentiated liposarcoma. Due to the
large variation in IHC scores, a natural log transformation after
adding 1 was made for each differentially expressed protein.
Table II reveals the individual
model of each marker for differentiating well-differentiated
liposarcoma from lipoma. These individual models can be used to
differentiate two types of tumor groups, and all considered markers
were found to be significantly associated with well-differentiated
liposarcoma. In the unadjusted analysis, the highest strength of
association with well-differentiated liposarcoma as compared with
lipoma was observed for HOXB7 (OR=20.590) followed by CIITA
(OR=14.963) and RGS2 (OR=11.509). Table III shows the cut-off value of each
marker to be used for differentiating the two groups. The highest
classification accuracy was obtained using CIITA (≥2) with
sensitivity 77.8% and specificity 90.1% followed by RGS2 (≥2) with
sensitivity 66.7% and specificity 81.8% and GLS2 (≥6) with
sensitivity 72.2% and specificity 72.7%. The overall performance of
each model was reported in Table
IV. The maximum AUC was obtained for the CIITA model (0.894)
followed by RGS2 (0.854). According to AUCs, the individual
performances of C4BPB, GLS2, RBBP5 and HOXB7 models were found to
be almost similar. The lowest AUC was obtained for EPHB2 model.
This indicates that the individual CIITA model can be reliably used
for differentiating the two types of lipomatous tumors followed by
the RGS2 model.
 | Table II.Individual model for differentiating
well differentiated liposarcoma from lipoma. |
Table II.
Individual model for differentiating
well differentiated liposarcoma from lipoma.
| Model | RC | 95% CI | P-value | OR |
|---|
| C4BPB |
2.169 | 0.329,
4.009 | 0.021 |
8.751 |
| _cons | −2.347 | −4.753, 0.059 | 0.056 |
|
| EPHB2 |
1.672 | 0.030,
3.314 | 0.046 |
5.321 |
| _cons | −2.459 | −5.423, 0.505 |
0.0505 |
|
| GLS2 |
1.992 | 0.415,
3.569 | 0.013 |
7.331 |
| _cons | −3.278 | −6.332,
−0.223 | 0.035 |
|
| HOXB7 |
3.025 | 0.753,
5.297 | 0.009 | 20.590 |
| _cons | −5.135 | −9.384,
−0.886 | 0.018 |
|
| RBBP5 |
1.520 | 0.246,
2.794 | 0.019 |
4.572 |
| _cons | −2.001 | −4.267, 0.265 | 0.084 |
|
| RGS2 |
2.443 | 0.633,
4.253 | 0.008 | 11.509 |
| _cons | −2.393 | −4.608,
−0.179 | 0.034 |
|
| CIITA |
2.706 | 0.641,
4.771 | 0.010 | 14.963 |
| _cons | −2.320 | −4.314,
−0.326 | 0.023 |
|
 | Table III.Individual threshold of each marker
for differentiating well-differentiated liposarcoma from
lipoma. |
Table III.
Individual threshold of each marker
for differentiating well-differentiated liposarcoma from
lipoma.
|
| Se (%) | Sp (%) | Correctly
classified | LR+log
threshold |
|---|
| C4BPB | 66.7 | 63.6 | 65.5%, 1.83 | 1.39 (3) |
| EPHB2 | 61.1 | 63.6 | 62.1%, 1.68 | 1.95 (6) |
| GLS2 | 72.2 | 72.7 | 72.4%, 2.65 | 1.95 (6) |
| HOXB7 | 72.2 | 63.6 | 69.0%, 1.99 | 1.95 (6) |
| RBBP5 | 66.7 | 72.7 | 69.0%, 2.44 | 1.95 (6) |
| RGS2 | 66.7 | 81.8 | 72.4%, 3.67 | 1.39 (3) |
| CIITA | 77.8 | 90.1 | 82.8%, 8.56 | 1.10 (2) |
 | Table IV.Overall performance of each marker in
differentiating well differentiated liposarcoma from lipoma. |
Table IV.
Overall performance of each marker in
differentiating well differentiated liposarcoma from lipoma.
| Model | AUC | 95% CI |
|---|
| C4BPB | 0.790 | 0.619, 0.962 |
| EPHB2 | 0.732 | 0.547, 0.918 |
| GLS2 | 0.798 | 0.625, 0.971 |
| HOXB7 | 0.828 | 0.676, 0.980 |
| RBBP5 | 0.803 | 0.641, 0.965 |
| RGS2 | 0.854 | 0.710, 0.997 |
| CIITA | 0.894 | 0.783, 1.000 |
We developed a multi-protein model of these markers
to increase discriminatory ability. Our analysis revealed the
combined model with Ciita and Rgs2 provides the highest ability
(AUC=0.93) to differentiate between lipomas and well-differentiated
liposarcomas (Table V). The
following equation can be used for predicting probability of
well-differentiated liposarcoma as compared to lipoma using Citta
and Rgs2 IHC scores: P(WDL) = exp[-4.822 + (2.183 ×
logCiita) + (1.404 × logRgs2)]/{1 +
exp[-4.822 + (2.183 × logCiita) + (1.404 ×
logRgs2)]} where P(WDL) is the probability of the tumor
being a well-differentiated liposarcoma, and Ciita and Rgs2 are the
IHC scores for each protein, respectively. The cut-off value for
the combined model differentiated the two tumor types with
sensitivity at 83.3% and specificity at 90.9%.
 | Table V.Individual model for differentiating
liposarcoma as compared to lipoma. |
Table V.
Individual model for differentiating
liposarcoma as compared to lipoma.
| Model | RC | 95% Cl | P-value | OR |
|---|
| Rbbp5 | 1.538 | −0.300, 3.377 | 0.101 | 4.657 |
| Ciita | 2.784 | 0.496, 5.072 | 0.017 | 16.179 |
| _cons | −4.822 | −9.022, −0.622 | 0.024 |
|
Discussion
Differentiation between lipoma and
well-differentiated liposarcoma can be challenging, particularly in
core biopsies where tumor tissue is sparse and where the
histological features are very similar. These issues lead to
increased diagnostic costs because of the need to send uncertain
biopsies to tertiary referral centers for correct classification
(20,21). A recent publication confirmed that
MDM2 amplification detected by FISH, which is considered the gold
standard for distinction of lipomas from well-differentiated
liposarcomas, showed high concordance rates with tumor types when
firm histological diagnoses were made and the tissues were not
considered equivocal diagnoses (22). Despite this finding, the authors of
that demonstrated that MDM2 amplification also occurs in other
types of liposarcomas (22).
Additionally, previous findings have shown that diverse soft tissue
sarcomas harbor MDM2 amplifications in up to 40% of cases (23–26).
MDM2 amplification occurs in 31.6% of ‘possible’
well-differentiated liposarcomas (22), in 28.6% of ‘probably’
well-differentiated liposarcomas (22), and in 5% of large and deep-seated
lipomas (18). Since FISH is both
labor and cost intensive, development of more reliable and/or
cost-effective diagnostic biomarkers for these lipomatous tumors
may reduce cost and speed of diagnosis.
Various methods have been utilized to identify
disease biomarkers, and within the past decade omics-based
technologies have allowed the knowledge-guided computational
identification of diagnostic and prognostic markers of disease
characterization, progression, outcome, and treatment
susceptibilities (27). Such studies
have been previously employed for soft tissue sarcomas. For
instance, bioinformatics analysis has been utilized on microarray
data from diverse sets of soft tissue sarcomas revealing
well-defined gene networks that may effectively classify sarcoma
subtypes and serve as a useful tool for rational taxonomy and
diagnosis of tumors (19,28–30). In
this study, we performed a meta-analysis on a panel of lipomatous
tumor samples that were previously classified based on gene
expression analysis. We identified 301 genes whose mRNA expression
differed at least 2-fold between lipomas and well-differentiated
liposarcomas. Of these identified genes, we validated our findings
at the protein level for 15 of them, finding that over half of the
expression patterns identified at the mRNA level in our
meta-analysis correlated well at the protein level. Indeed, our
most reliable multiprotein model exhibited sensitivity at 83.3% and
specificity at 90.9% with regard to differentiating lipomas from
well-differentiated liposarcoma tumors.
References
|
1
|
Nguyen T and Zuniga R: Skin conditions:
benign nodular skin lesions. FP Essent. 407:24–30. 2013.PubMed/NCBI
|
|
2
|
Bancroft LW, Kransdorf MJ, Peterson JJ and
O'Connor MI: Benign fatty tumors: classification, clinical course,
imaging appearance, and treatment. Skeletal Radiol. 35:719–733.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Paunipagar BK, Griffith JF, Rasalkar DD,
Chow LTC, Kumta SM and Ahuja A: Ultrasound features of deep-seated
lipomas. Insights into Imaging. 1:149–153. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Miao C, Liu D, Zhang F, Wang Y, Zhang Y,
Yu J, Zhang Z, Liu G, Li B, Liu X and Luo C: Association of FPGS
genetic polymorphisms with primary retroperitoneal liposarcoma. Sci
Rep. 5:90792015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zagars GK, Goswitz MS and Pollack A:
Liposarcoma: outcome and prognostic factors following conservation
surgery and radiation therapy. Int J Radiat Oncol Biol Phys.
36:311–319. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Clay MR, Martinez AP, Weiss SW and Edgar
MA: MDM2 amplification in problematic lipomatous tumors: Analysis
of FISH testing criteria. Am J Surg Pathol. 39:1433–1439. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dei Tos AP: Liposarcoma: New entities and
evolving concepts. Ann Diagn Pathol. 4:252–266. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Brisson M, Kashima T, Delaney D, Tirabosco
R, Clarke A, Cro S, Flanagan AM and O'Donnell P: MRI
characteristics of lipoma and atypical lipomatous
tumor/well-differentiated liposarcoma: Retrospective comparison
with histology and MDM2 gene amplification. Skeletal Radiol.
42:635–647. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gaskin CM and Helms CA: Lipomas, lipoma
variants, and well-differentiated liposarcomas (atypical lipomas):
Results of MRI evaluations of 126 consecutive fatty masses. AJR Am
J Roentgenol. 182:733–739. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
O'Donnell PW, Griffin AM, Eward WC,
Sternheim A, White LM, Wunder JS and Ferguson PC: Can experienced
observers differentiate between lipoma and well-differentiated
liposarcoma using only MRI? Sarcoma. 2013.982784PubMed/NCBI
|
|
11
|
Hasegawa T, Yamamoto S, Nojima T, Hirose
T, Nikaido T, Yamashiro K and Matsuno Y: Validity and
reproducibility of histologic diagnosis and grading for adult
soft-tissue sarcomas. Hum Pathol. 33:111–115. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nilbert M, Rydholm A, Mitelman F, Meltzer
PS and Mandahl N: Characterization of the 12q13-15 amplicon in soft
tissue tumors. Cancer Genet Cytogenet. 83:32–36. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pilotti S, Torre G Della, Lavarino C, Di
Palma S, Sozzi G, Minoletti F, Rao S, Pasquini G, Azzarelli A,
Rilke F, et al: Distinct mdm2/p53 expression patterns in
liposarcoma subgroups: Implications for different pathogenetic
mechanisms. J Pathol. 181:14–24. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shimada S, Ishizawa T, Ishizawa K,
Matsumura T, Hasegawa T and Hirose T: The value of MDM2 and CDK4
amplification levels using real-time polymerase chain reaction for
the differential diagnosis of liposarcomas and their histologic
mimickers. Hum Pathol. 37:1123–1129. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sirvent N, Coindre JM, Maire G, Hostein I,
Keslair F, Guillou L, Ranchere-Vince D, Terrier P and Pedeutour F:
Detection of MDM2-CDK4 amplification by fluorescence in situ
hybridization in 200 paraffin-embedded tumor samples: Utility in
diagnosing adipocytic lesions and comparison with
immunohistochemistry and real-time PCR. Am J Surg Pathol.
31:1476–1489. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sozzi G, Minoletti F, Miozzo M, Sard L,
Musso K, Azzarelli A, Pierotti MA and Pilotti S: Relevance of
cytogenetic and fluorescent in situ hybridization analyses in the
clinical assessment of soft tissue sarcoma. Hum Pathol. 28:134–142.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Weiss SW: Lipomatous tumors. Monogr
Pathol. 38:207–239. 1996.PubMed/NCBI
|
|
18
|
Wong DD, Low IC, Peverall J, Robbins PD,
Spagnolo DV, Nairn R, Carey-Smith RL and Wood D: MDM2/CDK4 gene
amplification in large/deep-seated ‘lipomas’: Incidence, predictors
and clinical significance. Pathology. 48:203–209. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nakayama R, Nemoto T, Takahashi H, Ohta T,
Kawai A, Seki K, Yoshida T, Toyama Y, Ichikawa H and Hasegawa T:
Gene expression analysis of soft tissue sarcomas: Characterization
and reclassification of malignant fibrous histiocytoma. Mod Pathol.
20:749–759. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Arbiser ZK, Folpe AL and Weiss SW:
Consultative (expert) second opinions in soft tissue pathology.
Analysis of problem-prone diagnostic situations. Am J Clin Pathol.
116:473–476. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Thway K and Fisher C: Histopathological
diagnostic discrepancies in soft tissue tumours referred to a
specialist centre. Sarcoma. 2009:7419752009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Thway K, Wang J, Swansbury J, Min T and
Fisher C: Fluorescence in situ hybridization for MDM2 amplification
as a routine ancillary diagnostic tool for suspected
well-differentiated and dedifferentiated liposarcomas: Experience
at a tertiary center. Sarcoma. 2015:8120892015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kimura H, Dobashi Y, Nojima T, Nakamura H,
Yamamoto N, Tsuchiya H, Ikeda H, Sawada-Kitamura S, Oyama T and Ooi
A: Utility of fluorescence in situ hybridization to detect MDM2
amplification in liposarcomas and their morphological mimics. Int J
Clin Exp Pathol. 6:1306–1316. 2013.PubMed/NCBI
|
|
24
|
Miura Y, Keira Y, Ogino J, Nakanishi K,
Noguchi H, Inoue T and Hasegawa T: Detection of specific genetic
abnormalities by fluorescence in situ hybridization in soft tissue
tumors. Pathol Int. 62:16–27. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nakayama T, Toguchida J, Wadayama B, Kanoe
H, Kotoura Y and Sasaki MS: MDM2 gene amplification in bone and
soft-tissue tumors: Association with tumor progression in
differentiated adipose-tissue tumors. Int J Cancer. 64:342–346.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Weaver J, Downs-Kelly E, Goldblum JR,
Turner S, Kulkarni S, Tubbs RR, Rubin BP and Skacel M: Fluorescence
in situ hybridization for MDM2 gene amplification as a diagnostic
tool in lipomatous neoplasms. Mod Pathol. 21:943–949. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Diamandis EP: Present and future of cancer
biomarkers. Clin Chem Lab Med. 52:791–794. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hadj-Hamou NS, Laé M, Almeida A, de la
Grange P, Kirova Y, Sastre-Garau X and Malfoy B: A transcriptome
signature of endothelial lymphatic cells coexists with the chronic
oxidative stress signature in radiation-induced post-radiotherapy
breast angiosarcomas. Carcinogenesis. 33:1399–1405. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Skubitz KM and Skubitz AP:
Characterization of sarcomas by means of gene expression. J Lab
Clin Med. 144:78–91. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tran D, Verma K, Ward K, Diaz D, Kataria
E, Torabi A, Almeida A, Malfoy B, Stratford EW, Mitchell DC, et al:
Functional genomics analysis reveals a MYC signature associated
with a poor clinical prognosis in liposarcomas. Am J Pathol.
185:717–728. 2015. View Article : Google Scholar : PubMed/NCBI
|