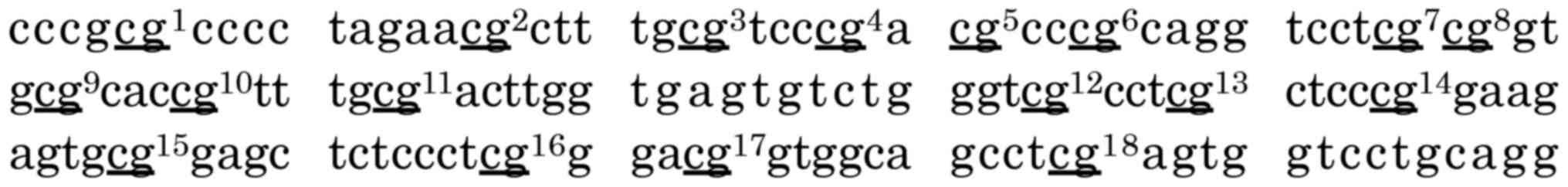Introduction
Medulloblastoma is a highly malignant brain tumor
that predominately affects children. The standard treatment of
medulloblastoma is surgery followed by radiotherapy and
chemotherapy using alkylating agents (1–3).
O6-methylguanine-DNA methyltransferase
(MGMT) is a DNA repair enzyme that plays an important role in tumor
resistance to alkylating agents. MGMT removes alkyl adducts from
the O6-position of guanine by inactivating itself. As
O6-alkylated guanine leads to double-strand breaks and
base mispairing, which eventually induces cell apoptosis, MGMT
protects normal cells as well as tumor cells from alkylating agents
(4). Expression of MGMT is
suppressed by methylation of CpG islands in the promoter region
(4–6). It was previously demonstrated that a
greater methylation status of the MGMT promoter region is
associated with favorable outcomes in adult and pediatric patients
with glioblastoma treated with alkylating agents, such as
temozolomide (7,8). However, only a limited number of
studies have investigated MGMT status in medulloblastoma and
its effect on disease outcome (5,9–11).
The aim of the present study was to determine the
methylation status of CpG sites in the MGMT promoter region
in tumor cells obtained from medulloblastoma patients and evaluate
the association between MGMT status and clinical
outcome.
Patients and methods
Patients
The records of pediatric patients with
medulloblastoma treated at Juntendo University Hospital (Tokyo,
Japan) between 1995 and 2012 were reviewed. Patients who underwent
institutional standard treatment for medulloblastoma (initial tumor
removal, craniospinal irradiation and chemotherapy) and who were
observed for at least 36 months after diagnosis, or who experienced
relapse of the disease after initiation of chemotherapy were
included in the study. Patients were excluded if their treatment
regimen deviated significantly from the standard treatment, such as
omission of radiotherapy, or underwent biopsy as the only surgical
intervention. Relevant clinical information, including current
disease status, was obtained from hospital charts. The study was
approved by the Juntendo University Ethics Committee, and written
informed consent was obtained from all the patients and/or their
legal guardians.
Analysis of MGMT status
Tumor tissues obtained at the first surgery for
tumor removal were used for analysis. Genomic DNA was extracted
from paraffin-embedded samples with deparaffinization solution and
the QIAamp DNA FFPE Tissue kit (Qiagen, Hilden, Germany). DNA from
each sample (300 mg) was treated with sodium bisulfite using the
Cells-to-CpG Bisulfite Conversion kit (Applied Biosystems, Foster
City, CA, USA).
The direct sequence method was used to analyze
bisulfite-treated DNA. MGMT promoter primers were designed
to cover 18 CpG sites (chr10:129467232-129467363 GenBank) by Methyl
Primer Express software v1.0 (Applied Biosystems) (Fig. 1). Two polymerase chain reaction (PCR)
products were made, namely product 1 (99 bp) and product 2 (89 bp).
Product 1 primers were as follows: Forward, GGA TAT GTT GGG ATA GTT
YG; and reverse, ACC CAA ACA CTC ACC AAA T. Product 2 primers were
as follows: Forward, ATT TGG TGA GTG TTT GGG; and reverse, ACR CCT
ACA AAA CCA CTC. PCR was performed by a two-step approach using
AmpliTaqGold 360 Master Mix (Applied Biosystems). PCR products were
sequenced on an ABI 3130 Genetic Analyzer (Applied Biosystems) with
the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied
Biosystems). Sequences were analyzed with SeqScape software v3.0
(Applied Biosystems). Genomic data were based on the GRCh38/hg38
assembly from the University of California Santa Cruz Genome
Browser (http://genome.ucsc.edu/), accessed
December 2013.
Statistical analysis
Differences between groups were analyzed by
Mann-Whitney U-test using GraphPad Prism 4 software (San Diego,
CA). P-values <0.05 were considered to indicate statistically
significant differences.
Results
Characteristics and clinical outcomes
of the patients
A total of 22 patients with available tumor tissue
and clinical data were identified. Of those, 7 patients were
excluded due to the following reasons: 3 were followed up for
<36 months after the diagnosis with no disease recurrence; 1 did
not receive radiotherapy due to being aged <24 months at
diagnosis; 2 received biopsy only as the initial surgical
intervention due to massive dissemination; and 1 experienced tumor
relapse prior to the initiation of radiotherapy and
chemotherapy.
A total of 15 patients were finally included in the
present study (Table I). The median
age at diagnosis was 9 years (range, 2–15 years) and the median
follow-up period was 41 months (range, 13–193 months). In all the
patients, the tumor was located in the fourth ventricle at the
midline of the cerebellum. The patients received multiple courses
of chemotherapy consisting of ifosfamide, cisplatin and etoposide
(n=8); cisplatin, vincristine and cyclophosphamide (n=3); or
ifosfamide, cisplatin, etoposide, vincristine and cyclophosphamide
(n=4). Following first-line treatment, 9 patients achieved complete
remission and 6 patients relapsed.
 | Table I.Patient characteristics and clinical
outcomes. |
Table I.
Patient characteristics and clinical
outcomes.
| Patient | Age at diagnosis
(years) | Sex | Pathological
classification | Dissemination | Surgery | Outcome after
first-line treatment |
|---|
| 1 | 11 | Female | Classic | − | GTR | CR |
| 2 | 7 | Female | Anaplastic | − | STR | CR |
| 3 | 5 | Female | Classic | − | STR | CR |
| 4 | 5 | Male | Desmoplastic | − | STR | CR |
| 5 | 4 | Male | Nodular | + | GTR | CR |
| 6 | 10 | Female | Anaplastic | + | GTR | CR |
| 7 | 11 | Male | Classic | − | STR | CR |
| 8 | 12 | Male | Anaplastic | − | STR | CR |
| 9 | 9 | Male | Classic | − | STR | CR |
| 10 | 10 | Male | Classic | − | STR | Relapse |
| 11 | 2 | Male | Anaplastic | − | STR | Relapse |
| 12 | 7 | Male | Classic | − | STR | Relapse |
| 13 | 15 | Female | Anaplastic | + | STR | Relapse |
| 14 | 11 | Male | Anaplastic | + | GTR | Relapse |
| 15 | 8 | Male | Classic | − | GTR | Relapse |
Analysis of MGMT status
The methylation status of 18 CpG sites of the
MGMT promoter region is shown in Table II. CpG sites with unmethylated
cytosine are displayed as thymine in the final sequences and are
indicated as ‘T’, whereas CpG sites with methylated cytosine are
indicated as ‘C’. Certain samples displayed mixed cytosine and
thymine signals and are indicated as ‘Y’. Among 270 CpG sites
analyzed, 59 (21.9%) were methylated and 170 (63.0%) were
unmethylated, whereas mixed signals were observed in 41 sites
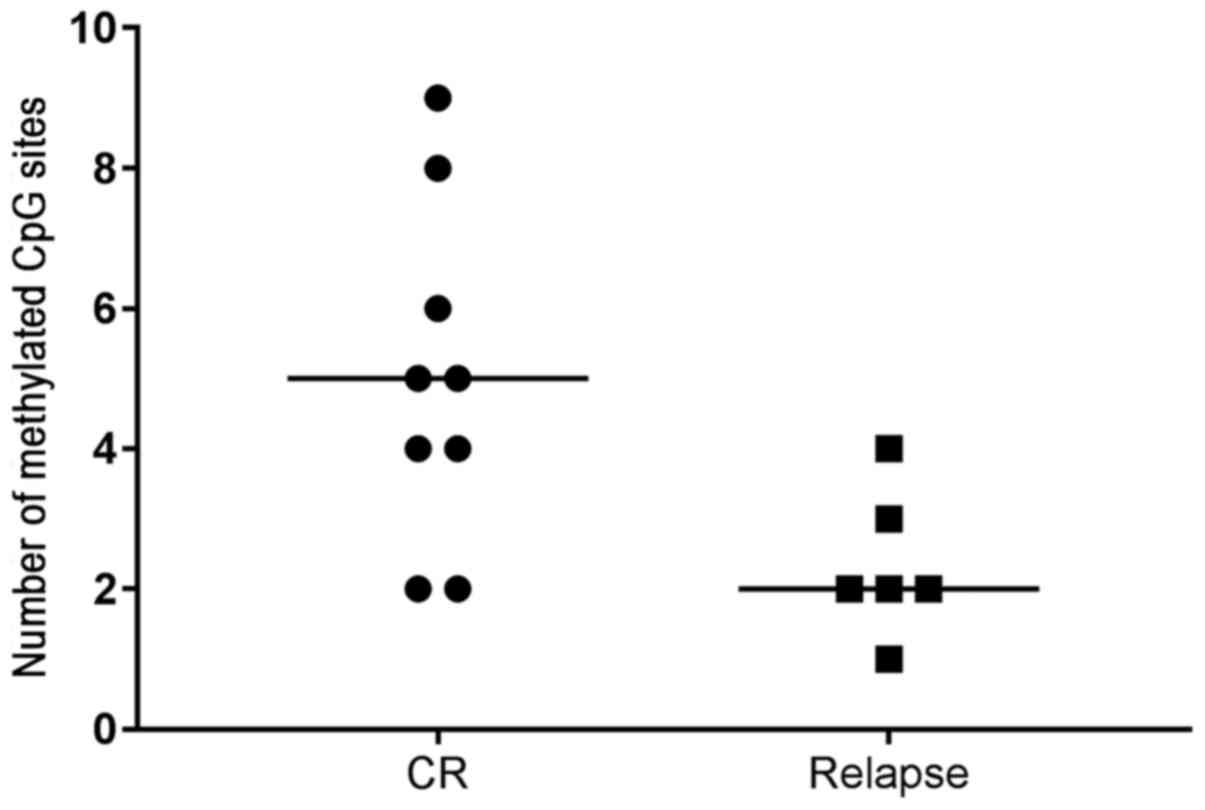
(15.2%). A higher number of methylated CpG sites was observed in
patients with complete remission compared with that in patients who
relapsed (P=0.041) (Fig. 2).
 | Table II.Methylation status of CpG sites in
the MGMT promoter region. |
Table II.
Methylation status of CpG sites in
the MGMT promoter region.
|
|
|
CpG
site |
|---|
|
|
|
|
|---|
| Patient | Outcome | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 |
|---|
| 1 | CR | T | Y | T | T | T | Y | C | Y | Y | Y | Y | C | C | C | T | T | T | T |
| 2 | CR | T | T | T | Y | C | T | Y | T | T | Y | T | T | T | T | T | T | T | C |
| 3 | CR | T | T | T | C | T | C | Y | T | T | Y | T | T | T | T | C | C | Y | C |
| 4 | CR | T | C | C | T | T | T | C | T | T | C | Y | T | T | T | T | T | T | T |
| 5 | CR | T | T | C | T | T | T | T | T | T | C | T | Y | T | C | T | C | C | T |
| 6 | CR | T | C | T | Y | T | T | T | Y | Y | C | Y | T | T | T | T | T | T | T |
| 7 | CR | T | T | T | T | T | C | C | C | C | T | Y | T | T | C | T | T | T | C |
| 8 | CR | C | T | C | T | T | C | C | C | C | T | T | T | T | C | T | C | Y | C |
| 9 | CR | C | C | C | T | C | C | T | T | T | C | T | T | T | T | T | C | C | T |
| 10 | Relapse | T | Y | T | Y | T | T | Y | T | Y | Y | Y | T | T | T | C | T | T | Y |
| 11 | Relapse | T | T | C | T | T | Y | Y | T | Y | Y | T | T | T | T | T | T | Y | C |
| 12 | Relapse | T | T | C | T | T | C | T | T | T | C | T | T | C | T | T | Y | T | T |
| 13 | Relapse | T | T | C | T | T | T | T | C | T | C | T | T | T | T | T | T | T | T |
| 14 | Relapse | C | T | T | T | T | T | T | C | T | T | T | T | T | T | T | T | T | T |
| 15 | Relapse | T | C | T | Y | Y | Y | T | Y | Y | Y | Y | T | T | Y | T | C | T | T |
Discussion
There was variability in the MGMT status
among medulloblastoma tumor samples, and an association was
observed between more extensive MGMT promoter methylation
and favorable clinical outcome of medulloblastoma. Previous studies
on the effect of MGMT status on the treatment of
medulloblastoma yielded conflicting results. Neben et al
reported that high levels of MGMT expression were associated
with unfavorable survival outcome using microarray-based screening
of 35 medulloblastomas (10). Bobola
et al observed that MGMT expression is a major
determinant of carmustine and temozolomide sensitivity in
medulloblastoma cell lines (12).
However, Faoro et al reported no association between
MGMT mRNA expression and progression-free or overall
survival of medulloblastoma patients (5).
These differences in the study results may be caused
in part by heterogeneity of chemotherapy regimens among studies. In
the study by Neben et al, the patients were treated with
lomustine, cisplatin and vincristine (10). In the study by Faoro et al,
the patients were treated as reported in the randomized trial
HIT'91, which consisted of two chemotherapy arms: One treated with
procarbazine, ifosfamide, etoposide, high-dose methotrexate,
cisplatin and cytarabine, and the other with cisplatin, lomustine
and vincristine. Recently, several genes were found to play
important roles in the pharmacokinetics or pharmacodynamics of
chemotherapy agents, such as the role of polymorphism of reduced
folate carrier 1 and methylenetetetrahydrofolate reductase in
high-dose methotrexate treatment (13). Therefore, the effect of MGMT
status on the survival of patients with medulloblastoma may vary
with different combinations of chemotherapy agents.
Another factor that may cause conflicting results
among studies is the complexity of determining MGMT status.
For example, a study using commercially available anti-MGMT
antibodies to determine MGMT expression reported major
interobserver variability (9,14).
Furthermore, as MGMT promoter methylation is inversely
correlated with MGMT expression (5,6),
methylation-specific PCR (MSP) with bisulfate-treated DNA has been
widely used to analyze MGMT promoter methylation status
(7–9). However, MSP is only able to detect a
limited number of methylated CpG sites in the primer region, and
recent studies report that the region commonly investigated by MSP
does not cover CpG sites that are most highly associated with
expression of MGMT, and that MSP may not be well-suited for
predicting the prognosis of patients with glioblastoma (15,16).
Direct sequencing and pyrosequencing are alternative methods for
quantitatively analyzing the methylation status of MGMT. As
pyrosequencing is effective for high-throughput screening, but is
quite costly, the direct sequence method was used to determine the
methylation status of selected CpG sites in the present study.
The MGMT promoter contains a 762-bp CpG
island with 98 CpG sites, with certain CpG regions reflecting
MGMT expression better than others. Everhard et al
reported CpG sites at +95, +113, +135 and +137 bp from the
transcriptional start site (TSS) (CpG 1, 3, 7 and 8 in our study,
respectively) and high concordance between methylation and
expression of MGMT in their analysis of 53 CpG sites in 54
glioblastoma samples (15). Malley
et al reported that individual or multiple consecutive
methylation of CpG sites at +153, +185, +195 and +213 bp from the
TSS (CpG 11, 14, 15 and 17 in our study, respectively) attenuated
the activity of the MGMT promoter in their study of 98 CpG
sites in xenografted glioblastoma samples and cell lines (6). Although we were unable to determine
whether methylation at specific CpG sites was more closely
associated with prognosis than others, due to our limited sample
size, our results suggest that the overall CpG methylation profile
of the targeted region in the present study is associated with the
outcome of medulloblastoma.
Recent rapid advances in genetic techniques
currently allow the subdivision of medulloblastoma into four
molecular subgroups with distinct demographics, clinical
presentations and clinical outcomes (17,18).
Unfortunately, the cases were not classified into molecular
subgroups at the time of diagnosis. However, Von Bueren et
al reported similar MGMT expression levels in the WNT and SHH
groups, that are higher compared with those of group 3 and group 4,
although there is wide variation even within groups (11). Considering that the prognosis of WNT
group patients is better compared with that of SHH group patients,
the MGMT methylation status may be an independent factor
affecting the prognosis of medulloblastoma.
The results of the present study should be
interpreted with caution. First, the methylation status of
MGMT was assessed in primary tumors resected prior to
initiation of radiotherapy and chemotherapy; however, radiotherapy
may affect MGMT methylation status and upregulate expression
of the gene (19). Chemotherapy may
also affect the MGMT status of the tumors; thus, relapsed
tumors may exhibit different methylation profiles. Temozolomide is
an alkylating agent that is increasingly used for relapsed
medulloblastoma (20–22). Although a significant correlation
between MGMT methylation status and temozolomide sensitivity
has been confirmed in glioblastoma (7,8),
sensitivity to temozolomide in relapsed medulloblastoma may not be
predicted from the MGMT status of primarily resected tumor
samples. Second, the small sample size prevented further analysis
of other clinical factors that may be associated with patient
outcome, such as molecular subtypes, pathological characteristics,
dissemination status and chemotherapy regimens.
In conclusion, there was variability in the
methylation status of the MGMT promoter region among tumor
samples from pediatric medulloblastoma patients using the direct
sequencing method. Our results indicate that a larger number of
methylated CpG sites in the MGMT promoter region is
associated with a favorable outcome of medulloblastoma.
Acknowledgements
The present study was supported by the Graduate
School Research Program of Juntendo University. The authors would
like to thank everyone who helped conduct this research at Juntendo
University and Juntendo University Hospital.
References
|
1
|
Packer RJ, Zhou T, Holmes E, Vezina G and
Gajjar A: Survival and secondary tumors in children with
medulloblastoma receiving radiotherapy and adjuvant chemotherapy:
Results of children's oncology group trial A9961. Neuro Oncol.
15:97–103. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jakacki RI, Burger PC, Zhou T, Holmes EJ,
Kocak M, Onar A, Goldwein J, Mehta M, Packer RJ, Tarbell N, et al:
Outcome of children with metastatic medulloblastoma treated with
carboplatin during craniospinal radiotherapy: A children's oncology
group phase I/II study. J Clin Oncol. 30:2648–2653. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gajjar A, Chintagumpala M, Ashley D,
Kellie S, Kun LE, Merchant TE, Woo S, Wheeler G, Ahern V, Krasin
MJ, et al: Risk-adapted craniospinal radiotherapy followed by
high-dose chemotherapy and stem-cell rescue in children with newly
diagnosed medulloblastoma (St Jude Medulloblastoma-96): Long-term
results from a prospective, multicentre trial. Lancet Oncol.
7:813–820. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gerson SL: MGMT: Its role in cancer
aetiology and cancer therapeutics. Nat Rev Cancer. 4:296–307. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Faoro D, von Bueren AO, Shalaby T,
Sciuscio D, Hürlimann ML, Arnold L, Gerber NU, Haybaeck J,
Mittelbronn M, Rutkowski S, et al: Expression of
O6-methylguanine-DNA methyltransferase in childhood
medulloblastoma. J Neurooncol. 103:59–69. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Malley DS, Hamoudi RA, Kocialkowski S,
Pearson DM, Collins VP and Ichimura K: A distinct region of the
MGMT CpG island critical for transcriptional regulation is
preferentially methylated in glioblastoma cells and xenografts.
Acta Neuropathol. 121:651–661. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hegi ME, Diserens AC, Gorlia T, Hamou MF,
de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani
L, et al: MGMT gene silencing and benefit from temozolomide in
glioblastoma. N Engl J Med. 352:997–1003. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Donson AM, Addo-Yobo SO, Handler MH, Gore
L and Foreman NK: MGMT promoter methylation correlates with
survival benefit and sensitivity to temozolomide in pediatric
glioblastoma. Pediatr Blood Cancer. 48:403–407. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rood BR, Zhang H and Cogen PH:
Intercellular heterogeneity of expression of the MGMT DNA repair
gene in pediatric medulloblastoma. Neuro Oncol. 6:200–207. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Neben K, Korshunov A, Benner A, Wrobel G,
Hahn M, Kokocinski F, Golanov A, Joos S and Lichter P:
Microarray-based screening for molecular markers in medulloblastoma
revealed STK15 as independent predictor for survival. Cancer Res.
64:3103–3111. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
von Bueren AO, Bacolod MD, Hagel C,
Heinimann K, Fedier A, Kordes U, Pietsch T, Koster J, Grotzer MA,
Friedman HS, et al: Mismatch repair deficiency: A temozolomide
resistance factor in medulloblastoma cell lines that is uncommon in
primary medulloblastoma tumours. Br J Cancer. 107:1399–1408. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bobola MS, Silber JR, Ellenbogen RG, Geyer
JR, Blank A and Goff RD: O6-methylguanine-DNA methyltransferase,
O6-benzylguanine, and resistance to clinical alkylators in
pediatric primary brain tumor cell lines. Clin Cancer Res.
11:2747–2755. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jabeen S, Holmboe L, Alnaes GI, Andersen
AM, Hall KS and Kristensen VN: Impact of genetic variants of RFC1,
DHFR and MTHFR in osteosarcoma patients treated with high-dose
methotrexate. Pharmacogenomics J. 15:385–390. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Preusser M, Charles Janzer R, Felsberg J,
Reifenberger G, Hamou MF, Diserens AC, Stupp R, Gorlia T, Marosi C,
Heinzl H, et al: Anti-O6-methylguanine-methyltransferase (MGMT)
immunohistochemistry in glioblastoma multiforme: Observer
variability and lack of association with patient survival impede
its use as clinical biomarker. Brain Pathol. 18:520–532.
2008.PubMed/NCBI
|
|
15
|
Everhard S, Tost J, El Abdalaoui H,
Crinière E, Busato F, Marie Y, Gut IG, Sanson M, Mokhtari K,
Laigle-Donadey F, et al: Identification of regions correlating MGMT
promoter methylation and gene expression in glioblastomas. Neuro
Oncol. 11:348–356. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kanemoto M, Shirahata M, Nakauma A,
Nakanishi K, Taniguchi K, Kukita Y, Arakawa Y, Miyamoto S and Kato
K: Prognostic prediction of glioblastoma by quantitative assessment
of the methylation status of the entire MGMT promoter region. BMC
Cancer. 14:6412014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Northcott PA, Korshunov A, Witt H,
Hielscher T, Eberhart CG, Mack S, Bouffet E, Clifford SC, Hawkins
CE, French P, et al: Medulloblastoma comprises four distinct
molecular variants. J Clin Oncol. 29:1408–1414. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Taylor MD, Northcott PA, Korshunov A,
Remke M, Cho YJ, Clifford SC, Eberhart CG, Parsons DW, Rutkowski S,
Gajjar A, et al: Molecular subgroups of medulloblastoma: The
current consensus. Acta Neuropathol. 123:465–472. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Grombacher T, Mitra S and Kaina B:
Induction of the alkyltransferase (MGMT) gene by DNA damaging
agents and the glucocorticoid dexamethasone and comparison with the
response of base excision repair genes. Carcinogenesis.
17:2329–2336. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cefalo G, Massimino M, Ruggiero A, Barone
G, Ridola V, Spreafico F, Potepan P, Abate ME, Mascarin M, Garrè
ML, et al: Temozolomide is an active agent in children with
recurrent medulloblastoma/primitive neuroectodermal tumor: An
Italian multi-institutional phase II trial. Neuro Oncol.
16:748–753. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nicholson HS, Kretschmar CS, Krailo M,
Bernstein M, Kadota R, Fort D, Friedman H, Harris MB, Tedeschi-Blok
N, Mazewski C, et al: Phase 2 study of temozolomide in children and
adolescents with recurrent central nervous system tumors: A report
from the Children's Oncology Group. Cancer. 110:1542–1550. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Grill J, Geoerger B, Gesner L, Perek D,
Leblond P, Cañete A, Aerts I, Madero L, de Toledo Codina JS,
Verlooy J, et al: Phase II study of irinotecan in combination with
temozolomide (TEMIRI) in children with recurrent or refractory
medulloblastoma: A joint ITCC and SIOPE brain tumor study. Neuro
Oncol. 15:1236–1243. 2013. View Article : Google Scholar : PubMed/NCBI
|