Introduction
MicroRNAs (miRNAs, miRs) are ~22-nucleotide long,
single-stranded non-coding RNA molecules (1). They were first discovered in the
nematode Caenorhabditis elegans with the identification of
the developmental regulator lin-4 (2). Thus far, there are ~2,588 annotated
miRNAs found in the human genome (3). With advances in research, it has been
demonstrated that miRNAs may play an important role in various
diseases. An increasing number of miRNAs were proven to participate
in crucial biological processes, such as cell proliferation,
migration, invasion and apoptosis (4–7), which
may enable use of the miRNA family in the diagnosis and treatment
of disease, due to the extensive alterations in miRNA expression in
different diseases.
miR-23a/24-2/27a encodes a ~2,159-nt pri-miRNA
transcript, which is located in chromosome 19p13.12 as an
intergenic miRNA cluster (8). The
profiling analysis suggested that the miR-23a/24-2/27a cluster was
significantly upregulated in hepatocellular carcinoma (HCC)
(9), pancreatic adenocarcinoma
(10) and breast cancer (11). There are several studies on the
association of the miR-23a/24-2/27a cluster with various types of
cancer. Thus, the present systematic review and meta-analysis were
designed to confirm whether miR-23a/24-2/27a may serve as a
diagnostic marker for cancer.
Data collection methods
Literature search
Two of the authors (Jing Quan and Suyue Liu)
independently conducted a search through PubMed, Medline and the
Cochrane Library to identify studies on miR-23a/24-2/27a and
cancer. The databases were searched from inception to September 26,
2016. In order to distinguish between miR-24-1 (also referred to as
miR-189 and miR-24-1*) and miR-24-2 (also referred to as miR-24
precursor-19 and miR-24-2*), the miRBase was searched. The
following search terms were used: (miR-23a or microRNA-23a or
has-miR-23a or miR-24 or microRNA-24 or has-miR-24 or miR-27a or
microRNA-27a or has-miR-27a) and (cancer or neoplasm or carcinoma
or tumor).
Inclusion and exclusion criteria
All the studies met the following criteria: i) They
were studies on miR-23a or miR-24 or miR-27a expression in cancer
patients; ii) they used tissue samples obtained from surgically
resected tumors and neighboring non-cancerous or normal tissues for
comparison; iii) the expression of miR-23a or miR-24 or miR-27a was
measured by quantitative polymerase chain reaction (qPCR) or
reverse transcription qPCR (RT-qPCR) analysis; iv) the association
between the expression level and survival outcome was clearly
demonstrated.
The exclusion criteria were as follows: i)
Duplicated studies; ii) studies without a control group; iii)
insufficient data; iv) meetings, reviews and meta-analysis articles
on animal and cell studies.
Data extraction and quality
assessment
Two independent authors (Jing Quan and Kangfu Dai)
extracted the following information from the studies: First author,
publication year, country, type of miRNA, type of cancer, role of
the gene, validation sample, number of the cases, survival
analysis, hazard ratio (HR), months of follow-up and quality
scores. The Newcastle-Ottawa Scale (NOS) was used to assess the
quality of the selected studies (a score of >5 was considered as
high quality).
Statistical analysis
The HR and associated 95% confidence interval (CI)
for each study was used to estimate the survival outcome of cancer
associated with miR-23a or miR-24 or miR-27a expression.
Heterogeneity of combined HRs was assessed by Cochran's Q test and
Higgin's I2 statistic. Heterogeneity was considered
statistically significant when P<0.05 or I2>50%.
In order to evaluate the association between miR-23a or miR-24 or
miR-27a expression and survival rate, a fixed-effects or
random-effects model was used to calculate the pooled HR. A
fixed-effects model (Mantel-Haenszel test) was applied in the
absence of between-study heterogeneity (P≥0.05 or
I2≤50%), while the random-effects model (Der Simonian
and Laird method) was applied if significant heterogeneity was
observed (P<0.05 or I2>50%). Stata 14.0 software
(StataCorp, College Station, TX, USA) was used to analyze the data
from the studies and construct the forest plot. The potential
publication bias among the included studies was assessed by Begg's
funnel plot and Egger's bias indicator test. P<0.05 in all the
two-sided statistical tests was considered to indicate
statistically significant differences.
Results
Basic information from the included
studies
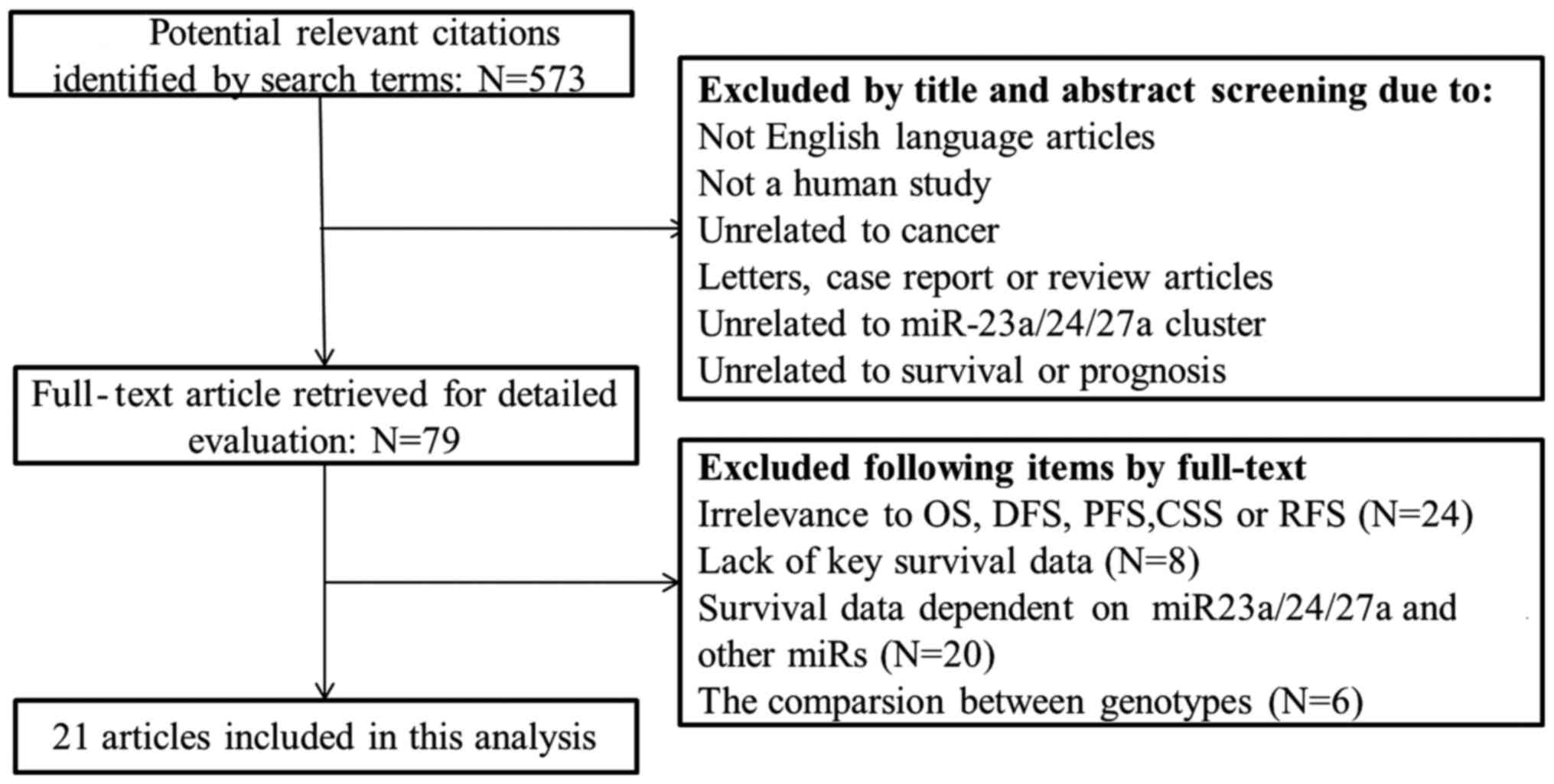
A search through PubMed, Medline and the Cochrane
Library identified 572 potentially relevant studies. A total of 79
full-text articles were selected for detailed evaluation following
exclusion of studies that were not in English, not human, unrelated
to cancer, letters, case reports or review articles, unrelated to
miR-23a/24-2/27a cluster, or unrelated to survival or prognosis.
Further selection depended on relevance to overall survival (OS),
recurrence-free survival (RFS), progression-free survival (PFS),
cancer-specific survival, key survival data and survival data on
miR-23a/24-2/27a, without any other miRNAs. Finally, 21 studies
were included in the analysis. The flow chart of the study
selection process is shown in Fig.
1.
The 21 studies included in this meta-analysis
included 8 studies on miR-23a, 6 on miR-24 and 8 on miR-27a
(12–32). A total of 1,974 cases were included,
including diffuse large B-cell lymphoma, laryngeal cancer,
non-small-cell lung cancer (NSCLC), prostate cancer, hepatocellular
carcinoma (HCC), colorectal cancer and renal cell carcinoma (RCC).
The NOS score was used by two independent authors to determine
study quality, and the scores of all the studies were >5. The
basic information from the 21 studies is summarized in Table I.
 | Table I.Basic characteristics of the 21
included studies. |
Table I.
Basic characteristics of the 21
included studies.
| First author | Year | Country | miR type | Cancer type | Gene role | Validation
sample | Sample size | Survival
analysis | Hazard ratios (95%
CI) | Follow-up
(months) | NOS | (Refs.) |
|---|
| Wang | 2014 | China | miR-23a | Diffuse large | Oncogene | Tissue | 104 | OS | 3.776
(1.10512.891)M | 34.1 | 7 | (30) |
|
|
|
|
| B-cell
lymphoma |
|
|
|
|
|
|
|
|
| Bao | 2014 | China | miR-23a | HCC | Oncogene | Tissue | 88 | OS, RFS | 2.286
(1.153–3.846)M | 60 | 8 | (12) |
|
|
|
|
|
|
|
|
|
| 2.205
(1.107–3.943)M |
|
|
|
| Ma | 2014 | China | miR-23a | Gastric cancer | Oncogene | Tissue | 84 | OS | 4.30
(1.5012.33)M | 72 | 6 | (19) |
| Zhang | 2015 | China | miR-23a | Laryngeal
cancer | Oncogene | Tissue | 52 | OS | 7.419
(2.561–21.491)U | 60 | 8 | (31) |
|
|
|
|
|
|
|
|
|
| 6.712
(2.076–21.700)M |
|
|
|
| Qu | 2015 | China | miR-23a | NSCLC | Oncogene | Tissue | 127 | OS | 3.558
(2.982–6.635)M | 60 | 7 | (26) |
| Cai | 2015 | China | miR-23a | Prostate
cancer | Anti-oncogene | Tissue | 123 | OS | 1.776
(1.116–2.829)M | 120 | 6 | (13) |
| Qu | 2015 | China | miR-23a | Nasopharyngeal
carcinoma | Oncogene | Tissue | 111 | DFS, OS | 0.384
(0.222–0.666)U | 80 | 7 | (25) |
|
|
|
|
|
|
|
|
|
| 0.435
(0.255–0.743)M |
|
|
|
|
|
|
|
|
|
|
|
|
| 0.357
(0.191–0.667)U |
|
|
|
|
|
|
|
|
|
|
|
|
| 0.392
(0.214–0.719)M |
|
|
|
| Liu | 2014 | China | miR-24 | HCC | Oncogene | Tissue | 207 | RFS, OS | 4.75
(2.66–8.47)M | 60 | 7 | (18) |
|
|
|
|
|
|
|
|
|
| 3.58
(2.34–5.46)M |
|
|
|
| Gao | 2014 | China | miR-24 | Colorectal
cancer | Anti-oncogene | Tissue | 95 | OS | 2.552
(1.647–3.956)U | 60 | 7 | (15) |
|
|
|
|
|
|
|
|
|
| 2.767
(1.203–6.364)M |
|
|
|
| Meng | 2014 | China | miR-24 | HCC | Oncogene | Serum | 72 | OS, DFS | 2.141
(1.158–3.960)M | 60 | 6 | (20) |
|
|
|
|
|
|
|
|
|
| 2.055
(1.114–3.792)M |
|
|
|
| Zhao | 2015 | China | miR-24 | NSCLC | Oncogene | Tissue | 53 | RFS | 1.77
(0.447.05)M | 20 | 8 | (32) |
|
|
|
|
|
|
| Serum |
|
| 1.85
(0.655.08)M | 23 |
|
|
| Organista-Nava | 2015 | Mexico | miR-24 | AL | Oncogene | Blood | 36 | OS | 1.83
(0.684.92)M | 72 | 7 | (23) |
| Mori | 2016 | Italy | miR-24 | Head and neck
squamous cell carcinomas | Oncogene | Tissue | 108 | OS | 1.80
(1.063.06)M | 72 | 6 | (21) |
| Eitan | 2009 | Israel | miR-23a | Ovarian cancer | Oncogene | Tissue | 26 | RFS | 2.70
(0.65–11.20)M | 60 | 7 | (14) |
|
|
|
| miR-27a |
| Oncogene |
|
|
| 3.83
(1.06–13.79)M |
|
|
|
| Han | 2011 | China | miR-27a | ALL | Anti-oncogene | Blood | 36 | RFS | 0.76
(0.0226.56)M | 36 | 7 | (16) |
| Huang | 2013 | China | miR-27a | Gastric cancer | Oncogene | Blood | 82 | OS | 1.91
(0.864.23)M | 15 | 7 | (17) |
| Taheriazam | 2015 | Iran | miR-27a | Osteosarcoma | Oncogene | Tissue | 53 | OS | 3.035
(1.731–9.897)M | 108 | 7 | (28) |
| Nakata | 2015 | Japan | miR-27a | ccRCC | Oncogene | Tissue | 183 | CSS, PFS | 1.21
(0.57–2.60)U | 120 | 7 | (22) |
|
|
|
|
|
|
|
|
|
| 2.33
(1.07–5.47)U |
|
|
|
|
|
|
|
|
|
|
|
|
| 2.71
(1.23–6.42)M |
|
|
|
| Tang | 2015 | China | miR-27a | Osteosarcoma | Oncogene | Serum | 166 | OS, DFS | 2.17
(1.303.62)M | 100 | 8 | (29) |
|
|
|
|
|
|
|
|
|
| 1.39
(0.464.22)M |
|
|
|
| Rivera-Daz | 2015 | Puerto | miR-27a | Glioblastoma | Anti- | Tissue | 35 | OS | 0.59
(0.20–1.77)M | 34 | 8 | (27) |
|
|
| Rico |
| multiforme | oncogene |
|
|
|
|
|
|
|
| Peng | 2015 | China | miR-27a | RCC | Oncogene | Tissue | 133 | OS, RFS | 0.91
(0.14–5.96)M | 36 | 7 | (24) |
|
|
|
|
|
|
|
|
|
| 1.61
(0.47–5.44)M |
|
|
|
Association of OS with the expression
of miR-23a
In order to elucidate the association between OS and
the expression of miR-23a, a survival analysis was performed. As
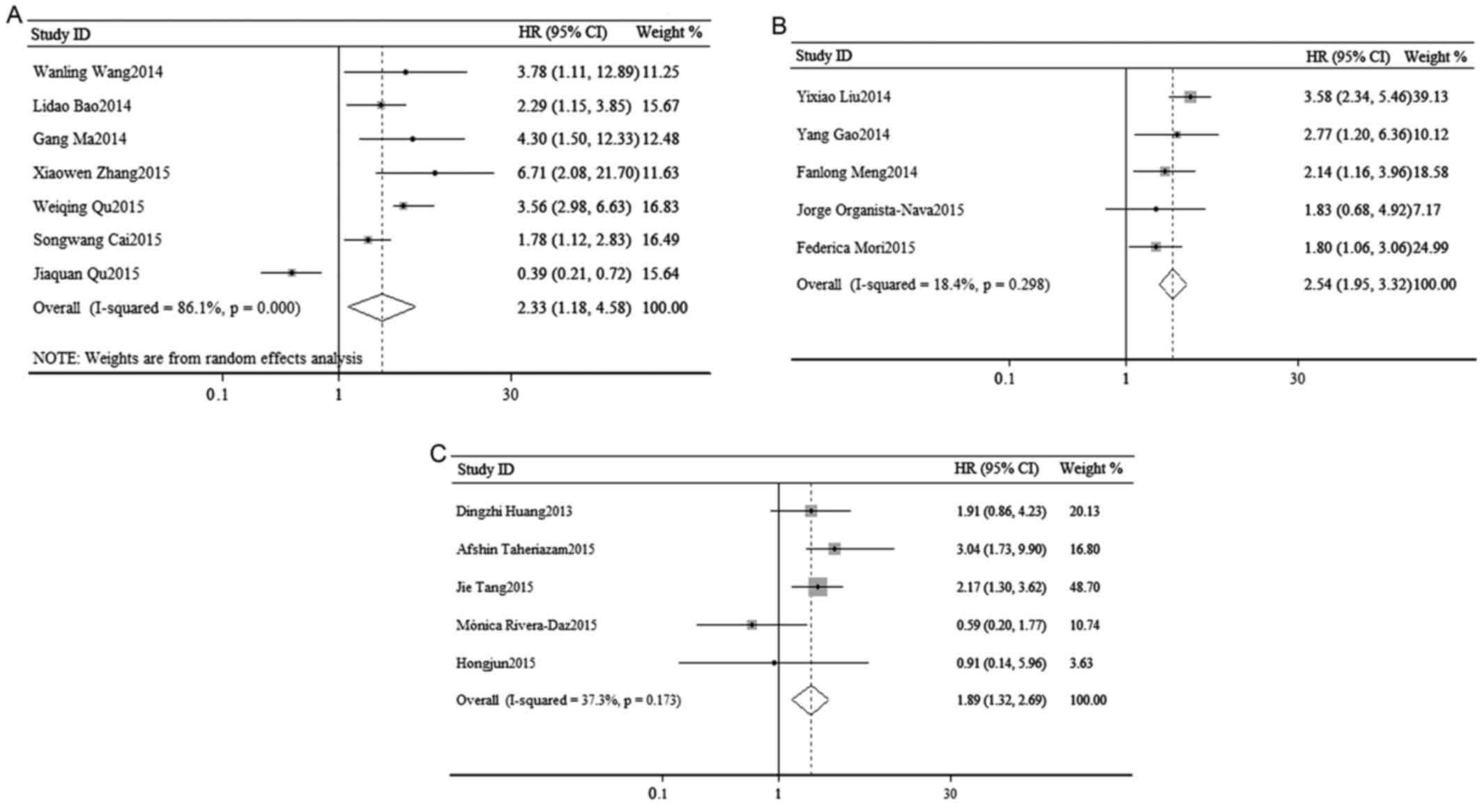
shown in Fig. 2A, a random-effects
analysis was used to calculate the pooled HR and its 95% CI due to
the relatively high heterogeneity in 7 cohorts
(I2=86.1%, P=0.000). However, the result indicated that
there was a statistically significant association between OS and
the expression of miR-23a (pooled HR=2.33, 95% CI: 1.18–4.58;
P=0.014).
To further elucidate this association, a stratified
analysis was performed (Table II).
In the subgroup analysis by cancer type, the result predicted that
a high expression level of miR-23a was associated with poorer OS in
digestive system cancers (pooled HR=2.69, 95% CI: 1.57–4.63;
P=0.034) by a random-effects model (I2=53.6%, P=0.142).
However, it failed to predict OS in respiratory system cancers
(pooled HR=2.03, 95% CI: 0.38–10.80; P=0.408) by a random-effects
model (I2=95%, P=0.000). In the subgroup analysis by
statistical methodology, there was a statistically significant
association between OS and the expression of miR-23a in the
multivariate analysis (pooled HR=2.33, 95% CI: 1.18–4.58; P=0.014)
by a random-effects model (I2=86.1%, P=0.000), while
there was no statistically significant association between OS and
the expression of miR-23a in the univariate analysis (pooled
HR=1.58, 95% CI: 0.08–30.81; P=0.76) by a random-effects model
(I2=95.7%, P=0.000). When stratified by dominant
ethnicity, a significant association was observed in Asians
(random-effects model; pooled HR=2.33, 95% CI: 1.18–4.58;
P=0.014).
 | Table II.Subgroup analysis and heterogeneity
analysis of miR-23a/24-2/27a. |
Table II.
Subgroup analysis and heterogeneity
analysis of miR-23a/24-2/27a.
|
|
|
|
| Test of
association | Test of
heterogeneity |
|---|
|
|
|
|
|
|
|
|---|
| Stratified
analysis |
|
| No. of studies | Pooled HR (95%
CI) | Z | P-value | Model | X2 | P-value | I2
(%) |
|---|
| miR-23a |
|
|
|
|
|
|
|
|
|
|
|
| OS | Overall | 7 | 2.33
(1.18–4.58) | 2.45 | 0.014 | R | 43.15 | 0.000 | 86.1 |
|
|
| Cancer types |
|
|
|
|
|
|
|
|
|
|
|
Digestive system | 2 | 2.46
(0.98–6.21) | 2.12 | 0.034 | R |
1.04 | 0.307 |
4.0 |
|
|
|
Respiratory system | 3 | 2.03
(0.38–10.80) | 0.83 | 0.408 | R | 39.85 | 0.000 | 95 |
|
|
| Statistical
methodology |
|
|
|
|
|
|
|
|
|
|
|
Univariate analysis | 2 | 1.58
(0.08–30.81) | 0.30 | 0.76 | R | 23.23 | 0.000 | 95.7 |
|
|
|
Multivariate analysis | 7 | 2.33
(1.18–4.58) | 2.45 | 0.014 | R | 43.15 | 0.000 | 86.1 |
|
|
| Ethnicity |
|
|
|
|
|
|
|
|
|
|
|
Asian | 7 | 2.33
(1.18–4.58) | 2.45 | 0.014 | R | 43.15 | 0.000 | 86.1 |
|
| DFS/RFS | Overall | 3 | 1.13
(0.37–3.44) | 0.21 | 0.836 | R | 13.18 | 0.000 | 84.8 |
| miR-24 |
|
|
|
|
|
|
|
|
|
|
|
| OS | Overall | 5 | 2.49
(1.84–3.37) | 6.91 | 0.000 | F |
4.90 | 0.298 | 18.4 |
|
|
| Sample |
|
|
|
|
|
|
|
|
|
|
|
Tissue | 3 | 2.74
(2.02–3.73) | 6.43 | 0.000 | F |
3.94 | 0.139 | 49.3 |
|
|
|
Blood | 2 | 2.05
(1.22–3.45) | 2.60 | 0.000 | F |
0.07 | 0.792 |
0.0 |
|
|
| Cancer types |
|
|
|
|
|
|
|
|
|
|
|
Digestive system | 3 | 2.99
(2.17–4.13) | 6.68 | 0.000 | F |
1.86 | 0.394 |
0.0 |
|
|
| Ethnicity |
|
|
|
|
|
|
|
|
|
|
|
Asian | 3 | 2.99
(2.17–4.13) | 6.68 | 0.000 | F |
1.86 | 0.394 |
0.0 |
|
|
|
Caucasian | 2 | 1.81
(1.13–2.88) | 2.48 | 0.013 | F |
0.00 | 0.977 |
0.0 |
|
| DFS/RFS | Overall | 4 | 2.85
(1.96–4.14) | 5.47 | 0.000 | F |
5.22 | 0.157 |
42.5 |
|
|
| Sample |
|
|
|
|
|
|
|
|
|
|
|
Tissue | 2 | 4.10
(2.40–7.00) | 5.18 | 0.000 | F |
1.66 | 0.198 | 39.7 |
|
|
|
Blood | 2 | 2.00
(1.18–3.38) | 2.58 | 0.01 | F |
0.03 | 0.863 |
0.0 |
| MiR-27a |
|
|
|
|
|
|
|
|
|
|
|
| OS | Overall | 5 | 1.89
(1.32–2.69) | 3.48 | 0.001 | F |
6.38 | 0.173 | 37.3 |
|
|
| Sample |
|
|
|
|
|
|
|
|
|
|
|
Tissue | 3 | 1.50
(0.79–2.69) | 1.24 | 0.214 | R |
5.60 | 0.061 | 64.3 |
|
|
|
Blood | 2 | 2.09
(1.36–3.22) | 3.36 | 0.001 | F |
0.07 | 0.792 |
0.0 |
|
|
| Cancer types |
|
|
|
|
|
|
|
|
|
|
|
Osteosarcoma | 2 | 2.37
(1.52–3.68) | 3.82 | 0.000 | F |
0.42 | 0.515 |
0.0 |
|
|
| Ethnicity |
|
|
|
|
|
|
|
|
|
|
|
Asian | 3 | 2.01
(1.32–3.05) | 3.25 | 0.001 | F |
0.79 | 0.675 |
0.0 |
|
|
|
Caucasian | 2 | 1.60
(0.81–3.17) | 1.36 | 0.175 | R |
5.29 | 0.021 | 81.1 |
|
| DFS/RFS/PFS | Overall | 5 | 2.19
(1.29–3.70) | 2.92 | 0.003 | F |
2.21 | 0.698 |
0.0 |
|
|
| Sample |
|
|
|
|
|
|
|
|
|
|
|
Tissue | 3 | 2.58
(1.41–4.72) | 3.07 | 0.002 | F |
0.95 | 0.623 |
0.0 |
|
|
|
Blood | 2 | 1.32
(0.46–3.80) | 0.51 | 0.608 | F |
0.10 | 0.753 |
0.0 |
|
|
| Cancer types |
|
|
|
|
|
|
|
|
|
|
|
RCC | 2 | 2.30
(1.16–4.57) | 2.39 | 0.017 | F |
0.48 | 0.49 |
0.0 |
|
|
| Ethnicity |
|
|
|
|
|
|
|
|
|
|
|
Asian | 4 | 1.95
(1.10–3.47) | 2.28 | 0.022 | F |
1.33 | 0.698 |
0.0 |
Association of OS with the expression
of miR-24
A survival analysis was performed to elucidate the
association between OS and the expression of miR-24. As shown in
Fig. 2B, a fixed-effects analysis
was used to calculate the pooled HR and its 95% CI in 5 cohorts
(I2=18.4%, P=0.298). The result indicated that the
dysregulation of miR-24 in various cancers may predict a poorer OS
(pooled HR=2.49, 95% CI: 1.84–3.37). The association was
statistically significant (P=0.000).
As shown in Table
II, further stratified analysis by detected samples suggested
that a poorer OS was associated with high expression level of
miR-24 in tissues (fixed-effects model, pooled HR=2.74, 95% CI:
2.02–3.73; P=0.000) as well as in the blood (fixed-effects model,
pooled HR=2.05, 95% CI: 1.22–3.45; P=0.000) sample. An obvious
statistically significant association was observed between OS and
the expression of miR-24 in digestive system cancers (pooled
HR=2.99, 95% CI: 2.17–4.13; P=0.000) by a fixed-effects model
(I2=0.0%, P=0.394). In the subgroup analysis of dominant
ethnicity, a significant association was observed in Asians
(fixed-effects model, pooled HR=2.99, 95% CI: 2.17–4.13; P=0.000)
as well as Caucasians (fixed-effects model, pooled HR=1.81, 95% CI:
1.13–2.88; P=0.013).
Association of OS with the expression
of miR-27a
A total of 5 studies evaluated OS and miR-27a. Due
to the relatively low significant heterogeneity, a fixed-effects
model was used to calculate the pooled HR and its 95% CI
(I2=37.3%, P=0.173). The result suggested that the
association between OS and the expression of miR-27a was
statistically significant (pooled HR=1.89, 95% CI: 1.32–2.69;
P=0.001; Fig. 2C).
Subgroup analyses failed to demonstrate a
significant association between poorer OS and high level of miR-27a
expression in the tissue subgroup (pooled HR=1.50, 95% CI:
0.79–2.69, P=0.214) by a random-effects model (I2=37.3%,
P=0.173), but revealed that a high level of miR-27a in the blood
was a significant predictor of poor OS (pooled HR=2.09, 95% CI:
1.36–3.22; P=0.001) by a fixed-effects model (I2=0.0%,
P=0.792). In addition, there was an obvious statistically
significant association between OS and the expression of miR-24 in
osteosarcoma (fixed-effects model, pooled HR=2.37, 95% CI:
1.52–3.68; P=0.000). In the subgroup analysis by dominant
ethnicity, a significant association was observed in Asians
(fixed-effects model, pooled HR=2.01, 95% CI: 1.32–3.05; P=0.001),
but not in Caucasians (random-effects model, pooled HR=1.60, 95%
CI: 0.81–3.17; P=0.175; Table
II).
Association of tumor progression
[disease-free survival (DFS)/recurrence-free survival (RFS)] with
the expression of miR-23a
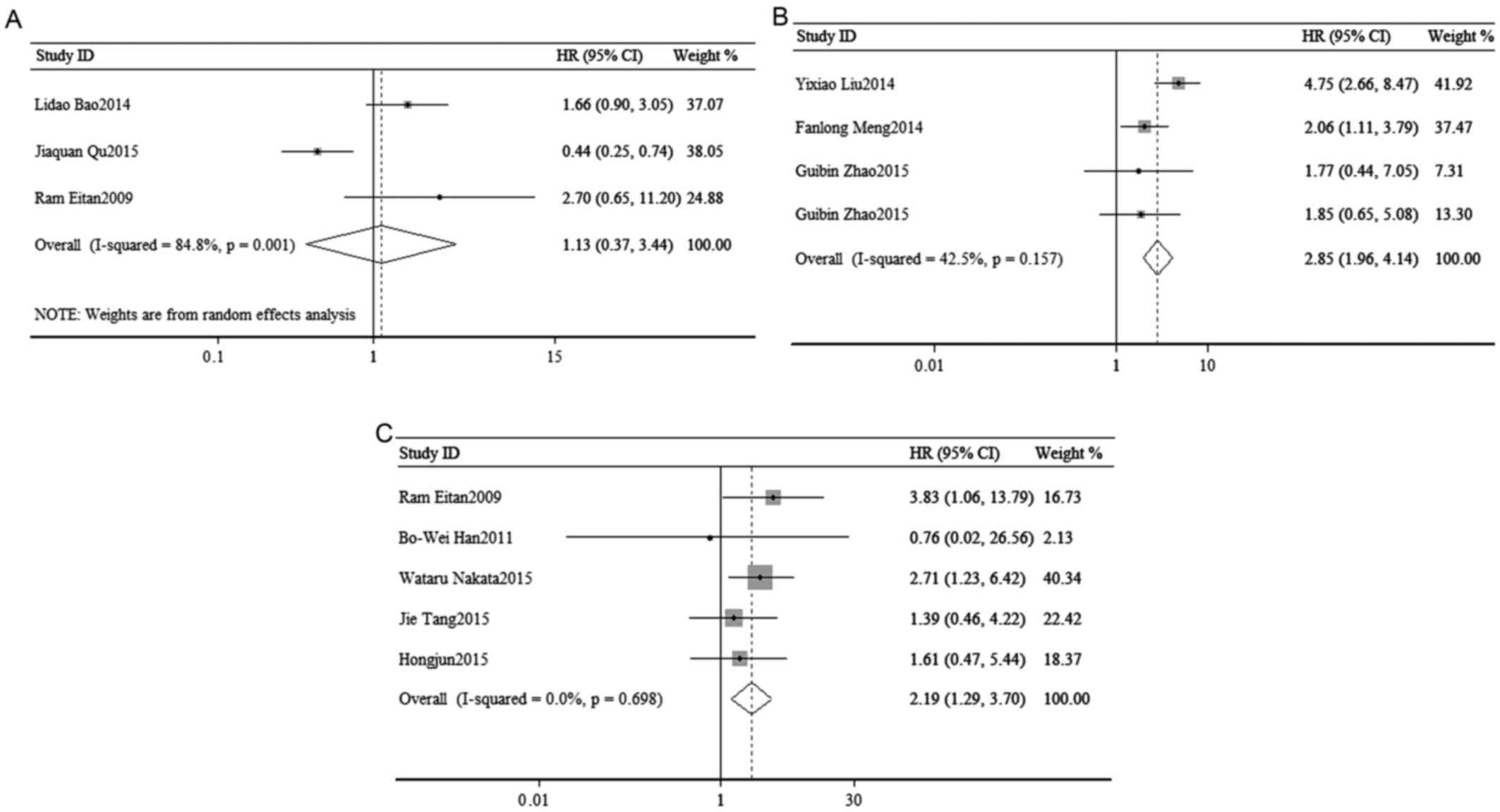
To access the association of tumor progression with
miR-23a expression, disease recurrence and metastasis were
assessed. As shown in the Fig. 5, a
random-effects model was used to calculate the pooled HR and its
95% CI in 3 cohorts (I2=84.8%, P=0.001), but failed to
show a significant association between the expression of miR-23a
and poor DFS/RFS (pooled HR=1.13, 95% CI: 0.37–3.44; P=0.836;
Fig. 3A).
Association of tumor progression
(DFS/RFS) with the expression of miR-24
Disease recurrence and metastasis were used to
access the association of tumor progression with miR-24 expression.
Due to the low heterogeneity, the pooled HR and its 95% CI were
calculated by a fixed-effects model (I2=42.5%, P=0.157)
and the meta-analysis result suggested that high expression of
miR-24 was significantly associated with poor DFS/RFS (pooled
HR=2.85, 95% CI: 1.96–4.14; P=0.000; Fig. 3B).
As shown in Table
II, further stratified analysis indicated that tumor
progression was associated with a high expression level of miR-24
in tissue samples (fixed-effects model, pooled HR=4.10, 95% CI:
2.40–7.00; P=0.000), as well as in the blood (fixed-effects model,
pooled HR=2.00, 95% CI: 1.18–3.38; P=0.01).
Association of tumor progression
[DFS/RFS/progression-free survival (PFS)] with the expression of
miR-27a
The association of tumor progression with miR-27a
expression was analyzed to combine disease recurrence and
metastasis. A total of 6 studies included a DFS/RFS/PFS analysis,
with significant heterogeneity (I2=0.00%, P=0.698), and
demonstrated a significant association between the expression of
miR-27a and poor DFS/RFS/PFS (pooled HR=2.19, 95% CI: 1.29–3.70;
P=0.003) (Fig. 3C).
In the subgroup analysis of the association of high
expression of miR-27a and tumor progression, a significant
association was observed for tissue samples (fixed-effects model,
pooled HR=2.58, 95% CI: 1.41–4.72; P=0.002). However, no
significant association was observed between tumor progression and
high expression of miR-27a in blood samples (fixed-effects model,
pooled HR=1.32, 95% CI: 0.46–3.80; P=0.608). Moreover, we found
that high expression of miR-27a was significantly associated with
poor DFS/RFS/PFS in RCC (fixed-effects model, pooled HR=2.30, 95%
CI: 1.16–4.67; P=0.017). No obvious heterogeneity was observed
(I2=0.00%, P=0.49) and the fixed-effects model was used.
A significant association was also observed in Asian patients
(fixed-effects model; pooled HR=1.95, 95% CI: 1.10–3.47;
P=0.022).
Heterogeneity analysis result
To assess OS for miR-23a (I2=86.1%),
miR-24 (I2=18.4%) and miR-27a (I2=37.3%), as
well as DFS/RFS/PFS for miR-23a (I2=84.8%), miR-24
(I2=42.5%) and miR-27a (I2=0.00%),
heterogeneity was analyzed among studies. In the subgroup analysis
by cancer type and miR-23a expression, significant heterogeneity
was observed in respiratory system cancers (I2=95%). In
the stratified analysis by statistical methodology and miR-23a
expression, significant heterogeneity was also observed in the
univariate analysis (I2=95.7%) as well as in the
multivariate analysis (I2=86.2%). However, in a subgroup
analysis, there was also significant heterogeneity among Asians (OS
for miR-23a: I2=86.1%), as well as Caucasians (OS for
miR-27a: I2=81.1%). In addition, significant
heterogeneity was observed in the subgroup of tissue samples (OS
for miR-27a: I2=64.3%), while no significant
heterogeneity was observed in other sample subgroups.
Publication bias and sensitivity
analysis
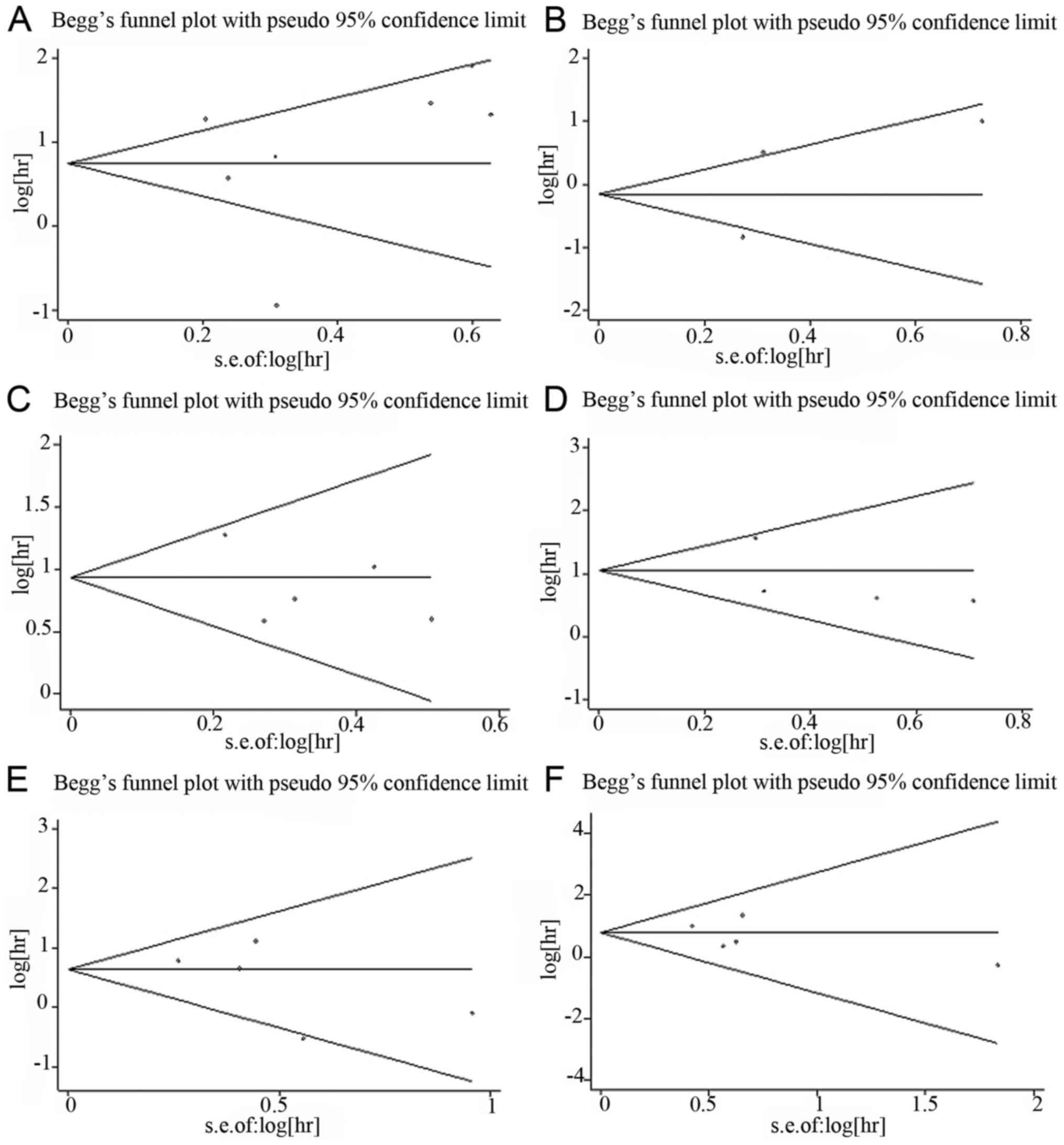
Begg's funnel plot and Egger's test were used to
assess the potential publication bias. For miR-23a, 7 cohorts
evaluating OS and 3 cohorts evaluating DFS/RFS were included. No
obvious asymmetry was observed in the Begg's funnel plot (Fig. 4A and B) and Egger's test indicated no
potential publication bias (OS: t=0.30, P=0.778; DFS/RFS: t=0.23,
P=0.854). For miR-24, 5 and 4 cohorts evaluating OS and DFS/RFS,
respectively, were included. There was no obvious asymmetry in the
Begg's funnel plot (Fig. 4C and D)
and Egger's test also indicated no potential publication bias (OS:
t=−0.99, P=0.395; DFS/RFS: t=−0.94, P=0.448). To evaluate OS and
DFS/RFS/PFS for miR-27a, 5 and 4 cohorts were included,
respectively. No obvious asymmetry was observed in the Begg's
funnel plot (Fig. 4E and F) and
Egger's test also indicated no potential publication bias (OS:
t=−1.21, P=0.312; DFS/RFS/PFS: t=−0.82, P=0.472).
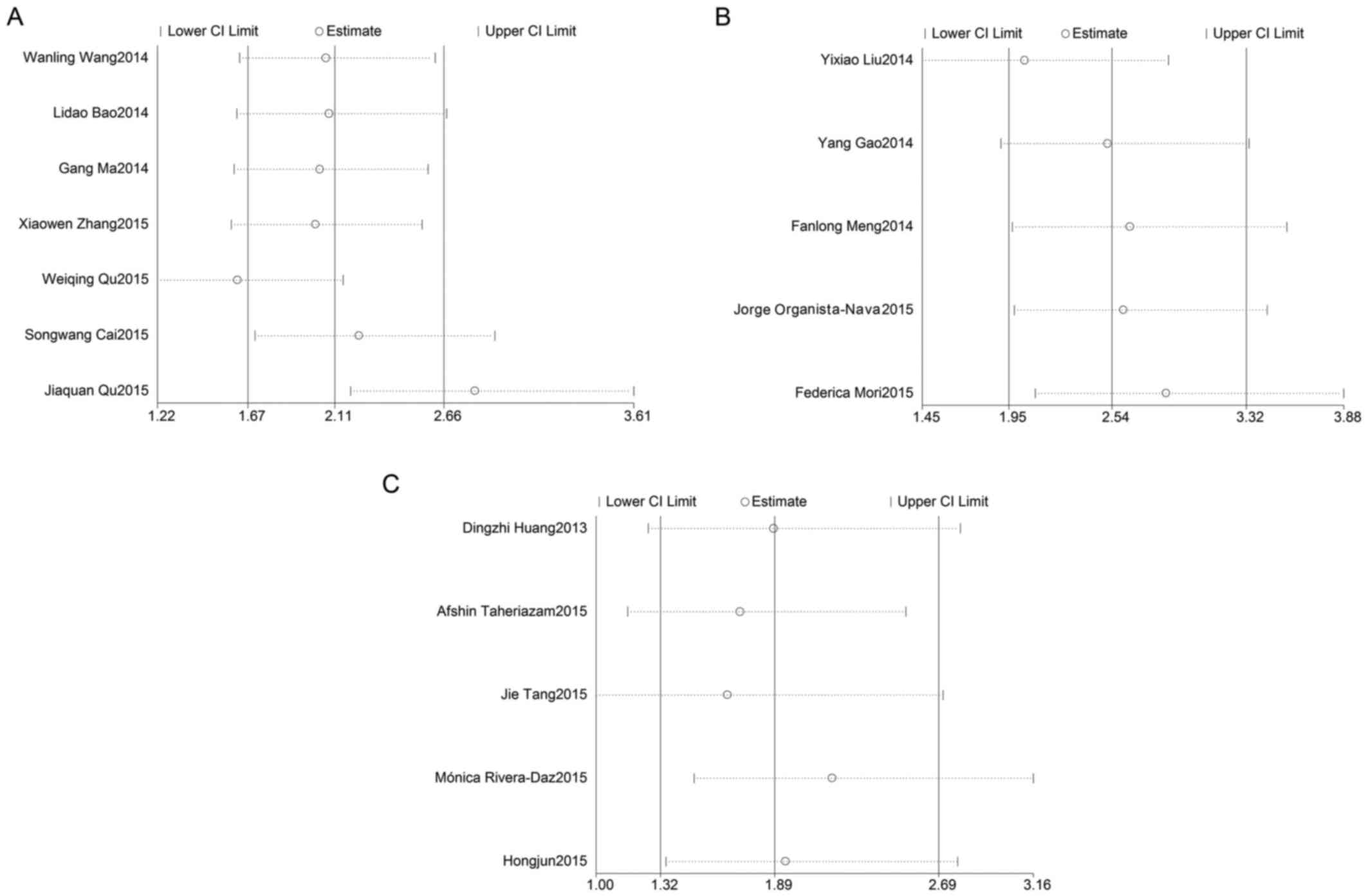
In order to assess the effect of any individual
study on the stability of the overall result, the sensitivity
analysis was used by omitting each study at a given time. As shown
in Fig. 5A, the result of
sensitivity analysis for miR-23a with OS was affected due to the
results of 2 studies: Qu et al (26) and Qu et al (25). The result of the sensitivity analysis
for miR-24 with OS (Fig. 5B)
reflected the stability of the studies, as did the result of the
sensitivity analysis for miR-27a with OS (Fig. 5C). Due to lack of a sufficient number
of studies, sensitivity analyses for miR-23a, miR-24 and miR-27a
with DFS/RFS were not preformed.
Discussion
To the best of our knowledge, this is the first
meta-analysis to investigate the association between the
miR-23a/24-2/27a cluster and various cancers. The miR-23a/24-2/27a
cluster exists in the vertebrate genome, and has been confirmed to
play an important role in cancer progression (33,34).
This cluster must be distinguished from the miR-23b/24-1/27b
cluster, as the latter is located in close proximity on the human
chromosome 9q22.32 region and also plays an important role in
cancer (35).
In the present meta-analysis, it was demonstrated
that a high expression level of miR-23a, as well as miR-24 and
miR-27a, was associated with worse OS in various types of cancers.
Particularly in cancers of the digestive system, high expression of
miR-23a and miR-24 indicated a worse prognosis. For cancers of the
respiratory system, there was no statistical significance in the
overall study sample, but significant relevance was observed in
certain individual studies. Therefore, more studies are required to
prove the association between high expression of miR-23a and OS in
respiratory system cancers. Furthermore, due to the lack of studies
on the association of high expression of miR-23a and OS in
Caucasians, the association of high expression level of miR-23a
with worse OS was only investigated in Asians. For miR-24, a
significant association was observed in Asians as well as
Caucasians. Unlike miR-23a and miR-24, a significant association
between a high expression level of miR-27a and OS was observed in
Asians, but not in Caucasians. A stratified analysis by detected
sample suggested that poorer OS was associated with high expression
levels of miR-24 and miR-27a in tissue as well as in blood samples.
And the results by tissue were more sensitive compared with the
results by blood for miR-24 and miR-27a. Moreover, there was a
statistically significant association between OS and the expression
of miR-24 in osteosarcoma.
In addition, there was a significant association
between the expression of miR-24, as well as miR-27a, and tumor
progression. However, there was no obvious association between the
expression of miR-23a and poor DFS/RFS. Further stratified analysis
indicated that a high expression level of miR-24, as well as
miR-27a, in tissue samples was associated with tumor progression.
The association persisted for miR-24, while no significant
association was observed for miR-27a expression in the blood.
Recently, a number of studies indicated that the
miR-23a/24-2/27 cluster plays an important role in the occurrence
and development of cancer. Huang et al demonstrated that a
high expression level of miR-23a/24/27a decreased transforming
growth factor-β-induced tumor-suppressive activity in a
Smad-dependent manner in HCC (9) and
lung cancer (36). Furthermore, the
cluster was also found to be involved in altering lymphoid cell
differentiation via the transcription factor PU.1 (37). An increasing number of studies
demonstrated that several miRNA clusters may regulate the
occurrence and development of cancer by cooperating with the c-Myc
oncogene (38–40). The association of miR-23a/24-2/27a
with c-Myc has also been investigated and the findings suggested
that the miR-23a/24-2/27a cluster was upregulated in breast cancer
and was correlated with cancer cell metastasis, migration and
invasion via c-Myc (34).
There were some limitations to this meta-analysis.
First, all the included studies were published in English;
therefore, English language bias may exist in this meta-analysis.
Second, the number of eligible studies was not sufficient, despite
the fact that no significant publication bias was detected in this
meta-analysis; furthermore, the subgroup analysis was limited by
the sample size, compromising the validity of the results. Third,
some HRs were estimated from survival curves and data were
extracted according to the Tierney's method (41). Finally, certain aspects were not
uniform among studies, including clinical characteristics and
follow-up time, which may lead to estimation errors.
In summary, the present meta-analysis demonstrated
that high expression levels of miR-23a, miR-24-2 and miR-27a were
associated with poor survival in various types of cancer. The
miR-23a/24-2/27a cluster may prove useful for monitoring the
progression and prognosis of cancer in clinical practice. However,
to verify the association between the expression of the
miR-23a/24-2/27a cluster and cancer, a larger number of relevant
studies are required.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81101922), the
Science and Technology Development Fund Project of Shenzhen (grant
nos. JCYJ20150403091443329 and JCYJ20170307111334308), the fund of
‘San-Ming’ project of medicine in Shenzhen and funds from the
Guangdong Key Medical Subject.
References
|
1
|
Hammond SM: An overview of microRNAs. Adv
Drug Deliv Rev. 87:3–14. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Horvitz HR and Sulston JE: Isolation and
genetic characterization of cell-lineage mutants of the nematode
Caenorhabditis elegans. Genetics. 96:435–454. 1980.PubMed/NCBI
|
|
3
|
Griffiths-Jones S, Grocock RJ, van Dongen
S, Bateman A and Enright AJ: miRBase: MicroRNA sequences, targets
and gene nomenclature. Nucleic Acids Res. 34:D140–D144. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hua K, Jin J, Zhang H, Zhao B, Wu C, Xu H
and Fang L: MicroRNA-7 inhibits proliferation, migration and
invasion of thyroid papillary cancer cells via targeting CKS2. Int
J Oncol. 49:1531–1540. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nip H, Dar AA, Saini S, Colden M, Varahram
S, Chowdhary H, Yamamura S, Mitsui Y, Tanaka Y, Kato T, et al:
Oncogenic microRNA-4534 regulates PTEN pathway in prostate cancer.
Oncotarget. 7:68371–68384. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jin L, Li Y, Liu J, Yang S, Gui Y, Mao X,
Nie G and Lai Y: Tumor suppressor miR-149-5p is associated with
cellular migration, proliferation and apoptosis in renal cell
carcinoma. Mol Med Rep. 13:5386–5392. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek
SH and Kim VN: MicroRNA genes are transcribed by RNA polymerase II.
EMBO J. 23:4051–4060. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang S, He X, Ding J, Huang S, He X, Ding
J, Liang L, Zhao Y, Zhang Z, Yao X, Pan Z, Zhang P, Li J, et al:
Upregulation of miR-23a approximately 27a approximately 24
decreases transforming growth factor-beta-induced tumor-suppressive
activities in human hepatocellular carcinoma cells. Int J Cancer.
123:972–978. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner
MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ and Schmittgen
TD: Expression profiling identifies microRNA signature in
pancreatic cancer. Int J Cancer. 120:1046–1054. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mattie MD, Benz CC, Bowers J, Sensinger K,
Wong L, Scott GK, Fedele V, Ginzinger D, Getts R and Haqq C:
Optimized high-throughput microRNA expression profiling provides
novel biomarker assessment of clinical prostate and breast cancer
biopsies. Mol Cancer. 5:242006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bao L, Zhao J, Dai X, Wang Y, Ma R, Su Y,
Cui H, Niu J, Bai S, Xiao Z, et al: Correlation between miR-23a and
onset of hepatocellular carcinoma. Clin Res Hepatol Gastroenterol.
38:318–330. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cai S, Chen R, Li X, Cai Y, Ye Z, Li S, Li
J, Huang H, Peng S, Wang J, et al: Downregulation of microRNA-23a
suppresses prostate cancer metastasis by targeting the PAK6-LIMK1
signaling pathway. Oncotarget. 6:3904–3917. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Eitan R, Kushnir M, Lithwick-Yanai G,
David MB, Hoshen M, Glezerman M, Hod M, Sabah G, Rosenwald S and
Levavi H: Tumor microRNA expression patterns associated with
resistance to platinum based chemotherapy and survival in ovarian
cancer patients. Gynecol Oncol. 114:253–259. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gao Y, Liu Y, Du L, Li J, Qu A, Zhang X,
Wang L and Wang C: Down-regulation of miR-24-3p in colorectal
cancer is associated with malignant behavior. Med Oncol.
32:3622015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Han BW, Feng DD, Li ZG, Luo XQ, Zhang H,
Li XJ, Zhang XJ, Zheng LL, Zeng CW, Lin KY, et al: A set of miRNAs
that involve in the pathways of drug resistance and leukemic
stem-cell differentiation is associated with the risk of relapse
and glucocorticoid response in childhood ALL. Hum Mol Genet.
20:4903–4915. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang D, Wang H, Liu R, Li H, Ge S, Bai M,
Deng T, Yao G and Ba Y: miRNA27a is a biomarker for predicting
chemosensitivity and prognosis in metastatic or recurrent gastric
cancer. J Cell Biochem. 115:549–556. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu YX, Long XD, Xi ZF, Ma Y, Huang XY,
Yao JG, Wang C, Xing TY and Xia Q: MicroRNA-24 modulates aflatoxin
B1-related hepatocellular carcinoma prognosis and tumorigenesis.
Biomed Res Int. 2014:4829262014.PubMed/NCBI
|
|
19
|
Ma G, Dai W, Sang A, Yang X and Gao C:
Upregulation of microRNA-23a/b promotes tumor progression and
confers poor prognosis in patients with gastric cancer. Int J Clin
Exp Pathol. 7:8833–8840. 2014.PubMed/NCBI
|
|
20
|
Meng FL, Wang W and Jia WD: Diagnostic and
prognostic significance of serum miR-24-3p in HBV-related
hepatocellular carcinoma. Med Oncol. 31:1772014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mori F, Ferraiuolo M, Santoro R, Sacconi
A, Goeman F, Pallocca M, Pulito C, Korita E, Fanciulli M, Muti P,
et al: Multitargeting activity of miR-24 inhibits long-term
melatonin anticancer effects. Oncotarget. 7:20532–20548. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nakata W, Uemura M, Sato M, Fujita K,
Jingushi K, Ueda Y, Kitae K, Tsujikawa K and Nonomura N: Expression
of miR-27a-3p is an independent predictive factor for recurrence in
clear cell renal cell carcinoma. Oncotarget. 6:21645–21654. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Organista-Nava J, Gómez-Gómez Y,
Illades-Aguiar B, Del Carmen Alarcón-Romero L, Saavedra-Herrera MV,
Rivera-Ramírez AB, Garzón-Barrientos VH and Leyva-Vázquez MA: High
miR-24 expression is associated with risk of relapse and poor
survival in acute leukemia. Oncol Rep. 33:1639–1649. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Peng H, Wang X, Zhang P, Sun T, Ren X and
Xia Z: miR-27a promotes cell proliferation and metastasis in renal
cell carcinoma. Int J Clin Exp Pathol. 8:2259–2266. 2015.PubMed/NCBI
|
|
25
|
Qu JQ, Yi HM, Ye X, Li LN, Zhu JF, Xiao T,
Yuan L, Li JY, Wang YY, Feng J, et al: MiR-23a sensitizes
nasopharyngeal carcinoma to irradiation by targeting IL-8/Stat3
pathway. Oncotarget. 6:28341–28356. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Qu WQ, Liu L and Yu Z: Clinical value of
microRNA-23a upregulation in non-small cell lung cancer. Int J Clin
Exp Med. 8:13598–13603. 2015.PubMed/NCBI
|
|
27
|
Rivera-Diaz M, Miranda-Roman MA, Soto D,
Quintero-Aguilo M, Ortiz-Zuazaga H, Marcos-Martinez MJ and
Vivas-Mejía PE: MicroRNA-27a distinguishes glioblastoma multiforme
from diffuse and anaplastic astrocytomas and has prognostic value.
Am J Cancer Res. 5:201–218. 2015.PubMed/NCBI
|
|
28
|
Taheriazam A, Bahador R, Karbasy SH,
Jamshidi SM, Torkaman A, Yahaghi E and Shakeri M: Down-regulation
of microRNA-26a and up-regulation of microRNA-27a contributes to
aggressive progression of human osteosarcoma. Diagn Pathol.
10:1662015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tang J, Zhao H, Cai H and Wu H: Diagnostic
and prognostic potentials of microRNA-27a in osteosarcoma. Biomed
Pharmacother. 71:222–226. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang WL, Yang C, Han XL, Wang R, Huang Y,
Zi YM and Li JD: MicroRNA-23a expression in paraffin-embedded
specimen correlates with overall survival of diffuse large B-cell
lymphoma. Med Oncol. 31:9192014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang XW, Liu N, Chen S, Wang Y, Zhang ZX,
Sun YY, Qiu GB and Fu WN: High microRNA-23a expression in laryngeal
squamous cell carcinoma is associated with poor patient prognosis.
Diagn Pathol. 10:222015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhao G, Liu L, Zhao T, Jin S, Jiang S, Cao
S, Han J, Xin Y, Dong Q, Liu X and Cui J: Upregulation of miR-24
promotes cell proliferation by targeting NAIF1 in non-small cell
lung cancer. Tumour Biol. 36:3693–3701. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhou Q, Gallagher R, Ufret-Vincenty R, Li
X, Olson EN and Wang S: Regulation of angiogenesis and choroidal
neovascularization by members of microRNA-23~27~24 clusters. Proc
Natl Acad Sci USA. 108:pp. 8287–8292. 2011; View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li X, Liu X, Xu W, Zhou P, Gao P, Jiang S,
Lobie PE and Zhu T: c-MYC-regulated miR-23a/24-2/27a cluster
promotes mammary carcinoma cell invasion and hepatic metastasis by
targeting Sprouty2. J Biol Chem. 288:18121–18133. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Goto Y, Kojima S, Nishikawa R, Enokida H,
Chiyomaru T, Kinoshita T, Nakagawa M, Naya Y, Ichikawa T and Seki
N: The microRNA-23b/27b/24-1 cluster is a disease progression
marker and tumor suppressor in prostate cancer. Oncotarget.
5:7748–7759. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cao M, Seike M, Soeno C, Mizutani H,
Kitamura K, Minegishi Y, Noro R, Yoshimura A, Cai L and Gemma A:
MiR-23a regulates TGF-β-induced epithelial-mesenchymal transition
by targeting E-cadherin in lung cancer cells. Int J Oncol.
41:869–875. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kong KY, Owens KS, Rogers JH, Mullenix J,
Velu CS, Grimes HL and Dahl R: MIR-23A microRNA cluster inhibits
B-cell development. Exp Hematol. 38:629–640 e1. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lu Y, Thomson JM, Wong HY, Hammond SM and
Hogan BL: Transgenic over-expression of the microRNA miR-17-92
cluster promotes proliferation and inhibits differentiation of lung
epithelial progenitor cells. Dev Biol. 310:442–453. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mehta A, Mann M, Zhao JL, Marinov GK,
Majumdar D, Garcia-Flores Y, Du X, Erikci E, Chowdhury K and
Baltimore D: The microRNA-212/132 cluster regulates B cell
development by targeting Sox4. J Exp Med. 212:1679–1692. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang CM, Chiba T, Brill B, Delis N, von
Manstein V, Vafaizadeh V, Oellerich T and Groner B: Expression of
the miR-302/367 cluster in glioblastoma cells suppresses
tumorigenic gene expression patterns and abolishes transformation
related phenotypes. Int J Cancer. 137:2296–2309. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tierney JF, Stewart LA, Ghersi D, Burdett
S and Sydes MR: Practical methods for incorporating summary
time-to-event data into meta-analysis. Trials. 8:162007. View Article : Google Scholar : PubMed/NCBI
|